Abstract
The Chlamydomonas anterograde intraflagellar transport motor, kinesin-2, is isolated as a heterotrimeric complex containing two motor subunits and a nonmotor subunit known as kinesin-associated polypeptide or KAP. One of the two motor subunits is encoded by the FLA10 gene. The sequence of the second motor subunit was obtained by mass spectrometry and sequencing. It shows 46.9% identity with the Fla10 motor subunit and the gene maps to linkage group XII/XIII near RPL9. The temperature-sensitive flagellar assembly mutants fla1 and fla8 are linked to this kinesin-2 motor subunit. In each strain, a unique single point mutation gives rise to a unique single amino acid substitution within the motor domain. The fla8 strain is named fla8-1 and the fla1 strain is named fla8-2. The fla8 and fla10 alleles show a chromosome loss phenotype. To analyze this chromosome loss phenotype, intragenic revertants of fla8-1, fla8-2, and fla10-14 were generated. The analysis of the mutants and the revertants demonstrates the importance of a pocket in the amino terminus of these motor subunits for both motor activity and for a novel, dominant effect on the fidelity of chromosome segregation.
INTRODUCTION
Kinesin-2 is the motor for anterograde intraflagellar transport (IFT) that serves to assemble and to maintain cilia and flagella in eukaryotic organisms. Cilia in many of these organisms are used for both motile and sensory functions. Defects that affect motile cilia/flagella in vertebrates cause randomization of the left-right axis of symmetry, chronic respiratory defects, and male sterility (McGrath and Brueckner, 2003; Snell et al., 2004). Defects in sensory cilia have been associated with polycystic kidney disease, retinal degeneration, smell and hearing loss, learning disabilities, obesity, and skeletal defects (Zhang et al., 2004). Genetic analysis of flagellar/ciliary assembly in Chlamydomonas, Caenorhabditis elegans, Drosophila, Tetrahymena, and mice show that these motors and proteins involved in IFT are conserved. Large particles (IFT particles) are moved from the basal body region to the tip of the cilia/flagella by kinesin-2 and returned to the cell body by a cytoplasmic dynein (Pazour et al., 1999; Porter et al., 1999).
Kinesin-2 is composed of two motor subunits and a third kinesin-associated protein known as KAP. This complex was first identified biochemically from sea urchin eggs (Cole et al., 1993; Wedaman et al., 1996). Its ciliary role was clarified by the identification of the gene product of the FLA10 gene as a kinesin-2 motor subunit (Walther et al., 1994), and observations that a mutant allele affected movement of particles in the flagella of Chlamydomonas (Kozminski et al., 1995). The fla10-1 allele is a temperature-sensitive mutation that lacks flagella at the restrictive temperature of 32°C (Huang et al., 1977; Adams et al., 1982; Lux and Dutcher, 1991). In this work, we show that the other motor subunit is encoded by the FLA8 gene (see below). The KAP subunit is encoded by the FLA3 gene (Mueller et al., 2005). Mutations in FLA8 and FLA3 also show temperature-sensitive defects in flagellar assembly (Adams et al., 1982).
Kinesin-2 also is required for ciliary assembly in C. elegans (Shakir et al., 1993), Tetrahymena (Brown et al., 1999), Drosophila (Han et al., 2003; Sarpal et al., 2003), sea urchins (Morris and Scholey, 1997), and mice (Yamazaki et al., 1995; Yang et al., 2001). In addition to roles in ciliary assembly, kinesin-2 has been implicated in other microtubule-based transport events. In Xenopus, kinesin-2 is involved in pigment granule dispersion (Tuma et al., 1998; Deacon et al., 2003); in endoplasmic reticulum-to-Golgi transport (Le Bot et al., 1998); and Vg1 mRNA localization (Betley et al., 2004) based on the introduction of dominant negative mutations. In Drosophila, neuronal transport of choline acetyltransferase (Ray et al., 1999) and transport of cell fate components during oogenesis require kinesin-2 (Pflanz et al., 2004). Kinesin-2 also seems to be involved in the establishment of neuronal polarity via association with the PAR-3 complex (Fan and Beck, 2004; Nishimura et al., 2004).
In ciliated cells, kinesin-2 localizes to the basal body region of cells and within the cilia/flagella (Vashishtha et al., 1996; Cole et al., 1998; Deane et al., 2001). During mitosis, kinesin-2 is found near centrioles and along the mitotic spindle (Henson et al., 1995; Vashishtha et al., 1996; Deane et al., 2001). Recently, Morris and colleagues showed in sea urchin blastulae using a green fluorescent protein GFP-KAP construct that KAP moves into the nucleus before nuclear envelope breakdown when the cilia disassemble and moves out of the nucleus after nuclear envelope reformation (Morris et al., 2004). Fan and Beck (2004) showed that KIF3B, a kinesin motor subunit, localizes to the midbody and that overexpression of the KIF3B tail results in cytokinesis defects. Previously, we had observed synthetic phenotypes in double mutants of fla10 and apm1 mutant alleles. The synthetic phenotypes were observed with three different alleles of fla10 (fla10-1, fla10-14, and fla10-15) in combination with the mutant apm1-122 allele (Lux and Dutcher, 1991). Mutations in the APM1 gene confer resistance to the microtubule-destabilizing drug ami-prophos-methyl or oryzalin (James et al., 1988), which suggests that the microtubules may be hyperstablized in this mutant background. The double mutant fla10-1; apm1-122 showed a cell cycle arrest at 21°C but growth at 32°C. The double mutants fla10-14; apm1-122 and fla10-15; apm1-122 show a slow growth phenotype at 21°C but show wild-type growth at 32°C. Given that no detectable Fla10 gene product is present at 32°C in fla10-1 cells (Cole et al., 1998), it is likely that these phenotypes arise via toxic interactions of the mutant gene products rather than by redundancy of the two genes. To ask whether the mutant gene products have other effects on the cell cycle, we have investigated the rates of chromosome loss and cytokinesis failures in several fla10 and fla8 alleles as well as in intragenic revertants of the fla8-1, fla8-2, and fla10-14 alleles.
MATERIALS AND METHODS
Cell Culture and Genetics
Cells were grown as described previously (Holmes and Dutcher, 1989). Matings were performed as described previously (Dutcher, 1995a). Revertants were isolated as described previously (Preble et al., 2001) but at 32°C.
Restriction Fragment Polymorphisms and Sequencing
DNA from fla10-14 and fla10-14 revertant cells was isolated and colony PCR was performed as described previously (Bowers et al., 2003). The coding region of FLA10 was amplified with overlapping primers that were designed with a melting temperature of 57°C with a spacing of 600 base pairs (sequences available upon request) and cloned into the pCR4-TOPO vector (TOPO TA cloning kit; Invitrogen, Carlsbad, CA). Transformed plasmids were prepared with Wizard Plus SV Minipreps DNA purification system (Promega, Madison, WI) and digested with EcoRI to verify the presence of an insert. Plasmid DNA was sequenced in conjunction with the Protein and Nucleic Acid Sequencing Laboratory (Washington University, St. Louis, MO) using M13 primers. All primers are given in the 5′-to-3′ direction. The E24K mutation was amplified using primers fla10-14F (CATCGTTGCAGCAAGCTTTA) and fla10-14R (ATGTAGACCGCTTCGCAGTC). A 207-base pair fragment from -53 to +155 in wild-type cells is not cut by MboII, whereas the E24K mutant is cut. The L196F mutation was amplified using primers fla10-3F (GTTGGGCGTGTGTGTTTATG) and fla10-3R (GACTCATAACCGACCGCTGT) and produced a 958-base pair fragment from +1417 to +2374. PpuMI cuts the wild-type PCR product into four fragments (512, 252, 149, and 45 base pairs), whereas the mutant is cut into three fragments (557, 252, and 149 base pairs). Primers fla10-5F (TTGGAATACCGCCATCT) and fla10–5R (CCAATCCCGTAAGGAAACAG) amplified the G331S mutation and produce a 894-base pair fragment from +3035 to +3928. The wild-type PCR product is cut into five fragments (270, 259, 153, 144, and 68 base pairs) with Sau96I, whereas the mutant is cut into four fragments (403, 270, 153, and 68 base pairs). The N329H mutation was amplified with the primers fla10-5F and fla10-5R, cloned into the TA vector, and sequenced because it does not have a restriction polymorphism with wild-type sequence.
A 242-base pair fragment was amplified from fla1, fla8, and CC1952 DNA with primers fla8-1F (TATCAAGCCCACGGGAGTAG) and fla8-1R (CGGATGTGTTACGAGTGTCG). The fla1 and fla8 PCR products were cut with RsaI to produce fragments of 178, 37, and 27 base pairs. The CC1952 DNA was cut to produce a fragment of 205 and 37 base pairs.
Cloning of a Kinesin-2 Motor Subunit
When Chlamydomonas expressed sequence tags (ESTs) first became available, BLAST searches using kinesin-2 motor sequences revealed a single 578-nucleotide EST (accession no. BE337430) that encoded part of a FLA10-related kinesin. This partial cDNA sequence was outside of the highly conserved kinesin motor domain, so it was suitable as a probe to screen both a cDNA library and a genomic BAC library (Clemson University Genomics Institute, Clemson, SC). Three BAC clones (28P7, 02I16, and 07F13) were identified. BAC clones and genomic DNA isolated directly from Chlamydomonas strains were used as PCR templates to amplify 4.8- to 5.2-kb products with high fidelity KOD XL DNA polymerase (Novagen, Madison, WI) supplemented with 2 M GC Melt (BD Biosciences Clontech, Palo Alto, CA). The sets of primers used were Kin2F1 (ACCCGCTGATTACGTTTCGAGTCTGG) and Kin2R1 5′-(GAAGCGGGTCTCAAGAGCGGGATAG) and Kin2F2 (GCGCGGCTGGAGCTGAAGACA) and Kin2R2 (GTAACGGCGACATACGGATGGAATAGACA). The 5-kb PCR products were cloned into TOPO blunt vectors (Invitrogen); the resulting plasmids were purified, and EcoRI digests were analyzed as described above. Sequencing primers were designed with a melting temperature of ∼55°C at an average spacing of ∼400 base pairs (sequences available upon request). An additional EST sequence (accession no. AV640755) corresponded to a cloned cDNA plasmid (#755) that was obtained from Kazusa DNA Research Institute (Chiba, Japan) (Asamizu et al., 1999; Asamizu et al., 2000). Plasmid #755 contained a 2.98-kbp cDNA insert with an open reading frame of 2304 base pairs that encodes a 768-amino acid protein. Sequencing of plasmid #755 confirmed the intron–exon structure determined from earlier analysis but it was found to contain a single base pair substitution at position 1721 of the open reading frame changing an A to a G transition. This produced a conservative amino acid substitution of arginine for lysine at residue 574. The Chlamydomonas reinhardtii strain used to generate clone #755 was C9 mt-, whereas all other sources of genomic and cDNA, including cDNA and BAC libraries, were of strains that were either 137c (CC-124 or CC-125) or mutants that were derived from the 137c strains. According to Harris (1989) C9 mt- and 137c arose from the same laboratory isolate but have been separated for 60 years. This length of time may account for the variation observed at amino acid residue 574.
Sequencing of Second Kinesin Motor Subunit Gene in fla Mutants and Revertants
Genomic DNA was isolated from Chlamydomonas fla1, fla3, fla8, and revertants by a modified procedure of (Koutoulis et al., 1997). The fla3 strain was chosen as a control. Chlamydomonas strains were plated onto solid medium and grown under continuous light for 7–9 d. Cells were removed from the plates and resuspended in 20 mM Tris, pH 7.5, 20 mM EDTA, 5% SDS, 1 mg/ml proteinase K and incubated overnight at 50°C. Phenol:chloroform (50:50) extraction was followed by chloroform extraction. Genomic DNA was precipitated with 7.5 M ammonium acetate in ice-cold ethanol followed by centrifugation for 5 min at 16,000 × g. The resulting DNA precipitate was washed two times with 80% ethanol. After drying, the DNA was resuspended in 10 mM Tris, 0.10 mM EDTA, pH 8.0, and stored at -28°C. PCR products (∼5 kb) containing the second motor subunit gene were amplified from each of three fla strains using KOD XL DNA polymerase with the Kin2F1, Kin2F2, Kin2R1, and Kin2R2 primers described above. Products from two different PCR reactions for each of the strains were cloned into TOPO Blunt vectors and resulting plasmids were purified as described above. For fla1, fla3, and fla8, the entire gene was sequenced with threefold coverage for at least two different PCR products. For revertant sequencing, the focus was on the motor domain of the kinesin with the primers Kin2F3 (CTTGCGCCCGAGAGCTGTACAGATTCAG) and Kin2R3 (CAGGGGCCATCAGCATCAATACACAGTTTG). The resulting 2254 base pairs PCR products were cloned into TOPO Blunt vectors (Invitrogen) and sequenced as described above.
Mass Spectrometry
Chlamydomonas kinesin-2 was purified from flagella essentially as described previously (Cole et al., 1998) with the exception that gel filtration was omitted. In brief, flagella were isolated from the wild-type CC-125 Chlamydomonas strain by pH shock followed by differential centrifugation (Witman et al., 1972) and storage as pellets at -80°C. The following steps were performed on ice or at 4°C. A 100-ml flagellar aliquot was extracted with 400 ml of 0.05% NP-40 in HMDEK buffer (10 mM HEPES, 5 mM MgSO4, 1.0 mM dithiothreitol, 0.5 mM EDTA, 25 mM KCl, pH 7.2) supplemented with 2.0 mM adenyl-5′-yl imidodiphosphate (AMPPNP) to keep kinesin-2 bound to the axonemal microtubules. After a 15-min centrifugation at 16,000 × g, the axonemal pellet was resuspended and extracted for 10 min with 10 mM MgATP in HMDEK buffer. After another 15-min centrifugation at 16,000 × g, the resulting supernatant was loaded onto a 13 ml of 10–25% sucrose density gradient (14 × 89-mm centrifuge tubes) and spun in an SW41-Ti rotor (Beckman Coulter, Fullerton, CA) for 12 h at 41,000 rpm. Fractions (0.5 ml) were analyzed by SDS-PAGE stained with Coomassie Blue. The heterotrimeric kinesin-2 was resolved at ∼10 S as three protein bands with relative mobilities of 86, 90, and 96 kDa. Each of the protein bands was carefully excised, digested with trypsin, and analyzed by matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectrometry to identify peptide masses. The peptides obtained for KRP86 were TFTFDNAFDWNVTQR, ASYLEIYNEEVR, SHSIFTITIETIEQTQAQPEGHIR, LNLVDLAGSER, AALEAEGGALPEGFATGPGGEIIVEK, ALDASFLEQMR, DMVIEAFIPPEEVQKVMK, AHWDDEREVWVLER, and FKSENILNLELDLPER. The peptides obtained for Fla10 were GLIPNTFR, YVFEIIAR, DLSQFVCK, LNLVDLAGSER, SSYLEIYNEEVR, and KGELADLQEQFQR. The peptides obtained for KAP were ATGMIAQLFR, LLHNLSFDHSLR, VVFYLVDLLQDK, SNVELLILATTFLK, AGQVEELLYQLQSR, NTPELIALAVNLTQNPR, and NTENFEVLLSHETLMQTLSR.
Chromosome Loss
Diploid strains were constructed by using either complementing arg7 alleles that had been taken through meiosis within 1 month of their use to ensure the absence of other mutations or by using complementation with nit2 and ac17 mutations in a trans-configuration. For chromosome loss on linkage group VI, one arm was marked with the recessive act2 mutation that confers resistance to 18 μg/ml cycloheximide. The other arm was marked with the recessive mating-type plus allele (mt+) and scored by mating or by a PCR reaction to identify the MID or FUS1 genes (Werner and Miergenhagen, 1998). Chromosome loss and mitotic recombination on linkage group III was assayed by the presence of maa7, nit2, and ac17 mutations. 5-Methylanthranilic acid or 5-aminobenzoic acid was used at 1.6 mM and maintained in yellow Lucite boxes to reduce breakdown (Palombella and Dutcher, 1998). One arm was marked by maa7 and nit2. The other arm is marked by ac17. Diploid cells were plated for single colonies on Sager and Granick rich medium with acetate and allowed to grow 5 d in the constant light at 21° or 32°C. Individual colonies were then transferred to liquid medium and grown for 24–48 h to a cell density of 106 cells/ml at 21°C to avoid clumps of cells that had been released from the mother cell wall. For experiments with fla10-2, the cells were treated with autolysin (Dutcher, 1995b) to release the cells from the mother cell to allow the plating of individual cells rather than groups of cells. Approximately 105 cells were inoculated onto cycloheximide or 5-methyl anthranilic acid (5-MAA) plates from each individual colony in four replicas. Approximately 3 × 102 cells were inoculated onto nonselective plates to determine cell number more precisely. After 10–14 d, the number of resistant colonies and the total number of viable cells on nonselective medium were determined. Because resistance could occur via spontaneous mutations, the rate of cycloheximide-resistant colonies in a haploid strain was determined. It occurred at <1 × 10-8 events/cell cycle. For linkage group VI, the mating type was determined and for linkage group III, the ability to grow on medium lacking acetate was determined.
Fluctuation analyses were performed on cultures, each derived from a single colony (Luria and Delbruck, 1943). For each fluctuation test, nine or 10 cultures were examined in parallel. The mean number of colonies was determined from the four replicas from each individual culture on selective medium and on nonselective medium. The number of distinct events rather than the number of resistant colonies was calculated through application of the “method of the median” (Lea and Coulson, 1949).
Modeling of Kinesin-2 onto KIF1A
The program Insight (Accelrys, Cambridge, MA) was used for modeling on a SGI workstation.
RESULTS
Identifying the Second Motor Subunit of Kinesin-2
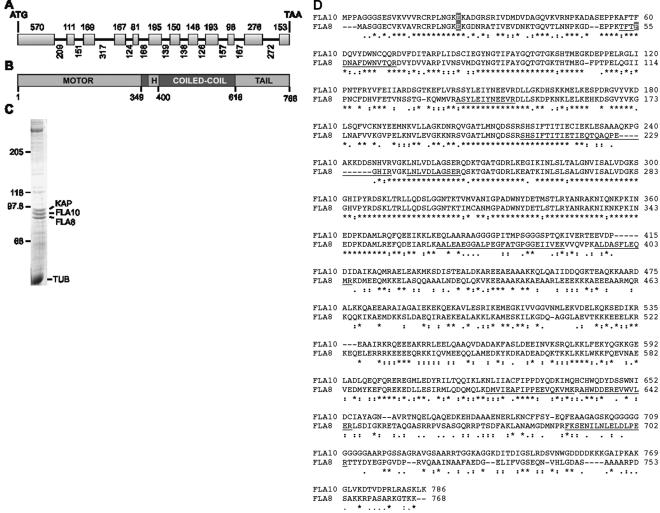
Chlamydomonas kinesin-2 was previously isolated from flagella and shown to be a heterotrimeric complex with two distinct motor subunits, one encoded by the FLA10 gene and a second motor subunit of unknown origin (Cole et al., 1998). As Chlamydomonas expressed sequence tags became available, a sequence closely related to the FLA10 gene was identified and used as a probe to screen libraries to identify both cDNA and genomic BAC clones for subsequent sequencing. cDNA and genomic DNA also were used directly as PCR template to amplify the gene for the purpose of sequencing. This kinesin-2 gene (KRP86) contains 12 exons with 11 introns (Figure 1A). The cDNA was determined to be 2926 base pairs with a 5′-untranslated region (UTR) of 141 base pairs and a 3′-UTR of 478 base pairs. The 2304-base pair open reading frame of KRP86 encodes a 768-amino acid kinesin with a predicted molecular mass of 86,056 Da and a pI of 6.59. The predicted protein was surprisingly similar to Fla10, displaying 46.9% identity throughout the protein and 71.7% identity within the NH2-terminal 360 amino acid motor domain (Figure 1B).
Figure 1.
The FLA8 gene encodes a kinesin-2 motor protein. (A) The FLA8 gene consists of 12 exons separated by 11 introns with the size of each shown. (B) The Fla8 protein is an amino-terminal kinesin predicted to form a 267-amino acid coiled coil domain broken by a 30-amino acid hinge (H). (C) Coomassie Blue-stained SDS-PAGE (7.5% acrylamide) of sucrose density gradient purified kinesin-2 used for MALDI-TOF mass spectrometry analysis. The subunits of 96, 90, and 86 kDa correspond to KAP, Fla10, and Fla8, respectively. TUB, tubulin. (D) Alignment of Fla10 and Fla8 protein sequences. Tryptic peptides identified by mass spectrometry are underlined. Amino acids that are changed in the fla8-1, fla8-2, or fla10-14 alleles are indicated by shading.
To verify that the KRP86 gene encoded the second motor subunit, kinesin-2 was removed from Chlamydomonas flagella by AMPPNP extraction followed by ATP release of the kinesin. The kinesin-2 complex was purified on a 10–25% sucrose gradient, and the three subunits were fractionated by SDS-PAGE as described previously (Figure 1C; Cole et al., 1998). MALDI-TOF mass spectrometry analysis of tryptic peptides of the 86-kDa kinesin-2 subunit generated 10 masses that matched nine predicted tryptic peptides for this kinesin (Figure 1D); one mass was unmatched. One of the nine sequences, LNLVDLAGSER, was not unique in that both Fla10 and the KRP86 protein both contain this conserved sequence. The kinesin-2 subunit with relative mobility of 90 kDa produced nine tryptic masses that matched exactly to five predicted Fla10 tryptic peptides (Figure 1D); four masses were unmatched. The 96-kDa protein generated 11 masses matched exactly eight predicted KAP tryptic peptides; three masses were unmatched (see Materials and Methods).
Identifying the FLA8 Locus
A restriction fragment length polymorphism (RFLP) for the kinesin-2 motor subunit was found previously and mapped to linkage group XII/XIII using meiotic progeny from a cross of a wild-type strain (137 or CC124) with a highly polymorphic strain (CC1952) (Gross et al., 1988). It is tightly linked to RPL9 in 40 progeny (Bowers et al., 2003). No known flagellar mutants mapped to this region. Although Adams et al. (1982) mapped many temperature-sensitive flagellar assembly mutants in Chlamydomonas, many of these were mapped incorrectly. Therefore, the mutant strains fla1and fla8 were chosen to examine because the anterograde-to-retrograde particle frequency ratio in these mutants was similar to the ratio for particles in the fla10-1 mutant flagella (Iomini et al., 2001). In addition, the flagellar IFT content of these mutants was biochemically similar to the patterns observed in the fla10-1 mutant (Cole et al., 1998). Thirty-nine progeny from a cross of fla1 by CC1952 and 45 progeny from a cross of fla8 by CC1952 were examined using the polymorphism used for mapping (see Materials and Methods). There was complete linkage between the temperature-sensitive flagellar assembly defect and the RFLP. As a control, no linkage was observed with the fla2 mutation in 25 progeny. The gene was sequenced from the fla1 strain and a single change was identified. The phenylalanine at position 55 is changed to a serine (F55 to S55) via a T-to-C transition. The gene from the fla8 strain was sequenced and a single change was identified. The glutamic acid at position 21 is changed to a lysine (E21 to K21) via a G-to-A transition. Therefore, we will refer to the KRP86 locus as FLA8; the fla8 allele (CC-1396) will be referred to as fla8-1. The fla1 allele (CC-1389) will be referred to as fla8-2.
Identifying the fla10-14 Mutation
Previously, we have shown that two of these mutations identified as temperature-sensitive flagellar assembly mutants in Chlamydomonas by Adams et al. (1982) were new alleles of fla10 (Lux and Dutcher, 1991). We sequenced the fla10-14 allele and found two changes between it and the published sequence (Walther et al., 1994). The glutamic acid at position 24 is changed to a lysine (E24 to K24) via a G-to-A transition and the leucine at position 196 is changed to a phenylalanine (L196-to-F196) via a C-to-T transition. This glutamic acid is in a similar position to the glutamic acid that is changed in the fla8-1 allele (Figure 1C).
Isolation of Revertants
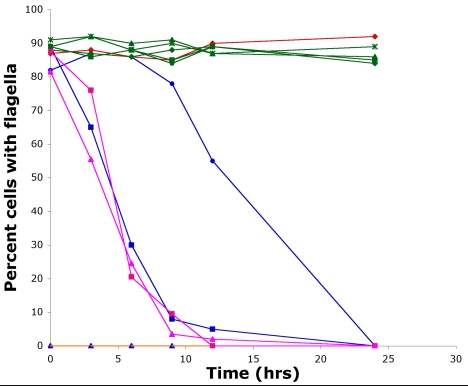
To determine whether these changes were responsible for the temperature-sensitive phenotypes, we mutagenized fla8-1, fla8-2, and fla10-14 cells with UV light and enriched for strains that regained the ability to assemble flagella and thus to swim in liquid medium at 32°C. The results with fla10-14 are described first and then the results with the two fla8 alleles. Nineteen fla10-14 revertants were obtained. Each was crossed by a wild-type strain to determine whether the event was linked or unlinked, and 20–32 tetrads were examined for each revertant. The reversion event in each of the 19 strains was linked to the original mutation. The flagellar phenotypes were determined by counting 200 cells at 21°C and at several time points for up to 24 h after shifting the cultures to 32°C (Figure 2). Cells with the fla10-14 mutation maintain flagella longer than in the fla10-1 allele (Lux and Dutcher, 1991). The cells with fla10-2 allele were aflagellate at both temperatures as reported previously (Matsuura et al., 2002). Each of the revertants tested was indistinguishable from wild-type cells in flagellar number at 21 and 32°C; five representative revertants are shown in Figure 2.
Figure 2.
Flagellar loss of fla8 and fla10 mutant alleles when shifted to the restrictive temperature. Compared with wild-type cells (red diamond), the fla10-1 (blue square), fla10-14 (blue circle), fla8-1 (pink square), and fla8-2 (pink triangle) alleles begin to lose their flagella after 3 h at 32°C, whereas fla10-2 (blue triangle) and fla10-14; fla3 (orange triangle) are aflagellate at all temperatures. The fla10-14 intragenic revertants, R16 (green square), R27 (green diamond), R10 (green circle), R8 (green asterisk), and R13 (green triangle) are identical to wild-type cells at the permissive and restrictive temperature.
To determine whether the mutations (E24K and L196F) were altered in the revertants, restriction fragment length polymorphisms were identified, and PCR fragments were amplified around the two mutations. A 207-base pair PCR product from wild-type cells is not cut by MboII, whereas the product from the E24K mutant is cut. PCR products from two of the 19 revertants were not cut by MboII and were sequenced. One, R27, was a true revertant that restores the sequence K23E24K25. The other revertant, R16, resulted in the sequence K23K24E25, which restores the KE motif, although it is one amino acid further into the protein (Table 1). A 958-base pairs PCR product from wild-type cells is cut three times by PpuM1, whereas PCR product from the L196F mutant is cut only twice. None of the revertants had an alteration at the sequence surrounding F196, based on this restriction enzyme difference. We verified the presence of F196 in four revertants (R8, R10, R13, and R27) by sequencing. This result together with the R27 revertant suggests that the L196F lesion has no detectable phenotypic effect on flagellar assembly. It is likely that the aflagellate phenotype arises from the change at glutamic acid to lysine at amino acid 24. The L196F change is likely to have been introduced by mutagenesis, because it is not present in the 137c parent.
Table 1.
Analysis of intragenic revertants of fla10-14
| Strain name | Amino acids of interest | Restriction fragment length polymorphism | Explanation | ||
|---|---|---|---|---|---|
| E24K25 | L196F | G331S | |||
| Wild-type | E24K25L196N329 | GGAGA | AGGTCCT | GGCCC | |
| G331 | Not cut by MboII | Cut by PpuMI | Cut by Sau96I | ||
| fla10-14 | E24K; F196 | GAAGA | AGGTCTT | GGCCC | Original mutations |
| Cut by MboII | Not cut by PpuMI | Cut by Sau96I | |||
| R27 | K24E; F196 | GGAGA | AGGTCTT | GGCCC | True revertant at position 24 |
| Not cut by MboII | Not cut by PpuMI | Cut by Sau96I | |||
| R16 | E24 K; K25E; F196 | GAAGG | AGGTCTT | GGCCC | Pseudorevertant |
| Not cut by MboII | Not cut by PpuMI | Cut by Sau96I | |||
| R10,R35 R36, R43 | E24K; F196; G331S | GAAGA | AGGTCTT | AGCCC | Pseudorevertant |
| Cut by MboII | Not cut by PpuMI | Not cut by Sau96 I | |||
| R8 | E24K; F196; N329H | GAAGA | AGGTCTT | GGCCC | Pseudorevertant |
| Cut by MboII | Not cut by PpuMI | Cut by Sau96I | |||
| R13 | E24K; F196; N329T | GAAGA | AGGTCTT | GGCCC | Pseudorevertant |
| Cut by MboII | Not cut by PpuMI | Cut by Sau96I | |||
Three of the remaining 17 revertants were picked at random and 7793 base pairs were sequenced to identify other reversion events. Three additional changes were found. Revertant R8 changes N329 to H329 via a transversion of an A to a C, R10 changes G331 to S331 via a transition of a G to an A, and R13 changes N329 to T329 via a transversion of an A to a C. An RFLP in an 894-base pair PCR product is observed with Sau96I in DNA from the G331 to S331 revertant. It cleaves the wild-type product into five bands and the mutant product into four bands. Using this polymorphism, three additional revertants were demonstrated to have the G331-to-S331 event. The remaining 11 revertants have not been sequenced at this time, and the change(s) is not known in these revertants.
Fifteen revertants of fla8-1 and 35 revertants of fla8-2 were obtained. Each was crossed by wild-type cells, and 20–32 tetrads were examined for each revertant. The reversion event in each of the strains was linked to the original mutation and no temperature-sensitive progeny were recovered, which suggests that these are intragenic revertants. The flagellar phenotypes were determined by counting 200 cells at 21°C and at several time points for up to 24 h after shifting the cultures to 32°C. All showed wild-type frequencies of flagella with wild-type lengths (our unpublished data).
The motor domain of the FLA8 gene from each revertant strain was sequenced (Table 2). For fla8-1, six true revertants that changed the serine back to phenylalanine were found. Nine other single or double mutants were found. The most frequent class (n = 6) changes M313 to an isoleucine. For fla8-2, three true revertants were found that changed K21 back to a glutamic acid. Thirty-two of the 35 revertants have a second-site alteration. Twenty-one of the strains change M313. Four change the methionine to a valine and the remaining ones change it to an isoleucine. Eight of the revertants change G314 to a serine, which was altered in three of the fla8-1 revertants and in four of the fla10-14 revertants.
Table 2.
Intragenic revertants of fla8-1 and fla8-2
| Strain | Amino acid change | Codon | Explanation |
|---|---|---|---|
| Wild-type | F55 | ||
| fla8-1 | F55S | TTC to TCC | Original mutation |
| R1, R5, R8, R19, R22, R27 | S55F | TCC to TTC | True revertant |
| R9 | S55Y | TCT to TAT | Pseudorevertant |
| R17 | F55S; N57I | AAT to ATT | Pseudorevertant |
| R29 | F55S; N42H | AAC to CAC | Pseudorevertant |
| R4, R7, R8, R18 | F55S; M313I | ATG to ATA | Pseudorevertant |
| R21, R24 | F55S; M313I; G314S | ATG GGC to ATA AGC | Pseudorevertant |
| fla8-2 | E21K | GAG to AAG | Original mutation |
| R17, R23, R36a | K21E | AAG to GAG | True revertant |
| R10 | E21K; N57Y | AAT to TAT | Pseudorevertant |
| R4, R8d | E21K; H100Y | CAT to TAT | Pseudorevertant |
| R29 | E21K; N312T | AAC to ACC | Pseudorevertant |
| R2, R19, R25, R26 | E21K; M313V | ATG to GTG | Pseudorevertant |
| R8a, R9, R10, R11, R13, R18, R20, R21, R28 | E21K; M313I | ATG to ATA | Pseudorevertant |
| R14, R15, R22, R24, R30, R33, R35 | E21K; M313I | ATG to ATT | Pseudorevertant |
| R16 | E21K; M313I; G314S | ATG GGC to ATA AGC | Pseudorevertant |
| R5, R6, R25, R27, R31, R32, R36b | E21K; G314S | GGC to AGC | Pseudorevertant |
Measuring Chromosome Loss and Mitotic Recombination in Chlamydomonas
The apm1-122 allele confers resistance to the microtubule depolymerizing agent, oryzalin, which binds to α-tubulin (Morrissette et al., 2004). The synthetic lethality of apm1-122 with the fla10 alleles and the dominance of this phenotype suggest that these alleles may interact with microtubules of the mitotic spindle in an aberrant way. We developed an assay for monitoring chromosome loss in Chlamydomonas.
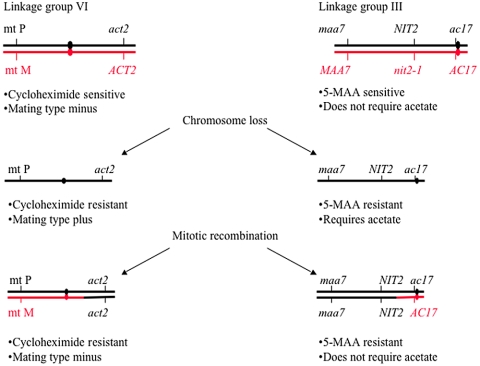
To determine the rate of chromosome loss in Chlamydomonas, we constructed diploid strains that are heterozygous for markers on either linkage groups VI or III. These marked strains allow the detection of chromosome loss compared with mitotic chromosome recombination by using recessive phenotypic markers on both arms of a linkage group. On linkage group VI, one arm was marked with the recessive act2 allele that confers resistance to cycloheximide and the other arm was marked by the mating-type locus. Cells heterozygous for the plus and minus mating-type alleles show a minus phenotype and cells that have lost the minus alleles show the plus mating-type phenotype. Chromosome loss is detected by resistance to cycloheximide and a mating-type plus phenotype. Mitotic recombination produces cycloheximide resistance and a mating-type minus phenotype. On linkage group III, one arm is marked with the NIT2 and maa7 alleles and the other arm is marked with the ac17 allele. The other homolog has the wild-type dominant alleles for MAA7 and AC17 and the recessive allele, nit2 (Figure 3). Loss of one homolog can be detected by cells that show resistance to 5-MAA, a suicide substrate for tryptophan synthetase, which is the product of the MAA7 gene (Palombella and Dutcher, 1998). These cells are able to grow on nitrite medium, but they require acetate. Mitotic recombination on this arm would result in cells that are resistant to 5-MAA but do not require acetate.
Figure 3.
Determining chromosome loss using markers on linkage group III and VI. An increased frequency of chromosome loss results in cells that are cycloheximide-resistant or mating-type plus- and 5-MAA–resistant and requires acetate for growth. Cells that have undergone mitotic recombination will be cyclohexamide resistant and mating-type minus and 5-MAA resistant and do not require acetate for growth.
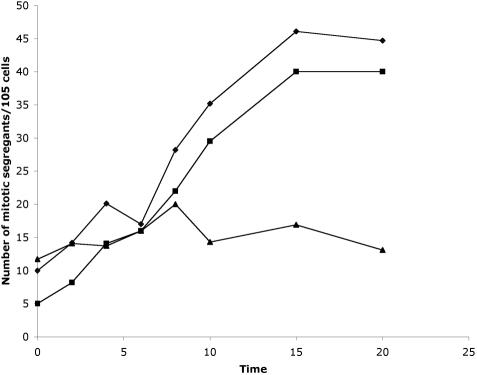
To verify that we could detect chromosome loss and mitotic recombination, we assayed untreated cells as well as cells treated with various agents. These agents include gamma irradiation, UV light, and addition of colchicine, a microtubule-destabilizing agent. Gamma irradiation at 500 mrem for 20 min increased the number of cycloheximide-resistant colonies from 10 per 105 to 45 per 105 cells (Figure 4). At the 20-min time point, 90% of the cycloheximide-resistant colonies showed a mating-type plus phenotype, which is consistent with chromosome loss. Given the starting frequency of chromosome loss events, it is likely that the increase in cells resistant to cycloheximide arises primarily from chromosome loss rather than mitotic recombination.
Figure 4.
Effect of UV irradiation on mitotic segregation of linkage group VI after various exposure times (in minutes) to UV irradiation. Mitotic recombination and chromosome loss of linkage group VI (diamond), chromosome loss of linkage group VI (square), and mitotic recombination of the left arm of linkage group VI (triangle).
UV irradiation had little effect on the rates of either mitotic recombination or chromosome loss. Treatment with colchicine resulted in only a twofold increase in cycloheximide resistance after treatment at 5 mM for 24 h, but the viability of the cultures decreased to <10% of the initial viability (our unpublished data). Wild-type cells show a similar rate of loss using linkage group III markers. Chromosome loss for linkage group III occurred at a rate of 6.9 per 105 cells (n = 6).
Mitotic Segregation in fla10 Alleles
The rate of mitotic segregation in three fla10 alleles that fail to assemble flagella as well as four of the intragenic revertant strains was determined using linkage group VI for the assay (Table 3). In wild-type diploid strains, mitotic recombination occurs with a rate of 5.3 events per 105 cells at 21°C and with a rate of 4.8 events per 105 cells at 32°C. Chromosome loss is slightly increased by temperature; the rates are 3.9 and 6.3 events per 105 cells at 21 and 32°C, respectively. Mitotic segregation in the two temperature-sensitive alleles, fla10-1 and fla10-14, was monitored both in heterozygous and homozygous diploid strains. Both alleles show an increased rate of loss compared with the wild-type cells in both heterozygous and homozygous strains at both temperatures. Thus, the chromosome loss phenotype is dominant. It is also dosage sensitive for both alleles as the homozygous strains show greater rate increases than the heterozygous strains (Table 3). The incubation at 32°C increases the loss for both alleles.
Table 3.
Rates of mitotic recombination and chromosome loss in wild-type, fla8, and fla10 diploid strains
| No. trials
|
21°C
|
32°C
|
||||
|---|---|---|---|---|---|---|
| Genotype | 21° | 32° | MR | CL | MR | CL |
| FLA10+/FLA10+; FLA8+/FLA8+ | 12 | 10 | 5.3 | 3.9 | 4.8 | 6.3 |
| FLA10/fla10-1 | 4 | 4 | 7.8 | 11.5 | 8.3 | 12.7 |
| fla10-1/fla10-1 | 2 | 8 | 4.9 | 19.2 | 7.8 | 21.5 |
| FLA10/fla10-14 | 3 | 3 | 6.1 | 13.8 | 5.7 | 15.9 |
| fla10-14/fla10-14 | 3 | 3 | 5.3 | 27.9 | 6.0 | 33.1 |
| FLA10/fla10-2 | 2 | 2 | 5.6 | 4.5 | 6.7 | 6.2 |
| fla10-2/fla10-2 | 2 | 2 | 5.9 | 6.3 | 6.3 | 5.9 |
| fla10-2/fla10-14 | 3 | 3 | 5.9 | 12.9 | 5.4 | 14.7 |
| fla10-14R16/fla10-14R16 | 2 | 2 | 4.9 | 4.6 | 4.3 | 5.7 |
| fla10-14R27/fla10-14R27 | 2 | 2 | 3.7 | 4.6 | 4.6 | 6.2 |
| fla10-14R10/fla10-14R10 | 2 | 2 | 4.6 | 5.1 | 3.6 | 4.9 |
| fla10-14R8/fla10-14R8 | 2 | 2 | 5.4 | 5.2 | 6.1 | 5.7 |
| fla10-14R13/fla10-14R13 | 2 | 2 | 5.2 | 5.9 | 6.3 | 6.0 |
| fla10-14/fla10-14; fla3-1/fla3-1 | 2 | 2 | 5.2 | 9.3 | 5.7 | 5.4 |
| fla10-14R16/fla10-14R16; fla3-1/fla3-1 | 2 | 2 | 4.9 | 5.5 | 5.3 | 4.6 |
| fla8-1/fla8-1 | 2 | 2 | 4.3 | 20.1 | 5.2 | 22.6 |
| fla8-2/fla8-2 | 2 | 2 | 4.7 | 17.5 | 3.9 | 18.4 |
| fla8-1/fla8-2 | 2 | 2 | 3.6 | 14.4 | 4.3 | 13.6 |
| fla8-1/FLA8 | 2 | 2 | 5.3 | 11.8 | 5.2 | 14.7 |
| fla8-2/FLA8 | 2 | 2 | 5.7 | 13.6 | 4.2 | 15.9 |
| fla8-1R17fla8-1R17 | 2 | 2 | 4.5 | 4.9 | 5.0 | 4.9 |
| fla8-1R29/fla8-2R29 | 2 | 2 | 4.7 | 5.3 | 4.5 | 4.1 |
| fla8-2R4/fla8-2R4 | 2 | 2 | 5.6 | 5.9 | 4.5 | 7.1 |
| fla8-2R14/fla8R14 | 2 | 2 | 3.3 | 6.0 | 3.7 | 6.9 |
| fla8-2R16/fla8R16 | 2 | 2 | 3.9 | 4.8 | 4.3 | 5.7 |
CL, chromosome loss; MR, mitotic recombination.
To ask whether the dominance results from a decrease in Fla10 gene product, the behavior of the fla10-2 strain, which has a null allele (Matsuura et al., 2002), was examined in both heterozygous and homozygous diploid strains. This allele shows no increase in chromosome loss at either temperature (Table 3). Furthermore, a diploid strain that is heteroallelic for the fla10-2 and fla10-14 alleles is similar to the fla10-14/FLA10 strain. Thus, the increased chromosome loss arises from an acquired feature of the mutant alleles.
Five different intragenic revertants were analyzed in homozygous diploid strains. None of the revertants show the loss phenotype (Table 3). Thus, both the flagellar assembly and the chromosome loss phenotypes were reverted by the second-site events. In addition, we monitored the rate of chromosome loss in the fla3-1 allele, which encodes KAP. This allele has no effect on chromosome segregation or mitotic recombination (Table 3).
Double mutants between fla10-14 and mutations in other kinesin-2 subunits were constructed. The fla10-14; fla8-1, fla10-14; fla8-2, and fla10-14; fla3–1 double mutants were aflagellate at 21 and 32°C (Figure 2). The double mutant fla10-14; fla3-1, shows no increase in chromosome loss compared with the wild-type strains (Table 3).
Mitotic Segregation in fla8 Alleles
We monitored chromosome loss and mitotic recombination in the two fla8 alleles to ask whether they resulted in increased rates of chromosome loss similar to the fla10 alleles. Both alleles show increased rates of loss (Table 3). We examined three intragenic revertants for both alleles and found they restored wild-type levels of chromosome loss (Table 3).
Monitoring Cytokinesis in fla8 and fla10 Cells
We monitored the fidelity of cytokinesis in wild-type, fla8-1, fla8-2, and fla10-14 cells by examining 21 and 32°C grown cells for the presence of binucleated cells, as determined by staining with the DNA-specific dye, 4,6-diamidino-2-phenylindole. No increase was observed (n = 200).
DISCUSSION
Identification of a Temperature-sensitive Flagellar Assembly Mutation in the Second Motor Subunit of Kinesin-2
The FLA10 gene encodes one of the two motor subunits that comprise the anterograde motor kinesin-2 (Walther et al., 1994; Kozminski et al., 1995). We have identified the second motor subunit by its homology to the Fla10 subunit among expressed sequence tags and then by sequencing cDNA and genomic DNA. Mass spectrometry of isolated kinesin-2 confirmed the identification. This subunit is encoded by the FLA8 gene in Chlamydomonas and sequencing of DNA from the fla8-1 and fla1 (renamed fla8-2) mutant strains revealed single amino acid changes in each. A phenylalanine is mutated to a serine (F55S) in fla8-1 and a glutamic acid is changed to a lysine (E21K) in fla8-2. These results agree with the suggestion of Iomini et al. (2001) that the fla8 and fla1 mutants are defective in assembly and transport of anterograde particles but that their retrograde particle assembly and transport are comparable with wild-type cells. Six additional temperature-sensitive alleles have been identified at the FLA8 locus, but the mutational changes have not been identified at this time (originally designated as fla1-2 to fla1-7) (Iomini et al., 2001). Unlike most of the loci identified with temperature-sensitive flagellar assembly defects (Adams et al., 1982; Iomini et al., 2001), multiple alleles are available for FLA8 (8 alleles) and for FLA10 (4 alleles). With the recent finding that the FLA3 locus encodes the KAP subunit (Mueller et al., 2005), conditional alleles are now available for the three subunits of kinesin-2 in Chlamydomonas. Double mutants between different temperature-sensitive alleles (fla10; fla8, fla10; fla3, and fla8; fla3) are unable to assemble flagella at any temperature, which suggests that the partial function of any two subunits result in the inability to assemble flagella.
Characterization of Intragenic Revertants of the fla8 and fla10 Mutants Reveals an Important Domain
In addition to sequencing the fla8 alleles, we identified two mutations in a second temperature-sensitive fla10 allele (fla10-14); a glutamic acid at position 24 is changed to a lysine (E24K) and a leucine at position 196 is changed to a phenylalanine (L196F). Both fla8-2 and fla10-14 have the same glutamic acid-to-lysine change, which suggests that this residue has an important function. This hypothesis is strengthened by characterization of the intragenic revertants of the three mutant alleles fla10-14, fla8-1, and fla8-2. The sequencing of independently isolated intragenic revertants of the fla10-14 allele revealed changes in amino acids 329 and 331. Because of a short insertion in FLA10 (Figure 1D), the amino acid sequences of FLA10 and FLA8 are offset; these amino acids of Fla10 correspond to amino acids 312 and 314 in Fla8. In the fla8-1 and fla8-2 intragenic revertants, the same or similar mutations are observed in the revertants. At amino acid 312, one fla8-2 revertant is changed from an asparagine to a threonine. Many fla8-1 and fla8-2 revertants change amino acid 313 from a methionine to either an isoleucine or a valine. Seven fla8-2 revertants have a glycine-to-serine change at amino acid 314. Three revertants have a dinucleotide change (GG to AA) that change positions 313 and 314; the methionine is mutated to an isoleucine and the glycine is changed to a serine. The fla10-1 mutation changes an asparagine residue to a lysine (N329K) (Walther et al., 1994), whereas changes to another polar amino acid revert the defects in two of the fla10-14 intragenic revertants (N329H) and in one of the fla8-2 intragenic revertants (N312T).
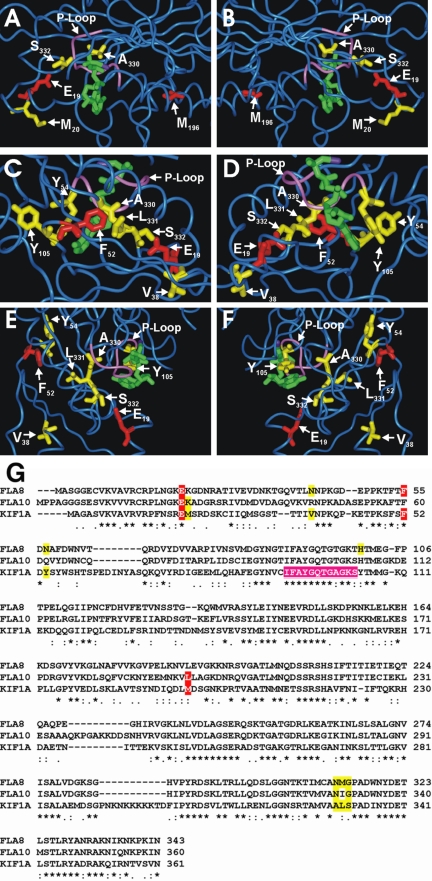
To ask whether the amino acids altered in the revertants lie near the original mutations, the Fla8 and Fla10 motor domain amino acid sequences were aligned with the KIF1A motor to allow the identification of amino acids that corresponded to the sites of interest in the fla10-14, fla8-1, and fla8-2 mutant and revertant proteins (Figure 5G). When the mutations of the intragenic revertants are positioned onto the structure (Nitta et al., 2004), most are found in proximity to the original mutations (Figure 5, A–F). One notable exception is the fla10-14 mutation, L196F, which corresponds to M196 in KIF1A. (Figure 5, A and B). Because reversion of the other mutation (E24K24E24) in the fla10-14 strain results in wild-type behavior, it seems that the L196F mutation does not affect activity of Fla10. These results are consistent with the conclusion that the L196F change may be a silent polymorphism.
Figure 5.
Location of amino acids altered in the fla8 and fla10 mutant and revertant alleles as modeled onto the crystal structure of KIF1A with AMPPNP (green) bound (PDB 1VFV). The P-loop is shown in purple. (A and B) Fla10 E24 and L196 correspond to KIF1A E19 and M196, respectively, and are shown in red. Revertant fla10-14 amino acids K25, N329, and G331 correspond to KIF1A M20, A330, and S332, respectively, and are shown in yellow. (C–F) Fla8 amino acids E21 and F55 correspond to KIF1A E19 and F52, respectively, and are shown in red. Revertant fla8-1 and fla8-2 amino acids N42, N57, H100, N312, M313, and G314 correspond to KIF1A V38, Y54, Y105, A330, L331, and S332, respectively, and are shown in yellow. (G) Alignment of Fla8, Fla10, and KIF1A in the regions of interest. Red shading indicates mutations in one of the kinesin-2 motor subunits, yellow shading indicates amino acids that are altered in the revertants, and purple shading indicates the P-loop residues.
Although KIF5 has the highest similarity with kinesin-2, KIF1A was chosen because it was crystallized with AMP-PNP and the positions of the nucleotide relative to the reversion events could be examined. The kinesin-2 mutations are not in the regions that change the interaction with the microtubule, but they are very close to the bound nucleotide and the strongly conserved P-loop (Figure 5, A–F). This suggests that reversion may result from a stabilization of the kinesin ATPase. The idea that these mutant kinesins may have reduced activity is supported by the observation that, even at the permissive temperature, the rate of anterograde IFT in fla8-2 strains is slower than the rate of anterograde IFT observed in wild-type strains (Iomini et al., 2001). Because the fla10-14 strain carries the same mutation as in fla8-2, we predict that anterograde IFT will move more slowly than in wild-type flagella. The anterograde IFT rate in the fla8-1 strain is close to wild type at the permissive temperature (Iomini et al., 2001). It is possible, however, that the rates of IFT in fla8-1, fla8-2, and fla10-14 could all dramatically decrease upon shifting to the restrictive temperature of 32°C.
Interestingly, the aromatic ring of Y105 of KIF1A is aligned with the nucleotide base so that these two are considered to be stacked (Figure 5, D and F). This is notable, in part, because two revertants of fla8-2, R4 and R8d, were rescued when a C-to-T transition changed the corresponding histidine at position 100 (H100) to a tyrosine. The N (M/I) G motif (A L S in KIF1A) is particularly close to the P-loop that wraps around the phosphate groups of the bound nucleotide (Figure 5). Without solving the motor domain structure of Fla8 or Fla10, we cannot be certain how the original and mutated amino acids might interact with the P-loop. However, it is tempting to speculate that this motif may be important for motor activity.
To determine whether these mutated residues are conserved in various orthologues or whether they are specific to the kinesin-2 subunits, BLAST searches were performed. The glutamic acid mutated to lysine in both the fla10-14 and fla8-2 mutants is conserved in all kinesin-2 subunits (n = 20). The phenylalanine, which is mutated in the fla8-1 mutant, is conserved only in kinesin-2 subunits with one conservative change to tyrosine in chicken. The N (M/I) G motif in FLA8 and FLA10 is not as well conserved among kinesins. Although the majority of the residues are identical in the kinesin-2 subunits, there are a few exceptions including Tetrahymena, which has an alanine, isoleucine, and serine at these positions. There is little sequence conservation of these residues in other kinesin family members. As such, it seems as though the mutated residues are specific to the kinesin-2 subunits and reinforce the hypothesis that they are critical to its proper function.
The fla8 and fla10 Mutant Alleles Produce Proteins with Aberrant Function
It was recently shown that overexpression of the KIF3B tail results in cytokinesis defects in mammalian cells (Fan and Beck, 2004). To determine whether the fla8 and fla10 alleles showed this phenotype, we assayed for the presence of binucleated cells. There is no cytokinesis defect observed in the fla10-14, fla8-1, and fla8-2 alleles; however, there is a small, but significant, increased frequency of chromosome loss in both homozygous and heterozygous diploids, which also was observed in the fla10-1 allele. These results indicate that the chromosome loss phenotype is dominant unlike the flagellar assembly defect, which is recessive. This phenotype was not observed in the fla10-2 allele, which is a null allele (Matsuura et al., 2002). Diploid strains heteroallelic for the fla10-2 and fla10-14 alleles also show an increased frequency of chromosome loss. Together, these data suggest that the fla10-1, fla10-14, fla8-1 and fla8-2 alleles show a gain of function phenotype and the mutant proteins have acquired an aberrant or toxic function(s). Previous studies showed that the Fla10 protein is found at the mitotic spindle poles as well as at the basal bodies during interphase in Chlamydomonas (Vashishtha et al., 1996; Deane et al., 2001). We hypothesize that these mutant kinesins could act at the spindle poles to alter centrosome separation or attachment. Alternatively, it is possible that one of the IFT polypeptides is required for chromosome segregation and the IFT polypeptide is mislocalized by the mutant kinesin-2.
Intriguingly, diploids homozygous for both fla10-14 and fla3-1 do not have a chromosome loss phenotype, which suggests that the phenotype relies on a functional Fla3 protein. Mueller and colleagues (2005) showed that the Fla10 protein does not localize to the basal body region properly in fla3 mutant cells. In sea urchins, it has been reported that the KAP subunit translocates into the nucleus before mitosis (Morris et al., 2004). In Chlamydomonas, the KAP subunit localizes to the basal body region in Chlamydomonas in wild-type interphase and mitotic cells, but it was not reported to be in the nucleus (Mueller el al., 2005). Regardless of its localization, the mutant motor subunits require wild-type Fla3 to affect chromosome segregation. The chromosome loss phenotype was not present in the fla10-14, fla8-1, or fla8-2 intragenic revertants, which suggests that these changes removed the toxic or aberrant interactions caused by the original mutations in the fla10 and fla8 alleles.
This aberrant function of mutant Fla10 protein could explain the genetic interaction between apm1-122 and various fla10 alleles that causes a synthetic cold-sensitive cell division phenotype (fla10-1) or a slow growth phenotype (fla10-14) at 21°C (Lux and Dutcher, 1991). Although the product of the APM1 gene is unknown, it may encode a microtubule-associated protein needed for spindle function.
Acknowledgments
We thank Dr. Gary Daughdrill for assistance with the modeling. Alex Edwards and Rona Robinson-Hill helped with the sequencing of the fla10-14 allele. They were supported by funds from Washington University and the Howard Hughes Medical Institute Award to Dr. Sarah Elgin. We thank Samantha Reed and Julie Sahalie for technical assistance cloning the FLA8 gene. They were supported by funds from the University of Idaho. This work was supported by National Institutes of Health Grants GM-32843 (to S.K.D.) and GM-61920 (to D.G.C.).
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05-05-0404) on June 8, 2005.
References
- Adams, G. M., Huang, B., Piperno, G., and Luck, D. J. (1982). Temperature-sensitive assembly-defective flagella mutants of Chlamydomonas reinhardtii. Genetics 100, 579-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asamizu, E., Miura, K., Kucho, K., Inoue, Y., Fukuzawa, H., Ohyama, K., Nakamura, Y., and Tabata, S. (2000). Generation of expressed sequence tags from low-CO2 and high-CO2 adapted cells of Chlamydomonas reinhardtii. DNA Res. 7, 305-307. [DOI] [PubMed] [Google Scholar]
- Asamizu, E., Nakamura, Y., Sato, S., Fukuzawa, H., and Tabata, S. (1999). A large scale structural analysis of cDNAs in a unicellular green alga, Chlamydomonas reinhardtii. I. Generation of 3433 non-redundant expressed sequence tags. DNA Res. 6, 369-373. [DOI] [PubMed] [Google Scholar]
- Betley, J. N., Heinrich, B., Vernos, I., Sardet, C., Prodon, F., and Deshler, J. O. (2004). Kinesin II mediates Vg1 mRNA transport in Xenopus oocytes. Curr. Biol. 14, 219-224. [DOI] [PubMed] [Google Scholar]
- Bowers, A. K., Keller, J. A., and Dutcher, S. K. (2003). Molecular markers for rapidly identifying candidate genes in Chlamydomonas reinhardtii. ERY1 and ERY2 encode chloroplast ribosomal proteins. Genetics 164, 1345-1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, J. M., Marsala, C., Kosoy, R., and Gaertig, J. (1999). Kinesin-II is preferentially targeted to assembling cilia and is required for ciliogenesis and normal cytokinesis in Tetrahymena. Mol. Biol. Cell 10, 3081-3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole, D. G., Chinn, S. W., Wedaman, K. P., Hall, K., Vuong, T., and Scholey, J. M. (1993). Novel heterotrimeric kinesin-related protein purified from sea urchin eggs. Nature 366, 268-270. [DOI] [PubMed] [Google Scholar]
- Cole, D. G., Diener, D. R., Himelblau, A. L., Beech, P. L., Fuster, J. C., and Rosenbaum, J. L. (1998). Chlamydomonas kinesin-II-dependent intraflagellar transport (IFT): IFT particles contain proteins required for ciliary assembly in Caenorhabditis elegans sensory neurons. J. Cell Biol. 141, 993-1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deacon, S. W., Serpinskaya, A. S., Vaughan, P. S., Lopez Fanarraga, M., Vernos, I., Vaughan, K. T., and Gelfand, V. I. (2003). Dynactin is required for bidirectional organelle transport. J. Cell Biol. 160, 297-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deane, J. A., Cole, D. G., Seeley, E. S., Diener, D. R., and Rosenbaum, J. L. (2001). Localization of intraflagellar transport protein IFT52 identifies basal body transitional fibers as docking sites for IFT particles. Curr. Biol. 11, 1586-1590. [DOI] [PubMed] [Google Scholar]
- Dutcher, S. K. (1995a). Mating and tetrad analysis in Chlamydomonas reinhardtii. Methods Cell Biol. 47, 531-540. [DOI] [PubMed] [Google Scholar]
- Dutcher, S. K. (1995b). Purification of basal bodies and basal body complexes from Chlamydomonas reinhardtii. Methods Cell Biol. 47, 323-334. [DOI] [PubMed] [Google Scholar]
- Fan, J., and Beck, K. A. (2004). A role for the spectrin superfamily member Syne-1 and kinesin II in cytokinesis. J. Cell Sci. 117, 619-629. [DOI] [PubMed] [Google Scholar]
- Gross, C. H., Ranum, L. P., and Lefebvre, P. A. (1988). Extensive restriction fragment length polymorphisms in a new isolate of Chlamydomonas reinhardtii. Curr. Genet. 13, 503-508. [DOI] [PubMed] [Google Scholar]
- Han, Y. G., Kwok, B. H., and Kernan, M. J. (2003). Intraflagellar transport is required in Drosophila to differentiate sensory cilia but not sperm. Curr. Biol. 13, 1679-1686. [DOI] [PubMed] [Google Scholar]
- Harris, E. H. (1989). The Chlamydonas Sourcebook: A Comprehensive Guide to Biology and Laboratory Use, San Diego: Academic Press. [DOI] [PubMed]
- Henson, J. H., Cole, D. G., Terasaki, M., Rashid, D., and Scholey, J. M. (1995). Immunolocalization of the heterotrimeric kinesin related protein KRP(85/95) in the mitotic apparatus of sea urchin embryos. Dev. Biol. 171, 182-194. [DOI] [PubMed] [Google Scholar]
- Holmes, J. A., and Dutcher, S. K. (1989). Cellular asymmetry in Chlamydomonas reinhardtii. J. Cell Sci. 94, 273-285. [DOI] [PubMed] [Google Scholar]
- Huang, B., Rifkin, M. R., and Luck, D. J. (1977). Temperature-sensitive mutations affecting flagellar assembly and function in Chlamydomonas reinhardtii. J. Cell Biol. 72, 67-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iomini, C., Babaev-Khaimov, V., Sassaroli, M., and Piperno, G. (2001). Protein particles in Chlamydomonas flagella undergo a transport cycle consisting of four phases. J. Cell Biol. 153, 13-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James, S. W., Ranum, L. P., Silflow, C. D., and Lefebvre, P. A. (1988). Mutants resistant to anti-microtubule herbicides map to a locus on the uni linkage group in Chlamydomonas reinhardtii. Genetics 118, 141-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutoulis, A., Pazour, G. J., Wilkerson, C. G., Inaba, K., Sheng, H., Takada, S., and Witman, G. B. (1997). The Chlamydomonas reinhardtii ODA3 gene encodes a protein of the outer dynein arm docking complex. J. Cell Biol. 137, 1069-1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozminski, K. G., Beech, P. L., and Rosenbaum, J. L. (1995). The Chlamydomonas kinesin-like protein FLA10 is involved in motility associated with the flagellar membrane. J. Cell Biol. 131, 1517-1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bot, N., Antony, C., White, J., Karsenti, E., and Vernos, I. (1998). Role of xklp3, a subunit of the Xenopus kinesin II heterotrimeric complex, in membrane transport between the endoplasmic reticulum and the Golgi apparatus. J. Cell Biol. 143, 1559-1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lea, D. E., and Coulson, C. A. (1949). The distribution of numbers of mutants in bacterial populations. J. Genet. 49, 264-285. [DOI] [PubMed] [Google Scholar]
- Luria, S. E., and Delbruck, M. (1943). Mutations of bacteria from virus sensitivity to virus resistance. Genetics 28, 491-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lux, F. G., 3rd, and Dutcher, S. K. (1991). Genetic interactions at the FLA10 locus: suppressors and synthetic phenotypes that affect the cell cycle and flagellar function in Chlamydomonas reinhardtii. Genetics 128, 549-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura, K., Lefebvre, P. A., Kamiya, R., and Hirono, M. (2002). Kinesin-II is not essential for mitosis and cell growth in Chlamydomonas. Cell Motil. Cytoskeleton 52, 195-201. [DOI] [PubMed] [Google Scholar]
- McGrath, J., and Brueckner, M. (2003). Cilia are at the heart of vertebrate left-right asymmetry. Curr. Opin. Genet. Dev. 13, 385-392. [DOI] [PubMed] [Google Scholar]
- Morris, R. L., English, C. N., Lou, J. E., Dufort, F. J., Nordberg, J., Terasaki, M., and Hinkle, B. (2004). Redistribution of the kinesin-II subunit KAP from cilia to nuclei during the mitotic and ciliogenic cycles in sea urchin embryos. Dev. Biol. 274, 56-69. [DOI] [PubMed] [Google Scholar]
- Morris, R. L., and Scholey, J. M. (1997). Heterotrimeric kinesin-II is required for the assembly of motile 9+2 ciliary axonemes on sea urchin embryos. J. Cell Biol. 138, 1009-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrissette, N. S., Mitra, A., Sept, D., and Sibley, L. D. (2004). Dinitroanilines bind alpha-tubulin to disrupt microtubules. Mol. Biol. Cell 15, 1960-1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller, J., Perrone, C. A., Bower, R., Cole, D. G., and Porter, M. E. (2005). The FLA3 KAP subunit is required for localization of kinesin-2 to the site of flagellar assembly and processive anterograde intraflagellar transport. Mol. Biol. Cell 16, 1341-1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitta, R., Kikkawa, M., Okada, Y., and Hirokawa, N. (2004). KIF1A alternately uses two loops t bind to microtubules. Science 305, 678-683. [DOI] [PubMed] [Google Scholar]
- Nishimura, T., Kato, K., Yamaguchi, T., Fukata, Y., Ohno, S., and Kaibuchi, K. (2004). Role of the PAR-3-KIF3 complex in the establishment of neuronal polarity. Nat. Cell Biol. 6, 328-334. [DOI] [PubMed] [Google Scholar]
- Palombella, A. L., and Dutcher, S. K. (1998). Identification of the gene encoding the tryptophan synthase beta-subunit from Chlamydomonas reinhardtii. Plant Physiol. 117, 455-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour, G. J., Dickert, B. L., and Witman, G. B. (1999). The DHC1b (DHC2) isoform of cytoplasmic dynein is required for flagellar assembly. J. Cell Biol. 144, 473-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pflanz, R., Peter, A., Schafer, U., and Jackle, H. (2004). Follicle separation during Drosophila oogenesis requires the activity of the Kinesin II-associated polypeptide Kap in germline cells. EMBO Rep. 5, 510-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter, M. E., Bower, R., Knott, J. A., Byrd, P., and Dentler, W. (1999). Cytoplasmic dynein heavy chain 1b is required for flagellar assembly in Chlamydomonas. Mol. Biol. Cell 10, 693-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preble, A. M., Giddings, T. H., Jr., and Dutcher, S. K. (2001). Extragenic bypass suppressors of mutations in the essential gene BLD2 promote assembly of basal bodies with abnormal microtubules in Chlamydomonas reinhardtii. Genetics 157, 163-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray, K., Perez, S. E., Yang, Z., Xu, J., Ritchings, B. W., Steller, H., and Goldstein, L. S. (1999). Kinesin-II is required for axonal transport of choline acetyltransferase in Drosophila. J. Cell Biol. 147, 507-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarpal, R., Todi, S. V., Sivan-Loukianova, E., Shirolikar, S., Subramanian, N., Raff, E. C., Erickson, J. W., Ray, K., and Eberl, D. F. (2003). Drosophila KAP interacts with the kinesin II motor subunit KLP64D to assemble chordotonal sensory cilia, but not sperm tails. Curr. Biol. 13, 1687-1696. [DOI] [PubMed] [Google Scholar]
- Shakir, M. A., Fukushige, T., Yasuda, H., Miwa, J., and Siddiqui, S. S. (1993). C. elegans osm-3 gene mediating osmotic avoidance behaviour encodes a kinesin-like protein. Neuroreport 4, 891-894. [DOI] [PubMed] [Google Scholar]
- Snell, W. J., Pan, J., and Wang, Q. (2004). Cilia and flagella revealed: from flagellar assembly in Chlamydomonas to human obesity disorders. Cell 117, 693-697. [DOI] [PubMed] [Google Scholar]
- Tuma, M. C., Zill, A., Le Bot, N., Vernos, I., and Gelfand, V. (1998). Heterotrimeric kinesin II is the microtubule motor protein responsible for pigment dispersion in Xenopus melanophores. J. Cell Biol. 143, 1547-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vashishtha, M., Walther, Z., and Hall, J. L. (1996). The kinesin-homologous protein encoded by the Chlamydomonas FLA10 gene is associated with basal bodies and centrioles. J. Cell Sci. 109, 541-549. [DOI] [PubMed] [Google Scholar]
- Walther, Z., Vashishtha, M., and Hall, J. L. (1994). The Chlamydomonas FLA10 gene encodes a novel kinesin-homologous protein. J. Cell Biol. 126, 175-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedaman, K. P., Meyer, D. W., Rashid, D. J., Cole, D. G., and Scholey, J. M. (1996). Sequence and submolecular localization of the 115-kD accessory subunit of the heterotrimeric kinesin-II (KRP85/95) complex. J. Cell Biol. 132, 371-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner, R., and Miergenhagen, D. (1998). Mating type determination of Chlamydomonas reinhardtii by PCR. Plant Mol. Biol. Rep. 16, 295-299. [Google Scholar]
- Witman, G. B., Carlson, K., Berliner, J., and Rosenbaum, J. L. (1972). Chlamydonas flagella. I. Isolation and electrophoretic analysis of microtubules, matrix, membranes, and mastigonemes. J. Cell Biol. 54, 507-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki, H., Nakata, T., Okada, Y., and Hirokawa, N. (1995). KIF3A/B: a heterodimeric kinesin superfamily protein that works as a microtubule plus end-directed motor for membrane organelle transport. J. Cell Biol. 130, 1387-1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Z., Roberts, E. A., and Goldstein, L. S. (2001). Functional analysis of mouse kinesin motor Kif3C. Mol. Cell. Biol. 21, 5306-5311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Q., Taulman, P. D., and Yoder, B. K. (2004). Cystic kidney diseases: all roads lead to the cilium. Physiology 19, 225-230. [DOI] [PubMed] [Google Scholar]