Abstract
The embryonic genome is formed by fusion of a maternal and a paternal genome. To accommodate the resulting diploid genome in the fertilized oocyte dramatic global genome reorganizations must occur. The higher order structure of chromatin in vivo is critically dependent on architectural chromatin proteins, with the family of linker histone proteins among the most critical structural determinants. Although somatic cells contain numerous linker histone variants, only one, H1FOO, is present in mouse oocytes. Upon fertilization H1FOO rapidly populates the introduced paternal genome and replaces sperm-specific histone-like proteins. The same dynamic replacement occurs upon introduction of a nucleus during somatic cell nuclear transfer. To understand the molecular basis of this dynamic histone replacement process, we compared the localization and binding dynamics of somatic H1 and oocyte-specific H1FOO and identified the molecular determinants of binding to either oocyte or somatic chromatin in living cells. We find that although both histones associate readily with chromatin in nuclei of somatic cells, only H1FOO is capable of correct chromatin association in the germinal vesicle stage oocyte nuclei. This specificity is generated by the N-terminal and globular domains of H1FOO. Measurement of in vivo binding properties of the H1 variants suggest that H1FOO binds chromatin more tightly than somatic linker histones. We provide evidence that both the binding properties of linker histones as well as additional, active processes contribute to the replacement of somatic histones with H1FOO during nuclear transfer. These results provide the first mechanistic insights into the crucial step of linker histone replacement as it occurs during fertilization and somatic cell nuclear transfer.
INTRODUCTION
The basic unit of chromatin is the nucleosome, which consists of 146 bp of DNA wrapped around an octamer of core histones. Single nucleosomes are connected by linker DNA to generate higher order chromatin structures. Members of the family of H1 linker histones bind in a sequence independent manner to the stretch of DNA connecting adjacent nucleosomes (Hansen, 2002; Bustin et al., 2005). Because association of linker histones leads to chromatin compaction, H1 is generally considered a transcriptional repressor both globally and in a gene-specific manner (Sera and Wolffe, 1998; Lee et al., 2004; Bustin et al., 2005). Binding of H1 to chromatin is dynamic with an estimated residence time of H1 molecules on chromatin of 2-3 min and other structural chromatin proteins, including the HMG proteins, dynamically competing for binding to H1 sites (Catez et al., 2002; Bustin et al., 2005).
Several linker histone variants exist in vertebrates (Khochbin, 2001). The variants are classified according to their tightly regulated expression pattern during embryonal development and cell differentiation (Khochbin, 2001). In mice the linker histones of the somatic lineage are the replication-dependent H1 isoforms, comprised of H1a through H1e, and the differentiation-dependent histone H1F0 (formerly referred to as H10). Linker histones are functionally redundant but expression levels of the variants are tightly controlled (Fan et al., 2001). All known H1 variants share a common domain structure. They consist of a short N-terminus, a central globular domain and a long C-terminal domain. The globular domain contains a helix-loop-helix motif and is believed to be responsible for specific positioning of the linker histone onto the nucleosome (Draves et al., 1992; Thomas et al., 1992; Ramakrishnan et al., 1993). The C-terminus, in contrast, appears unstructured and is highly negatively charged, thus possibly aiding the nonspecific binding of H1 to DNA. Both the globular domain and the C-terminus are essential for efficient binding and normal dynamic exchange (Hansen, 2002; Misteli et al., 2000). How H1 is positioned on nucleosomes in vivo is unknown.
In addition to the somatic linker histones, germline-specific linker histones are found in both sexes. Although the testis-specific linker histone H1t is expressed in pachytene spermatocytes (Meistrich et al., 1985; Churikov et al., 2004), the oocyte-specific linker histone H1FOO is expressed exclusively during development from fully meiotically competent GV-stage oocytes until the late two-cell stage embryo. H1FOO is the predominant, if not sole, linker histone in primary follicle 8 d postnatally and it is the only linker histone expressed at significant levels during these stages (Tanaka et al., 2001, 2003). H1FOO shares the general domain structure of the somatic linker histones (Tanaka et al., 2001). The protein is most closely related to the oocyte-specific cleavage stage histone (cs-H1) of sea urchin and the B4/H1M histone of Xenopus laevis, but shows only very limited sequence identity with somatic linker histones (Tanaka et al., 2003). Despite the substantial overall sequence differences, H1FOO shares an almost invariant octapeptide sequence with the somatic linker histones within the globular domain. H1FOO exists as two alternatively spliced forms (α and β) that originate from a single copy gene (Tanaka et al., 2004). The two proteins differ in the length of their C-terminal domain, with H1FOOα being the longer version. No functional differences between the two H1FOO isoforms are known.
The existence of an oocyte-specific linker histone suggests a dedicated function for this variant in oocyte maturation or fertilization. An obvious task for a specialized linker histone might arise from the requirement for the newly fertilized oocyte to remodel sperm chromatin. On entry of the sperm head, sperm-specific chromatin components are rapidly lost and H1FOO is integrated into the paternal chromosomes (Tanaka et al., 2001; Gao et al., 2004). This process requires dramatic global alterations of chromatin structure and appears to be accomplished solely by oocytic factors without the requirement for paternal components (McLay and Clarke, 2003). A similar extensive and rapid exchange of somatic linker histone with H1FOO is observed during somatic cell nuclear transfer (SCNT) in chromatin of an injected somatic nucleus in the process of cloning (Gao et al., 2004; Teranishi et al., 2004). Given the prominent role of H1FOO during fertilization and nuclear reprogramming in SCNT and its presumed functional involvement in global chromatin remodeling, we sought to analyze the dynamics of H1FOO binding in vivo. Here we characterize the differential behavior of somatic and oocyte-specific linker histone variants in living somatic cells and oocytes and we identify the intrinsic properties of H1FOO that determine its differential binding to either somatic or oocyte chromatin.
MATERIALS AND METHODS
Generation of Expression Vectors
The H1FOOα-GFP and H1FOOβ-GFP constructs were generated by insertion of the corresponding mouse cDNAs into the CT-GFP-TOPO vector (Invitrogen, Carlsbad, CA) as previously described (Tanaka et al., 2004). To generate the H1F0-GFP construct, the corresponding cDNA was PCR amplified from MTH10 GFPneo (Misteli et al., 2000). The PCR fragments were introduced into the pcDNA3.1/CT-GFP-TOPO vector (Invitrogen, Carlsbad, CA), following the TOPO T/A cloning protocol (Invitrogen). Primers to amplify H1F0 were as follows: ATGACCGAGAACTCCACCTC; TCTTCTTCTTGGCCTTCTTG.
To generate the H1FOOΔNG-GFP construct, the sequence coding for amino acids 134-346 of H1FOO α was PCR amplified using H1FOO-GFP as template. The PCR fragment was introduced into the pcDNA3.1/CT-GFP-TOPO vector (Invitrogen), following the TOPO T/A cloning protocol (Invitrogen). The primers were engineered to contain a transcription start site at the 5′ end. Primers (artificial start site underlined) were as follows: AGAAAGCCGGCAGGGGAGCTGCAGGTGCCA; GGCCTGAGTGTTCTCAGGGGTCTTTG.
To generate the H1FOOΔC-GFP construct the sequence comprising amino acids 1-152 of H1FOO was PCR amplified using H1FOO-GFP as template. The PCR fragment was introduced into the pcDNA3.1/CT-GFP-TOPO vector (Invitrogen), following the TOPO T/A cloning protocol (Invitrogen). Primers were as follows: GCCATGGCTCCTGGGAGTGTCTCCAGT; CCAATCCAGATTTTCTTGGAGCCTGTCT.
To generate the H1FOO(H1F0C)-GFP construct the sequence comprising the sequence coding for amino acids 101-194 of H1F0 was PCR amplified using H1F0-GFP as template. The primers were engineered to contain a 5′ NotI site and a 3′ XbaI site. The fragment was NotI/XbaI digested and introduced into the NotI/XbaI linearized H1FOOΔNG plasmid. The resulting H1FOO(H1F0C) construct encodes a chimeric protein consisting of amino acids 1-152 of H1FOOα, followed by aa 101-194 of H1F0.
To generate the H1FOO(H1F0G) plasmid the globular domain of H1F0 spanning amino acids 22-100 was PCR amplified. To allow subcloning of the fragment the primers were engineered to contain a 5′ NotI site and a 3′ Asp718 site. Primers (restriction sites underlined) were as follows: GGCGGCCGCTCCACGGACCACCCCAAGTAT; CCCCGGTACCTCATCGCCCTTGGCCAGCCTGAAGGA. The PCR fragment was digested with NotI and Asp718 and inserted into the corresponding sites of the pBluescript vector. A second fragment corresponding to amino acids 1-39 of H1FOO was generated by PCR amplification. For subcloning of this fragment the primers were engineered to contain a 5′ SacI, followed by an Asp718 site and a 3′ NotI site. Primers were as follows: GAGCTCGGTACCCATGGCTCCTGGGAGT; TGCGGCCGCTTCTGCGGCAACTTGG. The fragment was digested with SacI and NotI and inserted 5′ of the globular domain of H1F0 into the SacI/NotI-digested pBluescript vector containing the H1F0 globular domain fragment. This step created a DNA sequence consisting of the N-terminal domain of H1FOO in frame with the globular domain of H1F0 (N/G mix domain). The N/G mix domain was excised from the pBluescript backbone using Asp718 and inserted into the Asp718 digested H1FOOΔNG plasmid. The resulting H1FOO(H1F0G) construct encodes a chimeric protein consisting of amino acids 1-39 of H1FOOα, amino acids 22-100 of H1F0, followed by amino acids 134-304 of H1FOO.
The H1F0CC-GFP construct was generated by PCR amplification of the carboxy terminal domain and concomitant introduction of convenient restriction enzyme sites on the ends. This fragment was then spliced into the parent MTH10GFPneo expression construct. Subsequently, the chimeric DNA encoding the H1F0 with a duplicated C-terminal domain was PCR amplified. The primers used for the amplification of H1F0 (see above) were used in the PCR reaction. This reaction yielded a short and a long product, with the long product corresponding to H1F0 with the duplicated C-terminus. This fragment was introduced into the pcDNA3.1/CT-GFP-TOPO vector (Invitrogen), following the TOPO T/A cloning protocol (Invitrogen). All constructs were confirmed by sequencing.
Live Cell Microscopy and Fluorescence Recovery after Photobleaching Analysis
3134 mouse cells were cultured in DMEM supplemented with 10% fetal bovine serum (FBS) at 37°C.
Twenty-four hours before transfection the cells were plated into 35-mm glass-bottom dishes (MatTEK, Ashland, MA) at a concentration of 2 × 105 cells per dish in Phenol Red-free DMEM supplemented with 10% FBS. Transient transfection was carried out using the Lipofectamine 2000 transfection procedure (Invitrogen) with 2 μg DNA. Eighteen hours after transfection, the cells were imaged in the same medium supplemented with 20 nM HEPES (pH 7.3).
For live oocyte imaging mRNAs corresponding to the indicated constructs were generated by in vitro transcription using the Ambion T7 mRNA Machine kit and then polyadenylated using the Ambion polyA tailing kit (Ambion, Austin, TX) according to the manufacturer's instructions. Oocytes were harvested from 21-d-old C57Bl/6J mice 45-48 h after pregnant mare's serum gonadotropin (PMSG) injection. The oocytes were subsequently cultured at 37°C in M2 medium supplemented with 10 μM milrinone. Microinjection of mRNAs was carried out by standard techniques. Intact living oocytes were imaged 18-20 h after microinjection.
Live cell imaging and fluorescence recovery after photobleaching (FRAP) analysis of 3134 cells and oocytes was carried out on a temperature-controlled stage using a Zeiss 510 Laser scanning confocal microscope (Carl Zeiss, Oberkochen, Germany) as previously described (Becker et al., 2002).
Nuclear Transfer Experiments
MII-stage oocytes were obtained from adult (B6D2)F1 8-12-wk-old females by superovulation, and nuclear transfers were performed by piezo-mediated injection as described (Gao et al., 2004). Briefly, embryonic stem cells (R1) expressing H1F0-GFP were injected into the ooplasm of an enucleated MII-stage oocyte and under the zona pellucida (control). The cloned constructs were incubated in CZB medium supplemented with glucose (CZB-G) at 37°C, 5% CO2, and transferred after 5 min, 1 h, or 3 h to small drops of phosphate-buffered saline (PBS) for confocal microscopy observation. Immunofluorescence procedure was performed on three different pools of R1 ES cell nuclei constructs that had been kept in CZB-G for 5 min, 1 h, or 3 h and then fixed in 4% paraformaldehyde (Sigma, St. Luis, MO) in PBS for 30 min at 37°C. The fixed constructs were permeabilized for 20 min in 0.5% Triton X-100 (Sigma), incubated 1 h in PBS + 0.5% Tween + 2% bovine serum albumin, and then overnight at 4°C with the primary antibody, a rabbit polyclonal antibodies against the somatic linker histone (H1F0) or the oocyte-specific linker histone (H1FOO). Constructs were incubated for 1 h at room temperature in FITC-conjugated donkey anti-rabbit IgG (Jackson ImmunoResearch Laboratories, West Grove, PA), DNA labeled with propidium iodide (PI, Trevigen, Gaithersburg, MD) for 30 min, deposited on slides, and mounted in Citifluor (AF1, Citifluor Products, Canterbury, United Kingdom). Confocal microscopy was performed on an inverted Olympus (Olympus America, Lake Success, NY) microscope equipped with the Fluorview confocal lasers scanning software (Olympus America).
RESULTS
Dynamics of Replacement of H1F0 by H1FOO during ES Cell Nuclear Transfer
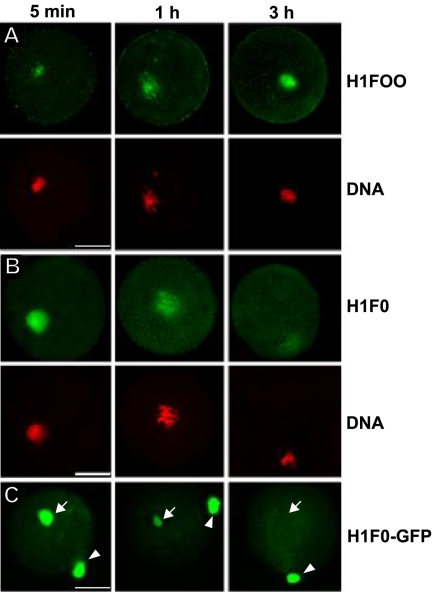
To monitor the kinetics of replacement of somatic linker histone with oocyte-specific H1FOO, we introduced isolated nuclei of R1 ES cells into stage II mouse oocytes and followed endogenous H1FOO and somatic H1 isoforms using indirect immunofluorescence (Figure 1, A and B). H1FOO rapidly populated the introduced ES cell nucleus. Traces of H1FOO were visible on the introduced R1 chromatin as soon as 5 min after nuclear transfer increasing over the next 3 h (Figure 1A). The association of H1FOO was paralleled by loss of the endogenous somatic linker histone from the introduced nucleus within 3 h with some residual levels visible at the end of this period (Figure 1B). This time course was somewhat slower than that observed after SCNT of cumulus and myoblast nuclei (Gao et al., 2004; Teranishi et al., 2004) but comparable to that observed after SCNT of embryonic fibroblast nuclei (Teranishi et al., 2004), suggesting that the kinetics of the histone exchange upon SCNT are cell-type dependent. As observed for endogenous H1F0, stably expressed H1F0-GFP was lost from injected R1 nuclei with similar kinetics and was almost undetectable 3 h after nuclear transfer (Figure 1C). This latter observation indicates that the exogenously expressed H1F0-GFP shares dynamic properties of the endogenous somatic linker histone in the oocytic environment and is well suited for dynamic studies within this model system.
Figure 1.
Exchange of somatic linker histones with oocyte-specific linker histones following transfer of R1 ES cell nuclei. (A-C) Cloned constructs were prepared as previously described (Gao et al., 2003, 2004). R1 ES cell nuclei were injected into enucleated MII-stage oocytes and then cultured as previously described (Gao et al., 2003, 2004) for the indicated time points. (A and B) Exchange of somatic linker histones with H1FOO in chromatin of injected R1 ES cell nuclei. Constructs were fixed and imaged for DNA content, and either oocyte-specific linker histone (A) or somatic linker histone (B) content as described (Gao et al., 2004). (C) H1F0-GFP expressed from a chromosomal locus in a R1 ES cell nucleus is removed from R1 chromatin with a kinetic similar to that of the endogenous somatic H1. To monitor fluorescence loss by bleaching a second nucleus was placed in the perivitelline space next to the ooplasm for comparison. Arrow indicates the injected nucleus; arrow head indicates the nucleus placed in the perivitelline space. Scale bars, 20 μm.
Differential Localization of H1 Variants in Somatic Cells and Oocytes
To characterize potential differences between H1F0 and H1FOO we compared the in vivo subcellular distribution of GFP-tagged versions of the two proteins in GV stage oocytes and in somatic 3134 mouse epithelial cancer cells. H1F0-GFP and H1FOOα-GFP showed similar subnuclear distributions in somatic cells (Figure 2). Both proteins were for the most part excluded from nucleoli and localized throughout the nucleus, but accumulated in several distinct foci (Figure 2, A and C). These foci correspond to heterochromatic regions as previously demonstrated by colocalization of somatic H1F0 and H1C with HP1 and centromere proteins (Misteli et al., 2000). The localization pattern of the two isoforms was identical to the reported distribution of endogenous H1C and H1F0 and GFP-fusion proteins in murine somatic cells (Misteli et al., 2000). The localization patterns of both proteins was largely independent of the transgene expression level as previously observed (Lever et al., 2000; Misteli et al., 2000). The proper localization of the oocyte-specific isoform in somatic cells suggests that despite the significant differences in sequence, H1FOO has all necessary determinants for correct distribution in somatic chromatin.
Figure 2.
Representative examples of the subnuclear distribution of H1F0-GFP and H1FOOα-GFP in somatic cells and oocytes. (A and C) Distribution of H1F0-GFP and H1FOOα-GFP in somatic 3134 nuclei. Both H1 isotypes show a distribution characteristic for linker histones. Cells were transiently transfected as described in Materials and Methods. (B and C) Distribution of H1F0-GFP and H1FOOα-GFP in GV stage oocytes. GV-stage oocytes were injected with in vitro-transcribed polyadenylated RNA as previously described (Tanaka et al., 2004). Only H1FOO-GFP shows the stage-specific surrounding nucleolus distribution (B), whereas H1F0-GFP shows aberrant predominantly nucleolar distribution and illuminates only a few nucleoplasmic foci (D). Live cells were monitored using confocal laser scanning microscopy as previously described (Becker et al., 2002). Arrowheads indicate nucleoli; the GV is indicated by a dashed line. Scale bars, 3 mm.
To ask whether the somatic H1 isoform is able to localize properly in oocyte chromatin, we expressed H1F0-GFP, and as a control H1FOO-GFP, in GV stage oocytes by microinjection of polyA-mRNA. The two isoforms localized in dramatically different patterns. H1FOO-GFP accumulated strongly in chromatin in perinucleolar regions. Only very little GFP fluorescence was observed throughout the nucleoplasm and within the nucleolus (Figure 2B). This distribution was identical to that of endogenous H1FOO (Tanaka et al., 2004) and reflected the typical chromatin organization in oocytes at this developmental stage in the form of nucleolar surrounding chromatin (Miyara et al., 2003). In contrast, H1F0 localized predominantly to the nucleolus with only a low amount of the protein accumulating in distinct foci (Figure 2, B and D). The difference in subnuclear distribution of the different H1 variants could not be attributed to differing expression levels, because increasing the amount of injected RNA by up to 50-fold consistently produced the described distribution for both constructs (unpublished data). In addition, injection of approximately the same amount of RNA into somatic cells yielded a distribution for both fusion proteins identical to the distribution seen in transfected cells (unpublished data), indicating that the in vitro-transcribed RNA was properly suited for translation. No differences in localization were observed for H1FOO α and β in both somatic cells and oocytes (unpublished data). These results demonstrate that the cellular localization of linker histones is isoform-specific and they imply functional specificity between somatic and oocyte-specific linker histones.
Localization Determinants in H1FOO
To define the regions in H1FOO and H1F0 responsible for their differential localization in oocytes and somatic cells, we generated a set of deletion mutants of H1FOO and H1F0 (Figure 3A).
Figure 3.
The globular/N-terminal domain of H1FOO is responsible for the correct subnuclear distribution. (A) Schematic representation of the analyzed H1FOO deletion and chimeric constructs. N, N-terminal domain; G, globular domain; C, C-terminal domain. (B-I) Subnuclear distribution of H1FOO mutants in oocytes (B, D, F, and H) and 3134 cells (C, E, G, and I). (D and E) The N-terminal and globular domain of H1FOO are sufficient to mediated correct localization in oocyte and somatic nuclei. (F and G) The C-terminal domain of H1F0 fused to the N-terminal and globular domain of H1FOO shows a subnuclear localization similar to wild-type H1FOO in oocyte and somatic nuclei. (B, C, H, and I) Neither the C-terminal domain of H1FOO alone (B and C), nor the globular domain of H1F0 fused to the N-terminal and the C-terminal domain of H1FOO (H and I) shows correct localization in oocyte and somatic cell nuclei. Arrowheads indicate nucleoli. Scale bars, 3 mm.
Expression of the GFP-tagged C-terminal domain of H1FOO yielded exclusive nucleolar localization of this protein in oocytes and somatic cells (Figure 3, B and C). Thus, the C-terminal domain of H1FOO is not sufficient to mediate correct localization of H1FOO. In contrast, a mutant lacking the C-terminal domain but containing the full-length N-terminal and globular domain was sufficient for wild-type subnuclear distribution both in oocytes and somatic cells (Figure 3, D and E; H1FOOΔC).
To ask which part of the H1FOO molecule conferred its differentiation localization in oocytes compared with somatic H1F0, we tested the localization of chimeric H1F0/H1FOO fusion proteins (Figure 3A). A chimera consisting of the N-terminus and the globular domain of H1FOO and the C-terminal domain of H1F0 (H1FOO(H1F0C)) yielded the same distribution as wild-type H1FOO in both cellular systems (Figure 3, F and G). This observation demonstrates that the localization of H1FOO is not dependent on its C-terminus. In contrast, replacement of the globular domain of H1FOO with that of H1F0 resulted in an intermediate distribution in both the oocyte and in somatic cells (Figure 3, H and I). A significant fraction of the protein localized to the nucleolus, whereas some of the protein surrounded the nucleolus and was found within the nuclear compartment. This distribution suggests that the globular domain of H1FOO is essential for proper localization. We conclude that the globular domain, possibly in conjunction with the N-terminus, contributes significantly the oocyte-specific localization of H1FOO.
Differential Binding Dynamics of H1 Variants in Oocytes and Somatic Cells
Somatic linker histones bind dynamically to chromatin with a residence time on the order of minutes (Lever et al., 2000; Misteli et al., 2000). Because the oocyte-specific H1FOO rapidly populates the paternal genome after fertilization and the genome of the transferred somatic nucleus after SCNT, we sought to determined the dynamic binding properties of somatic and oocyte-specific linker histones in living somatic cells and oocytes. To this end we used FRAP because the FRAP recovery kinetics of chromatin proteins are directly related to the proteins' binding properties (Phair et al., 2004).
In living oocytes, H1FOO showed significantly slower FRAP recovery kinetics than somatic H1F0. Upon bleaching of a fraction of the chromatin signal, the half-time (τ1/2) of recovery for H1FOO was 31.8 s, whereas that of the nucleoplasmic H1F0 was ∼5.8 s (Figure 4C). This observation suggests tighter association of the oocyte-specific H1FOO with chromatin than H1F0 (Figure 4, A-C). The exchange of H1F0 inside the nucleolus was rapid with τ1/2 < 3 s (unpublished data). In somatic cells, recovery kinetics were similar with τ1/2 of ∼30 s for H1FOO and τ1/2 ∼6 s for H1F0 (Figure 5, A-C). The fact that H1FOO showed significantly slower recovery kinetics in the somatic nucleus than H1F0 demonstrates that the tighter binding is a property of H1FOO and does not depend on the chromatin.
Figure 4.
Dynamic exchange of linker histone isoforms and mutants with oocyte chromatin. (A and B) The nuclei of oocytes expressing either H1F0-GFP (A) or H1FOO-GFP (B) were imaged before and after photobleaching of chromatin foci at the indicated time points. The recovery of the fluorescent signal was monitored by time-lapse microscopy. A segment magnification of the bleached area indicated by the red rectangle is shown in false color below the corresponding panels. (C and D) Quantitation of recovery kinetics. For quantitation, at least four oocytes from two independent experiments were used. Scale bars, 3 mm.
Figure 5.
Dynamic exchange of linker histone isoforms and mutants with somatic chromatin. (A and B) 3134 cells expressing either H1F0-GFP (A) or H1FOO-GFP (B) were imaged before and after photobleaching of chromatin. The recovery of the fluorescent signal was monitored by time-lapse microscopy. (C and D) Quantitation of recovery kinetics. For quantitation, at least 10 cells from two independent experiments were used. Scale bars, 3 mm.
Because H1FOO was more tightly bound to chromatin than H1F0, it was possible that the differential localization of H1FOO and H1F0 in the oocytes nucleus was a direct consequence of the differential binding dynamics. To test this possibility, we determined the binding dynamics of all mutants and chimeras in order to correlate them with their localization patterns. The properly localized C-terminal deletion construct H1FOOΔC showed an exchange rate significantly faster (τ1/2 = 8 s) than the wild-type H1FOO and its binding dynamics were similar to those of H1F0 in oocyte chromatin (Figure 4D). The properly localized H1FOO-(H1F0C) showed even slower exchange dynamics (τ1/2 = 90 s) than the wild-type H1FOO, whereas the aberrantly localized H1FOO(H1F0G) was found to exchange more rapidly (τ1/2 = 3.5 s) than H1F0 (Figure 4D). Similar recovery kinetics of mutants and chimeras were observed in somatic nuclei (Figure 5D). These observations, and especially the rapid exchange dynamics of the properly localized H1FOOΔC, demonstrate that the aberrant localization of H1F0 in oocytes is not simply due to its reduced binding affinity to oocytic chromatin.
Localization and Replacement of a High-affinity H1F0
To test directly whether proper localization of H1FOO in oocytes was related to tighter association of H1FOO with chromatin, we engineered a somatic linker histone with increased binding affinity. We duplicated the C-terminal domain of H1F0 to generate an H1F0-CC mutant (Figure 6A). Increased binding of this mutant was confirmed by FRAP analysis in somatic cells, where H1F0-CC showed dramatically reduced recovery with τ1/2 <180 s, compared with 6 s for the wild-type protein (Figure 6B). When introduced into oocytes H1F0-CC revealed a similarly dramatically reduced exchange rate compared with either H1F0 or H1FOO (Figure 6C). However, the tightly binding H1F0-CC did not assume the distribution pattern of H1FOO in oocytes, localizing instead with an intermediate distribution of the mutant very similar to that found for H1FOO(H1F0G) (Figure 6E). We conclude that, although increased binding of the somatic H1 appears to aid in properly localizing the somatic linker histone in the oocyte, strong binding to chromatin alone is not sufficient to mediate the oocyte-specific distribution of H1FOO.
Figure 6.
Reduced dynamic exchange of H1F0CC-GFP is not sufficient for H1FOO-like localization in oocytes and does not block removal from R1 nuclei during SCNT. (A) Schematic representation of the H1F0CC-GFP chimeric protein. N, N-terminal domain; G, globular domain; C, C-terminal domain. (B and C) Quantitation of recovery kinetics of H1F0CC-GFP and H1F0-GFP in somatic cells (B) and oocytes (C). For quantitation of recovery kinetics in oocytes, four oocytes from two independent experiments were used. For quantitation of recovery kinetics in somatic cells, 10 cells from at least two independent experiments were used. Scale bar, 3 mm. (D and E) Representative example of the distribution of H1F0CC-GFP in 3134 nuclei (D) and oocyte nuclei (E). (F) R1 nuclear transfer experiment as described in Figure 1 (also see Materials and Methods). H1F0CC-GFP expressed from a chromosomal locus in a R1 ES cell nucleus is removed from R1 chromatin with a kinetic similar to that of the endogenous somatic H1 (compare Figure 1, B and C). To monitor fluorescence loss by bleaching a second nucleus was placed in the perivitelline space next to the ooplasm for comparison. Arrow indicates the injected nucleus; arrowhead indicates the nucleus placed in the perivitelline space. Scale bar, 20 mm.
The availability of the tighter binding H1F0-CC mutant also allowed us to test whether the loss of somatic H1 upon SCNT is simply due to competition between the more tightly binding H1FOO compared with the somatic H1F0 or involves active processes in the oocyte. We therefore transplanted nuclei from R1 ES cells expressing H1F0-CC-GFP into MII stage mouse oocytes. H1F0-CC-GFP was lost rapidly from the introduced nucleus with kinetics similar or somewhat faster than H1F0-wt-GFP (Figure 6F). This observation suggests that replacement of somatic H1 is an active process and that oocytic factors contribute to removing somatic H1s.
DISCUSSION
Fertilization and SCNT are characterized by dramatic, global chromatin remodeling events, including erasure and de novo establishment of DNA methylation patterns, global changes in histone modifications and the exchange of histones (Vignon et al., 2002). These events are largely, and possibly exclusively, driven by oocytic factors (McLay and Clarke, 2003). Here we have compared the cellular behavior of somatic and oocyte-specific linker histones in living oocytes and somatic cells. We find significant differences in the localization and binding properties of somatic and oocyte-specific histone H1 variants. We also find preliminary evidence for the existence of active mechanisms that mediate H1 linker histone transitions in the oocyte or upon SCNT.
Dynamic population of sperm chromatin during fertilization and of somatic chromatin during SCNT by H1FOO is a well-established phenomenon. On fertilization by intracytoplasmic sperm injection (ICSI) the paternal chromatin is populated within minutes by H1FOO and reaches significant levels within 60 min (Gao et al., 2004). An analogous event occurs during normal fertilization (Gao et al., 2004). Similarly, upon introduction of cumulus or myoblast nuclei during SCNT somatic histone H1 is lost from the somatic nucleus and replaced within 60 min by oocyte-specific H1 (Gao et al., 2004). We find that exchange of endogenous and exogenously introduced somatic H1 in R1 ES cells takes somewhat longer and is complete within 3 h. This difference suggests that the reprogramming and transformation of the somatic genome might differ among cell types. It will be interesting to determine systematically whether different somatic cell types undergo morphological changes and reprogramming of their gene expression profiles with different kinetics upon SCNT and whether these differences might be related to global chromatin organization.
Although somatic and oocyte-specific linker histones localize similarly in chromatin of somatic cells, we find significant differences in oocytes. In particular, the somatic H1 is unable to associate effectively with the oocyte chromatin and to assume an oocyte-specific distribution. Our analysis of a set of deletion mutants and isoform chimeras suggests that the N-terminus and the globular domains of histone H1 determine this localization. This conclusion is supported by the observation that the C-terminus of H1FOO alone gives aberrant localization and that a mutant including the N-terminal and globular domains is sufficient to give normal localization in oocytes. In addition, a chimeric protein containing the N-terminal and C-terminal domain of H1FOO but the globular domain of H1F0 was aberrantly localized in a similar manner as the full-length H1F0. Taken together, these observations show that the N and globular domains of both isoforms contribute significantly to their localization.
Somatic and oocyte-specific linker histones also differ significantly in their binding dynamics to chromatin in vivo. We find based on FRAP experiments that H1FOO binds significantly more tightly to chromatin than H1F0 in both somatic cells and in oocytes. Efficient binding of H1FOO in somatic cells and oocytes required all protein domains. As was observed for localization, the globular and N-terminal domain of H1FOO appear to contribute most strongly to the increased binding of the H1FOO, because replacement of the H1FOO globular domain with that of H1F0 resulted in dramatically reduced binding. This finding is in line with the requirement of both the C-terminal and the globular domain of mouse H1F0 and H1FC for full binding activity in somatic cells, where the globular domain appears to make a stronger contribution to binding than the C-terminal domain (Misteli et al., 2000). This behavior is consistent with the observation that both the N- and the C-terminus are involved in binding of human H1.1 (Hendzel et al., 2004). Interestingly, for H1.1 the C-terminus makes a stronger contribution than the N-terminal and globular domain (Hendzel et al., 2004), suggesting isoform dependent differences in the contribution of the individual subdomains to DNA binding.
The effect of the globular domain on binding strength was observed both in somatic cells and in oocytes, suggesting that the differential binding characteristics mediated by the globular domain are independent of the chromatin and are an intrinsic property of the globular domain. Despite significant overall differences, the highest degree of homology between H1FOO and other somatic linker histones including H1F0 is found within the globular domain. Structural analysis suggests that the classical “hist1-fold” motif of a three-helix bundle and a C-terminal β-sheet hairpin within the globular domain is the most likely conformation for the H1FOO globular domain (Kihara and Adashi, unpublished data). However, the globular domain of H1FOO is more highly positively charged than other somatic linker histones and this higher positive charge may facilitate a stronger electrostatic interaction with DNA and therefore stabilize H1FOO binding to the chromatosome. Although binding of human H1.1, and possibly other H1 isoforms, is modulated by phosphorylation of specific residues in the C-terminus (Hendzel et al., 2004; Lu and Hansen, 2004), we conclude that for H1FOO, H1FC, and H1F0 the major binding contribution appears to come from the globular and/or N-terminal domains. This observed strong contribution of the globular domain to binding is consistent with its proposed function in the specific binding of linker histones to the nucleosomes (Zhou et al., 1998). Whereas, in contrast, the highly charged C-terminus is likely involved in nonspecific interactions (Thomas, 1999).
Comparison of the effect of mutations on localization and dynamics showed that the dynamic binding behavior of the linker histones does not automatically determine the observed overall localization pattern. The exchange rate of the properly localized H1FOOΔC and H1FOO(H1F0C) was significantly faster or slower than the wild-type H1FOO, respectively. The most direct evidence for additional components in determining the distribution of the linker histones is the observation that a mutant containing a duplication of the somatic C-terminus, which results in greatly increased binding still does not localize properly in the oocyte. The fact that the more tightly binding somatic H1F0 mutant localizes more similar to the oocyte-specific H1FOO than wild-type H1F0 suggest, however, that binding strength might contribute to the localization pattern, but certainly does not determine it. Taken together these observations indicate that other factors than binding dynamics contribute to localization.
Although the higher binding affinity of H1FOO relative to somatic H1 isoforms likely contributes to rapid replacement of somatic histones with oocyte-specific histones upon fertilization or in SCNT, our observation that oocyte-specific H1FOO can still displace H1F0-CC with rapid kinetics strongly suggest that somatic linker histones are removed by an active process in the oocyte. Consistent with the existence of an active component to this transition, it was previously shown that the ability of the oocyte to mediate this transition is developmentally regulated (Gao et al., 2004). Although it is not clear what this process is, protein degradation might be one of its components because treatment of SCNT embryos with the proteasome inhibitor MG132 partially inhibits the H1 to H1FOO transition (Gao et al., 2005). Also, the observation that H1F0, which has a half-life on the order of several hours, is entirely lost from somatic nuclei within 1-3 h strongly implies active protein degradation in this process. Therefore it appears that, although the increased binding of H1FOO to chromatin may facilitate the replacement of somatic linker histones, at least one, and possibly additional oocyte-specific factors or processes contribute to the orchestrated loss of histones from somatic chromatin during SCNT. These same processes or factors may promote the H1 to H1FOO transition during normal fertilization as well, although comparison between SCNT into enucleated oocytes and normal fertilization is complicated by the possibility that critical chromatin associated proteins are removed during the enucleation process and the H1 transition observed in SCNT may not fully correspond to the event during normal fertilization. The identification of the factors involved in histone exchange will lead to a better understanding of the function of H1 in chromatin structure and should provide important insights into the processes of oogenesis, embryonic genome formation, and nuclear reprogramming during cloning.
Acknowledgments
We thank Paola Scaffidi and Lisa Garrett-Beal for their assistance with microinjection experiments and Tatiana Karpova for imaging support. Imaging was done at the National Cancer Institutes Fluorescence Imaging Facility. We are grateful to Hannes Drexler for critical review of the manuscript. This study was supported in part by a grant from the National Institutes of Health/National Institute of Child Health and Human Development (HD43092) to K.L. and from the National Science Foundation to D.T.B. T.M. is a Fellow of the Keith R. Porter Endowment for Cell Biology.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05-04-0350) on June 8, 2005.
References
- Becker, M., Baumann, C., John, S., Walker, D. A., Vigneron, M., McNally, J. G., and Hager, G. L. (2002). Dynamic behavior of transcription factors on a natural promoter in living cells. EMBO Rep. 3, 1188-1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin, M., Catez, F., and Lim, J. H. (2005). The dynamics of histone h1 function in chromatin. Mol. Cell 17, 617-620. [DOI] [PubMed] [Google Scholar]
- Catez, F., Brown, D. T., Misteli, T., and Bustin, M. (2002). Competition between histone H1 and HMGN proteins for chromatin binding sites. EMBO Rep. 3, 760-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churikov, D., Zalenskaya, I. A., and Zalensky, A. O. (2004). Male germline-specific histones in mouse and man. Cytogenet. Genome Res. 105, 203-214. [DOI] [PubMed] [Google Scholar]
- Draves, P. H., Lowary, P. T., and Widom, J. (1992). Co-operative binding of the globular domain of histone H5 to DNA. J. Mol. Biol. 225, 1105-1121. [DOI] [PubMed] [Google Scholar]
- Fan, Y., Sirotkin, A., Russell, R. G., Ayala, J., and Skoultchi, A. I. (2001). Individual somatic H1 subtypes are dispensable for mouse development even in mice lacking the H1(0) replacement subtype. Mol. Cell. Biol. 21, 7933-7943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, S., Chung, Y. G., Parseghian, M. H., King, G. J., Adashi, E. Y., and Latham, K. E. (2004). Rapid H1 linker histone transitions following fertilization or somatic cell nuclear transfer: evidence for a uniform developmental program in mice. Dev. Biol. 266, 62-75. [DOI] [PubMed] [Google Scholar]
- Gao, S., Han, Z., Kihara, M., Adashi, E., and Latham, K. E. (2005). Protease inhibitor MG132 in cloning: no end to the nightmare. Trends Biotechnol. 23, 66-68. [DOI] [PubMed] [Google Scholar]
- Hansen, J. C. (2002). Conformational dynamics of the chromatin fiber in solution: determinants, mechanisms, and functions. Annu. Rev. Biophys. Biomol. Struct. 31, 361-392. [DOI] [PubMed] [Google Scholar]
- Hendzel, M. J., Lever, M. A., Crawford, E. and Th'ng, J. P. (2004). The C-terminal domain is the primary determinant of histone H1 binding to chromatin in vivo. J. Biol. Chem. 279, 20028-20034. [DOI] [PubMed] [Google Scholar]
- Khochbin, S. (2001). Histone H1 diversity: bridging regulatory signals to linker histone function. Gene 271, 1-12. [DOI] [PubMed] [Google Scholar]
- Lee, H., Habas, R., and Abate-Shen, C. (2004). MSX1 cooperates with histone H1b for inhibition of transcription and myogenesis. Science 304, 1675-1678. [DOI] [PubMed] [Google Scholar]
- Lever, M. A., Th'ng, J. P., Sun, X., and Hendzel, M. J. (2000). Rapid exchange of histone H1.1 on chromatin in living human cells. Nature 408, 873-876. [DOI] [PubMed] [Google Scholar]
- Lu, X., and Hansen, J. C. (2004). Identification of specific functional subdomains within the linker histone H10 C-terminal domain. J. Biol. Chem. 279, 8701-8707. [DOI] [PubMed] [Google Scholar]
- McLay, D. W., and Clarke, H. J. (2003). Remodelling the paternal chromatin at fertilization in mammals. Reproduction 125, 625-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meistrich, M. L., Bucci, L. R., Trostle-Weige, P. K., and Brock, W. A. (1985). Histone variants in rat spermatogonia and primary spermatocytes. Dev. Biol. 112, 230-240. [DOI] [PubMed] [Google Scholar]
- Misteli, T., Gunjan, A., Hock, R., Bustin, M., and Brown, D. T. (2000). Dynamic binding of histone H1 to chromatin in living cells. Nature 408, 877-881. [DOI] [PubMed] [Google Scholar]
- Miyara, F. et al. (2003). Chromatin configuration and transcriptional control in human and mouse oocytes. Reprod. Dev. 64, 458-470. [DOI] [PubMed] [Google Scholar]
- Phair, R. D., Scaffidi, P., Elbi, C., Vecerova, J., Dey, A., Ozato, K., Brown, D. T., Hager, G., Bustin, M., and Misteli, T. (2004). Global nature of dynamic protein-chromatin interactions in vivo: three-dimensional genome scanning and dynamic interaction networks of chromatin proteins. Mol. Cell. Biol. 24, 6393-6402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan, V., Finch, J. T., Graziano, V., Lee, P. L., and Sweet, R. M. (1993). Crystal structure of globular domain of histone H5 and its implications for nucleosome binding. Nature 362, 219-223. [DOI] [PubMed] [Google Scholar]
- Sera, T., and Wolffe, A. P. (1998). Role of histone H1 as an architectural determinant of chromatin structure and as a specific repressor of transcription on Xenopus oocyte 5S rRNA genes. Mol. Cell. Biol. 18, 3668-3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, M., Hennebold, J. D., Macfarlane, J., and Adashi, E. Y. (2001). A mammalian oocyte-specific linker histone gene H1oo: homology with the genes for the oocyte-specific cleavage stage histone (cs-H1) of sea urchin and the B4/H1M histone of the frog. Development 128, 655-664. [DOI] [PubMed] [Google Scholar]
- Tanaka, M. et al. (2004). H1FOO is coupled to the initiation of oocytic growth. Biol. Reprod. 72, 135-142. [DOI] [PubMed] [Google Scholar]
- Tanaka, M., Kihara, M., Meczekalski, B., King, G. J., and Adashi, E. Y. (2003). H1oo: a pre-embryonic H1 linker histone in search of a function. Mol. Cell. Endocrinol. 202, 5-9. [DOI] [PubMed] [Google Scholar]
- Teranishi, T., Tanaka, M., Kimoto, S., Ono, Y., Miyakoshi, K., Kono, T., and Yoshimura, Y. (2004). Rapid replacement of somatic linker histones with the oocyte-specific linker histone H1foo in nuclear transfer. Dev. Biol. 266, 76-86. [DOI] [PubMed] [Google Scholar]
- Thomas, J. O. (1999). Histone H1, location and role. Curr. Opin. Cell Biol. 11, 312-317. [DOI] [PubMed] [Google Scholar]
- Thomas, J. O., Rees, C., and Finch, J. T. (1992). Cooperative binding of the globular domains of histones H1 and H5 to DNA. Nucleic Acids Res. 20, 187-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignon, X., Zhou, Q., and Renard, J. P. (2002). Chromatin as a regulative architecture of the early developmental functions of mammalian embryos after fertilization or nuclear transfer. Cloning Stem Cells 4, 363-377. [DOI] [PubMed] [Google Scholar]
- Zhou, Y. B., Gerchman, S. E., Ramakrishnan, V., Travers, A., and Muyldermans, S. (1998). Position and orientation of the globular domain of linker histone H5 on the nucleosome. Nature 395, 402-405. [DOI] [PubMed] [Google Scholar]