Abstract
Rab GTPases and SNARE fusion proteins direct cargo trafficking through the exocytic and endocytic pathways of eukaryotic cells. We have used steady state mRNA expression profiling and computational hierarchical clustering methods to generate a global overview of the distribution of Rabs, SNAREs, and coat machinery components, as well as their respective adaptors, effectors, and regulators in 79 human and 61 mouse nonredundant tissues. We now show that this systems biology approach can be used to define building blocks for membrane trafficking based on Rab-centric protein activity hubs. These Rab-regulated hubs provide a framework for an integrated coding system, the membrome network, which regulates the dynamics of the specialized membrane architecture of differentiated cells. The distribution of Rab-regulated hubs illustrates a number of facets that guides the overall organization of subcellular compartments of cells and tissues through the activity of dynamic protein interaction networks. An interactive website for exploring datasets comprising components of the Rab-regulated hubs that define the membrome of different cell and organ systems in both human and mouse is available at http://www.membrome.org/.
INTRODUCTION
Understanding the molecular basis for the organization of the exocytic and endocytic membrane trafficking pathways in the eukaryotic cell remains a formidable challenge. The foundation of these pathways is the lipid bilayer that separates different subcellular compartments, their distinguishing features encoded by phospholipid composition, and unique sets of integral and peripheral membrane proteins. By harnessing and regulating the fundamental processes of membrane fission and fusion through the action of protein complexes, the lipid bilayer can be exploited to produce a variety of distinct subcellular compartments with unique chemical environments that play essential roles in cell and organ function. Moreover, it is now evident that these subcellular compartments are dynamic structures in continuous and specific communication through carrier vesicles and tubules that mobilize cargo to specific destinations. They can be disassembled and reassembled in a remarkably facile manner in response to cell signaling pathways, mitosis, or by simple chemical perturbants.
Implicit in these dynamic pathways is the need to systematically and reversibly regulate protein interactions. Although traditional phylogenetic analyses provided significant insights into the diversity of components that direct membrane traffic (Pereira-Leal and Seabra, 2000; Chen and Scheller, 2001; Pereira-Leal and Seabra, 2001), our understanding of the basic cellular building blocks that organize this diversity into contiguous pathways is still fragmentary. Reductionist approaches using biochemical and molecular tools also provide important insights into specific steps of a pathway. However, understanding the global interconnectivities of complex biological pathways, such as cargo trafficking, will require new approaches utilizing modern computational tools to organize cell biological data in a manner that provides a more integrated systems biology view.
It is now well established that cargo movement between subcellular compartments involves transport containers whose formation is directed by evolutionary conserved coat complexes. These include the coat protein complex II (COPII) involved in endoplasmic reticulum (ER) export (Antonny and Schekman, 2001; Barlowe, 2003) and the coat protein complex I (COPI; Nickel et al., 2002), clathrin (Lafer, 2002), and caveolin (Williams and Lisanti, 2004) coat families involved in the subsequent steps defined by the exocytic and endocytic pathways. In some cases, these coats are known to be linked to cargo through families of adaptors to ensure efficient cargo selection and coordination with vesicle and tubule formation. Moreover, cargo capture is necessarily coupled to cellular components that direct the transport of vesicles to their unique destinations. Two large protein families that contribute significantly to vesicle targeting are the Rab GTPases (Pfeffer, 2001; Seabra et al., 2002; Deneka et al., 2003; Spang, 2004) and the soluble N-ethyl-maleimide-sensitive factor attachment protein receptor (SNARE) family of docking/fusion proteins (Chen and Scheller, 2001; Gerst, 2003; Ungar and Hughson, 2003).
Rab proteins are molecular switches that regulate the dynamic assembly and disassembly of multiprotein scaffolds involved in vesicle traffic (Miaczynska and Zerial, 2002; Pfeffer, 2003). The number of Rab family members found in the cell strictly correlates evolutionarily with increasing membrane complexity (Pereira-Leal and Seabra, 2000, 2001): Schizosaccharomyces pombe (7 members), Saccharomyces cerevisiae (11 members), Caenorhabditis elegans (29 members), Drosophila melanogaster (29 members), Arabidopsis thaliana (57 members), and Homo sapiens (63 members). Thus, the nearly fivefold increase in the number of Rab family members in the mammalian genome over that found in yeast may reflect the larger number of specialized trafficking pathways in the differentiated cell types forming mammalian organ systems, although no systematic approach has so far been applied to understand the organization of these pathways.
Because Rabs lack efficient intrinsic guanine nucleotide exchange and hydrolysis activity, their interactions with effectors are regulated by guanine nucleotide exchange factors (GEFs) and GTPase-activating proteins (GAPs) that promote the cyclical assembly and disassembly of Rab containing protein complexes (Pfeffer, 2001; Bernards, 2003; Spang, 2004). Indeed, recent results suggest that sequential Rab function may be regulated by the activity of GEFs (Wang and Ferro-Novick, 2002). Effector complexes formed in response to Rab activation can perform a variety of functions. They couple membranes to the cytoskeleton through the recruitment of kinesin- and myosin-based motors (Hammer and Wu, 2002; Karcher et al., 2002), direct the recruitment of tethering factors to initiate transport container docking (Allan et al., 2000; Moyer et al., 2001), potentially facilitate the function of proteins that alter membrane lipid composition (Czech, 2003; Gruenberg, 2003), and may organize the activity of SNARE components that mediate membrane fusion (Pfeffer, 2001; Gerst, 2003; Spang, 2004).
The SNARE family consists of a cognate group of integral and peripheral membrane proteins required for bilayer recognition and fusion (Chen and Scheller, 2001; Gerst, 2003; Ungar and Hughson, 2003). SNARE family members are divided into Q- and R-SNARE subfamilies based on their contribution to the reversible assembly of quaternary docking-fusion complexes (Ungar and Hughson, 2003). Each Q- and R-SNARE family member is believed to contribute differentially to docking and fusion by providing specific information that correctly directs the close juxtaposition of two membrane bilayers at specific steps of the exocytic and endocytic pathways. Like the Rab GTPases, bilayer docking/fusion mediated by SNARE complexes is highly regulated by a variety of pathway-specific effectors that either promote (matchmakers) or prevent (matchbreakers) SNARE assembly pathways (Gerst, 2003). However, unlike the Rab GTPases, the number of evolutionarily divergent members in the SNARE family has increased only modestly (1.5-fold) with expanding developmental complexity (Chen and Scheller, 2001), therefore raising the possibility that SNARE interactions are organized by Rab-based molecular switches that contribute significantly to the membrane complexity in higher eukaryotes.
How Rab GTPases, SNARE proteins, and their associated effectors and regulators confer membrane identity and coordinate the dynamics of cargo flux through sequential compartments to define the highly distinct subcellular organizations found in different mammalian cell and organ systems remains largely unknown. Given the hypothesis that the marked increase in Rab diversity during evolution of higher eukaryotes reflects ongoing specialization of membrane trafficking pathways, we explored the use of tissue-specific mRNA expression profiling and hierarchical clustering methods to organize components involved in membrane trafficking into groups of activity that direct specific membrane composition and transport networks within highly divergent cell types. A systems biology approach involving expression profiling of 79 human and 61 mouse nonredundant tissues (Su et al., 2004) leads us to propose that membrane trafficking events are orchestrated by Rab-regulated protein hubs. We find that these hubs can be linked to biochemically characterized components of the coat, targeting, tethering, and fusion machineries, validating the use of computational methods to extend our current knowledge base. We refer to this collection of interacting components that define the specific membrane architectures of a given cell type as the membrome network. We have compiled human and mouse membrome datasets comprising the known components of the membrome networks of different cell and organ systems. These membrome datasets are available online (http://www.membrome.org/) through the SymAtlas web application. Considering the fragmentary nature of current reductionist approaches in elucidating trafficking component functions, the membrome datasets provide a systems biology or top-down perspective that not only complements our current understanding of transport in complex tissues but now provides a more integrated view of Rab activity in controlling membrane architecture.
MATERIALS AND METHODS
Microarray Data
Microarray data used in this study were from the Human and Mouse Gene Atlases (Version 2) of The Genomics Institute of the Novartis Research Foundation (GNF). These define the steady state mRNA expression profiles of 79 human and 61 mouse nonredundant tissues (Su et al., 2004). These gene atlases are publicly available online and can be accessed through the SymAtlas web application (http://symatlas.gnf.org) for searching and visualization by keyword, accession number, gene symbol, genome interval, sequence, expression pattern, and coregulation. We have also compiled human and mouse membrome datasets that currently comprise ∼470 human/mouse proteins corresponding to known trafficking components within the cell. These datasets can be accessed through the Membrome homepage (http://www.membrome.org) for direct searching and visualization using the SymAtlas web application as described above. They may also be accessed by selecting either human or mouse membrome dataset from the pulldown menu available on any given SymAtlas web page. Further directions for using the SymAtlas web application and Membrome datasets are presented in the Supplementary Methods.
Hierarchical Clustering and Display of mRNA Expression Profiles
Hierarchical clustering and graphical representation of microarray data were carried out using the Cluster 3.0 and TreeView 1.6 programs, respectively (Eisen et al., 1998; de Hoon et al., 2004). Briefly, median-scaling was performed on both genes (probe sets) and arrays so that both genes and arrays had on average median and variance equal to zero and one. Hierarchical clustering of genes was then performed using an uncentered correlation distance metric and complete linkage aggregation. The initial PostsSript (PS) images produced by TreeView were edited using the Adobe Illustrator software (San Jose, CA), first by moving and splicing the dendrogram with the corresponding list of genes and then by separately scaling the list of genes, tissues, arrays, and the dendrogram, while still preserving the original alignments between the individual elements. To facilitate rapid browsing through the manuscript figures (e.g., to locate a favorite gene among up to 500 other genes), we took a novel approach at their presentation. Figures have been prepared in a vector-based graphics format that is still preserved in the online portable document format (PDF) version of the manuscript figures. When these PDF documents are opened with the Adobe Acrobat Reader (freely available for download at http://www.adobe.com/products/acrobat/readermain.html), the manuscript figures readily become interactive, keyword searchable databases. Figures can also be scaled up and down without any loss in resolution, such as by using the dynamic zoom function of the Acrobat Reader. A search can be implemented by clicking the search icon available on the Acrobat toolbar and entering the common name for a given gene (e.g., Rab3A) in the “Search PDF” window. If the given gene is present in the manuscript figures, a clickable list of search results will then appear within the same Search PDF window, wherein all the different occurrences of the given gene name will be listed that will be hyperlinked to the precise locations of the given gene within the document. Acrobat Reader will display the document page on which the given gene is first mentioned in the list and also highlight in gray the gene name on this page. Clicking on a different result in the Search PDF window will highlight the given gene name on the document. Clicking on any gene in the figures (e.g., Rab3A in Figure 1B) will also launch the default internet browser on user's computer and open the corresponding bioentry page at SymAtlas (Su et al., 2004). A list of the actual probe sets used corresponding to the human and mouse genes shown in the manuscript figures is provided in the Supplementary Table 1. This table is also fully keyword searchable when opened with the Acrobat Reader, and each probe set/accession number given can be directly queried at SymAtlas for more information.
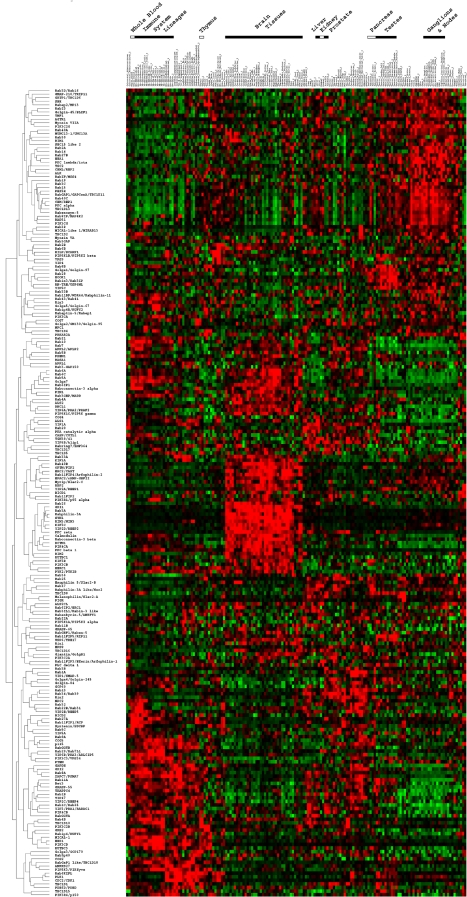
Figure 1.
Human rabome. (A) Seventy-nine human tissues were profiled in duplicate as described in Materials and Methods. Displayed in numerical order are the expression profiles of individual Rabs. (B) Hierarchical clustering was used to group Rabs (left column) based on the similarity of their expression profiles across the entire tissue array (top panel). A dendrogram is provided where lengths of the branches leading up to each node directly reflect the degree of correlation between the expression profiles as assessed by the pairwise similarity function described in Materials and Methods (also see Supplementary Methods for further discussion and a list of the actual correlation values used for the construction of this dendrogram). The online full text/PDF versions of this manuscript provide links to enhanced figure files that are keyword searchable using the search menu of the Adobe Acrobat Reader, and the blue highlighted components (gene names) are also hyperlinked to their corresponding entries at the SymAtlas web application (see Materials and Methods for further directions for accessing the interactive features of this manuscript's figures). (C) Highlighted are the expression profiles of Rab1, Rab5, Rab3, and Rab27 isoforms.
RESULTS
The Rabome
Although phylogenetic analysis of proteins in a gene family is commonly used to identify potential functional relationships to other family members (Pereira-Leal and Seabra, 2000, 2001), computational approaches applying hierarchical clustering algorithms to systematic tissue profiling can complement this annotation by providing insights into the physiological activity of close and distant family members and to different gene families in different cell types (Panda et al., 2002; Su et al., 2004; Walker et al., 2004). This is particularly useful in order to begin to elucidate the function of diverse members of the Rab, SNARE, and coat families where the molecular and biochemical basis for their functional integration is still largely unknown.
Given the hypothesis that the marked increase in Rab diversity during the evolution of higher eukaryotes reflects ongoing specialization of membrane trafficking pathways, we first examined the differential role of the Rab GTPase family members in these pathways (Pereira-Leal and Seabra, 2001) using a database of mRNA expression profiles derived from 79 human (Figure 1A, right panel) and 61 mouse (Supplementary Figures S1–S7) nonredundant tissues (Su et al., 2004). In human tissues, expression profiles of 51 of 63 currently known Rabs were available, and these were found to vary considerably between different tissues (Figure 1A). This is depicted by a heat map of colors comprising shades of red and green that correspond to up- and down-regulation events, respectively, relative to the median expression level (black) across the entire atlas. The brightest red and green arrays correspond to maximal values of up- and down-regulation events, respectively. The median expression level varied considerably between different Rabs, possibly reflecting their specialized function(s) in distinct tissues (see Supplementary Methods for instructions for accessing individual expression profiles at http://symatlas.gnf.org/).
To establish an order to the fluctuations observed in the Rab expression (Figure 1A), we used well-established hierarchical clustering methods (Eisen et al., 1998; Quackenbush, 2001; de Hoon et al., 2004) to position each Rab on the array with respect to all other Rabs in our dataset, based on the similarities in their individual tissue expression profiles (Figure 1B, right panel). In this way, we were able to generate an initial view of the relationships between Rab protein expression and developmentally similar and divergent cellular trafficking pathways. Here, the branch lengths of the dendrogram provided (Figure 1B, left panel) directly reflect the degree of correlation between the expression profiles of the Rab genes as assessed by the pairwise similarity function described in the Materials and Methods (Eisen et al., 1998; see Supplementary Methods for a further discussion and a list of actual correlation values used for the construction of the dendrogram shown in Figure 1B). As can be readily observed, Rabs cluster in unique groups based on similarities in their expression patterns across different tissues. Using data available from reductionist approaches that define the individual functions of a number of Rabs based on genetic and biochemical methods (Miaczynska and Zerial, 2002; Seabra et al., 2002; Pfeffer, 2003; Prekeris, 2003; Pfeffer and Aivazian, 2004; Spang, 2004), we can validate the utility of computational approaches for ordering Rab activities into physiologically relevant clusters and develop a systems biology perspective of both known and unknown function(s) in the context of cellular differentiation pathways.
Housekeeping Rabs. Rabs can be divided into several categories, beginning with those that perform generic functions to maintain the normal operation of the constitutive exocytic and endocytic pathways. These housekeeping Rabs include, among others, Rab1 isoforms involved in ER-to-Golgi transport in the exocytic pathway, and Rab4, 5, 7, and 11 isoforms involved in early and late endosome function (Pfeffer, 2001). These Rabs families are phylogenetically divergent with <35% (amino acid sequence) identity (Pereira-Leal and Seabra, 2000, 2001). Surprisingly, despite their purported generic housekeeping functions, each of these Rab shows highly variable expression profiles and do not necessarily cluster together (Figure 1B; e.g., click here to see Rab1A expression profile at SymAtlas). Elevated levels of Rab1 isoforms relative to the mean were found in lung, liver, kidney, intestines, testes, and immune system lineage tissues (Figure 1, B and C). Given their secretory function, these tissues are likely to require extensive amounts of ER and Golgi compartments for normal function. Although no tissue examined lacked Rab1 expression, there are three Rab1 isoforms (A, B, and C/Rab35; A has 93 and 54% identity with B and C, respectively; Tisdale et al., 1992), and their expression was generally found to be relatively tissue-specific. Although human Rab1B and Rab1C/Rab35 are up-regulated in lung and immune system lineages (i.e., thymus), Rab1A is down-regulated in these cells, but is highly expressed in cardiac and smooth muscle. Because the latter are not secretory tissues, these observations suggest unanticipated functions for Rab1A, perhaps in the maintenance of the extensive sarcoplasmic reticulum in muscle cells by Golgi-linked transport pathways (Wu et al., 2001). Similar to Rab1, housekeeping Rab5 isoforms A-C that are critical for the function of the endosomal pathway (Bucci et al., 1994) showed highly variable expression profiles across different tissues. Up-regulation of a given Rab5 isoform (Figures 1, B and C) occurred in developmentally distinct cell and tissue types, suggesting that the trafficking properties of the early endosome in different cell types is tuned to endocytic pathway specialization. A similar conclusion can be reached for Rabs involved in movement between early and late endosomes (Rab7) and between the early endosome and the cell surface through recycling endosomes (Rab4 and Rab11; Figure 1B).
Use of a computational approach to organize Rab function demonstrates that variations in the expression of housekeeping Rabs reflect tissue/cell-specific specialization of even the most basic trafficking events underlying the general constitutive functions of the exocytic and endocytic pathways. This suggests that the steady state levels and dynamics of mainstream organelles (ER, Golgi, endosomes, and lysosomes) are highly variable and specialized in response to housekeeping Rab-regulated activity. This could reflect differences in the basic demands by different classes of cargo that need to flow through these pathways in different cell types, a point not directly evident using traditional methods.
Specialized Rabs. Given the limited number of Rabs in lower eukaryotes (7–11), one possibility is that most Rabs in higher eukaryotes contribute to the function of more evolved membrane architectures and specialized subcellular trafficking pathways. Computational approaches involving hierarchical clustering methods have the potential to help understand the organization and function of these specialized Rabs.
One of the most notable and well characterized of the specialized Rabs is Rab3A, which is active in the regulated secretory pathway in the neuron (Schluter et al., 2002). Rab3A has been extensively studied in neurotransmitter release given its high abundance in the brain and presence on synaptic vesicles. Rab3A is believed to play an important role in the tethering and docking of synaptic vesicles in preparation for fusion (Lonart et al., 1998). Consistent with the biochemical studies, Rab3A is highly up-regulated in all brain tissues examined (Figures 1, B and C). Relative to the median expression level of housekeeping Rabs, Rab3A expression is low or absent in most other tissues (<1–2 copies/cell; click here to see Rab3A expression profile at SymAtlas). Thus, Rab3A is spatially restricted to a subset of specialized trafficking events at the brain synapse. Moreover, three other Rab3 isoforms, B, C, and D, show strikingly different expression profiles that contrast with the distribution of Rab3A (Figures 1, B and C). Although not mutually exclusive, these results are consistent with a recent analysis of Rab3 isoform expression and function in a number of different tissues based on immunoblotting (Schluter et al., 2002) that demonstrated a high degree of subspecialization of Rab3 isoform functions during the development of regulated secretory pathways.
In contrast to the activity Rab3 isoforms in the regulated secretory pathway, the phylogenetically distinct Rab27 family (<35% identity to Rab3A; Pereira-Leal and Seabra, 2000, 2001) has been implicated genetically and biochemically in the function of a variety of specialized subcellular organelles forming the secretory-lysosome pathway (Izumi et al., 2003; Tolmachova et al., 2004). These include toxic granules in cytotoxic T-lymphocytes, antigen-processing compartments involved in MHC class II presentation, platelet granules, and melanosomes in melanocytes (Seabra et al., 2002; Izumi et al., 2003; Tolmachova et al., 2004). We found that, consistent with these data, Rab27A is largely absent from brain tissue, but is markedly up-regulated in the immune system lineages (myeloid, T-cells, and NK cells) and whole blood samples rich in platelets containing these pathways (Figures 1, B and C). Interestingly, human Rab27B is down-regulated in these tissues, but up-regulated in a subset of ganglion and skeletal/uterine muscle tissues, suggesting an anticipated function of the secretory-lysosome pathway in these tissues.
A variety of other uncharacterized Rab proteins and their subfamily members exhibit profiles that suggest tissue-specific expression. Tissue-specific distributions of housekeeping and specialized Rabs suggest the existence of Rab “hubs” that direct highly defined trafficking pathways.
Rab Clusters Define Coupled Steps in Tissue-specific Pathways. Although individual Rab expression profiles highlight the cellular differences in the activity of even closely related Rab hubs, it is also evident that groups of divergent Rabs may function as a cohort to provide identity to tissue-specific pathways (Figure 1B). For example, in brain tissue, up-regulation of Rab3A correlates strongly with that of Rab40B (30% identity), Rab26 (55% identity), and Rab33A (34% identity), as indicated by the corresponding dendrogram branch lengths (Figure 1B, left panel). Because these Rabs are phylogenetically divergent from one another, it is likely that each has a unique function at the synapse. The biological functions of Rab40A and B are currently unknown, although Rab40C has been recently associated with endocytic traffic in oligodendrocytes (Rodriguez-Gabin et al., 2004). Although Rab40A and C are up-regulated in neuronal ganglions and nodes, up-regulation of Rab40B is confined to brain tissue with an expression profile nearly identical to that of Rab3A (Figure 1B). One possibility is that Rab40B may define an as yet uncharacterized Rab-regulated hub modulating a linked step in the Rab3A-dependent synaptic vesicle cycle in the brain. Alternatively, Rab40B could direct a comparably active postsynaptic dynamic pathway cycling neurotransmitter receptors given that expression profiling of tissues can highlight activities that are linked across cell boundaries.
Unlike Rab40B that is unique to brain tissue, Rab26 was originally cloned from pancreas (Wagner et al., 1995), a highly active secretory tissue that contains both zymogen and insulin granules, and is localized to secretory granules (Yoshie et al., 2000). Consistent with this observation, expression profiling reveals that Rab26 is strongly up-regulated in pancreatic, liver, and several other secretory tissues (Figure 1B). These results suggest that brain and pancreatic tissues may have common elements in the organization of protein interactions directing regulated secretion by several different Rab hubs.
Although Rab26 may control a late stage in regulated secretion, Rab33A is thought to facilitate endosome to Golgi transport and/or retrograde transport from late to early Golgi compartments. Consistent with the expression profiling data, Northern blot analyses of Rab33A showed prominent expression in brain and immune system lineages (Zheng et al., 1998). Thus, Rab33A in brain tissue may direct an unanticipated strong link between synaptic Rab3A/Rab40A/Rab26 function and endosome-Golgi recycling pathways.
In addition to the Rab3A, 26, 33A, 40B cluster, Rab4B and 11B are also up-regulated in brain tissue and cocluster with the Rab3A-regulated hub (Figure 1B), reinforcing the link between Rab3A function and recycling pathways. This can be contrasted with a different Rab cluster consisting of Rab4A, 5A, 6A, 6C, 7, and 10, which, although also up-regulated in brain tissue, segregates away from the Rab3A hub because of differences in overall expression profiles. These results suggest that this unlinked cluster, although important in brain tissue, is also likely to function as a hub cluster in the endocytic pathways of other cell and tissue types that are divergent from the highly specialized synaptic pathway(s).
Consistent with the need for specialized Rab cohorts in membrane trafficking events, a different Rab cluster is found in cells of immune lineage including Rab9A, Rab21, Rab27A, and Rab29/Rab7L1 (Figure 1B). Rab9A is required for lysosomal biogenesis via recycling pathways involving the trans-Golgi network (Riederer et al., 1994) and, as discussed above, Rab27A has a general role in the secretory-lysosome pathway (Izumi et al., 2003; Tolmachova et al., 2004). Although Rab21 has been suggested to be involved in the regulation of vesicular transport in polarized intestinal epithelial cells (Opdam et al., 2000), the function of Rab29/Rab7L1 remains unknown. One possibility is that this Rab cluster contributes to coupled trafficking pathway(s) related to MHC class I and II antigen processing and presentation. In the case of cytotoxic T-lymphocytes, Rab linkages could define sequential hub activities leading to the generation and fusion of cytotoxic granules that are targeted to the immune synapse (Trambas and Griffiths, 2003).
In general, hierarchical clustering reveals that Rab isoforms define tissue-specific subspecializations of a given pathway, whereas evolutionarily divergent Rabs form clusters of activity to facilitate linked membrane-trafficking pathways in divergent cell types.
Rab Regulators. Rab GTPases do not function in isolation. Using hierarchical clustering methods, we examined the tissue-specific distribution of known and putative Rab regulators. For example, Rab3A coclusters with the Rab3-interacting proteins calmodulin (Park et al., 1997); GDI1(α) (Sudhof, 2004); RIM2, RIM3, Rabphilin-3A (Fukuda, 2003); and synapsin (Syn1; Giovedi et al., 2004a, b; Figure 2). Regulators also include a diverse group of GEFs and GAPs that coordinate Rab activation and inactivation, respectively (Pfeffer, 2001; Bernards, 2003; Spang, 2004). For example, Rab3A clusters with HERC1 (Rosa et al., 1996) and the potential GEF Syn1 (Giovedi et al., 2004a). Although the Rab3A GEF Rab3GEP and GAP Rab3GAP have more divergent distributions suggestive of interaction with other Rab GTPases, the Rab3A-hub did contain the Rab3GEP-interacting protein Rabconnectin-3 beta (Kawabe et al., 2003), which may provide added specificity to Rab3A GEF recognition in vivo. As a second example, clustering of ALS2 (Otomo et al., 2003) with Rab5A; syntenin (Tomoda et al., 2004) with Rab5C; and RASA1, APPL1, and APPL2 (Miaczynska et al., 2004) with Rab5B (Figure 2) is also in good agreement with well-established biochemical data and highlights potential strong differences in isoform activity in different tissues. Thus, these examples indicate the utility of a systems biology approach to establish linkages between functional components. It should be emphasized that not all biochemical interactions reported in the literature will be necessarily seen by the hierarchical clustering of mRNA expression profiles (i.e., there are some false negatives in our data; see Supplementary Methods for further discussion), possibly reflecting tissue specific properties of expression and regulation not recapitulated by reductionist approaches.
Figure 2.
Human rabome including Rab regulators, effectors, and related kinases/phosphatases. Expression profiling was performed as that described in Figure 1 (see the online full text/PDF versions of the manuscript to access high-resolution, keyword searchable, and hyperlinked version of this figure).
Although Rab5 GEFs Rabex-5 (Horiuchi et al., 1997) and Rin1 (Tall et al., 2001) exhibit similar expression profiles, their tissue distribution differs from that of the Rab5 isoforms, suggestive of their additional functions in endocytic recycling pathways. Interestingly, Rabex-5 and Rin1 cluster with Rab11B, along with a known Rab11-interacting protein, Rab11FIP5 (Prekeris, 2003). This raises the possibility that the Rab5 GEFs Rabex-5 and Rin1 may also function as Rab11B GEFs, hence coupling the Rab5- and Rab11-regulated endocytic recycling activities (Bucci et al., 1994; Schlierf et al., 2000), as has been observed for yeast GEFs linking exocytic Rabs (Wang and Ferro-Novick, 2002). Similarly, the expression profile of Rabaptin-5 (Ohya et al., 1998) showed a stronger correlation with Rab33B than with Rab5 isoforms (Figure 2), consistent with the recent biochemical evidence that Rabaptin-5 also interacts with Rab33B (Valsdottir et al., 2001). We also noted that TRAPPC4, a mammalian homologue to a component of the Ypt1/Rab1 specific yeast GEF Trapp complex (Jones et al., 2000), clustered with Rab1B/C, suggestive of an evolutionary conserved function in the Rab1 hub in early exocytic pathways of mammalian cells.
Rab Effectors. It is now clear that Rabs can interact with multiple effectors to regulate the spatiotemporal function of organelles in membrane traffic (Deneka et al., 2003). Effectors interacting with activated Rabs include proteins that may direct vesicle tethering, docking, and fusion (Segev, 2001; Zerial and McBride, 2001; Deneka et al., 2003; Spang, 2004). Known tethers for Rab1 include Golgin-84 (Satoh et al., 2003), Grasp-55 (Shorter et al., 1999), Grasp-65 (Moyer et al., 2001), and p115 (Allan et al., 2000). Hierarchical clustering methods reveal that Grasp-55 clusters closely with the Rab1B and Rab1C/Rab35 isoforms, whereas Golgin-84 clustering is closest to that of Rab1A and Grasp-65 and p115 exhibit broader expression profiles (Figure 2). Intriguingly, Grasp-65 closely coclusters with Rab11B instead, suggesting that Rab11B may either utilize Grasp-65 as an effector and/or that Grasp65 has an important role in linking the endocytic recycling pathways to Grasp-65-dependent Golgi function(s) (Schlierf et al., 2000).
Rabs also couple membranes to motors directing movement along the cytoskeleton. For example, Rab27A has been shown to use melanophilin as a linker to Myosin Va (Fukuda et al., 2002). Although the expression profile of melanophilin does not directly correlate with that of Rab27A, the former clusters very closely with pIgR, the polymeric immunoglobulin (Ig) receptor, which interacts directly with Rab3D to control ligand-stimulated transcytosis (van IJzendoorn et al., 2002; Figure 2). Furthermore, the Rab3 GAP Rab3IL1 (Luo et al., 2001) also clusters with melanophilin and pIgR. Thus, melanophilin may interact with multiple Rabs not detected by traditional biochemical approaches, linking these pathways to the actin cytoskeleton.
A family of functionally diverse, kinesin-related motor subunits (KIF3A, B, and C) associate to form the kinesin-II motor complex in which KIF3C and KIF3B are alternative partners for KIF3A (Navone et al., 2001). Kinesin-II motor complex can mediate anterograde membrane traffic in neurons and melanosomes, ER-to-Golgi transport, and the mobility of protein complexes within cilia and flagella required for their morphogenesis (Hirokawa, 2000). Notably, we also observe close clustering of KIF3A and C with Rab33A and Rab3A, respectively, whereas the KIFB expression profile is also closest to that of Rab3A. Tight correlation of KIF3C with Rab3A suggests unanticipated function at the synapse in conjunction with KIF3B.
A universal effector required for Rab recycling is GDI (Wu et al., 1996). In humans, GDI has two isoforms. As mentioned earlier, GDI1(α) distribution correlates strongly with the Rab3A, an interaction already well established biochemically (Sudhof, 2004). In contrast, GDI2(β) has a broader distribution, although it is clearly specialized in cells of immune lineage (Figure 2). Distribution of GDI12(β) is suggestive of a more housekeeping role in the constitutive pathways than GDI1(α), but it is also evident that GDI2(β) has, in addition, a specialized role(s) for Rab GTPases in immune tissues that have extensive endocytic pathways.
Although retrieval involves GDI, delivery of Rabs to membranes is associated with the members of the YIP family (Pfeffer and Aivazian, 2004). Indeed, consistent with the biochemical data that the yeast Yip1p (Heidtman et al., 2003) and the mammalian YIP1 (Tang et al., 2001) are involved in the regulation of ER-to-Golgi traffic, human YIP1 clusters tightly with Rab1A (Figure 2). Furthermore, YIP3/PRA1 promotes Rab dissociation from GDI during loading onto Golgi membranes (Hutt et al., 2000; Sivars et al., 2003) and has an important role in ER-to-Golgi and Golgi transport (Hutt et al., 2000; Abdul-Ghani et al., 2001). YIP3/PRA1 exhibits close clustering with Rab1B, Rab1C/Rab35, and Grasp-55. Recent studies suggest that when overexpressed or down-regulated in heterologous expression systems, YIP3/PRA1 activity is also linked to the function of Rab9, an endosomal Rab required for the recycling of the mannose-6-phosphate receptor between the late endosome and the trans-Golgi network (Sivars et al., 2003). Given the observation that Yip family members form heterocomplexes (Calero and Collins, 2002; Calero et al., 2002; Pfeffer and Aivazian, 2004), cell-specific complexes between YIP family members may augment the site and specificity of YIP3/PRA1 function in different cell types to accommodate GDI recycling of Rabs from multiple compartments. Interestingly, we also observe that hitherto uncharacterized mammalian YIP2C and YIP2D isoforms (Pfeffer and Aivazian, 2004) have expression profiles very similar to Rab1B and Rab3A, respectively (Figure 2).
In general, application of hierarchical clustering methods to Rab regulators and effectors begins to provide insight into the tissue-specific organization of Rab hubs to broaden our perspective on their potential interactions with other Rabs and linked pathways.
Lipid Kinases and Phosphatases. Given the observation that many Rab effectors contain PH and FYVE domains involved in binding to phosphoinositides and that different phosphoinositides are localized to different compartments and involved in membrane targeting (Simonsen et al., 2001), we profiled known phosphatidylinositol (PI) kinases and phosphatases as possible direct or indirect effectors of Rab function (Figure 2).
Members of the PIP5KI family are known to be involved in trafficking at the cell surface (Martin, 2001). Indeed, we observe that the PIP5KIA and PIP5KIC isoforms exhibit similar expression profiles as that of the endocytic Rabs, Rab22A and Rab5A, respectively. In particular, Rab22A regulates the recycling of membrane proteins by a clathrin-independent pathway (Weigert et al., 2004). The PIK4 family is critical for the maintenance of the structural and functional organization of the Golgi complex (Audhya et al., 2000; De Matteis and Godi, 2004). PIK4CB/PI4KIIIβ appears in a distinct cluster with Rab1C/Rab35 and YIP3/PRA1, hence predicting a potential role for PIK4CB/PI4KIIIβ in ER-to-Golgi and Golgi transport regulated by the Rab1C/Rab35 hub. We also noted that PIK4CB/PI4KIIIβ clusters with Rab11A associated with the apical recycling endosomes (Wang et al., 2000), confirming a recent finding that the former is requisite for the functional association of the latter with the Golgi complex (de Graaf et al., 2004).
Cellular PI(3) kinase activities need to be balanced by counteracting PI(3) phosphatase activities. Indeed, PIK3C3/VPS34, which is essential for internal vesicle formation within multivesicular endosomes (Futter et al., 2001), clusters with PTEN, a PI(3) phosphatase (Maehama et al., 2001). Other lipid phosphatase and kinase isoforms show corresponding tissue-specific distributions. Thus, hierarchical clustering supports the concept that PH/FYVE domain containing Rab effectors and lipid modifying enzymes are used to coordinate membrane lipid composition with specific protein traffic patterns controlled by Rab hubs.
The SNAREome
Hierarchical Clustering of SNAREs. Although Rab function diverged significantly as higher eukaryote cell pathways became more specialized, only a modest diversification took place for SNARE components involved in membrane targeting and fusion (Chen and Scheller, 2001). This led us to suggest that Rab proteins function as the primary diversification element of membrane trafficking pathways by altering the combinatorial potential for protein interactions through their effector interacting (switch) domains and GTPase activity. We find that hierarchical clustering of SNAREs alone is consistent with this conclusion and that it provides further insight to the more global features of SNARE pairing pathways (Figure 3). For example, the brain profile is remarkably distinctive with most brain-derived structures up-regulating the SNAREs VAMP2 (R), Syntaxin 1A (Q), and SNAP-25 (Q). This reflects their well-established role in synaptic vesicle fusion and neurotransmitter release by Rab3A-regulated pathways (Chen and Scheller, 2001; Gerst, 2003; Sollner, 2003; Ungar and Hughson, 2003). Even within the spectrum of brain tissues analyzed, the various Q/R isoforms show unique distributions. Thus, hierarchical clustering emphasizes the possibility that specific quaternary fusion complexes involved in synaptic transmission are optimized for different neuronal cell types. In contrast to the activity of specialized SNAREs involved in neural function, the distributions of other SNAREs (Figure 3) clearly reflect specialized activity within exocytic/endocytic membrane systems found in other tissue-specific pathways. These results are in accord with the current view that SNAREs participate in specific membrane targeting and fusion events (Gerst, 2003; Ungar and Hughson, 2003).
Figure 3.
Human SNAREome. Expression profiling was performed as that described in Figure 1 (see the online full text/PDF versions of the manuscript to access high-resolution, keyword searchable, and hyperlinked version of this figure).
SNARE Regulators and Effectors. SNARE complex assembly and disassembly is highly regulated (Gerst, 2003). SNARE regulators include NSF and SNAP isoforms, and more specialized components including, among others, synaptotagmins, tomosyns, complexins, and the Munc18/Sec1 and Munc13 families (Chen and Scheller, 2001; Sollner, 2002, 2003; Gerst, 2003; Ungar and Hughson, 2003). Although NSF is considered a general component, its distribution (Figure 4) is most tightly correlated with that of the SNAREs VAMP2, syntaxin 1A, and SNAP-25, undoubtedly reflecting the intense level of SNARE activity in the brain tissue that directs neurotransmission and, potentially, synaptic remodeling.
Figure 4.
Human SNAREome including SNARE regulators and effectors. Expression profiling was performed as that described in Figure 1 (see the online full text/PDF versions of the manuscript to access high-resolution, keyword searchable, and hyperlinked version of this figure).
The synaptotagmin family is believed to couple signaling pathways to neurotransmitter release through Ca2+ signaling (Sudhof, 2002; Bai and Chapman, 2004). As expected from biochemical studies, Synaptotagmin I (Koh and Bellen, 2003) and its binding protein STXBP1/Munc18–1 (Swanson et al., 1998), along with another key mediator of neurotransmission, synaptophysin (Calakos and Scheller, 1994), are highly up-regulated in nearly all brain tissues examined, and cluster closely with Syntaxin 1A, VAMP2, and SNAP-25 (Figure 4). Other synaptotagmins show very different tissue distributions. Thus, the expression of synaptotagmins correlates with specific subsets of Q- and R-SNAREs, leading to the conclusion that synaptotagmin regulation of SNARE function is tissue and cell type specific.
Similarly, distribution of other SNARE regulator family members such as Munc18/Sec1 (e.g., Munc18–1/2/3, Sly1, VPS33A/B, and VPS45), Munc13 (e.g., Unc13A/B), tomosyn (e.g., STXBP5), and complexin (e.g., CPLX1/2; Gerst, 2003) suggests that they are also specialized for a particular cell type. Expression of these proteins could reflect the need for an altered balance in the negative and positive regulation of SNARE assembly/disassembly pathways reflecting cargo activity and integration with extracellular signaling events.
Coat Machineries
In general, the function of a Rab-regulated hub is to segregate cargo in transit from “resident” components that define the identity of exocytic and endocytic compartments. To date, three basic types of general coat polymers (clathrin, COPI, and COPII) are known to select cargo for transport between specific compartments (Kirchhausen, 2000; Antonny and Schekman, 2001; Lee et al., 2004). Figure 5 depicts hierarchical clustering of the known coat components and their adaptors and regulators exclusive of the Rab-SNARE machinery.
Figure 5.
Human coat components, adaptors, regulators, and related kinases/phosphatases. Expression profiling was performed as that described in Figure 1 (see the online full text/PDF versions of the manuscript to access high-resolution, keyword searchable, and hyperlinked version of this figure).
COPII mediates anterograde transport from ER, where the small GTPase Sar1 is responsible for the recruitment of Sec23/24 (Lee et al., 2004), a protein complex that has a well-established function in cargo selection (Aridor et al., 1998; Kuehn et al., 1998). Considering that the four Sec24 isoforms largely exhibit differential expression profiles independent of the two Sec23 isoforms (Figure 5), distinct Sec23/24 complexes are likely to be functionally specialized for tissue-specific export of cargo from the ER, consistent with the observation in yeast wherein the various Sec24 isoforms recruit different cargo from the ER (Lee et al., 2004). Nevertheless, the expression profiles of the Sec13R and Sec31A are near identical, in close agreement with the biochemical data that the two proteins form a functional complex (Sec13/31) requisite for COPII coat polymerization (Lederkremer et al., 2001; Gurkan and Balch, 2005). In fact, Sec13R and Sec31A seed a “COPII cluster” that gradually expands to include Sec24C; the Sar1-specific GEF, Sec12 (Weissman et al., 2001); the putative phospholipid modifying/Sec23-interacting protein Sec23IP/P125 (Tani et al., 1999); and finally Sec24A, with a decreasing degree of correlation in the given order. Here, hierarchical clustering of expression profiles highlights an integrated unit of function for the early secretory pathway. Furthermore, we observe isolated coclustering of Sec23A and Sec24D that may comprise a tissue-specialized Sec23/24 complex. Finally, the expression profiles of Sar1B and its GAP Sec23B (Yoshihisa et al., 1993) are also highly correlated, independent of the other COPII components, suggesting a unique function in ER export more strongly correlated with specific types of cargo and Sec24 isoforms. This interpretation is supported by the recent observation that Sar1B loss-of-function leads to a unique genetic deficiency in chylomicron trafficking from the ER (Shoulders et al., 2004).
In contrast to interaction of cargo with the Sec24 subunit of the COPII machinery, interaction of cargo in the Golgi involves the whole COPI complex (Nickel et al., 2002). COPI has been characterized in vivo and in vitro and appears to be synthesized from subcomplexes with the 7 subunit holocomplex being considered a stable structure (Lowe and Kreis, 1995). The near identical expression profiles of the COPI subunits α and ζ1 clearly places them in a larger cluster that also includes COPI subunits β, γ, and β2 (Figure 5). On the other hand, the COPI subunits ζ2 and ε exhibit different expression profiles raising the possibility that there may be unanticipated tissue-specific heterogeneity in the composition of COPI coats. Interestingly, the expression profile of the COPI subunit ε exhibits a very high level of correlation with that of Arf1. This is consistent with in vitro biochemical data on the importance of its interaction with this GTPase and tissue-specific functions (Eugster et al., 2000).
Adaptor protein (AP) complexes select cargo for inclusion into coated vesicles in the late secretory and endocytic pathways (Robinson, 2004). AP1 components γ2 and σ2, and AP2 components α2, μ1, and σ1 are strongly up-regulated in the immune lineages, whereas the AP3 components β2 and μ2 are up-regulated in brain tissues, consistent with their known biochemical functions in the neurons (Hinners and Tooze, 2003; Figure 5). The AP4 components ε1 and σ1 are up-regulated in neuronal ganglions and nodes, suggesting an enhanced function for AP4-mediated pathways in these tissues (Yap et al., 2003). AP complex clusters also include other components with which biochemical interactions have already been established (Lafer, 2002; Hinners and Tooze, 2003). For example, AP1 (γ2 and σ2) clusters with the AP1 subunit γ binding protein 1, AP1γBP1, the small GTPase Arf6, and Arf6 GAPs GIT2 and Centaurin β1. On the other hand, the AP2 (α2, μ1, and σ1) cluster includes Dynamin II, Arf1, Arf1 GAPs GIT1/p95-APP1 and PSCD2/ARNO, and the clathrin-binding protein HRS/HGS. These results raise the possibility that AP1 function might be tightly integrated with ARF6 function in the endocytic pathway. Conversely, linkage between AP2 and Arf1 raises the surprising possibility of Arf1 function in cell surface trafficking events. Alternatively, this could reflect a strong link between Arf1-mediated Golgi trafficking pathways with more distal AP2-mediated endocytic trafficking occurring at the plasma membrane.
Golgi-associated, γ-ear-containing, Arf-binding proteins (GGAs) constitute a family of monomeric adaptors for clathrin (Robinson, 2004). An interesting cluster is observed around the muscle-specific clathrin heavy polypeptide like 1 (CLH22; Sirotkin et al., 1996; Doray and Kornfeld, 2001) consisting of the AP1γ1 subunit, GGA1 (Doray et al., 2002), the transcription factor regulating Arf1 levels, APA1/ZFP410 (Benanti et al., 2002), and the adaptor-like protein Stonin-1 (Martina et al., 2001), suggesting a cargo-specific activity associated with CLH22 function. Other GGAs (Lafer, 2002; Hinners and Tooze, 2003) exhibit differential expression profiles reflecting tissue-specific roles.
Although the cargo selection mechanisms for COPII, COPI and clathrin are becoming clear (Nickel et al., 2002; Barlowe, 2003; Bonifacino and Glick, 2004; Spang, 2004), the basis for cargo recruitment by uncoated tubules emanating from Golgi compartments, from endosomal recycling compartments, or from invaginations formed by caveolin-coated membranes (Williams and Lisanti, 2004) remains to be defined. In the endocytic pathway, expression profiles of Cav1 and Cav2 exhibit very high correlation, whereas Cav3 has a unique distribution (Figures 5). The tight coclustering of Cav1/2 is consistent with the observation that they form a functional complex (Cohen et al., 2004).
In general, like Rabs and SNAREs, coat components involved in cargo selection are tightly linked to tissue-specific expression patterns, undoubtedly reflecting a close link between the type and extent of specialized cargo and their requisite trafficking pathways in different cell types.
The Membrome
Hierarchical clustering of the Rab, SNARE, and coat complex components (Figures 1, 2, 3, 4, 5) provides parallel snapshots of the basic machineries driving the overall membrane traffic. To begin integrating these activities, we clustered Rabs and SNAREs exclusive of their respective regulators and effectors to highlight these two fundamental components of membrane trafficking (Figure 6). Strikingly, we observed tight clustering of known components involved at the synapse including Rab3A, SNAP-25, VAMP2, and Syntaxin 1A (Figure 6; Chen and Scheller, 2001; Gerst, 2003; Sollner, 2003; Ungar and Hughson, 2003), validating the utility of linking the activities of different protein families using expression profiling.
Figure 6.
Human Rabome and SNAREome. Expression profiling was performed as that described in Figure 1 (see the online full text/PDF versions of the manuscript to access high-resolution, keyword searchable and hyperlinked version of this figure).
Considering the requisite cooperativity between Rab and SNARE components and their regulators and effectors, we extended the hierarchical clustering analysis to encompass all known membrane trafficking components (Figure 7). Although biochemical and genetic evidence linking Rabs directly to coat components, tethers, or SNAREs using reductionist approaches is limited in scope, and in many cases indirect, the results from computational clustering methods provide for the first time a systems biology view of their linked activities. Significantly, in this inclusive hierarchical clustering analysis (Figure 7), the initial distributions defined by Rabs and/or SNAREs alone (Figures 1, 3, and 6) are not only largely maintained, but actually enhanced with additional components that are consistent with biochemical and genetic studies. Again, the Rab3A-regulated hub of the synapse provides the best validation of the approach. We observe tight coclustering of Rab3A with three distinct groups: 1) Rab regulators and effectors including GDI1(α) (Sudhof, 2004), RIM3, Rabphilin-3A (Fukuda, 2003), calmodulin (Park et al., 1997), Synapsin (Syn1; Giovedi et al., 2004a, b), Rab3 GEP-binding protein Rabconnectin-3 beta (Kawabe et al., 2003), and a potential RabGAP, RUTBC1 (Katoh, 2004); 2) SNARE system components including VAMP2, NSF (Chen and Scheller, 2001; Gerst, 2003; Sollner, 2003; Ungar and Hughson, 2003), Synaptotagmin I (Koh and Bellen, 2003), and Synaptophysin (Calakos and Scheller, 1994); and finally 3) coat machinery components AP3 μ2 (Hinners and Tooze, 2003), DNJC6/Auxilin, and Dynamin III (Lafer, 2002). Although Dynamin I has a broad tissue distribution and has a distinct clustering profile, the striking linkage of Dynamin III to Rab3A suggests a central, if not specialized, role in the synapse (Gray et al., 2003). The ability of synapse components to dominate the hierarchical clustering profile suggest that synaptic function throughout the brain is a very high maintenance process even when compared with housekeeping Rab activities associated with neurons, or the highly abundant glial component. Moreover, in the all inclusive profile, subspecialization of Rab hub activity within the brain becomes more evident.
Figure 7.
Human membrome including currently known components involved in exocytic and endocytic membrane traffic. Key sections of the dendrogram that are mentioned in the manuscript text are shown in blue and they are also magnified on the left hand side. Expression profiling was performed as that described in Figure 1 (see the online full text/PDF versions of the manuscript to access high-resolution, keyword searchable, and hyperlinked version of this figure).
Although the synapse provides an example of the Rab3A hub defining function of the membrane architecture of the regulated secretory pathway, coat assembly in constitutive pathways must also be linked to membrome components that direct the transport container to the correct target membrane. In the inclusive array (Figure 7), we note a Rab1A-hub comprising further known components involved in the regulation of ER-to-Golgi traffic, such as YIP1 (Tang et al., 2001), the small GTPase Sar1A and its specific GAP, Sec23A (Yoshihisa et al., 1993). This Rab1A-hub also contains ER-to-Golgi membrane traffic SNAREs VAMP3 and Ykt6 (Bonifacino and Glick, 2004).
Arf1, a GTPase involved in COPI coat assembly and recycling from Golgi compartments to the ER (Balch et al., 1992), clusters not only with the COPI subunit ε as discussed above (Eugster et al., 2000), but also with Rab1B, which modulates COPI recruitment to pre-Golgi and Golgi compartments (Figure 7; Tisdale et al., 1992; Alvarez et al., 2003). Curiously, Rab11A, which is associated with the apical recycling endosomes (Wang et al., 2000), also clusters with Arf1 along with a network of coat machinery components that include Dynamin II, clathrin light chain A (CLTA), the clathrin-binding protein HRS/HG, HRS/HG-interacting protein TSG101 (Lu et al., 2003), and TSG101-interacting protein EAP30 (von Schwedler et al., 2003). Further significant members of this Arf1 cluster include Rab1B/C along with the GEF TRAPPC4 (Jones et al., 2000) and known Rab1 tether Grasp-55 (Shorter et al., 1999), TIP47 (Diaz and Pfeffer, 1998), and the COPII components Sec13R and Sec24C. Interestingly, the various subunits of the COG protein complex, which is necessary for normal Golgi morphology and function (Ungar et al., 2002), are widely scattered throughout the profile, suggesting that different subunits contribute differentially to COG function in a tissue specific manner. This conclusion is consistent with the recent observation that COG complexes are involved in trafficking of Golgi glycosylation enzymes that have variable tissue distributions. With respect to Golgi adaptors, the AP1γ1/CLH22/GGA1/APA1 cluster discussed in the previous section (Figure 5), now expands to include Rab8B, which regulates AP1-dependent basolateral transport in Madin Darby canine kidney cells (Ang et al., 2003), the Golgi- and endosome-associated SNAREs GOSR2/Membrin (Lowe et al., 1997), and Syntaxin 8 (Subramaniam et al., 2000), respectively (Figure 7). Thus, hierarchical clustering highlights features of Golgi organization that may function in concert to balance anterograde and retrograde lipid/protein flow.
In endocytic clathrin-based pathways, we observe many correlations between distributions of adaptors with specific Rabs and SNAREs, suggesting linkage for tissue-specialized recycling (Figure 7). Interestingly, Cav1/2 now clusters tightly with Rab13, raising the possibility that this Rab may be a component of caveosomes or required for targeting of Cav1/2 derived vesicles to the ER/Golgi in a variety of cell types. Potentially consistent with this interpretation, Rab13 has also been implicated in endocytic recycling of occludin, a tight junction integral membrane protein (Morimoto et al., 2005).
Given the many strong correlations between hierarchial clustering and biochemical interactions in both exocytic and endocytic pathways, we propose that Rab-centric hierarchical coding systems regulate specific membrane interactions and cargo flow through the exocytic and endocytic pathways. We refer to this general system of Rab-regulated hubs of protein interactions as the membrome for a given cell type or transport activity. In this view, Rabs and SNAREs form the minimal core components of the membrome. Their activity is regulated by cohub components that will also include Rab/SNARE regulators and effectors that directly or indirectly interact with coat components to define cargo trafficking pathways. For a given cell type, the membrome will vary substantially reflecting the unique expression profiles of its components, thereby dictating unique membrane architectures. These results raise the provocative possibility that the membrome may also include collections of Rab hubs that define closely linked pathways that are coupled across cellular barriers where cross-talk between cells is critical for normal function, such as at the synapse. Indeed, given that the composition of the synaptic membrome seen in brain tissues (Figure 7) is consistent with all biochemical evidence regarding the critical role of each component in synaptic transmission, it is evident that the hierarchical clustering has predictive value for defining the interactions comprising other uncharacterized Rab-regulated hubs.
DISCUSSION
We have applied a systems biology approach to help elucidate functional relationships of eukaryotic membrane trafficking components by using hierarchical clustering of mRNA expression profiles of 79 human and 61 mouse tissues. Although we focused on the analysis of human tissue profiles, similar results were also obtained during the analysis of mouse tissue profiles (Supplementary Figures S1–S7). Differences between mouse and human profiles highlight the possibility that mouse and human physiologies are evolutionary divergent and require novel specializations of Rab-regulated hubs.
Alternatively, using the “profile neighborhood” feature of the SymAtlas web application, a search can be carried out against the entire GNF Human and Mouse Gene Atlases (http://symatlas.gnf.org) or the human and mouse membrome datasets (http://www.membrome.org) to identify potential interacting partners for a given protein based on the correlation of their expression profiles (see Supplementary Material for further directions). Because the membrome datasets are limited to currently known components compiled from the literature, they allow a more focused view of trafficking interactions. On the other hand, searches carried out against the entire GNF Human and Mouse Gene Atlases benefit from potential identification of unanticipated interactions that may help expand our understanding of trafficking pathways.1
Lessons Learned from Hierarchical Clustering
Gene expression profiling has a number of known limitations that may influence the interpretation of the membrome datasets (see Supplementary Methods). Despite these limitations, hierarchical clustering still provides for the first time a more systems biology view of membrane architecture of higher eukaryotes. Such a top-down view provides a number of insights into more general features of the Rab-regulated trafficking networks. First, even housekeeping Rabs exhibit tissue specialization, with differing levels of expression reflecting the intensity of a particular constitutive exocytic or endocytic trafficking pathway. Housekeeping Rab isoforms define further levels of subspecialization of constitutive pathways. Second, clustering helps to identify Rabs whose hub activities define unique membrane trafficking pathways characteristic of differentiated cell types. This is particularly important for understanding the function of the majority (∼50) of Rab hub functions whose activities are currently poorly characterized. Third, clustering provides potential insight into the organization of linked Rab hubs defining specific trafficking pathways (Wang and Ferro-Novick, 2002). Fourth, a systems biology approach provides insight into potential biochemical interactions where such interactions would not be readily apparent using reductionist approaches. Fifth, a systems biology approach highlights the combinatorial nature of Rab regulation of distinct subsets of regulators and effectors and coat machineries in establishing tissue-specific pathways. By highlighting unanticipated connections from a more global perspective, hierarchical clustering methods have predictive value.
The Membrome
Given the hypothesis that the marked increase in Rab diversity during evolution of higher eukaryotes largely reflects the ongoing specialization of membrane trafficking pathways, we propose that a Rab-centric perspective provides a rational approach to begin to organize the membrane architecture of eukaryotic cells. This coding system, referred to as the membrome, containing Rabs and SNAREs, their regulators and effectors in conjunction with cargo adaptors and coat components, provides a foundation for understanding the multitude of trafficking events that dictate the distinct physiologies of differentiated cells.
Although biochemical and genetic approaches have so far been instrumental in elucidating the basic Rab GTPase function, it is now clear that reductionist techniques have more limited use in elucidating the intricate protein interaction networks driving overall membrane architecture of the cell. Indeed, we found that the tissue-specific expression levels of Rab GTPases and their cognate regulators and effectors are highly variable. In the case of Rabs, this was true for both housekeeping Rabs involved in the function of constitutive exocytic and endocytic compartments, as well as developmentally specialized members. Even highly related Rab isoforms that correspond to Rab subfamily members show tissue/cell specialization. In yeast, only two Rabs are essential for growth—Rab1/Ypt1 involved in ER-to-Golgi transport and Sec4 for transfer from the Golgi to the cell surface (Pereira-Leal and Seabra, 2001). Moreover, only a small number of Rabs are required to maintain overall constitutive function of the exocytic and endocytic pathways in S. pombe (7 Rabs) and S. cerevisiae (11 Rabs; Pereira-Leal and Seabra, 2001). In contrast, mammalian cells have at least 63 Rab proteins (Pereira-Leal and Seabra, 2000). Therefore, 50–55 Rabs found in mammalian cells have unique function not found in lower eukaryotes. A similar divergent group of Rabs defines the organization of plants (Rutherford and Moore, 2002) and other multicellular organisms including D. melanogaster (Satoh et al., 1997), C. elegans (Bock et al., 2001), and various parasites (Field and Field, 1997; Quevillon et al., 2003).
Given that Rab interaction with regulators and effectors largely occurs through the switch domains (Pfeffer, 2001; Spang, 2004), it is apparent that membrane trafficking pathways have utilized the sequence and folding flexibility of these domains to generate highly specialized responses to GTP loading and hydrolysis. This reflects the evolutionary need to develop pathways that integrate not only the unique properties of a particular Rab-regulated hub, but also link the trafficking activity of each cell type with that of other cells comprising different tissue types. Thus, we suggest that even the basic 5–10 Rab-regulated hubs that direct the constitutive exocytic and endocytic pathways of lower eukaryotes are likely to have many functional components and links that are unique to the membrane architecture of higher eukaryotes. It is apparent that it will be important to study the function of a particular Rab and its effector in the correct cellular context.
What is a common theme that defines a general role for Rab hubs from a systems biology perspective? Although SNAREs direct late events leading to membrane fusion, it is becoming increasingly apparent that Rab proteins initiate targeting through recruitment of membrane-oriented tethering components that forge links with fusion factors to coordinate cargo transport with membrane flow. Moreover, the ability of Rab GTPases to couple transport containers to motor proteins can be conceptually viewed as a “tethering” function that establishes the distribution of organelles within the cytoskeletal network. Such links will rely heavily on the unique tissue distribution of GEFs and GAPs as well as general modulators such as GDIs that facilitate recycling. Thus, a Rab-centric hub coordinates targeting and fusion through integrating diverse activities required for defining the overall membrane architecture of the differentiated cell.
The Rab World
By developing a Rab-centric view, components comprising the membrome datasets can be used to define more limited subsets of functionally linked pathways (e.g., the synaptic membrome) or to elucidate links between different Rab-regulated hubs (e.g., to coordinate antigen presentation in immune cell lineages). As such global insight is not available from traditional classification schemes based simply on sequence or biochemical homologies, we suggest that the Rab world perspective provided by hierarchical clustering may present a systems biology framework to outline the protein and lipid organization of cellular membranes within higher eukaryotes.
Supplementary Material
Acknowledgments
We thank Andre Bernards for providing access to the Ras GTPase superfamily database. These studies are supported by grants from the National Institutes of Health (GM33301, GM42336, and EY11606) to W.E.B. C.G. is a Cystic Fibrosis Foundation Postdoctoral Research Fellowship recipient. This is TSRI Manuscript No. 16969-CB.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05-01-0062) on June 8, 2005.
Abbreviations used: AP, adaptor protein complex; COPI, coat protein complex I; COPII, coat protein complex II; ER, endoplasmic reticulum; GAPs, GTPase-activating proteins; GEFs, guanine nucleotide exchange factors; GGAs, Golgi-associated, γ-ear-containing, Arf-binding proteins; PI, phosphatidylinositol; SNARE, soluble N-ethyl-maleimide-sensitive factor attachment protein receptor
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
Footnotes
To ensure complete annotation of the known mammalian membrane trafficking components comprising the online human and mouse membrome datasets, these will be periodically updated. We encourage readers to submit (membrome@scripps.edu) gene accession numbers corresponding to the proteins currently not listed (see Supplementary Table 1).
References
- Abdul-Ghani, M., Gougeon, P. Y., Prosser, D. C., Da-Silva, L. F., and Ngsee, J. K. (2001). PRA isoforms are targeted to distinct membrane compartments. J. Biol. Chem. 276, 6225-6233. [DOI] [PubMed] [Google Scholar]
- Allan, B. B., Moyer, B. D., and Balch, W. E. (2000). Rab1 recruitment of p115 into a cis-SNARE complex: programming budding COPII vesicles for fusion. Science 289, 444-448. [DOI] [PubMed] [Google Scholar]
- Alvarez, C., Garcia-Mata, R., Brandon, E., and Sztul, E. (2003). COPI recruitment is modulated by a Rab1b-dependent mechanism. Mol. Biol. Cell 14, 2116-2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang, A. L., Folsch, H., Koivisto, U. M., Pypaert, M., and Mellman, I. (2003). The Rab8 GTPase selectively regulates AP-1B-dependent basolateral transport in polarized Madin-Darby canine kidney cells. J. Cell Biol. 163, 339-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonny, B., and Schekman, R. (2001). ER export: public transportation by the COPII coach. Curr. Opin. Cell Biol. 13, 438-443. [DOI] [PubMed] [Google Scholar]
- Aridor, M., Weissman, J., Bannykh, S., Nuoffer, C., and Balch, W. E. (1998). Cargo selection by the COPII budding machinery during export from the ER. J. Cell Biol. 141, 61-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audhya, A., Foti, M., and Emr, S. D. (2000). Distinct roles for the yeast phosphatidylinositol 4-kinases, Stt4p and Pik1p, in secretion, cell growth, and organelle membrane dynamics. Mol. Biol. Cell 11, 2673-2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai, J., and Chapman, E. R. (2004). The C2 domains of synaptotagmin— partners in exocytosis. Trends Biochem. Sci. 29, 143-151. [DOI] [PubMed] [Google Scholar]
- Balch, W. E., Kahn, R. A., and Schwaninger, R. (1992). ADP-ribosylation factor is required for vesicular trafficking between the endoplasmic reticulum and the cis-Golgi compartment. J. Biol. Chem. 267, 13053-13061. [PubMed] [Google Scholar]
- Barlowe, C. (2003). Signals for COPII-dependent export from the ER: what's the ticket out? Trends Cell Biol. 13, 295-300. [DOI] [PubMed] [Google Scholar]
- Benanti, J. A., Williams, D. K., Robinson, K. L., Ozer, H. L., and Galloway, D. A. (2002). Induction of extracellular matrix-remodeling genes by the senescence-associated protein APA-1. Mol. Cell. Biol. 22, 7385-7397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernards, A. (2003). GAPs galore! A survey of putative Ras superfamily GTPase activating proteins in man and Drosophila. Biochim. Biophys. Acta 1603, 47-82. [DOI] [PubMed] [Google Scholar]
- Bock, J. B., Matern, H. T., Peden, A. A., and Scheller, R. H. (2001). A genomic perspective on membrane compartment organization. Nature 409, 839-841. [DOI] [PubMed] [Google Scholar]
- Bonifacino, J. S., and Glick, B. S. (2004). The mechanisms of vesicle budding and fusion. Cell 116, 153-166. [DOI] [PubMed] [Google Scholar]
- Bucci, C., Wandinger-Ness, A., Lutcke, A., Chiariello, M., Bruni, C. B., and Zerial, M. (1994). Rab5a is a common component of the apical and basolateral endocytic machinery in polarized epithelial cells. Proc. Natl. Acad. Sci. USA 91, 5061-5065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calakos, N., and Scheller, R. H. (1994). Vesicle-associated membrane protein and synaptophysin are associated on the synaptic vesicle. J. Biol. Chem. 269, 24534-24537. [PubMed] [Google Scholar]
- Calero, M., and Collins, R. N. (2002). Saccharomyces cerevisiae Pra1p/Yip3p interacts with Yip1p and Rab proteins. Biochem. Biophys. Res. Commun. 290, 676-681. [DOI] [PubMed] [Google Scholar]
- Calero, M., Winand, N. J., and Collins, R. N. (2002). Identification of the novel proteins Yip4p and Yip5p as Rab GTPase interacting factors. FEBS Lett. 515, 89-98. [DOI] [PubMed] [Google Scholar]
- Chen, Y. A., and Scheller, R. H. (2001). SNARE-mediated membrane fusion. Nat. Rev. Mol. Cell. Biol. 2, 98-106. [DOI] [PubMed] [Google Scholar]
- Cohen, A. W., Hnasko, R., Schubert, W., and Lisanti, M. P. (2004). Role of caveolae and caveolins in health and disease. Physiol. Rev. 84, 1341-1379. [DOI] [PubMed] [Google Scholar]
- Czech, M. P. (2003). Dynamics of phosphoinositides in membrane retrieval and insertion. Annu. Rev. Physiol. 65, 791-815. [DOI] [PubMed] [Google Scholar]
- de Graaf, P. et al. (2004). Phosphatidylinositol 4-kinasebeta is critical for functional association of rab11 with the Golgi complex. Mol. Biol. Cell 15, 2038-2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Hoon, M. J., Imoto, S., Nolan, J., and Miyano, S. (2004). Open source clustering software. Bioinformatics 20, 1453-1454. [DOI] [PubMed] [Google Scholar]
- De Matteis, M. A., and Godi, A. (2004). Protein-lipid interactions in membrane trafficking at the Golgi complex. Biochim. Biophys. Acta 1666, 264-274. [DOI] [PubMed] [Google Scholar]
- Deneka, M., Neeft, M., and van der Sluijs, P. (2003). Regulation of membrane transport by rab GTPases. Crit. Rev. Biochem. Mol. Biol. 38, 121-142. [DOI] [PubMed] [Google Scholar]
- Diaz, E., and Pfeffer, S. R. (1998). TIP 47, a cargo selection device for mannose 6-phosphate receptor trafficking. Cell 93, 433-443. [DOI] [PubMed] [Google Scholar]
- Doray, B., Ghosh, P., Griffith, J., Geuze, H. J., and Kornfeld, S. (2002). Cooperation of GGAs and AP-1 in packaging MPRs at the trans-Golgi network. Science 297, 1700-1703. [DOI] [PubMed] [Google Scholar]
- Doray, B., and Kornfeld, S. (2001). Gamma subunit of the AP-1 adaptor complex binds clathrin: implications for cooperative binding in coated vesicle assembly. Mol. Biol. Cell 12, 1925-1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen, M. B., Spellman, P. T., Brown, P. O., and Botstein, D. (1998). Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95, 14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eugster, A., Frigerio, G., Dale, M., and Duden, R. (2000). COP I domains required for coatomer integrity, and novel interactions with ARF and ARF-GAP. EMBO J. 19, 3905-3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field, H., and Field, M. C. (1997). Tandem duplication of rab genes followed by sequence divergence and acquisition of distinct functions in Trypanosoma brucei. J. Biol. Chem. 272, 10498-10505. [DOI] [PubMed] [Google Scholar]
- Fukuda, M. (2003). Distinct Rab binding specificity of Rim1, Rim2, rabphilin, and Noc2. Identification of a critical determinant of Rab3A/Rab27A recognition by Rim2. J. Biol. Chem. 278, 15373-15380. [DOI] [PubMed] [Google Scholar]
- Fukuda, M., Kuroda, T. S., and Mikoshiba, K. (2002). Slac2-a/melanophilin, the missing link between Rab27 and myosin Va: implications of a tripartite protein complex for melanosome transport. J. Biol. Chem. 277, 12432-12436. [DOI] [PubMed] [Google Scholar]
- Futter, C. E., Collinson, L. M., Backer, J. M., and Hopkins, C. R. (2001). Human VPS34 is required for internal vesicle formation within multivesicular endosomes. J. Cell Biol. 155, 1251-1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerst, J. E. (2003). SNARE regulators: matchmakers and matchbreakers. Biochim. Biophys. Acta 1641, 99-110. [DOI] [PubMed] [Google Scholar]
- Giovedi, S., Darchen, F., Valtorta, F., Greengard, P., and Benfenati, F. (2004a). Synapsin is a novel Rab3 effector protein on small synaptic vesicles. II. Functional effects of the Rab3A-synapsin I interaction. J. Biol. Chem. 279, 43769-43779. [DOI] [PubMed] [Google Scholar]
- Giovedi, S., Vaccaro, P., Valtorta, F., Darchen, F., Greengard, P., Cesareni, G., and Benfenati, F. (2004b). Synapsin is a novel Rab3 effector protein on small synaptic vesicles. I. Identification and characterization of the synapsin I-Rab3 interactions in vitro and in intact nerve terminals. J. Biol. Chem. 279, 43760-43768. [DOI] [PubMed] [Google Scholar]
- Gray, N. W., Fourgeaud, L., Huang, B., Chen, J., Cao, H., Oswald, B. J., Hemar, A., and McNiven, M. A. (2003). Dynamin 3 is a component of the postsynapse, where it interacts with mGluR5 and Homer. Curr. Biol. 13, 510-515. [DOI] [PubMed] [Google Scholar]
- Gruenberg, J. (2003). Lipids in endocytic membrane transport and sorting. Curr. Opin. Cell Biol. 15, 382-388. [DOI] [PubMed] [Google Scholar]
- Gurkan, C., and Balch, W. E. (2005). Recombinant production in baculovirus-infected insect cells and purification of the mammalian Sec13/Sec31 complex. Methods Enzymol. (in press). [DOI] [PubMed]
- Hammer, J. A., 3rd, and Wu, X. S. (2002). Rabs grab motors: defining the connections between Rab GTPases and motor proteins. Curr. Opin. Cell Biol. 14, 69-75. [DOI] [PubMed] [Google Scholar]
- Heidtman, M., Chen, C. Z., Collins, R. N., and Barlowe, C. (2003). A role for Yip1p in COPII vesicle biogenesis. J. Cell Biol. 163, 57-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinners, I., and Tooze, S. A. (2003). Changing directions: clathrin-mediated transport between the Golgi and endosomes. J. Cell Sci. 116, 763-771. [DOI] [PubMed] [Google Scholar]
- Hirokawa, N. (2000). Stirring up development with the heterotrimeric kinesin KIF3. Traffic 1, 29-34. [DOI] [PubMed] [Google Scholar]
- Horiuchi, H., Lippe, R., McBride, H. M., Rubino, M., Woodman, P., Stenmark, H., Rybin, V., Wilm, M., Ashman, K., Mann, M., and Zerial, M. (1997). A novel Rab5 GDP/GTP exchange factor complexed to Rabaptin-5 links nucleotide exchange to effector recruitment and function. Cell 90, 1149-1159. [DOI] [PubMed] [Google Scholar]
- Hutt, D. M., Da-Silva, L. F., Chang, L. H., Prosser, D. C., and Ngsee, J. K. (2000). PRA1 inhibits the extraction of membrane-bound rab GTPase by GDI1. J. Biol. Chem. 275, 18511-18519. [DOI] [PubMed] [Google Scholar]
- Izumi, T., Gomi, H., Kasai, K., Mizutani, S., and Torii, S. (2003). The roles of Rab27 and its effectors in the regulated secretory pathways. Cell Struct. Funct. 28, 465-474. [DOI] [PubMed] [Google Scholar]
- Jones, S., Newman, C., Liu, F., and Segev, N. (2000). The TRAPP complex is a nucleotide exchanger for Ypt1 and Ypt31/32. Mol. Biol. Cell 11, 4403-4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karcher, R. L., Deacon, S. W., and Gelfand, V. I. (2002). Motor-cargo interactions: the key to transport specificity. Trends Cell Biol. 12, 21-27. [DOI] [PubMed] [Google Scholar]
- Katoh, M. (2004). Characterization of RUSC1 and RUSC2 genes in silico. Oncol. Rep. 12, 933-938. [PubMed] [Google Scholar]
- Kawabe, H., Sakisaka, T., Yasumi, M., Shingai, T., Izumi, G., Nagano, F., Deguchi-Tawarada, M., Takeuchi, M., Nakanishi, H., and Takai, Y. (2003). A novel rabconnectin-3-binding protein that directly binds a GDP/GTP exchange protein for Rab3A small G protein implicated in Ca(2+)-dependent exocytosis of neurotransmitter. Genes Cells 8, 537-546. [DOI] [PubMed] [Google Scholar]
- Kirchhausen, T. (2000). Three ways to make a vesicle. Nat. Rev. Mol. Cell. Biol. 1, 187-198. [DOI] [PubMed] [Google Scholar]
- Koh, T. W., and Bellen, H. J. (2003). Synaptotagmin I, a Ca2+ sensor for neurotransmitter release. Trends Neurosci. 26, 413-422. [DOI] [PubMed] [Google Scholar]
- Kuehn, M. J., Herrmann, J. M., and Schekman, R. (1998). COPII-cargo interactions direct protein sorting into ER-derived transport vesicles. Nature 391, 187-190. [DOI] [PubMed] [Google Scholar]
- Lafer, E. M. (2002). Clathrin-protein interactions. Traffic 3, 513-520. [DOI] [PubMed] [Google Scholar]
- Lederkremer, G. Z., Cheng, Y., Petre, B. M., Vogan, E., Springer, S., Schekman, R., Walz, T., and Kirchhausen, T. (2001). Structure of the Sec23p/24p and Sec13p/31p complexes of COPII. Proc. Natl. Acad. Sci. USA 98, 10704-10709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, M. C., Miller, E. A., Goldberg, J., Orci, L., and Schekman, R. (2004). Bi-directional protein transport between the ER and Golgi. Annu. Rev. Cell Dev. Biol. 20, 87-123. [DOI] [PubMed] [Google Scholar]
- Lonart, G., Janz, R., Johnson, K. M., and Sudhof, T. C. (1998). Mechanism of action of rab3A in mossy fiber LTP. Neuron 21, 1141-1150. [DOI] [PubMed] [Google Scholar]
- Lowe, M., and Kreis, T. E. (1995). In vitro assembly and disassembly of coatomer. J. Biol. Chem. 270, 31364-31371. [DOI] [PubMed] [Google Scholar]
- Lowe, S. L., Peter, F., Subramaniam, V. N., Wong, S. H., and Hong, W. (1997). A SNARE involved in protein transport through the Golgi apparatus. Nature 389, 881-884. [DOI] [PubMed] [Google Scholar]
- Lu, Q., Hope, L. W., Brasch, M., Reinhard, C., and Cohen, S. N. (2003). TSG101 interaction with HRS mediates endosomal trafficking and receptor down-regulation. Proc. Natl. Acad. Sci. USA 100, 7626-7631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, H. R., Saiardi, A., Nagata, E., Ye, K., Yu, H., Jung, T. S., Luo, X., Jain, S., Sawa, A., and Snyder, S. H. (2001). GRAB: a physiologic guanine nucleotide exchange factor for Rab3A, which interacts with inositol hexakisphosphate kinase. Neuron 31, 439-451. [DOI] [PubMed] [Google Scholar]
- Maehama, T., Taylor, G. S., and Dixon, J. E. (2001). PTEN and myotubularin: novel phosphoinositide phosphatases. Annu. Rev. Biochem. 70, 247-279. [DOI] [PubMed] [Google Scholar]
- Martin, T. F. (2001). PI(4,5)P(2) regulation of surface membrane traffic. Curr. Opin. Cell Biol. 13, 493-499. [DOI] [PubMed] [Google Scholar]
- Martina, J. A., Bonangelino, C. J., Aguilar, R. C., and Bonifacino, J. S. (2001). Stonin 2, an adaptor-like protein that interacts with components of the endocytic machinery. J. Cell Biol. 153, 1111-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miaczynska, M., Christoforidis, S., Giner, A., Shevchenko, A., Uttenweiler-Joseph, S., Habermann, B., Wilm, M., Parton, R. G., and Zerial, M. (2004). APPL proteins link Rab5 to nuclear signal transduction via an endosomal compartment. Cell 116, 445-456. [DOI] [PubMed] [Google Scholar]
- Miaczynska, M., and Zerial, M. (2002). Mosaic organization of the endocytic pathway. Exp. Cell Res. 272, 8-14. [DOI] [PubMed] [Google Scholar]
- Morimoto, S., Nishimura, N., Terai, T., Manabe, S., Yamamoto, Y., Shinahara, W., Miyake, H., Tashiro, S., Shimada, M., and Sasaki, T. (2005). Rab13 mediates the continuous endocytic recycling of occludin to the cell surface. J. Biol. Chem. 280, 2220-2228. [DOI] [PubMed] [Google Scholar]
- Moyer, B. D., Allan, B. B., and Balch, W. E. (2001). Rab1 interaction with a GM130 effector complex regulates COPII vesicle cis-Golgi tethering. Traffic 2, 268-276. [DOI] [PubMed] [Google Scholar]
- Navone, F., Consalez, G. G., Sardella, M., Caspani, E., Pozzoli, O., Frassoni, C., Morlacchi, E., Sitia, R., Sprocati, T., and Cabibbo, A. (2001). Expression of KIF3C kinesin during neural development and in vitro neuronal differentiation. J. Neurochem. 77, 741-753. [DOI] [PubMed] [Google Scholar]
- Nickel, W., Brugger, B., and Wieland, F. T. (2002). Vesicular transport: the core machinery of COPI recruitment and budding. J. Cell Sci. 115, 3235-3240. [DOI] [PubMed] [Google Scholar]
- Ohya, T., Sasaki, T., Kato, M., and Takai, Y. (1998). Involvement of Rabphilin3 in endocytosis through interaction with Rabaptin5. J. Biol. Chem. 273, 613-617. [DOI] [PubMed] [Google Scholar]
- Opdam, F. J., Kamps, G., Croes, H., van Bokhoven, H., Ginsel, L. A., and Fransen, J. A. (2000). Expression of Rab small GTPases in epithelial Caco-2 cells: Rab21 is an apically located GTP-binding protein in polarised intestinal epithelial cells. Eur. J. Cell Biol. 79, 308-316. [DOI] [PubMed] [Google Scholar]
- Otomo, A. et al. (2003). ALS2, a novel guanine nucleotide exchange factor for the small GTPase Rab5, is implicated in endosomal dynamics. Hum. Mol. Genet. 12, 1671-1687. [DOI] [PubMed] [Google Scholar]
- Panda, S., Antoch, M. P., Miller, B. H., Su, A. I., Schook, A. B., Straume, M., Schultz, P. G., Kay, S. A., Takahashi, J. S., and Hogenesch, J. B. (2002). Coordinated transcription of key pathways in the mouse by the circadian clock. Cell 109, 307-320. [DOI] [PubMed] [Google Scholar]
- Park, J. B., Farnsworth, C. C., and Glomset, J. A. (1997). Ca2+/calmodulin causes Rab3A to dissociate from synaptic membranes. J. Biol. Chem. 272, 20857-20865. [DOI] [PubMed] [Google Scholar]
- Pereira-Leal, J. B., and Seabra, M. C. (2000). The mammalian Rab family of small GTPases: definition of family and subfamily sequence motifs suggests a mechanism for functional specificity in the Ras superfamily. J. Mol. Biol. 301, 1077-1087. [DOI] [PubMed] [Google Scholar]
- Pereira-Leal, J. B., and Seabra, M. C. (2001). Evolution of the Rab family of small GTP-binding proteins. J. Mol. Biol. 313, 889-901. [DOI] [PubMed] [Google Scholar]
- Pfeffer, S. (2003). Membrane domains in the secretory and endocytic pathways. Cell 112, 507-517. [DOI] [PubMed] [Google Scholar]
- Pfeffer, S., and Aivazian, D. (2004). Targeting Rab GTPases to distinct membrane compartments. Nat. Rev. Mol. Cell. Biol. 5, 886-896. [DOI] [PubMed] [Google Scholar]
- Pfeffer, S. R. (2001). Rab GTPases: specifying and deciphering organelle identity and function. Trends Cell Biol. 11, 487-491. [DOI] [PubMed] [Google Scholar]
- Prekeris, R. (2003). Rabs, Rips, FIPs, and endocytic membrane traffic. Sci. World J. 3, 870-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quackenbush, J. (2001). Computational analysis of microarray data. Nat. Rev. Genet. 2, 418-427. [DOI] [PubMed] [Google Scholar]
- Quevillon, E., Spielmann, T., Brahimi, K., Chattopadhyay, D., Yeramian, E., and Langsley, G. (2003). The Plasmodium falciparum family of Rab GTPases. Gene 306, 13-25. [DOI] [PubMed] [Google Scholar]
- Riederer, M. A., Soldati, T., Shapiro, A. D., Lin, J., and Pfeffer, S. R. (1994). Lysosome biogenesis requires Rab9 function and receptor recycling from endosomes to the trans-Golgi network. J. Cell Biol. 125, 573-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, M. S. (2004). Adaptable adaptors for coated vesicles. Trends Cell Biol. 14, 167-174. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Gabin, A. G., Almazan, G., and Larocca, J. N. (2004). Vesicle transport in oligodendrocytes: probable role of Rab40c protein. J. Neurosci. Res 76, 758-770. [DOI] [PubMed] [Google Scholar]
- Rosa, J. L., Casaroli-Marano, R. P., Buckler, A. J., Vilaro, S., and Barbacid, M. (1996). p619, a giant protein related to the chromosome condensation regulator RCC1, stimulates guanine nucleotide exchange on ARF1 and Rab proteins. EMBO J. 15, 4262-4273. [PMC free article] [PubMed] [Google Scholar]
- Rutherford, S., and Moore, I. (2002). The Arabidopsis Rab GTPase family: another enigma variation. Curr. Opin. Plant Biol. 5, 518-528. [DOI] [PubMed] [Google Scholar]
- Satoh, A., Wang, Y., Malsam, J., Beard, M. B., and Warren, G. (2003). Golgin-84 is a rab1 binding partner involved in Golgi structure. Traffic 4, 153-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh, A. K., Tokunaga, F., and Ozaki, K. (1997). Rab proteins of Drosophila melanogaster: novel members of the Rab-protein family. FEBS Lett. 404, 65-69. [DOI] [PubMed] [Google Scholar]
- Schlierf, B., Fey, G. H., Hauber, J., Hocke, G. M., and Rosorius, O. (2000). Rab11b is essential for recycling of transferrin to the plasma membrane. Exp. Cell Res. 259, 257-265. [DOI] [PubMed] [Google Scholar]
- Schluter, O. M., Khvotchev, M., Jahn, R., and Sudhof, T. C. (2002). Localization versus function of Rab3 proteins. Evidence for a common regulatory role in controlling fusion. J. Biol. Chem. 277, 40919-40929. [DOI] [PubMed] [Google Scholar]
- Seabra, M. C., Mules, E. H., and Hume, A. N. (2002). Rab GTPases, intracellular traffic and disease. Trends Mol. Med. 8, 23-30. [DOI] [PubMed] [Google Scholar]
- Segev, N. (2001). Ypt and Rab GTPases: insight into functions through novel interactions. Curr. Opin. Cell Biol. 13, 500-511. [DOI] [PubMed] [Google Scholar]
- Shorter, J., Watson, R., Giannakou, M. E., Clarke, M., Warren, G., and Barr, F. A. (1999). GRASP55, a second mammalian GRASP protein involved in the stacking of Golgi cisternae in a cell-free system. EMBO J. 18, 4949-4960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoulders, C. C., Stephens, D. J., and Jones, B. (2004). The intracellular transport of chylomicrons requires the small GTPase, Sar1b. Curr. Opin. Lipidol. 15, 191-197. [DOI] [PubMed] [Google Scholar]
- Simonsen, A., Wurmser, A. E., Emr, S. D., and Stenmark, H. (2001). The role of phosphoinositides in membrane transport. Curr. Opin. Cell Biol. 13, 485-492. [DOI] [PubMed] [Google Scholar]
- Sirotkin, H., Morrow, B., DasGupta, R., Goldberg, R., Patanjali, S. R., Shi, G., Cannizzaro, L., Shprintzen, R., Weissman, S. M., and Kucherlapati, R. (1996). Isolation of a new clathrin heavy chain gene with muscle-specific expression from the region commonly deleted in velo-cardio-facial syndrome. Hum. Mol. Genet. 5, 617-624. [DOI] [PubMed] [Google Scholar]
- Sivars, U., Aivazian, D., and Pfeffer, S. R. (2003). Yip3 catalyses the dissociation of endosomal Rab-GDI complexes. Nature 425, 856-859. [DOI] [PubMed] [Google Scholar]
- Sollner, T. H. (2002). Vesicle tethers promoting fusion machinery assembly. Dev. Cell 2, 377-378. [DOI] [PubMed] [Google Scholar]
- Sollner, T. H. (2003). Regulated exocytosis and SNARE function (Review). Mol. Membr. Biol. 20, 209-220. [DOI] [PubMed] [Google Scholar]
- Spang, A. (2004). Vesicle transport: a close collaboration of Rabs and effectors. Curr. Biol. 14, R33-34. [DOI] [PubMed] [Google Scholar]
- Su, A. I. et al. (2004). A gene atlas of the mouse and human protein-encoding transcriptomes. Proc. Natl. Acad. Sci. USA 101, 6062-6067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniam, V. N., Loh, E., Horstmann, H., Habermann, A., Xu, Y., Coe, J., Griffiths, G., and Hong, W. (2000). Preferential association of syntaxin 8 with the early endosome. J. Cell Sci. 113(Pt 6), 997-1008. [DOI] [PubMed] [Google Scholar]
- Sudhof, T. C. (2002). Synaptotagmins: why so many? J. Biol. Chem. 277, 7629-7632. [DOI] [PubMed] [Google Scholar]
- Sudhof, T. C. (2004). The synaptic vesicle cycle. Annu. Rev. Neurosci. 27, 509-547. [DOI] [PubMed] [Google Scholar]
- Swanson, D. A., Steel, J. M., and Valle, D. (1998). Identification and characterization of the human ortholog of rat STXBP1, a protein implicated in vesicle trafficking and neurotransmitter release. Genomics 48, 373-376. [DOI] [PubMed] [Google Scholar]
- Tall, G. G., Barbieri, M. A., Stahl, P. D., and Horazdovsky, B. F. (2001). Ras-activated endocytosis is mediated by the Rab5 guanine nucleotide exchange activity of RIN1. Dev Cell 1, 73-82. [DOI] [PubMed] [Google Scholar]
- Tang, B. L., Ong, Y. S., Huang, B., Wei, S., Wong, E. T., Qi, R., Horstmann, H., and Hong, W. (2001). A membrane protein enriched in endoplasmic reticulum exit sites interacts with COPII. J. Biol. Chem. 276, 40008-40017. [DOI] [PubMed] [Google Scholar]
- Tani, K., Mizoguchi, T., Iwamatsu, A., Hatsuzawa, K., and Tagaya, M. (1999). p125 is a novel mammalian Sec23p-interacting protein with structural similarity to phospholipid-modifying proteins. J. Biol. Chem. 274, 20505-20512. [DOI] [PubMed] [Google Scholar]
- Tisdale, E. J., Bourne, J. R., Khosravi-Far, R., Der, C. J., and Balch, W. E. (1992). GTP-binding mutants of rab1 and rab2 are potent inhibitors of vesicular transport from the endoplasmic reticulum to the Golgi complex. J. Cell Biol. 119, 749-761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolmachova, T., Anders, R., Stinchcombe, J., Bossi, G., Griffiths, G. M., Huxley, C., and Seabra, M. C. (2004). A general role for Rab27a in secretory cells. Mol. Biol. Cell 15, 332-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomoda, T., Kim, J. H., Zhan, C., and Hatten, M. E. (2004). Role of Unc51.1 and its binding partners in CNS axon outgrowth. Genes Dev. 18, 541-558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trambas, C. M., and Griffiths, G. M. (2003). Delivering the kiss of death. Nat. Immunol. 4, 399-403. [DOI] [PubMed] [Google Scholar]
- Ungar, D., and Hughson, F. M. (2003). SNARE protein structure and function. Annu. Rev. Cell Dev. Biol. 19, 493-517. [DOI] [PubMed] [Google Scholar]
- Ungar, D., Oka, T., Brittle, E. E., Vasile, E., Lupashin, V. V., Chatterton, J. E., Heuser, J. E., Krieger, M., and Waters, M. G. (2002). Characterization of a mammalian Golgi-localized protein complex, COG, that is required for normal Golgi morphology and function. J. Cell Biol. 157, 405-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valsdottir, R., Hashimoto, H., Ashman, K., Koda, T., Storrie, B., and Nilsson, T. (2001). Identification of rabaptin-5, rabex-5, and GM130 as putative effectors of rab33b, a regulator of retrograde traffic between the Golgi apparatus and ER. FEBS Lett. 508, 201-209. [DOI] [PubMed] [Google Scholar]
- van IJzendoorn, S. C., Tuvim, M. J., Weimbs, T., Dickey, B. F., and Mostov, K. E. (2002). Direct interaction between Rab3b and the polymeric Ig receptor controls ligand-stimulated transcytosis in epithelial cells. Dev. Cell 2, 219-228. [DOI] [PubMed] [Google Scholar]
- von Schwedler, U. K. et al. (2003). The protein network of HIV budding. Cell 114, 701-713. [DOI] [PubMed] [Google Scholar]
- Wagner, A. C., Strowski, M. Z., Goke, B., and Williams, J. A. (1995). Molecular cloning of a new member of the Rab protein family, Rab 26, from rat pancreas. Biochem. Biophys. Res. Commun. 207, 950-956. [DOI] [PubMed] [Google Scholar]
- Walker, J. R., Su, A. I., Self, D. W., Hogenesch, J. B., Lapp, H., Maier, R., Hoyer, D., and Bilbe, G. (2004). Applications of a rat multiple tissue gene expression data set. Genome Res. 14, 742-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, W., and Ferro-Novick, S. (2002). A Ypt32p exchange factor is a putative effector of Ypt1p. Mol. Biol. Cell 13, 3336-3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X., Kumar, R., Navarre, J., Casanova, J. E., and Goldenring, J. R. (2000). Regulation of vesicle trafficking in madin-darby canine kidney cells by Rab11a and Rab25. J. Biol. Chem. 275, 29138-29146. [DOI] [PubMed] [Google Scholar]
- Weigert, R., Yeung, A. C., Li, J., and Donaldson, J. G. (2004). Rab22a regulates the recycling of membrane proteins internalized independently of clathrin. Mol. Biol. Cell 15, 3758-3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman, J. T., Plutner, H., and Balch, W. E. (2001). The mammalian guanine nucleotide exchange factor mSec12 is essential for activation of the Sar1 GTPase directing endoplasmic reticulum export. Traffic 2, 465-475. [DOI] [PubMed] [Google Scholar]
- Williams, T. M., and Lisanti, M. P. (2004). The caveolin proteins. Genome Biol. 5, 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, G. et al. (2001). Increased myocardial Rab GTPase expression: a consequence and cause of cardiomyopathy. Circ. Res. 89, 1130-1137. [DOI] [PubMed] [Google Scholar]
- Wu, S. K., Zeng, K., Wilson, I. A., and Balch, W. E. (1996). Structural insights into the function of the Rab GDI superfamily. Trends Biochem. Sci. 21, 472-476. [DOI] [PubMed] [Google Scholar]
- Yap, C. C., Murate, M., Kishigami, S., Muto, Y., Kishida, H., Hashikawa, T., and Yano, R. (2003). Adaptor protein complex-4 (AP-4) is expressed in the central nervous system neurons and interacts with glutamate receptor delta2. Mol. Cell Neurosci. 24, 283-295. [DOI] [PubMed] [Google Scholar]
- Yoshie, S., Imai, A., Nashida, T., and Shimomura, H. (2000). Expression, characterization, and localization of Rab26, a low molecular weight GTP-binding protein, in the rat parotid gland. Histochem. Cell Biol. 113, 259-263. [DOI] [PubMed] [Google Scholar]
- Yoshihisa, T., Barlowe, C., and Schekman, R. (1993). Requirement for a GTPase-activating protein in vesicle budding from the endoplasmic reticulum. Science 259, 1466-1468. [DOI] [PubMed] [Google Scholar]
- Zerial, M., and McBride, H. (2001). Rab proteins as membrane organizers. Nat. Rev. Mol. Cell. Biol. 2, 107-117. [DOI] [PubMed] [Google Scholar]
- Zheng, J. Y., Koda, T., Fujiwara, T., Kishi, M., Ikehara, Y., and Kakinuma, M. (1998). A novel Rab GTPase, Rab33B, is ubiquitously expressed and localized to the medial Golgi cisternae. J. Cell Sci. 111(Pt 8), 1061-1069. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.