Abstract
Constructing a mitotic spindle requires the coordinated actions of several kinesin motor proteins. Here, we have visualized the dynamics of five green fluorescent protein (GFP)-tagged mitotic kinesins (class 5, 6, 8, 13, and 14) in live Drosophila Schneider cell line (S2), after first demonstrating that the GFP-tag does not interfere with the mitotic functions of these kinesins using an RNA interference (RNAi)-based rescue strategy. Class 8 (Klp67A) and class 14 (Ncd) kinesin are sequestered in an active form in the nucleus during interphase and engage their microtubule targets upon nuclear envelope breakdown (NEB). Relocalization of Klp67A to the cytoplasm using a nuclear export signal resulted in the disassembly of the interphase microtubule array, providing support for the hypothesis that this kinesin class possesses microtubule-destabilizing activity. The interactions of Kinesin-5 (Klp61F) and -6 (Pavarotti) with microtubules, on the other hand, are activated and inactivated by Cdc2 phosphorylation, respectively, as shown by examining localization after mutating Cdc2 consensus sites. The actions of microtubule-destabilizing kinesins (class 8 and 13 [Klp10A]) seem to be controlled by cell cycle-dependent changes in their localizations. Klp10A, concentrated on microtubule plus ends in interphase and prophase, relocalizes to centromeres and spindle poles upon NEB and remains at these sites throughout anaphase. Consistent with this localization, RNAi analysis showed that this kinesin contributes to chromosome-to-pole movement during anaphase A. Klp67A also becomes kinetochore associated upon NEB, but the majority of the population relocalizes to the central spindle by the timing of anaphase A onset, consistent with our RNAi result showing no effect of depleting this motor on anaphase A. These results reveal a diverse spectrum of regulatory mechanisms for controlling the localization and function of five mitotic kinesins at different stages of the cell cycle.
INTRODUCTION
The formation of a bipolar spindle precedes the segregation of chromosomes to the two daughter cells during cell division. The attachment of microtubules to sister kinetochores and cross-linking of microtubules emanating from opposite poles (antiparallel microtubules) are both critical events for establishing bipolarity of mitotic and meiotic spindles (Rieder and Salmon, 1998; Compton, 2000; Scholey et al., 2003). The creation of the metaphase spindle and the subsequent segregation of sister chromatids in anaphase also require the actions of dynein and several members of the kinesin superfamily (Scholey et al., 2003). A major challenge in understanding cell division has been to define the precise roles and regulation of these various molecular motors. Because an anti-kinesin drug has entered clinical trials for chemotherapy (Sakowicz et al., 2004), a better understanding of the cell biological mechanisms of mitotic kinesins also generates new insights into future therapy.
Several kinesin motor classes play important roles in mitosis in numerous eukaryotes. A critical and universally conserved motor is the Kinesin-5 (BimC/Eg5) (nomenclature defined by Lawrence et al., 2004), a bipolar tetrameric motor that pushes apart antiparallel microtubules, which causes separation of spindle poles during prophase and establishes spindle bipolarity after nuclear envelope breakdown (NEB) (Sharp et al., 2000b). Minus end-directed kinesins (Kinesin-14; e.g., Ncd) and cytoplasmic dynein help to focus microtubules to spindle poles (Endow et al., 1994; Heald et al., 1997), and Ncd also helps to generate inward force upon antiparallel microtubules (Sharp et al., 2000a). Several kinesins attached to chromosome arms (e.g., chromokinesin [Kinesin-4] and Kid [Kinesin-10]) or kinetochores (CENP-E [Kinesin-7] and Kin I/MCAK [Kinesin-13]) are involved in attaching chromosomes to the spindle and aligning them at the metaphase (Theurkauf and Hawley, 1992; Vernos et al., 1995; Yucel et al., 2000; Kline-Smith et al., 2004). The Kin I (Kinesin-13) microtubule-destabilizing kinesins regulate overall microtubule length and are thought to depolymerize microtubules at centrosomes and kinetochores to generate centrosome-directed spindle flux and anaphase A motion, respectively (Rogers et al., 2004). The Kip3 (Kinesin-8) also may be involved in microtubule destabilization, although the evidence has been indirect and mainly involves the observation of long microtubules after motor mutation or depletion (Garcia et al., 2002; West et al., 2002; Goshima and Vale, 2003; Gandhi et al., 2004; Savoian et al., 2004). Finally, one or more “cytokinetic” kinesins (Kinesin-6) function in the late phases of mitosis in higher eukaryotes to create the central spindle and likely transport molecules that are needed for initiating the cytokinesis furrow (Adams et al., 1998; Minestrini et al., 2002).
To better understand the roles of motor proteins in mitosis, we previously undertook a systematic analysis of the contributions of all 25 kinesin genes in cell division in the Drosophila S2 cell line by RNA interference (RNAi) (Goshima and Vale, 2003). Strong phenotypes were observed for single RNAi of five kinesins: monopolar spindles for Klp61F [BimC/Eg5; Kinesin-5], unfocused poles for Ncd [Kin C; Kinesin-14], monopolar and monastral bipolar spindles with long astral microtubules for Klp10A [Kin I; Kinesin-13], monopolar and monastral bipolar spindles with long kinetochore microtubules for Klp67A [Kip3; Kinesin-8], and central spindle formation defects and cytokinesis failure for Pav [MKLP1; Kinesin-6] (phenotypes are summarized in Supplemental Figure 1). Three kinesin genes (CENP-meta [CENP-E; Kinesin-7], Klp3A [chromokinesin; Kinesin-4], and Nod [Kid, Kinesin-10]) generated partial defects in metaphase chromosome alignment.
The localization and dynamics of motor proteins also can generate valuable insight into function and regulation, which complements phenotypic information derived by RNAi or genetics. Although localization information has been reported for many mitotic motors, some of the data have been obtained by immunofluorescence or high-level green fluorescent protein (GFP) expression, which can be subject to potential artifacts of fixation and mislocalization of overexpressed proteins, respectively. Relatively few studies have investigated dynamics using live cell imaging, and no single study has compared the localization and dynamics of several mitotic motors all within the same animal cell type. Here, using an inducible expression system and an RNAi-based rescue assay, we have determined the localization and dynamics of the five kinesins essential for mitotic spindle formation in S2 cells by live cell GFP imaging and examined how cell cycle transitions affect localization and function.
MATERIALS AND METHODS
Cell Culture and RNA Interference
Drosophila Schneider cell line (S2) was cultured, and RNAi was performed according the methods of Clemens et al. (2000). Templates for in vitro transcription were generated by PCR using the primers listed in the Supplemental Table. The synthesized double-stranded RNA (dsRNA) was added to cell culture in 96-well or 24-well plates in amounts of 1 or 5 μg, respectively. UTR sequences are predictable from the full-length cDNA sequences available in the database. At the end of the RNAi (days 3–7; Supplemental Table), cells were resuspended and replated on concanavalin A (ConA)-coated coverslips typically for 2 h before imaging or fixation (Rogers et al., 2002). In the rescue experiment for Pav-GFP, cells were put on ConA-coated coverslips for 20 min before fixation. CuSO4 was added for induction of gene expression (for low level expression, 100 μM for 4 h or 0–10 μM for >1 d; and for rescue assay and overexpression, 100–500 μM addition for >2 d). For microtubule disruption, cells were incubated in the presence of 50 μM colchicines for 16 h.
Plasmids and Transfection
GFP was fused onto the C termini of kinesins by introduction of a NotI site. A KpnI site was introduced at the N terminus of kinesins. The resulting kinesin-GFP sequences were inserted into pMT plasmid (Invitrogen, Carlsbad, CA), which has a copper-inducible promoter. The following cDNAs (Open Biosystems, Huntsville, AL) containing full-length kinesin were used for the PCR template: LD15641 (Klp61F), LD29208 (Klp10A), RE52076 (Klp67A), LD29131 (Ncd), and RE22456 (Pav). The S929 residue of Klp61F was mutated to Leu during cloning, but this substitution did not change their function as shown by rescue experiment. For NES construct, an established nuclear export signal (NELALKLAGLDINK; Roth et al., 2003) was attached to the C terminus of GFP. Site-directed mutagenesis was performed by QuikChange PCR (Stratagene, La Jolla, CA) using mutation-containing oligo DNAs. Transfection was performed using Cellfectin reagent (Invitrogen), and stably expressing cells were selected by hygromycin. Most of the cells (usually >70%) in the stable cell express GFP. Note, however, that the expression level of GFP is variable among cells, because we did not perform clonal selection of the cell lines.
Immunofluorescence Microscopy and Immunoblotting
For microtubules and Cid, cells were fixed by 3 or 6% paraformaldehyde for 15 min and stained by anti-tubulin antibody (DM1A; 1:500), anti-Cid antibody (Henikoff et al., 2000), and Hoechst 33342 (0.5 μg/ml) after 0.1% Triton treatment. The signals of GFP-kinesins were well maintained during this procedure. Klp10A, Klp67A, and Ncd were stained after methanol fixation (Rogers et al., 2002, 2004). In brief, cells were fixed by prechilled methanol solution (90% methanol, 3% paraformaldehyde, and 5 mM sodium bicarbonate, pH 9) for 15 min, followed by 0.1% Triton treatment. Specimens were imaged by a cooled charge-coupled device (CCD) camera Sensicam mounted on a Zeiss Axiovert microscope. Immunoblotting was performed with a rabbit polyclonal anti-Klp61F (Sharp et al., 1999), anti-Klp10A (Rogers et al., 2004), and anti-Ncd (a gift from Jonathan Scholey, University of California–Davis, Davis, CA). Rabbit anti-Klp67A antiserum was produced using C-terminal fragment of Klp67A (346–781 a.a.) fused with glutathione S-transferase (GST) and was used for immunoblot (1:100). For immunofluorescence, affinity purification was further performed using shorter GST-Klp67A-C fragment (602–781 a.a.).
Live Imaging of GFP-tagged Kinesins
For time-lapse imaging of kinesins, images from low-expressing GFP-kinesin cells were collected at 1- to 5-s intervals with 500-ms exposure times at room temperature (23–25°C) using a cooled CCD camera Orca-ER (Hamamatsu, Bridgewater, NJ) attached to a Yokogawa spinning-disk confocal scanhead (PerkinElmer Life and Analytical Sciences, Boston, MA) that was mounted on Nikon TE2000 inverted microscope equipped with excitation and emission filter wheels (Sutter Instrument, Novato, CA). Camera and filter wheels were controlled by MetaMorph software (Universal Imaging, Downingtown, PA; Molecular Devices, Sunnyvale, CA) on a PC computer. For GFP-tubulin, images were acquired every 5–10 s with 50- to 150-ms exposure time by Orca-ER-II (Hamamatsu) attached to an Axiovert microscope (Carl Zeiss, Thornwood, NY). For imaging at fixed time point of living cells, cells on ConA were supplied with 0.1 ng/μl Hoechst 33342 for 30 min, and images were acquired by a cooled CCD camera Sensicam mounted on an Axiovert microscope.
RESULTS
Rescue of Mitotic Defects by GFP-tagged Kinesins
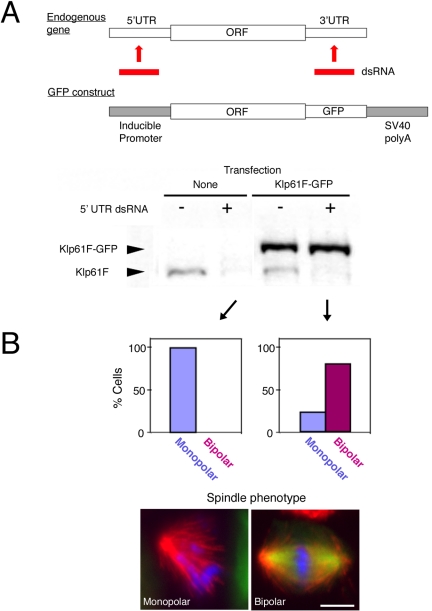
In Drosophila S2 cells, five kinesins are essential for proper spindle formation during mitosis: Klp61F [BimC/Eg5; Kinesin-5], Klp10A [Kin I; Kinesin-13], Klp67A [Kip3; Kinesin-8], Ncd [Kin C; Kinesin-14], and Pav [MKLP1; Kinesin-6] (Supplemental Figure 1). To determine the intracellular localization and dynamics of these kinesins, we tagged each kinesin at its C terminus with GFP. To ensure that the GFP fusion proteins were functional, a strategy commonly used in yeast or in whole animal (Endow and Komma, 1996) but generally lacking for higher eukaryotic cells in culture, we developed an RNAi-based rescue assay. RNAi is usually performed using dsRNA homologous to the open reading frame in Drosophila (Clemens et al., 2000). However, we tested whether dsRNA homologous to either the 5′UTR or 3′UTR also efficiently interferes with protein expression and whether an ectopically expressed gene lacking the UTR sequences is resistant to this dsRNA treatment (Figure 1A). We constructed a cell line stably maintaining the Klp61F-GFP gene under the control of an inducible promoter and containing 3′ SV40 poly-A sequences but lacking endogenous UTR sequences. After treating S2 cells with dsRNA corresponding to the 5′UTR of Klp61F, a significant reduction of the endogenous motor protein was observed. However, the induced Klp61F-GFP protein level was not affected (Figure 1A, bottom). Phenotypic analysis showed that Klp61F-GFP expression efficiently rescued the monopolar spindle defect induced by 5′UTR RNAi (Figure 1B and Table 1). This result indicates that Klp61F-GFP can functionally replace endogenous Klp61F. Similarly, Klp10A-GFP, Klp67A-GFP, Ncd-GFP, and Pav-GFP rescued RNAi phenotypes produced by UTR RNAi of these motors (Table 1 and Supplemental Figure 2). This method provides a means of ensuring that the incorporation of GFP does not interfere with protein function.
Figure 1.
Rescue assay in S2 cells. (A) Top, schematic representation of a rescue assay in S2 cells. dsRNA targeting 5′UTR or 3′UTR is used for RNAi knockdown of an endogenous gene. An exogenous gene lacking UTR sequences, such as GFP-fusion gene shown here, can be expressed and is resistant to the dsRNA. Bottom, immunoblotting with anti-Klp61F against control untransfected cells and a Klp61F-GFP–expressing cell line after 5′UTR RNAi (lanes 2 and 4) or no RNA treatment (lanes 1 and 3). 5′UTR dsRNA specifically knocked down endogenous Klp61F but not the expressed Klp61F-GFP. (B) The RNAi rescue assay shows that Klp61F-GFP is functional. Top, phenotypic analysis shows that monopolar spindles were observed after RNAi of untransfected cells (left), whereas bipolar spindles were found predominantly in the Klp61F-GFP–expressing cell line (right). Note that some Klp61F-GFP cells (22%) do not express GFP, and those cells formed monopolar spindles. Bottom, immunofluorescence examples of a monopolar spindle after 5′UTR-based RNAi (left) and a bipolar spindle after rescued by Klp61F-GFP expression (right). Blue, DNA; green, GFP; and red, tubulin. Bar, 5 μm.
Table 1.
Rescue experiment for GFP-tagged mitotic kinesins
| Klp61F UTR RNAi | % Monopolar spindles (n) |
| No transfection | 100 (37) |
| Klp61F-GFP | 0 (23) |
| Klp61F [T933A]-GFP | 100 (20) |
| Ncd UTR RNAi | % Unfocused/multipolar spindlesa (n) |
| No transfection | 84 (31) |
| Ncd-GFP | 23 (27) |
| Klp10A UTR RNAi | % Spindles with abnormally long MTs (n) |
| No transfection | 100 (31) |
| Klp10A-GFP | 0 (36) |
| Klp67A UTR RNAi | % Spindles with abnormally long/curled MTs (n) |
| No transfection | 100 (17) |
| Klp67A-GFP | 0 (26) |
| Pav UTR RNAi | % Interphase cells with >2 nuclei (n)b |
| No transfection | 56 (200) |
| Pav-GFP | 11 (200) |
| Pav [T7A/T15A/T458A/T467A]-GFP | 9 (200) |
Each phenotype is described in Goshima and Vale (2003) (also see Supplemental Figure 2 for representative cell images). Only GFP-expressing cells were scored for spindle phenotypes of transfected cell lines.
Note that ∼20% of wild-type S2 cells have this type of spindles
Approximately 10% of wild-type S2 cells have more than two nuclei
Localization and Dynamics of Mitotic Kinesins during the Cell Cycle
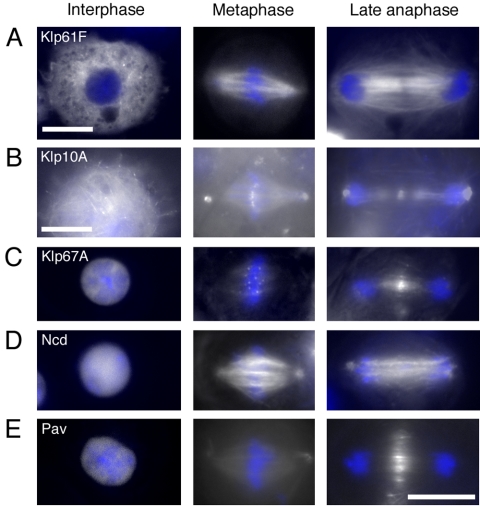
Figure 2 shows unfixed GFP images of five kinesins taken by wide-field microscopy in interphase, metaphase, and late anaphase after RNAi knockdown of endogenous kinesins. Several of our results with GFP-tagged kinesins are consistent with previous publications, but several novel observations were obtained as well. Because identical GFP localization pattern was observed in the presence of endogenous kinesin (Supplemental Figure 3), we performed most of the further localization analysis (e.g., time-lapse observation) in the presence of endogenous proteins. Results for Pav-GFP [MKLP1; Kinesin-6] and Klp61F-GFP [BimC/Eg5; Kinesin-5] are largely consistent with previous studies and thus will only be mentioned briefly (Figure 2, A and E). Pav, which is essential for central spindle bundling during cytokinesis, was localized to the nucleus during interphase, was primarily diffusely distributed during metaphase and accumulated in central spindle region after anaphase (Adams et al., 1998; Minestrini et al., 2002). Klp61F-GFP was localized diffusely throughout the cytoplasm during interphase, exhibited a strong signal at centrosomes in prophase before NEB (our unpublished data), and localized in the interior regions of the bipolar spindle, including kinetochore microtubule bundles (K-fibers) and nonkinetochore microtubules from metaphase to anaphase, which is largely in agreement with previous findings (Barton et al., 1995; Sharp et al., 1999). However, we could not see nuclear Klp61F-GFP before prometaphase, which is detected in embryonic nuclei (Sharp et al., 1999). The spindle association of Klp61F is dependent on microtubules, because no spindle-like localization was detected after microtubules were depolymerized with colchicine, and cells were arrested in prometaphase (Supplemental Figure 4A).
Figure 2.
Localization of GFP-tagged mitotic kinesins in the S2 cell cycle. Localization of five GFP-tagged mitotic kinesins in interphase (left), metaphase (middle), and anaphase (right). Endogenous kinesins were knocked down by selective RNAi using UTR sequences (see Figure 1), and cells moderately expressing kinesin-GFP (white) were put on a ConA plate to spread, and images were taken without fixation by wide-field microscope. DNA was stained by Hoechst 33342 (blue). See text for explanations. Bar, 10 μm.
Ncd-GFP [Kinesin-14] was localized to the nucleus during interphase, which was undetected by previous immunofluorescence studies of S2 cells (Hatsumi and Endow, 1992). However, using another recently generated polyclonal antibody, nuclear enrichment also was confirmed by immunofluorescence microscopy (Supplemental Figure 5A). In metaphase, Ncd-GFP signals were detected on centrosomes and uniformly along spindle microtubules by a wide-field microscope (Figure 2D). However, time-lapse imaging by spinning-disk confocal microscope also revealed a previously unreported enrichment of Ncd-GFP at the tips of growing mitotic microtubules emanating from centrosomes (Goshima and Vale, unpublished data). Similar to Klp61F-GFP, colchicine treatment led to diffused distribution of Ncd-GFP in mitosis (Supplemental Figure 4B).
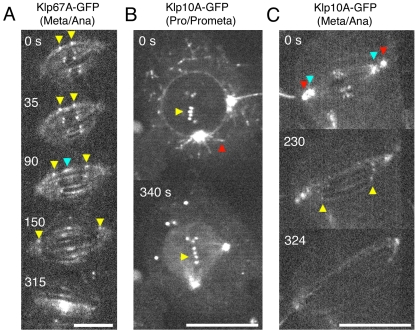
The localization and dynamics of Klp67A-GFP implicated new information on its function. During interphase, Klp67A-GFP was sequestered in the nucleus (Figure 2C); mitochondrial localization (previously shown in embryo by immunofluorescence using polyclonal antibodies; Pereira et al., 1997) was not detected in this study. During metaphase, Klp67A-GFP was localized clearly to kinetochores and also faintly to spindle microtubules, but it was not concentrated at centrosomes (see Supplemental Figure 6A for costaining with Cid, a centromere protein). Double visualization of Klp67A-GFP and anti-Klp10A [Kin I; Kinesin-13] immunostaining showed that Klp67A has distinct localization from Klp10A on metaphase chromosomes (Supplemental Figure 6B). In late anaphase, Klp67A-GFP was absent from the kinetochore and was concentrated in the central region of the spindle (Figure 2C). To unambiguously show that the above-mentioned localization is not the artifact of GFP-tagging and/or overexpression, immunostaining of Klp67A was performed for wild-type and RNAi-treated cells. anti-Klp67A antibody staining was identical to Klp67A-GFP throughout the wild-type cell cycle; the signals were greatly diminished after RNAi, confirming the specificity of the staining (Supplemental Figure 5B). Time-lapse imaging revealed that enrichment at the central spindle began after the onset of anaphase spindle elongation (Figure 3A, blue; Movie 1) and that kinetochore localization markedly diminished from midanaphase. However, this microscopy also detected some kinetochore signals retained until the completion of anaphase A (Figure 3A, yellow).
Figure 3.
Time-lapse imaging of Klp67A-GFP and Klp10A-GFP during mitosis. (A) Dynamics of Klp67A-GFP from metaphase to late anaphase. Clear punctate signals are visible at kinetochores and fainter signals are along K-fibers. Kinetochore signals (yellow arrowheads) are clearly seen during metaphase and diminish during anaphase A (90–150 s). Central spindle accumulation (blue arrowhead) begins at the onset of anaphase spindle elongation. Bar, 5 μm. See also Movie 1. (B) Dynamics of Klp10A-GFP from prophase to prometaphase. Clear punctate signals are visible on the astral microtubules emanating from centrosomes (red) and at centromeres (yellow) during prophase, whereas astral microtubule staining was dramatically reduced after NEB (340 s). Aggregated GFP signals are temporarily visible in cytoplasm, the function of which is unclear. Bar, 10 μm. See also Movie 2. (C) Dynamics of Klp10A-GFP from metaphase to late anaphase. Clear signals are visible at centrosomes (red), pole regions where minus ends of K-fibers are focused (blue) and at centromeres. Centromere signals are seen during metaphase and persist until the end of anaphase A (yellow). Nearly uniform spindle localization is detected during late anaphase spindle elongation. Bar, 10 μm. See also Movie 3.
For Klp10A-GFP, we observed localization at spindle poles and centromeres during metaphase (Figure 2B), consistent with previous immunofluorescence results (Rogers et al., 2004). Accumulated signals were occasionally detected at central spindle region in anaphase, but the enrichment was not as dramatic as for of Klp67A or Pav. We also confirmed microtubule plus end-tracking during interphase (Mennella et al., 2005) by time-lapse imaging (our unpublished data). Interestingly, plus end tracking on astral microtubules was clearly visible during prophase but abruptly diminished after NEB and bipolar spindle formation (Figure 3B and Movie 2). In metaphase, plus end tracking was much less obvious, and instead, the majority of Klp10A-GFP was localized at centrosomes, centromeres and also the pole region interior to centrosomes (Figure 3C and Movie 3). We also observed that centromere signals were clearly maintained during anaphase A (Figure 3C, yellow arrows) and that Klp10A-GFP localized fairly uniformly along interpolar microtubules in late anaphase in addition to centrosomes.
Roles of Cdc2 Kinase for Klp61F and Pav Function
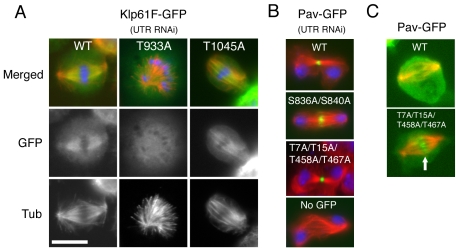
We next investigated whether kinesin localization is affected by Cdc2 kinase (cyclin-dependent kinase) (Figure 4). To investigate the effects of Cdc2 phosphorylation, we first searched for consensus Cdc2 phosphorylation sites in the mitotic kinesin sequences. The most favorable motif for Cdc2 phosphorylation is S/T P x K/R, although the S/T P motif can occasionally be used as a substrate (“imperfect consensus”). Recent genome-wide screening of Cdc2 substrates in yeast suggested that phosphorylation of these imperfect consensus sites often occurs when they are located near a perfect consensus sequence (Ubersax et al., 2003). In the five kinesins studied here, we found six perfect consensus sites: two in Klp61F [BimC/Eg5; Kinesin-5] and four in Pav [MKLP1; Kinesin-6]) and four imperfect sequences within 40 a.a. of the perfect ones (two in Klp61F and two in Pav). We constructed 15 plasmids in which single, double, triple, or quadruple Ser or Thr in these consensus sites were mutated to Ala (Klp61F: T933A, S949A, T1045A, S1050A/S1054A, T1045A/S1050A/S1054A; Pav: T7A, T15A, T7A/T15A, T458A, T467A, T458A/T467A, S836A, S840A, S836A/S840A, T7A/T15A/T458A/T467A). Stable cell lines expressing these alanine mutant proteins with GFP-tags were made, because stable cell lines allowed more reliable tests of mitotic rescue than transient transfections. We then determined whether expression of these mutant kinesins could rescue the Klp61F or Pav mitotic defects after UTR RNAi. Of these 15 constructs, only one construct, Klp61F [T933A], failed to rescue the mitotic defect (monopolar spindles after 5′UTR RNAi of Klp61F) (Table 1). Moreover, Klp61F [T933A]-GFP did not localize to the spindle and instead was diffusely distributed throughout the cell in mitosis (Figure 4A).
Figure 4.
Requirement of Cdc2 kinase sites for mitotic kinesin localization. GFP-kinesins localization (green) and immunofluorescence of tubulin (red) after rescue assay (DNA; blue). (A) The mutation T933A in a Cdc2 phosphorylation site of Klp61F-GFP caused mislocalization of Klp61F-GFP and abolished its rescue ability. Control wild-type Klp61F-GFP and another Cdc2 site mutant T1045A rescued the monopolar phenotype. (B) Alanine mutations in Cdc2 sites in Pav did not affect central spindle localization and bundling activity of Pav during cytokinesis, and cells would initiate cleavage furrow formation. (C) A quadruple mutant (T7A/T15A/T458A/T467A) precociously localized to central spindle region in metaphase (arrow). See Table 1 for quantitation. Bar, 10 μm.
In contrast, all of the alanine mutants for Pav, including the quadruple mutant, still localized to the central spindle (Figure 4B) and clearly rescued the Pav UTR RNAi-generated cytokinesis defect (Table 1). This result is surprising, given previous results showing that a mutation of a conserved Cdc2 site in ZEN-4 (the Caenorhabditis elegans ortholog of Pav) abolishes its ability to rescue the cytokinesis-defective zen-4 null mutant (Mishima et al., 2004). Although normal for cytokinesis function, we found that the localization patterns of two Pav mutants were altered in metaphase. Specifically, the quadruple mutant (T7A/T15A/T458A/T467A) localized prematurely to the central region of the spindle before anaphase (>90% of preanaphase bipolar spindle [n = 50]; Figure 4C, arrow). A similar localization of T7A/T15A double mutant also was frequently but not always observed (60%), and the accumulation in the central spindle was weaker than the quadruple mutant (our unpublished data). These data suggest that Cdc2 phosphorylations near catalytic regions prevent precocious association of Pav with central spindle.
Effect of Nuclear Export of Ncd-GFP, Klp67A-GFP, and Pav-GFP during Interphase
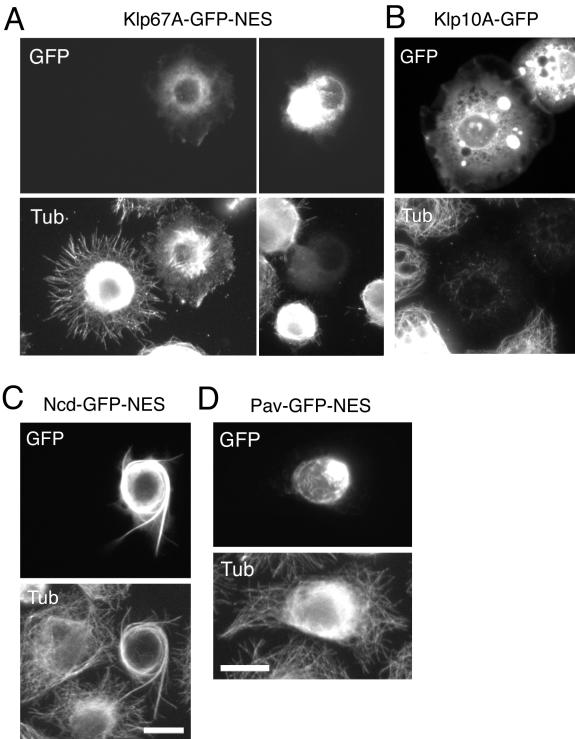
Three mitotic kinesins (Klp67A, Ncd, and Pav) were localized in the nucleus during interphase (Figure 2). To understand the role of nuclear import during interphase, we artificially exported these kinesins to the cytoplasm by addition of a nuclear export signal (NES). We found that Klp67A-GFP-NES indeed localized to the cytoplasm and that this change in distribution resulted in the destabilization of microtubules (Figure 5A). This was particularly dramatic in cells expressing high levels of this construct, where a virtually complete loss of the cytoplasmic microtubule network was observed (Figure 5A, right). This effect was directly brought by ATPase domain of Klp67A, because expression of N terminus motor domain alone (358 a.a.) induced similar microtubule destabilization in cells (our unpublished data). Previous results have shown an increase in microtubule length after Klp67A RNAi or genetic loss of function of equivalent motor types in yeast and fly (Garcia et al., 2002; West et al., 2002; Goshima and Vale, 2003; Gandhi et al., 2004; Savoian et al., 2004), leading to the speculation that this motor might destabilize microtubules. However, our results showing that Klp67A expression leads to microtubule destabilization provide more direct evidence for this hypothesis. These results in interphase cells also suggest that the microtubule-destabilizing activity of Klp67A is constitutive and not specifically regulated by the cell cycle. Overexpression of Klp10A destabilizer similarly destroyed the cytoplasmic microtubule network (Figure 5B), and this motor was recently shown to play a role in regulating cytoplasmic microtubule dynamics in this cell line (Mennella et al., 2005). Ncd-GFP-NES, in contrast, localized to cytoplasmic microtubules. At lower expression levels, Ncd-GFP-NES accumulated at the growing tips of microtubules (Goshima and Vale, unpublished data). Overexpression caused abnormal bundling of cytoplasmic microtubules (Figure 5C). In contrast, Pav-GFP-NES, a microtubule cross-linker during cytokinesis, made filamentous aggregates at perinuclear region and did not affect global microtubule network after overproduction (Figure 5D), suggesting that additional mitotic regulation, such as phosphorylation by Polo kinase, is required for its function.
Figure 5.
Effects of cytoplasmic activation of Klp67A and Ncd. (A) Destabilization of cytoplasmic microtubules (bottom) by expression of Klp67A-GFP-NES (top). Klp67A-GFP-NES construct was transfected and transiently overexpressed, and the cells were fixed and stained with an anti-tubulin antibody. The cell with higher GFP intensity has fewer microtubules. (B) Klp10A-GFP overexpression also led to microtubule destabilization. (C) Excessive bundling of microtubules (bottom) after Ncd-GFP-NES (top) overexpression in the cytoplasm. (D) The cytoplasmic microtubule network was not destroyed after overexpression of Pav-GFP-NES.
Roles of Klp67A and Klp10A Microtubule Destabilizers in Kinetochore Microtubule Depolymerization during Anaphase A
At the onset of anaphase A, the sister chromatids are separated and then segregated to opposite poles by the depolymerization of kinetochore microtubules. The localization of Klp67A and Klp10A to kinetochores led us to test whether these microtubule-destabilizing kinesins might contribute to anaphase A movement. We first quantitatively examined the anaphase spindle elongation and K-fiber shortening in control GFP-tubulin cells. Because spindle poles and K-fibers were both detectable by GFP-tubulin from metaphase to anaphase, we could monitor pole separation as well as K-fiber shortening by this single fluorophore imaging approach (Figure 6, A and E, and Movie 4). Observations of >30 control cells from metaphase or early anaphase through to telophase revealed that K-fiber shortening always occurs in the middle of spindle elongation (190–225 s in Figure 6A). Almost all of the analyzed cells (17/18) started K-fiber shortening within 250 s after the onset of spindle elongation, although the duration is very variable among cells (60–310 s, average 140 ± 60 s [n = 18]). The rate of K-fiber shortening was fairly uniform from cell to cell (2.6 ± 0.4 μm/min [n = 12]).
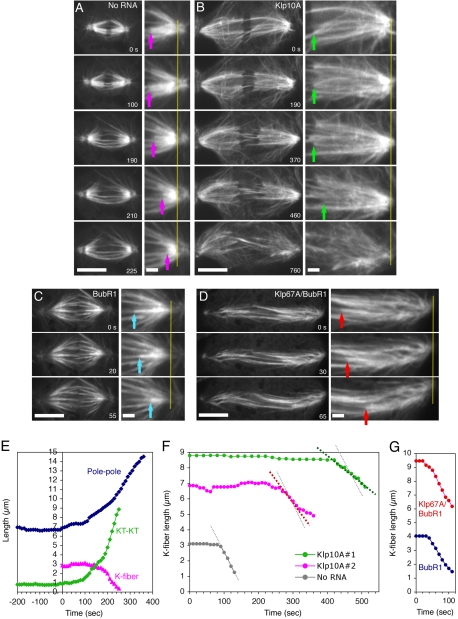
Figure 6.
Effects of Klp10A or Klp67A RNAi on anaphase A K-fiber shortening. (A–D) Time-lapse GFP-tubulin images from metaphase to late anaphase in a control cell (no RNA addition) (A), a Klp10A RNAi-treated cell (day 7) (B), a BubR1 RNAi-treated cell (day 3) (C), and a Klp67A/BubR1 double RNAi-treated cell (day 3) (D). See also Movies 4–7 corresponding to these treatments. In the right panels (2× enlarged images), spindle poles are vertically aligned, and tracking of kinetochores (arrow) relative to poles (yellow line) is shown. Bars, 10 μm (left) and 2 μm (right). (E) Pole-to-pole distance, sister kinetochore distance (KT-KT; distance between plus ends of K-fiber) and K-fiber length (KT-pole distance of a selected K-fiber) of the cell in A are plotted. Mean velocity of K-fiber shortening was 2.6 ± 0.4 μm/min (n = 12). (F) K-fiber length change in two Klp10A RNAi cells and a control untreated cell is plotted (Klp10A #1 corresponds to the cell in B). Another example is also shown as #2. Mean velocity of K-fiber shortening was 23% slower than control (2.0 ± 0.8 μm/min [n = 15]), and one-half of the cells took >250 s to start K-fiber shortening after the onset of anaphase. K-fiber could not be traced after certain short length, because abundant microtubules from centrosomes masked the K-fiber. (G) K-fiber length change for the single BubR1 and double Klp67A/BubR1 RNAi cells in C and D is plotted. Mean velocity of the K-fiber shortening is similar between single BubR1 (2.1 ± 0.4 μm/min [n = 16]) and double Klp67A/BubR1 RNAi (2.2 ± 0.5 μm/min [n = 13]).
After Klp10A RNAi, anaphase entry was not blocked, and we observed that the average velocity of K-fiber shortening was 23% slower (2.0 ± 0.8 μm/min [n = 15]; p < 0.025), and five of 15 cells exhibited velocities <1.5 μm/min, which was never observed in 12 control cells (Figure 6, B and F, and Movie 5). Furthermore, one-half of the cells showed significant delay (>250 s) in the onset of K-fiber shortening (observed in only 1 of 18 control cells). These observations strongly suggest that Klp10A plays important role in anaphase A chromosome motility in S2 cells, as was seen in the Drosophila embryo (Rogers et al., 2004).
Klp67A RNAi leads to chromosome misalignment in the spindle and blocks anaphase entry in S2 cells (Goshima and Vale, 2003). Thus, to assess the contribution of Klp67A in anaphase K-fiber shortening, we observed GFP-tubulin after simultaneous knockdown of Klp67A and a spindle checkpoint protein BubR1. First, however, we evaluated anaphase A chromosome movements in cells treated singly with BubR1 RNAi. BubR1 RNAi caused precocious entry into anaphase before chromosome congression, which made it difficult to detect the onset of anaphase. Thus, the duration of anaphase could not be determined. However, the rate of K-fiber shortening that occurs after mid-anaphase could be measured (Figure 6, C and G, and Movie 6). The mean velocity (2.1 ± 0.4 μm/min [n = 16]) was 20% slower than control cells. The basis of this reduction is uncertain, although perhaps the precocious entry into anaphase might limit the full activity of microtubule depolymerizers. We then investigated the rate of anaphase A in double Klp67A/BubR1 RNAi cells. The long spindle microtubules (characteristic of Klp67A RNAi) and precocious anaphase entry (characteristic of BubR1 RNAi) indicated the knockdown of both proteins in the cells examined by microscopy. The chromosome misalignment was more severe than with single BubR1 RNAi, and accordingly, kinetochore microtubules were not easily detected as bundles. Nevertheless, we could trace K-fiber(s) in about ∼30% of the observed cells from early anaphase to mid-/late anaphase (Figure 6D and Movie 7). These K-fibers shortened at a similar velocity to single BubR1 RNAi (2.2 ± 0.5 μm/min [n = 13]; Figure 6G). This result suggests that Klp67A is not involved in K-fiber shortening during anaphase A (see Discussion).
DISCUSSION
Strategies for Regulating Mitotic Kinesin Activity
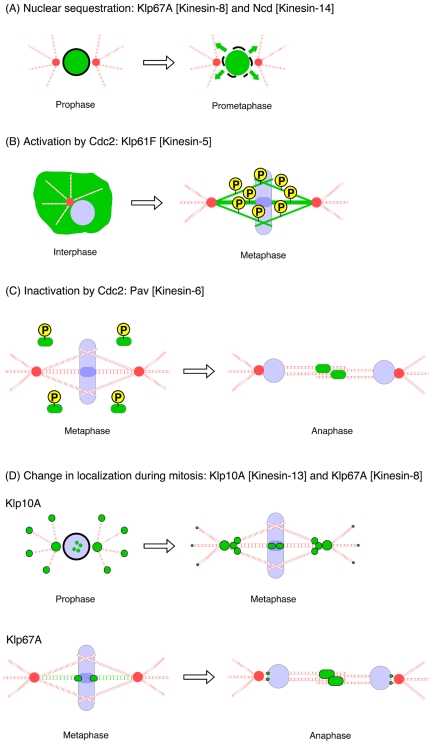
Our studies reveal a diversity of mechanisms for controlling the mitotic kinesins during the cell cycle (Figure 7). The motor activities of two mitotic kinesins, Klp67A and Ncd, are not controlled by cell cycle kinases but are “sequestered” by localization to the nucleus during interphase, shielding interphase microtubules from undesirable effects (destabilization by Klp67A and bundling by Ncd). NEB at prometaphase enables these motors to “meet” and perform their actions on microtubules. Nuclear accumulation of vertebrate Ncd also occurs in cells (Kuriyama et al., 1995; Walczak et al., 1997), implicating the evolutional conservation of the motor inactivation mechanism. Nuclear import might require the importin protein, because tail domain of a Xenopus Kinesin-14 is shown to interact with importin to down-regulate microtubule binding activity of this domain (Ems-McClung et al., 2004).
Figure 7.
Strategies to regulate mitotic kinesin activity in the cell cycle. (A) Nuclear sequestration of Klp67A or Ncd protects cytoplasmic microtubules from the undesired depolymerizing or cross-linking activities of these motors. Nuclear envelope breakdown enables the motors to perform their actions on microtubules. (B) Phosphorylation of Thr 933 residue by Cdc2 kinase is essential for Klp61F targeting to microtubules during mitosis. (C) Cdc2 phosphorylations around the motor domain of Pav prevent this motor for prematurely binding to the central spindle during metaphase. Dephosphorylation of these sites is required for central spindle targeting after anaphase. (D) Change of localization of Klp10A and Klp67A depolymerases after mitotic progression. Klp10A-GFP shows clear microtubule plus end tracking in interphase. Plus end tracking is still clearly detected during prophase, while centrosome/centromere localization also is seen. After nuclear envelope breakdown, plus end tracking becomes much less evident, and the majority of Klp10A localizes to centrosomes, centromeres, and interior pole regions. Klp67A enriched at the outer region of kinetochores during metaphase, followed by central spindle accumulation immediately upon anaphase onset.
In contrast, Klp61F (and related Kinesin-5 motors) is constitutively inactive in the cytoplasm and requires activation by Cdc2 phosphorylation at a single site. Mislocalization of the T933A mutant agrees with studies of unphosphorylated Klp61F in the fly embryo (Sharp et al., 1999) and results of alanine substitution of the corresponding residue in Xenopus and human Eg5 (Blangy et al., 1995; Sawin and Mitchison, 1995). However, these previous studies did not demonstrate the necessity of this phosphorylation event for mitotic function. Our results extend these studies by showing that other Cdc2 sites are not needed for function and that phosphorylation of T933 is required not only for spindle localization but also for this motor's function in bipolar spindle formation. The mechanism by which incorporation of a single phosphate in the tail domain activates microtubule binding in the motor domain remains a fascinating issue. On the other hand, Pav (and related Kinesin-6 motors) is inactivated by Cdc2 kinase before anaphase; its activation seems to require dephosphorylation of Cdc2 sites. Independently, Mishima et al. (2004) examined and recently published the localization of MKLP1 [T8A/T450A], a Cdc2 mutant of human Kinesin-6. Their results show premature localization to the spindle as well, but they reported a diffuse localization to the entire spindle and not confined to the overlapping microtubules in the central region of the spindle as shown here. Our localization is more consistent with a mechanism of localization involving antiparallel microtubules.
Microtubule plus end-tracking of Klp10A is regulated at the point of NEB. Plus end-tracking in interphase is mediated by the interaction with EB1 protein that is enriched at microtubule plus ends throughout the cell cycle (Rogers et al., 2002; Mennella et al., 2005). This implicates that NEB releases a factor, possibly derived from chromatin, which dissociates Klp10A from EB1 in prometaphase. The molecular mechanism of this dissociation is unknown. Klp67A relocalization, from kinetochores to central spindle, occurs during anaphase. The localization of Klp67A to the central spindle in late anaphase also is likely to serve a functional role, because mutations in the Klp67A gene cause cytokinesis failure in meiosis by central spindle deformation (Gandhi et al., 2004; Savoian et al., 2004).
The requirement of the above-mentioned regulatory mechanisms for mitotic kinesin function also seem to vary. Our results show that Cdc2 phosphorylation of Klp61F is absolutely required for function, because mutation of this phosphorylation site generates a monopolar phenotype as severe as the motor RNAi depletion. On the other hand, interphase nuclear import of Pav, Klp67A, and Ncd are not essential for the subsequent mitotic functions of these motors, because the addition of a NES to these motors allows full rescue of RNAi phenotype (our unpublished data). The regulation requirements for Kinesin-6 are more difficult to establish from this study and those in the literature. In S2 cells, interference with Cdc2 phosphorylation of Pav causes premature mislocalization to overlap zones of metaphase spindles, but this does not seem to cause problems in bipolar spindle formation, alignment of chromosomes, or cytokinesis. In contrast, Mishima et al. (2004) showed that mutations of two analogous Cdc2 phosphorylation sites (T8 and T450) in human Kinesin-6 (MKLP1) caused chromosome missegregation. The same group also showed that the T9A mutation of the C. elegans Kinesin-6 (ZEN-4) cannot rescue zen-4 null mutant (Mishima et al., 2004). It is unlikely that Cdc2 phosphorylation of Pav is required for cytokinesis in any of these systems, because Cdc2 activity drops precipitously at the metaphase/anaphase transition. Rather, the above-mentioned differences may be due to the fact that premature spindle localization may cause dominant negative effects in some cell types but not in others. However, another possibility is that Drosophila Kinesin-6 has additional phosphorylation sites that contribute to the complete down-regulation of the motor activity.
Distinct Roles for Kinetochore-associated Microtubule Depolymerases: Klp67A [Kinesin-8] and Klp10A [Kinesin-13]
Chromosome-to-pole movement during anaphase A has been hypothesized to be driven by microtubule depolymerization (Inoue and Salmon, 1995; Kline-Smith and Walczak, 2004). Two sites of depolymerization of kinetochore microtubules seem to contribute to this movement: minus end depolymerization at the centrosome region, leading to poleward flux of microtubule subunits; and plus end depolymerization at the kinetochore. The depolymerization of microtubules by Klp10A and other Kinesin-13 family members has been well established through in vitro studies. Studies in Drosophila embryos show that Klp10A drives microtubule depolymerization at the centrosome and that another Kinesin-13 member (Klp59C) is localized to the kinetochore and is responsible for depolymerization driving anaphase A movement (Rogers et al., 2004). The class 8 kinesins not only have microtubule-translocating activity (Pereira et al., 1997) but also have been implicated in microtubule destabilization based upon the finding that longer than normal mitotic microtubules arise after genetic mutation or RNAi depletion of these kinesins (Garcia et al., 2002; West et al., 2002; Goshima and Vale, 2003; Gandhi et al., 2004; Savoian et al., 2004). Unlike Klp10A, the length increase is specific for kinetochore microtubules and noncentrosomal interpolar microtubules (Goshima and Vale, 2003). However, such an effect upon microtubules could be indirect. Here, we show for the first time that expression of Klp67A in the interphase cytoplasm leads to a profound destabilization of the microtubule network. This result strongly suggests that the Kinesin-8 motors are microtubule-destabilizing proteins.
Savoian et al. (2004) recently found that anaphase chromosome-to-pole movement in meiosis I of a hypomorphic Klp67A mutant is significantly slower than wild type, demonstrating that this motor contributes to chromosome movement in the anaphase spindle. However, the slow movement could also be the consequence of unstable kinetochore–microtubule interaction in the Klp67A mutant (Savoian et al., 2004). We showed by time-lapse imaging that subpopulation of Klp67A-GFP persistently localized at the kinetochore, perhaps at outer region of the kinetochore where most of the K-fibers terminate, throughout anaphase A, which was undetected by immunofluorescent microscopy (Savoian et al., 2004; this study). These results led us to hypothesize that Klp67A might drive chromosome-to-pole movement by depolymerizing the plus ends of kinetochore microtubules. However, the rate of K-fiber shortening was unchanged after RNAi knockdown of Klp67A. To interpret this “negative” result, it may be noted that RNAi method does not completely deplete the endogenous proteins and therefore some residual proteins might be sufficient to execute the depolymerizing function. However, Klp67A RNAi cells analyzed in this study had much longer K-fibers and interpolar microtubules than normal, which indicates that any residual small amount of Klp67A is insufficient to constrain metaphase microtubule length. Release of large population of Klp67A from kinetochores after mid-anaphase also may result in the little contribution of this motor-to-anaphase A depolymerization, which occurs from the middle of the anaphase in this cell line. Together, we suggest that Klp67A is an essential preanaphase K-fiber depolymerase that controls the length of K-fibers but that is not involved in K-fiber shortening during anaphase, at least in S2 cells. The residual population of Klp67A in the anaphase kinetochore might be counterbalanced by kinetochore microtubule polymerase such as Mast/Orbit (CLASP) (Maiato et al., 2005) or inactivated by certain mechanisms such as phosphorylation/dephosphorylation.
Regarding Klp10A, we show that in one-half of the Klp10A RNAi cells, K-fiber shortening did not take place for >5 min after sister chromatid separation and that shortening occurred with ∼25% reduced velocity. These results indicate that Klp10A is a major contributor of anaphase A chromosome movement in S2 cells, in agreement with studies in embryo by Rogers et al. (2004), although we cannot confirm here that its site of action is at the centrosome or kinetochore. Residual Klp10A might be contributing to the shortening in these cells, or other proteins also may participate in this process. Klp59C is the most reasonable candidate to depolymerize K-fiber from its plus end as shown in embryos (Rogers et al., 2004). It also is reported that knockdown of this motor in S2 cells elevates the frequency with which lagging chromosomes are observed in anaphase (Rogers et al., 2004). However, we found no defects in the rate of K-fiber depolymerization in anaphase A of Klp59C RNAi cells (our unpublished data; however, antibodies were not available to evaluate the efficiency of RNAi knockdown). Other mechanisms also might contribute to chromosome-to-pole movement, such as inactivation of microtubule-stabilizing proteins (e.g., Mast/Orbit [CLASP]), other destabilizers (e.g., katanin) or dynein-dependent transport of kinetochores on microtubules. Such mechanisms might be possible to explore in S2 cells depleted of Klp10A by RNAi.
Supplementary Material
Acknowledgments
We are grateful to E. D. Salmon, T. Mitchison, and other Cell Division Group members at Marine Biological Laboratory for the use of microscopes; to D. Sharp and V. Mennella for communicating results before publication; to D. Sharp, G. Rogers, D. Buster, J. Scholey, and S. Henikoff for antibodies; and to N. Stuurman for technical assistance and S. Rogers for helpful discussions. G. G. is the recipient of a postdoctoral fellowship from Human Frontier Science Program Organization.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05-02-0118) on June 15, 2005.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Adams, R. R., Tavares, A. A., Salzberg, A., Bellen, H. J., and Glover, D. M. (1998). pavarotti encodes a kinesin-like protein required to organize the central spindle and contractile ring for cytokinesis. Genes Dev. 12, 1483-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton, N. R., Pereira, A. J., and Goldstein, L. S. (1995). Motor activity and mitotic spindle localization of the Drosophila kinesin-like protein KLP61F. Mol. Biol. Cell 6, 1563-1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blangy, A., Lane, H. A., d'Herin, P., Harper, M., Kress, M., and Nigg, E. A. (1995). Phosphorylation by p34cdc2 regulates spindle association of human Eg5, a kinesin-related motor essential for bipolar spindle formation in vivo. Cell 83, 1159-1169. [DOI] [PubMed] [Google Scholar]
- Clemens, J. C., Worby, C. A., Simonson-Leff, N., Muda, M., Maehama, T., Hemmings, B. A., and Dixon, J. E. (2000). Use of double-stranded RNA interference in Drosophila cell lines to dissect signal transduction pathways. Proc. Natl. Acad. Sci. USA 97, 6499-6503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compton, D. A. (2000). Spindle assembly in animal cells. Annu. Rev. Biochem. 69, 95-114. [DOI] [PubMed] [Google Scholar]
- Ems-McClung, S. C., Zheng, Y., and Walczak, C. E. (2004). Importin α/β and Ran-GTP regulate XCTK2 microtubule binding through a bipartite nuclear localization signal. Mol. Biol. Cell 15, 46-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endow, S. A., Chandra, R., Komma, D. J., Yamamoto, A. H., and Salmon, E. D. (1994). Mutants of the Drosophila ncd microtubule motor protein cause centrosomal and spindle pole defects in mitosis. J. Cell Sci. 107, 859-867. [DOI] [PubMed] [Google Scholar]
- Endow, S. A., and Komma, D. J. (1996). Centrosome and spindle function of the Drosophila Ncd microtubule motor visualized in live embryos using Ncd-GFP fusion proteins. J. Cell Sci. 109, 2429-2442. [DOI] [PubMed] [Google Scholar]
- Gandhi, R., Bonaccorsi, S., Wentworth, D., Doxsey, S., Gatti, M., and Pereira, A. (2004). The Drosophila kinesin-like protein KLP67A is essential for mitotic and male meiotic spindle assembly. Mol. Biol. Cell 15, 121-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia, M.A., Koonrugsa, N., and Toda, T. (2002). Two kinesin-like Kin I family proteins in fission yeast regulate the establishment of metaphase and the onset of anaphase A. Curr. Biol. 12, 610-621. [DOI] [PubMed] [Google Scholar]
- Goshima, G., and Vale, R. D. (2003). The roles of microtubule-based motor proteins in mitosis: comprehensive RNAi analysis in the Drosophila S2 cell line. J. Cell Biol. 162, 1003-1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatsumi, M., and Endow, S. A. (1992). The Drosophila ncd microtubule motor protein is spindle-associated in meiotic and mitotic cells. J. Cell Sci. 103, 1013-1020. [DOI] [PubMed] [Google Scholar]
- Heald, R., Tournebize, R., Habermann, A., Karsenti, E., and Hyman, A. (1997). Spindle assembly in Xenopus egg extracts: respective roles of centrosomes and microtubule self-organization. J. Cell Biol. 138, 615-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff, S., Ahmad, K., Platero, J. S., and van Steensel, B. (2000). Heterochromatic deposition of centromeric histone H3-like proteins. Proc. Natl. Acad. Sci. USA 97, 716-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue, S., and Salmon, E. D. (1995). Force generation by microtubule assembly/disassembly in mitosis and related movements. Mol. Biol. Cell 6, 1619-1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kline-Smith, S. L., Khodjakov, A., Hergert, P., and Walczak, C. E. (2004). Depletion of centromeric MCAK leads to chromosome congression and segregation defects due to improper kinetochore attachments. Mol. Biol. Cell 15, 1146-1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kline-Smith, S. L., and Walczak, C. E. (2004). Mitotic spindle assembly and chromosome segregation: refocusing on microtubule dynamics. Mol. Cell 15, 317-327. [DOI] [PubMed] [Google Scholar]
- Kuriyama, R., Kofron, M., Essner, R., Kato, T., Dragas-Granoic, S., Omoto, C. K., and Khodjakov, A. (1995). Characterization of a minus end-directed kinesin-like motor protein from cultured mammalian cells. J. Cell Biol. 129, 1049-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence, C. J., et al. (2004). A standardized kinesin nomenclature. J. Cell Biol. 167, 19-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiato, H., Khodjakov, A., and Rieder, C. L. (2005). Drosophila CLASP is required for the incorporation of microtubule subunits into fluxing kinetochore fibres. Nat. Cell Biol. 7, 42-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mennella, V., Rogers, G. C., Rogers, S. L., Buster, D. W., Vale, R. D., and Sharp, D. J. (2005). Functionally distinct kinesin-13 family members cooperate to regulate microtubule dynamics during interphase. Nat. Cell Biol. 7, 235-245. [DOI] [PubMed] [Google Scholar]
- Minestrini, G., Mathe, E., and Glover, D. M. (2002). Domains of the Pavarotti kinesin-like protein that direct its subcellular distribution: effects of mislocalisation on the tubulin and actin cytoskeleton during Drosophila oogenesis. J. Cell Sci. 115, 725-736. [DOI] [PubMed] [Google Scholar]
- Mishima, M., Pavicic, V., Gruneberg, U., Nigg, E. A., and Glotzer, M. (2004). Cell cycle regulation of central spindle assembly. Nature 430, 908-913. [DOI] [PubMed] [Google Scholar]
- Pereira, A. J., Dalby, B., Stewart, R. J., Doxsey, S. J., and Goldstein, L. S. (1997). Mitochondrial association of a plus end-directed microtubule motor expressed during mitosis in Drosophila. J. Cell Biol. 136, 1081-1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder, C. L., and Salmon, E. D. (1998). The vertebrate cell kinetochore and its roles during mitosis. Trends Cell Biol. 8, 310-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers, G. C., Rogers, S. L., Schwimmer, T. A., Ems-McClung, S. C., Walczak, C. E., Vale, R. D., Scholey, J. M., and Sharp, D. J. (2004). Two mitotic kinesins cooperate to drive sister chromatid separation during anaphase. Nature 427, 364-370. [DOI] [PubMed] [Google Scholar]
- Rogers, S. L., Rogers, G. C., Sharp, D. J., and Vale, R. D. (2002). Drosophila EB1 is important for proper assembly, dynamics, and positioning of the mitotic spindle. J. Cell Biol. 158, 873-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth, P., Xylourgidis, N., Sabri, N., Uv, A., Fornerod, M., and Samakovlis, C. (2003). The Drosophila nucleoporin DNup88 localizes DNup214 and CRM1 on the nuclear envelope and attenuates NES-mediated nuclear export. J. Cell Biol. 163, 701-706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakowicz, R., et al. (2004). Antitumor activity of a kinesin inhibitor. Cancer Res. 64, 3276-3280. [DOI] [PubMed] [Google Scholar]
- Savoian, M. S., Gatt, M. K., Riparbelli, M. G., Callaini, G., and Glover, D. M. (2004). Drosophila Klp67A is required for proper chromosome congression and segregation during meiosis I. J. Cell Sci. 117, 3669-3677. [DOI] [PubMed] [Google Scholar]
- Sawin, K. E., and Mitchison, T. J. (1995). Mutations in the kinesin-like protein Eg5 disrupting localization to the mitotic spindle. Proc. Natl. Acad. Sci. USA 92, 4289-4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholey, J. M., Brust-Mascher, I., and Mogilner, A. (2003). Cell division. Nature 422, 746-752. [DOI] [PubMed] [Google Scholar]
- Sharp, D. J., Brown, H. M., Kwon, M., Rogers, G. C., Holland, G., and Scholey, J. M. (2000a). Functional coordination of three mitotic motors in Drosophila embryos. Mol. Biol. Cell 11, 241-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp, D. J., McDonald, K. L., Brown, H. M., Matthies, H. J., Walczak, C., Vale, R. D., Mitchison, T. J., and Scholey, J. M. (1999). The bipolar kinesin, KLP61F, cross-links microtubules within interpolar microtubule bundles of Drosophila embryonic mitotic spindles. J. Cell Biol. 144, 125-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp, D. J., Rogers, G. C., and Scholey, J. M. (2000b). Microtubule motors in mitosis. Nature 407, 41-47. [DOI] [PubMed] [Google Scholar]
- Theurkauf, W. E., and Hawley, R. S. (1992). Meiotic spindle assembly in Drosophila females: behavior of nonexchange chromosomes and the effects of mutations in the nod kinesin-like protein. J. Cell Biol. 116, 1167-1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubersax, J. A., Woodbury, E. L., Quang, P. N., Paraz, M., Blethrow, J. D., Shah, K., Shokat, K. M., and Morgan, D. O. (2003). Targets of the cyclin-dependent kinase Cdk1. Nature 425, 859-864. [DOI] [PubMed] [Google Scholar]
- Vernos, I., Raats, J., Hirano, T., Heasman, J., Karsenti, E., and Wylie, C. (1995). Xklp1, a chromosomal Xenopus kinesin-like protein essential for spindle organization and chromosome positioning. Cell 81, 117-127. [DOI] [PubMed] [Google Scholar]
- Walczak, C. E., Verma, S., and Mitchison, T. J. (1997). XCTK 2, a kinesin-related protein that promotes mitotic spindle assembly in Xenopus laevis egg extracts. J. Cell Biol. 136, 859-870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West, R. R., Malmstrom, T., and McIntosh, J. R. (2002). Kinesins klp5(+) and klp6(+) are required for normal chromosome movement in mitosis. J. Cell Sci. 115, 931-940. [DOI] [PubMed] [Google Scholar]
- Yucel, J. K., Marszalek, J. D., McIntosh, J. R., Goldstein, L. S., Cleveland, D. W., and Philp, A. V. (2000). CENP-meta, an essential kinetochore kinesin required for the maintenance of metaphase chromosome alignment in Drosophila. J. Cell Biol. 150, 1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.