Abstract
The transactivator responsive region (TAR) present in the 5′-NTR of the HIV-1 genome represents a potential target for antiretroviral intervention and a model system for the development of specific inhibitors of RNA–protein interaction. Earlier, we have shown that an anti-TAR polyamide nucleotide analog (PNATAR) conjugated to a membrane transducing (MTD) peptide, transportan, is efficiently taken up by the cells and displays potent antiviral and virucidal activity [B. Chaubey, S. Tripathi, S. Ganguly, D. Harris, R. A. Casale and V. N. Pandey (2005) Virology, 331, 418–428]. In the present communication, we have conjugated five different MTD peptides, penetratin, tat peptide, transportan-27, and two of its truncated derivatives, transportan-21 and transportan-22, to a 16mer PNA targeted to the TAR region of the HIV-1 genome. The individual conjugates were examined for their uptake efficiency as judged by FACScan analysis, uptake kinetics using radiolabeled conjugate, virucidal activity and antiviral efficacy assessed by inhibition of HIV-1 infection/replication. While FACScan analysis revealed concentration-dependent cellular uptake of all the PNATAR–peptide conjugates where uptake of the PNATAR–penetratin conjugate was most efficient as >90% MTD was observed within 1 min at a concentration of 200 nM. The conjugates with penetratin, transportan-21 and tat-peptides were most effective as an anti-HIV virucidal agents with IC50 values in the range of 28–37 nM while IC50 for inhibition of HIV-1 replication was lowest with PNATAR–transportan-27 (0.4 μM) followed by PNATAR–tat (0.72 μM) and PNATAR–penetratin (0.8 μM). These results indicate that anti-HIV-1 PNA conjugated with MTD peptides are not only inhibitory to HIV-1 replication in vitro but are also potent virucidal agents which render HIV-1 virions non-infectious upon brief exposure.
INTRODUCTION
HIV-1 infection is spreading at an alarming rate and despite the best efforts at prevention where statistical data confirm a steady increase in the number of new HIV-1 infected patients. In 2004, around five million new infections were reported (http://www.unaids.org/wad2004/EPIupdate2004_html_en/epi04_02_en.htm) with >13 000 new infections occurring daily. The rate of new infection depicts the failure of existing and approved methods to control the spread of the deadly virus. Highly active antiretroviral therapy (HAART) continues to demonstrate increased clinical efficacy due to the continuous introduction of new antiretroviral agents (1). However, the efficacy of new treatments does not assist in preventing the spread of the disease and the cost of drug therapy which is prohibitive for a vast majority of acquired immunodeficiency syndrome (AIDS) patients globally. Given this bleak scenario, strategies should be rapidly developed to reduce the number of newly infected individuals. Developing new virucidal agents to be used as topical formulations during sexual contact, the primary route of new infection, is an essential part of any such strategy (2). Recently, enfuvirtide (Fuzeon™) was approved by FDA as an HIV-1 entry inhibitor. This 36mer peptide directed against the HIV-1 glycoprotein gp41 (3) is administered by subcutaneous injection and is currently the only AIDS drug which exerts its action outside the target cell prior to infection. Besides this, a number of topical microbicides of the NNRTI class (4,5) and surfactants (6) are reported to have virucidal activity but none of them have yet been approved by the FDA for clinical use.
This situation warrants the exploration of alternative strategies for combating HIV infection, one of which involves an in-depth critical analysis of non-mutable, highly conserved regulatory regions of the HIV genome resistant to mutational changes. The targeting of such regions by inhibitory agents logically constitutes a feasible strategy for halting viral replication. The unique 5′ (U5) non-translated region (1–333 nt) of the HIV-1 genome comprises a number of regulatory regions essential for replication. These critical domains include (i) the primer-binding site (183–201 nt), essential for -primed initiation of reverse transcription (7–9), (ii) the A-loop (168–173 nt) located upstream of the primer binding site (PBS) and essential for the selection and interaction of primer (10), (iii) the LTR sequences at the 5′ and 3′ ends of the genome which are essential for viral transcription and integration (11) and (iv) the transactivation responsive region (TAR) essential for gene expression via transcriptional activation (12,13) and which also plays an additional role in the initiation of reverse transcription (14).
Many approaches have been used in attempts to arrest retroviral replication and prominent among them is the antisense strategy (15,16). In the quest for attractive antisense agents, Nielsen et al. (17) first introduced polyamide nucleic acids (PNA) in 1991 and the PNAs were subsequently shown to exhibit improved target affinity, increased duplex stability, nuclease resistance and superior pharmacokinetic properties compared with other antisense agents. We have previously used PNA to target two critical sites within the 5′-untranslated region of the HIV-1 genome. We have reported that PNA as well as PNA–DNA chimera complementary to the primer-binding site of the HIV genome can completely block priming by (18–20). We also targeted another equally important non-mutable region, TAR, and have shown that anti-PNATAR inhibits the tat-mediated transactivation of HIV-1 LTR transcription by efficient sequestration of TAR (21). Additionally, we were able to prove that a 16mer PNA completely blocks the loop and bulge region of TAR in contrast to its shorter analogs, viz. 15, 13 and 12mer, which were also found to be effective in blocking transactivation, albeit to a lesser degree (22). Although PNAs have enormous potential as antisense agents, the success of PNA-mediated gene therapy requires efficient cellular uptake, which has hitherto presented a major barrier to success in strategies employing the use of PNAs as potential therapeutic agents. In order to improve cellular uptake of PNA targeted to the TAR region of the HIV-1 genome we have earlier conjugated an anti-TAR PNA with Transportan, a 27 amino acid membrane transducing (MTD) peptide. We also demonstrated successful in vitro inhibition of HIV-1 replication using this conjugate (23,24).
In the present communication, a 16mer anti-TAR PNA complementary to the minimal functional TAR sequence, comprising the apical stem-loop and bulge regions, has been conjugated with five different carrier peptides. The conjugated PNA effectively permeates the cellular membrane and is able to inhibit HIV-1 replication in infected cells in vitro. In addition, the PNATAR–peptide conjugates are able to penetrate HIV-1 virion particles, resulting in effective inactivation of viral infectivity (24). Since, our main goal is to develop various derivatives of anti-HIV-1 PNAs as effective antiviral/virucidal agents for intervention of HIV-1 infection, all important aspects related to drug delivery of this class of compounds need to be explored extensively. One of the main obstacles to the success of PNA as clinically relevant agents is their poor cellular uptake. Therefore, this issue needs to be addressed before designing experiments for animal models. We have used five different cell-penetrating peptides as delivery vectors. All the peptides besides transportan are shorter than transportan. The use of shorter peptides as an effective cargo for PNA biodelivery will be desirable as they are expected to elicit little or no immunologic response when used in animal models. Penetratin, a well-studied cell-penetrating peptide derived from the homeodomain, is common to all homeoproteins (25,26). We have also used the highly basic 13mer tat-peptide (48–60 amino acids) derived from the 86 amino acid HIV-1 tat protein. This peptide has been shown to actively traverse membranes of various cell types (27). The sequence contains six arginine and two lysine residues, conferring a highly cationic character to this peptide. Transportan-27, is a chimeric peptide containing a 13 amino acid peptide derived from galanin, and a 14 amino acid peptide derived from mastoparan (wasp venom) connected via a lysine residue (23,24). Two transportan analogs, one having a deletion in the galanin arm (Transportan-21) (28) and the other having a deletion in the mastoparan arm (Transportan-22), are included in this study. While Transportan-21, has been described earlier (28), Transportan-22, was designed and synthesized for this study to examine the effect of deleting the cationic domain comprising the six C-terminal amino acids of the mastoparan peptide. The antiviral and virucidal properties, as well as cellular uptake kinetics of these five conjugates were evaluated extensively.
MATERIALS AND METHODS
Designing of PNAs and PNA conjugates
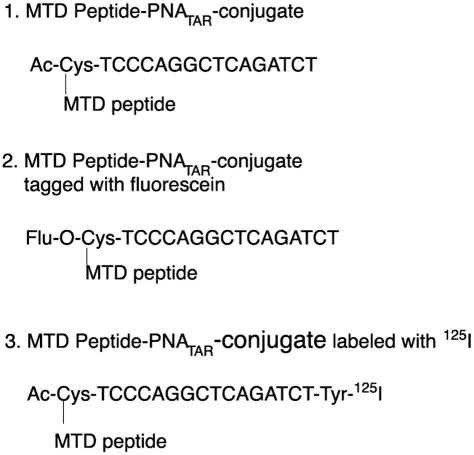
Sequences of PNATAR, PNATAR–MTD peptides and fluorescein labeled PNATAR–MTD peptides are shown in Figure 1. The 16mer penetratin (RQIKIWFQNRRMKWKK) and 13mer tat-peptide (GRKKRRQRRRPPQ) were purchased from BACHEM Biosciences Inc. Transportan-27 (GWYLNSAGYLLGK(e-Cys)INLKALAALAKKIL) and its truncated derivatives Transportan-21 (AGYLLGK(e-Cys)INLKALAALAKKIL) and Transportan-22 (GWYLNSAGYLLGK(e-Cys)INLKALAAL) were obtained from Applied Biosystems. A cysteine residue was incorporated at the N-terminus of the peptides and the –SH group was activated with 3-nitro pyridine sulphenyl groups (Scheme 1). The peptides with 3-nitro-2-pyridinesulfenyl (NPYS) group are reported to react rapidly with thiols to form disulfide linkage (29). Three PNAs purchased from Applied Biosystems were Fluorenylmethyloxycarbonyl (Fmoc)-O-Cys-TCC-CAG-GCT-CAG-ATC-T, Ac-Cys-TCC-CAG-GCT-CAG-ATC-T, and Ac-Cys-TCC-CAG-GCT-CAG-ATC-T-Tyr. The Fmoc-O-Cys-TCC-CAG-GCT-CAG-ATC-T was used to attach fluorescein with 8-amino-2, 4-dioxaoctanoic acid (egl) linker at the 5′ N-terminus following removal of the Fmoc protecting group. Ac-Cys-TCC-CAG-GCT-CAG-ATC-T-Tyr was used for introducing radioiodine in the aromatic ring of Tyr residue at the C-terminus. The Ac-Cys-TCC-CAG-GCT-CAG-ATC-T is designed for the cell culture studies. All the three forms of PNAs had Cys residue at the N-terminus, which provides an activated –SH group for conjugation with transporter peptides.
Figure 1.
Sequence of anti-PNATAR used in the present study along with the variations: (i) PNATAR–MTD peptide conjugate, (ii) Fluorescein-tagged PNATAR–MTD peptide and (iii) I125-labeled PNATAR–MTD peptide.
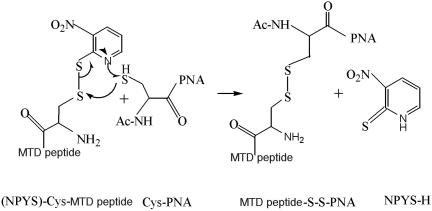
Scheme 1.
Mechanism of conjugation chemistry for PNA–MTD peptides. The peptide moiety has cysteine at its N-terminus and it is activated by incorporating an -NPYS group. The PNA has a terminal cysteine at its 5′ end. In a redox reaction, the S-S linkage is formed which is stable in cell culture media but is easily degraded in the reductive intracellular environment. This degradation is critical for the peptide moiety to complete its task as a transporting vector, releasing the PNA inside the cell. Upon reduction of the disulfide bond, the PNA is released and migrates towards its target.
Fluorescein labeling of PNA
Fmoc-O-Cys-TCC-CAG-GCT-CAG-ATC-T sequence was used for linking fluorescein with egl linker. The fluorescein was attached to the PNA molecule while still immobilized on MBHA resin (polystyrene beads carrying 4-methyl benzhydryl amine). First, the Fmoc group was removed by treating with 20% piperidine in N,N′-Dimethylformamide (DMF) (1 ml) for 5 min. After washing the column with DMF (3× 1 ml), 300 μl of DMF containing 1.66% (w/v) 5-carboxy fluorescein succinimidyl ester (Molecular Probes, Eugene, OR) and 8.3% diisopropyl ethylamine (v/v) was added to the resin and incubated for 1 h with occasional shaking. The labeling solution was discarded and the resin was washed with DMF and finally with dichloromethane (DCM). The resin was further treated with 10% trifluoro acetic acid (TFA) and 2% m-cresol in DCM (100 μl) to remove the trityl group from the terminal cysteine as well as the Bhoc (benzhydrylcarbonyl) protecting groups from PNA. The fluorescein labeled PNA was released from the resin by treating with TFA: m-cresol (4:1) for 2 h and the fluoresceinated PNA was precipitated by the addition of 5-fold excess of dry cold ether. The fluorescein-tagged PNA was further conjugated with different MTD peptides as described below.
Synthesis of PNA-peptide conjugates
All the solvents used were degassed prior to use by bubbling helium gas for 10 min. PNA (1 μmol, 1 eq.) dissolved in 1:1 NMP-H2O to a final concentration of 1 mM (1.0 ml) was supplemented with 1.3 eq. of NPYS-peptide dissolved in N-methyl pyrrolidinone (NMP) to a final concentration of 10 mM. Finally, 100 μl of 1 M sodium acetate buffer (pH 5.0) was added to it and the solution was vortexed and incubated at 40°C for 3 h in the dark. The reaction was quenched by addition of 5% degassed aqueous TFA (final conc. 650 mM) and the conjugate was purified by high-performance liquid chromatography using YMC guardpak ODS-AQ C18 column (S-3 μm, 12 nm, 4× 50 mm) using water: acetonitrile gradient (containing 0.5% acetic acid v/v) with the column temperature maintained at 45°C. The fractions containing the conjugate were pooled and lyophilized and stored at −20°C. During the purification the trifluoroacetate is converted to acetate salt to eliminate the toxicity related with trifluoroacetate. The quantitation of PNA was done by measuring OD at 260 nm divided by molar extinction coefficient [150.6 ml/(μmol × cm)] for PNA. The purity of the conjugated product was examined by mass spectroscopy on a Perspective biosystems voyager DE matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) system using sinapinic acid matrix.
Preparation of 32P-labeled RNA-TAR
The plasmid pEM-7 encoding the HIV-1 TAR under the control of the T7 promoter (30) was used for preparing the RNA transcript representing wild-type RNA-TAR. The plasmid pEM-7 was linearized with HindIII and the 32P-labeled run-off transcript was prepared using [α32P]CTP and T7 RNA polymerase kit according to manufacturer's protocol (Roche Molecular Biochemicals). After the transcription reaction, the plasmid DNA was digested with 25 U of DNase I and the labeled transcript was purified by phenol–chloroform extraction followed by alcohol precipitation.
Gel retardation assay
The affinity and specificity of the PNATAR–peptide conjugate for its target was evaluated by gel electrophoretic mobility shift analysis (EMSA). The 32P-labeled TAR- RNA (20 nM; 10 000 Cerenkov c.p.m.) was incubated with increasing concentrations of PNATAR–peptide conjugate (penetratin and tat) in a buffer containing 30 mM Tris–HCl (pH 8.0), 75 mM KCl, 5.5 mM MgCl2, 1.3 mM DTT, 0.01% NP-40 and 500 ng of poly r(I-C) in a final volume of 15 μl. After 30 min of incubation at 37°C, samples were subjected to gel electrophoresis on 6% polyacrylamide gel using the Tris–borate buffer system. The RNA–PNA-peptide complex was resolved at a constant voltage of 150 V at r.t. for 3 h and subjected to phosphorImager analysis (Molecular Dynamics).
Reverse transcription of TAR-RNA
Reverse transcription catalyzed by HIV-1 RT was carried out in the presence of individual PNATAR–peptide conjugates (penetratin and tat-peptides) using TAR-RNA template primed with 5′ 32P-labeled 18mer DNA primer as described before (22). The individual PNATAR–peptide conjugates at the indicated concentrations were pre-incubated with 10 nM of the annealed template primer at room temperature in a reaction buffer containing 50 mM Tris–HCl (pH 7.8), 10 mM DTT, 100 μg/ml BSA, 60 mM KCl and 5 mM MgCl2 and 100 μM each of the 4 dNTPs. Reverse transcription was initiated by the addition of 50 nM of HIV-1 RT. The reactions were performed at 37°C and terminated by the addition of equal volume of Sanger's gel loading dye (31). The products were resolved on 8% polyacrylamide denaturing urea gel and visualized by phosphoimaging (Molecular Dynamics).
Cell culture, transfection of 293T cells and purification of HIV-1 virions
The CEM CD4+ lymphocytes 12D7 (32) were grown in RPMI-1640 medium supplemented with 10% fetal calf serum (FCS), 4 mM l-glutamine at 37°C in 5% CO2 containing humidified air. Early-mid log phase cells were harvested and washed with an equal volume of PBS with Ca2+ and Mg2+. For making pseudotyped HIV-1, 293T cells grown in complete DMEM were co-transfected with pHIV-1JR-CSF-lucenv(−) (33) and pVSV-G using the calcium phosphate transfection system (Life Technologies). The culture supernatant was saved at 24, 48 and 72 h post-transfection and analyzed for p24 antigen using the enzyme-linked immunosorbent assay p24 antigen kit (Abbott Laboratories). The pseudotyped HIV-1 virions were isolated from the culture supernatant by filtration through a 0.45 μm pore size PVDF membrane (Millipore) followed by ultracentrifugation at 70 000 g for 45 min. The viral pellet was re-suspended in 2 ml of PBS and re-centrifuged through 5 ml of 20% sucrose/PBS cushion for 30 min at 100 000 g.
Cellular uptake studies of PNA-peptides
CEM cells grown in complete RPMI medium containing 10% fetal bovine serum (FBS) were harvested, washed with PBS and re-suspended in the same medium containing 2% FBS at a cell density of 0.5 × 106 cells/ml. Cells were aliquoted in a 12-well costar microtiter plate at 0.5 × 106 cells per well and incubated at 37°C with varying concentrations of fluorescein-tagged PNATAR–MTD peptide conjugates. At varying time points the cells were harvested and washed with PBS buffer and re-suspended in complete RPMI medium. The fluoresceinated cells were counted by FACS analysis (FACScan) on a Beckton Dickinson flow cytometer. All the sets were scanned in the presence of 1 μg of propidium iodide to ensure that the uptake occurred in live cells only. Cell Quest Pro software (Beckton Dickinson) was used to acquire and analyze 104 events detected by the FL1 detector (for FITC), which excluded FL3 detector (propidium iodide).
Luciferase assay
Luciferase assays were performed using the Promega luciferase assay kit and manufacturer's protocol (Luciferase Reporter assay system, Promega Corp., Madison, WI). Briefly, the cells were harvested, washed once with PBS and lysed by addition of 250 μl of the 1× reporter lysis buffer (Promega). Following incubation at 25°C for 15 min on a rocking shaker, the lysates were centrifuged at 15 000 r.p.m. for 10 min and the supernatants were collected in fresh tubes. Luciferase assay was performed by adding 100 μl of the luciferase substrate to 25 μl of each samples in a 96-well Fluorotec plate, and the light emission was measured using a Packard top count fluorescence counter. The total light units were normalized by the total protein content in the extract.
Uptake kinetics
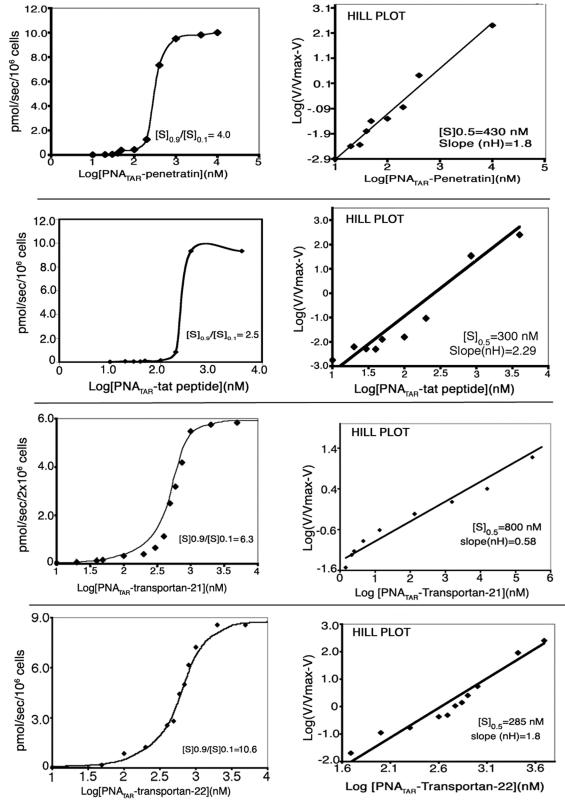
The PNATAR–peptide conjugates and PNATAR were labeled with [125I] using the Na[125I] and the chloramine-T labeling kit from ICN. For radiolabeling, 0.5 nmol of the conjugate and 0.28 nmol of [125I] (0.5 mCi) were reacted in the presence of 2.8 nmol of chloramine-T for 1 min and quenched by addition of 62 nmol of sodium metabisulphite according to manufacturer's protocol. The labeled conjugate was purified using NAP-10 gel filtration (Amersham-Pharmacia) and C18 reverse phase columns. The final product was quantified by absorption at 260 nm and the specific radioactivity was adjusted to desired levels by adding unlabeled conjugate. For uptake experiments, 1 × 106 CEM cells were incubated with varying concentrations of the radiolabeled conjugate at room temperature. After 30 s of incubation, each individual sample was filtered through a glass fiber filter (GF-B), washed extensively with PBS and the amount of radiolabeled PNA conjugate internalized by the cells was determined by gamma counting. The [S]0.9/[S]0.1 ratio and cooperativity index of the substrate at 0.9 Vmax and 0.1 Vmax was determined by plotting the uptake versus substrate concentration. Hill coefficient (nH) was determined from the Hill plot of uptake data.
Anti-HIV-1 activity in cell culture
The effect of anti-TAR PNA on HIV-1 production was monitored by infecting CEM cells with pseudotyped HIV-1 virions expressing the firefly luciferase reporter gene, described above. Briefly, pseudotyped HIV-1 virions (100 ng p24) in the presence of 10 μg of polybrene/ml were added to 0.5 × 106 CEM cells in a final volume of 1.0 ml to achieve multiplicities of infection (MOI) of 0.003. The cell cultures were incubated at 37°C for 2 h, and spun down at 1250 r.p.m. The cells were washed twice with 1× PBS and re-suspended in 1 ml of RPMI medium containing 10% FBS and varying amounts of PNATAR–peptide conjugate, anti-TAR PNA, or peptide alone. At 48 h post-transfection, the cells were harvested and the level of infection/viral gene expression was quantitated by luciferase assay. In another experiment, the cells were first incubated with the PNATAR–peptides, anti-PNATAR, or peptide alone for 2 h. The cells were then spun down at 1250 r.p.m. and the cells were washed and infected with the pseudovirus, as described above. After 2 h the cells were again harvested and washed with PBS and re-suspended in culture medium. The cells were incubated for 48 h, harvested and processed as described above.
Determination of virucidal activity PNATAR–peptide conjugates
To examine the virucidal activity of the conjugate, 150 μl of HIV-1 pseudovirus (14 ng, p24 eq.) was incubated with different concentrations of PNATAR–peptide conjugates at 37°C for 1 h. The mixture was then ultracentrifuged at 100 000 g for 1 h and the viral pellet was re-suspended in RPMI medium containing CEM cells (0.5 × 106) and 10 μg polybrene in a final volume of 1 ml. The cells were incubated on a rocking shaker at 37°C for 2 h, harvested, washed once with 1× PBS and finally re-suspended in 1 ml fresh complete RPMI media in a 12-well tissue culture plate. The cells were incubated in a 5% CO2-containing humidified incubator at 37°C for 48 h and then harvested and washed with PBS. The washed cells were lysed in 250 μl cell lysis buffer provided with the luciferase assay kit (Promega) and the cell lysates were assayed for firefly luciferase activity. The level of luciferase activity was normalized by protein content of the extract (BCA protein assay, Pierce). The data from three independent experiments were evaluated for the determination of dose median-effect using the Calcusyn software (Biosoft).
Calculation of median dose effect
The effect of PNATAR–MTD peptide concentrations on the luciferase expression were analyzed using the Calcusyn software (Biosoft) based on the median-effect. This software is based on a median-effect equation developed by Chou (34,35) for determining dose–effect relationship. The IC50 corresponds to median-effect dose which is given with 95% confidence in the results. The equation used is fa/fu = (D/Dm)m, where D is the dose of drug; Dm is the median-effect dose signifying the potency determined from the x-intercept of the median-effect plot; fa is the fraction effected by the dose while fu is the unaffected fraction (fu = 1 − fa); m is an exponent signifying the sigmoidicity of the dose–effect curve. The IC50 corresponds to median-effect dose which is given with its 95% confidence limit in the result table.
RESULTS
Binding specificity of the PNATAR–penetratin and PNATAR–tat-peptide to TAR-RNA
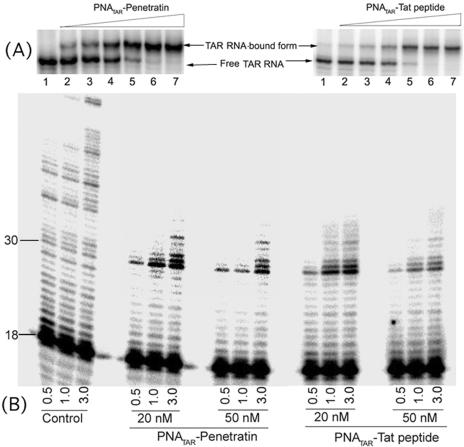
In order to evaluate the binding affinity of the PNATAR–peptide conjugates to their target sequence, we performed a gel mobility shift assay with internally 32P-labeled 82 bp long TAR-RNA transcript (20 nM) and increasing concentrations of PNATAR–peptide (penetratin and tat-peptides) conjugates (Figure 2A). At an equimolar ratio of PNATAR–peptide to TAR-RNA, ∼90% shift was achieved (lane 5). The same binding pattern was observed in a gel shift assay with the unconjugated PNATAR (data not shown), suggesting that the conjugation with the transporter peptide does not inhibit binding to its target. Binding experiments were done for PNATAR–transportan and its truncated derivatives and the results revealed a pattern similar to those exhibited by the PNATAR– penetratin and PNATAR–tat conjugates (data not shown).
Figure 2.
(A) Specificity of interaction of PNATAR–penetratin or PNATAR–tat-peptide with its target sequence on the HIV-1 genomic RNA. The binding affinity of anti-PNATAR to the TAR-RNA was assessed by EMSA. PNATAR–peptide conjugates at varying concentrations (2.5, 5, 10, 20, 30 and 40 nM) were incubated with 20 nM of 32P-labeled TAR-RNA for 30 min at 37°C in binding buffer and subjected to native PAGE. The RNA–PNA complexes were resolved at a constant voltage of 150 V at r.t. and visualized by phosphorImaging. (B) Primer extension assay with TAR-RNA template in the presence PNATAR–penetratin or PNATAR–tat-peptide. The TAR-RNA template was first annealed with a 5′ 32P-labeled 18mer DNA and then incubated with indicated concentrations of PNATAR–peptide at 37°C for 30 min. Reverse transcription reactions were initiated under standard reaction condition as described in Materials and Methods. Reactions were carried out at 37°C and stopped after indicated time points (min) by addition of 2× Sanger's gel loading dye. Reaction products were resolved on an 8% denaturing polyacrylamide-urea gel. Control samples represent reverse transcription in the absence of PNATAR–peptide. The positions of the labeled 18 and 30mer extension product are indicated. The 30mer position corresponds to the beginning of the loop region of the TAR-RNA, which is selectively targeted by the antisense PNA.
Inhibition of reverse transcription of TAR-RNA in the presence of PNATAR–peptide
Since reverse transcription of the viral RNA is blocked by PNA bound to the template downstream of the reverse transcription site (18,19), similar inhibition of reverse transcription was expected with PNA–peptide conjugates. To examine this contention, the 82mer TAR-RNA primed with 32P-labeled 18mer DNA primer was incubated in the absence and presence of increasing concentrations (20 and 50 nM) of individual PNATAR–peptide conjugates at 37°C for 30 min, followed by initiation of reverse transcription by HIV-1 RT (Figure 2B). In the control lanes, (no PNATAR–peptide) primer extension was not inhibited and complete synthesis product was formed within 3 min. However, in presence of PNATAR–peptide, a distinct pause was evident at position 30 of the RNA-TAR loop just prior to the PNA binding site (22). These results clearly demonstrate that conjugation of PNA with the MTD peptide(s) does not reduce its target efficacy or the halting of reverse transcription at the site of binding.
Cellular uptake of PNATAR–peptide
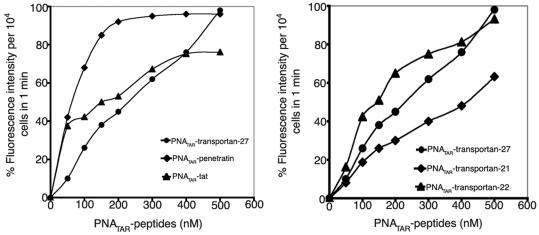
We have previously shown that PNA conjugated with transportan is efficiently taken up by cells (24). In order to explore the suitability of other carrier peptides to further improve the uptake of PNA, we conjugated anti-TAR PNA with four different MTD peptides and evaluated their uptake efficiency by FACScan analysis. Each of the conjugates contained a fluorescein tag attached to the PNA molecule. The unconjugated naked PNA with a similar fluorescein tag was used as a control. The lymphocyte CEM cells were incubated with varying concentrations (50–500 nM) of individual fluorescein-tagged PNATAR–peptide conjugates and the fluorescence signals per 10 000 cells were recorded by FACScan. Figure 3 displays differential uptake patterns as judged by fluorescence intensity in the presence of individual PNATAR–peptide conjugates. For instance, the PNATAR–penetratin conjugate exhibited ∼85% cellular uptake at 150 nM while only 40–50% of cells were found to be fluorescence positive at similar concentrations of PNATAR–tat and PNATAR–transportan conjugates (Figure 3A). The cellular uptake of the penetratin conjugate was the most efficient of the tested conjugates as a sharp linear increase in the fluorescence intensity was consistently noted at lower concentrations (50–100 nM) with ∼99% uptake at 200 nM within 1 min of incubation. Transportan was the second most efficient peptide carrier as PNATAR–transportan exhibited a linear increase in the uptake at all the concentrations (50–500 nM) ranging from 10 to 15% uptake at 50 nM to ∼99% at 500 nM. Its truncated derivative, transportan-22, was equally efficient while transportan-21 displayed relatively poor cellular uptake as compared to its parent peptide (Figure 3B). This suggests that the hydrophobic domain of the galparan unit is indispensable and its deletion changes its uptake pattern relative to the parent transportan peptide. Extending the incubation period up to 5 min had no impact on the fluorescence intensity, indicating that cellular uptake is very rapid for all the PNATAR–peptide conjugates.
Figure 3.
Comparison of uptake patterns of PNATAR–peptide conjugates in CEM cells. In panel A, PNATAR–Penetratin, PNATAR–tat, and PNATAR–transportan-27 are compared. In panel B, PNATAR–transportan-22, PNATAR–transportan-21, and PNATAR–transportan-27 are compared. Cells (0.5 × 106) were incubated with varying concentrations of fluorescein-tagged PNATAR–peptide conjugates in RPMI medium with 2% FCS at room temperature. Uptake of conjugate within 1 min was evaluated by FACS analysis.
It was further confirmed that the incubation temperature had no influence on the cellular uptake of each of the PNA–peptide conjugates as fluorescence intensity per 10 000 cells did not change significantly when incubation was carried out at 4, 25 and 37°C (data not shown). Within 1 min of incubation the uptake of PNATAR–peptide reaches a maximum at all the tested temperatures, suggesting that the uptake does not occur via receptor-mediated endocytosis, which is largely inhibited at lower temperatures. To rule out the possibility that the PNA–peptide conjugates adhere non-specifically to the cell surface, falsely elevating fluorescence readings, a parallel experiment was carried out. Following incubation with the individual fluorescein-tagged PNA–peptide conjugates, cells were washed once with PBS containing 0.01% trypsin, then twice with PBS and finally re-suspended in RPMI media for FACScan. An 8–10% decrease in the fluorescence intensities was seen following the trypsin wash while the overall uptake pattern remained unchanged, suggesting that the major portion of the fluorescence intensity observed in the FACScan was due to cellular uptake of the individual conjugates and not non-specific association with the cell membrane (data not shown).
Uptake kinetics
The cellular internalization of penetratin, tat-peptide and transportan has been reported to be uninhibited at 4°C suggesting that the uptake mechanism does not involve receptor-mediated endocytosis (25,26,36,37). In addition, the rapid cellular internalization of penetratin as well as transportan has been reported to remain unsaturated (25,26,37). PNATAR–penetratin, having a net positive charge, is reported to enter the hydrophobic membrane lipid environment following interaction with negatively charged molecules on the cell surface through a two-step process, phase transfer followed by tryptophan-dependent translocation (38). A similar pattern is expected when these peptide carriers are used as the vehicle to deliver PNA into the cells. To determine this, the PNA moiety of the individual PNA–peptide conjugates was labeled with radioiodine (125I) and used for quantitative evaluation of their cellular uptake. Briefly, 0.5 × 106 cells (lymphocytes) were incubated with varying concentrations of radiolabeled PNATAR–peptide conjugate at room temperature for 30 s followed by rapid filtration through the GF-B. The filters were washed with PBS and the radioactivity within the cells was determined by gamma counting. The results shown in Figure 4 represent the uptake kinetics obtained with individual PNA–peptide conjugates. All the PNA–peptide conjugates yielded a sigmoidal curve (Figure 4, left panel) suggesting a cooperative interaction between the conjugate and the cellular membrane. The cooperativity index (ratio of [S]0.9/[S]0.1) obtained with individual conjugates suggest the amount of conjugate that would be required for increasing the uptake velocity from 0.1 to 0.9 Vmax. The cooperativity indices varied from 2.5 to 10.6 with PNA–tat conjugate having the lowest and the PNATAR–transportan-22 having the highest. The PNATAR–penetratin and PNATAR–transportan-27 conjugates displayed intermediate values of 4.0 and 6.33, respectively. A Hill plot of the uptake data was generated to determine the nH values for individual PNA–peptide conjugates (Figure 4, right panel). The [S]0.5 value as well as nH for individual PNA–peptide conjugate was determined from this plot. The [S]0.5 values indicate the concentration of the conjugate at which uptake velocity is half of Vmax. The [S]0.5 values in the range of 285–430 nM were obtained with PNATAR–tat, PNATAR–transportan-22 and PNATAR–penetratin conjugates while conjugate with transportan and its truncated derivative transportan-21, exhibited [S]0.5 values in the range of 0.8–1.5 μM. The nH values determined from the slope of the Hill plot varied from 0.58 for PNATAR–transportan-21 to 2.29 for PNATAR–tat conjugate with an intermediate value of 1.8 obtained for both PNATAR–penetratin and PNATAR–transportan-22 conjugates. The nH >1 for PNATAR–tat, PNATAR–penetratin and PNATAR–transportan-22 suggest that these conjugates may have more than one interaction site on the cellular membrane.
Figure 4.
Uptake kinetics studies of PNATAR–penetratin, PNATAR–tat-peptide, PNATAR–transportan-21 and PNATAR–transportan-22 in CEM cells. The 125I-labeled conjugate was incubated with CEM cells and the amount of conjugate taken up by the cells was determined from specific radioactivity of the conjugate as described in the Materials and Methods. The uptake of the conjugate was plotted against the log of conjugate concentration and the cooperativity index ([S]0.9/[S] 0.1) was determined (left panels). Hill coefficient (nH) was determined from the slope of the Hill plot of the uptake data shown in the right panels. Cooperativity index represents the ratio of the substrate at 0.9 Vmax and 0.1 Vmax while nH represents the concentration of the conjugate at which the velocity of the uptake is equal to 0.5 Vmax. PNATAR–transportan data (24) were provided for comparison.
Inhibition of HIV-1 replication in vivo in the presence of anti-PNATAR analogs
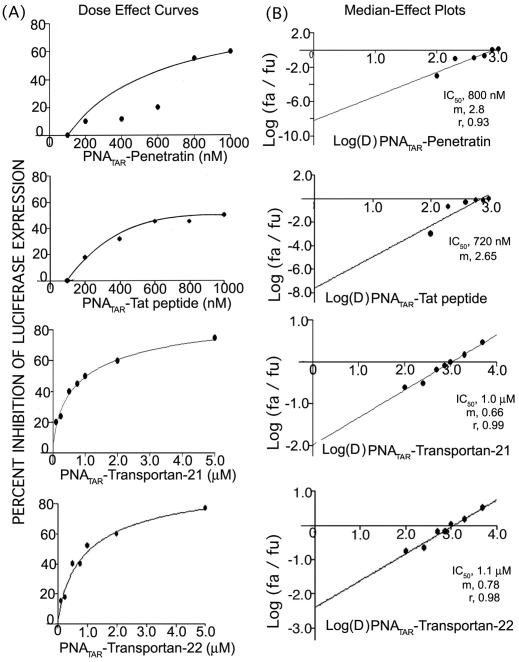
In order to evaluate the in vitro antiviral efficacy of the individual PNATAR–peptide conjugates, the growth media for CEM cells infected with pseudotyped HIV-1 was supplemented with the conjugates. In this experiment, CD4+ lymphocytes infected with highly infectious VSV-G pseudotyped HIV-1 virus (carrying the firefly luciferase reporter gene) were incubated with varying concentrations of individual PNATAR–peptide conjugates. Similar experiments were performed with unconjugated anti-PNATAR and peptide alone as controls. Experiments were carried out for 48 h to assess the inhibition of viral replication during this interval. Following the incubation, the cells were lysed and the cell extract was normalized for total protein content and analyzed for quantitative levels of luciferase expression. The luciferase activity obtained in the infected cells in the absence of PNA was assigned an arbitrary value of 100 while the other samples were given values relative to this control value. A substantial decrease in viral replication was observed as the concentration of PNA conjugate was increased. Figure 5 shows the dose–effect curves (left panel; Figure 5A) and the median-effect plot (right panel; Figure 5B) obtained with individual conjugates. Addition of 100 nM PNATAR–penetratin to the culture media did not inhibit HIV-1 replication but higher concentrations did effectively inhibit viral replication. At 800 nM PNATAR–penetratin, ∼50% inhibition is achieved. The same pattern was achieved when the cells were first incubated with PNATAR–penetratin for 2 h prior to infection with the virus demonstrating that the uptake of the PNA conjugate and binding to its target sequence is rapid and effective (data not shown). The pattern with the other PNATAR–peptides was found to be the same and the IC50 values were calculated using Calcusyn software are tabulated in Table 1.
Figure 5.
(A) Dose–effect curve for antiviral efficacy of PNATAR–penetratin, PNATAR–tat-peptide PNATAR–transportan-21 and PNATAR–transportan-22 conjugates. CEM cells were infected with VSV-G pseudotyped S1 strain of HIV-1 and grown in the presence of increasing concentrations of PNATAR–penetratin and PNATAR–tat-peptide (0–1000 nM). Cells were harvested, lysed and assayed for luciferase expression 48 h post infection. The effect PNATAR–peptide concentrations on the luciferase expression was analyzed using the Calcusyn software (Biosoft). (B) Median-effect Plot: The IC50 values determined from the ratio of log of luciferase expression in treated cells (fa) and untreated cells (fu) as a function of the log concentration of PNATAR–penetratin, PNATAR–tat-peptide, PNATAR–transportan-21 and PNATAR–transportan-22 conjugates were 0.8, 0.70, 1.0 and 1.1 μM, and the linear correlation coefficients, r, were 0.93, 0.87, 0.99 and 0.98, respectively.
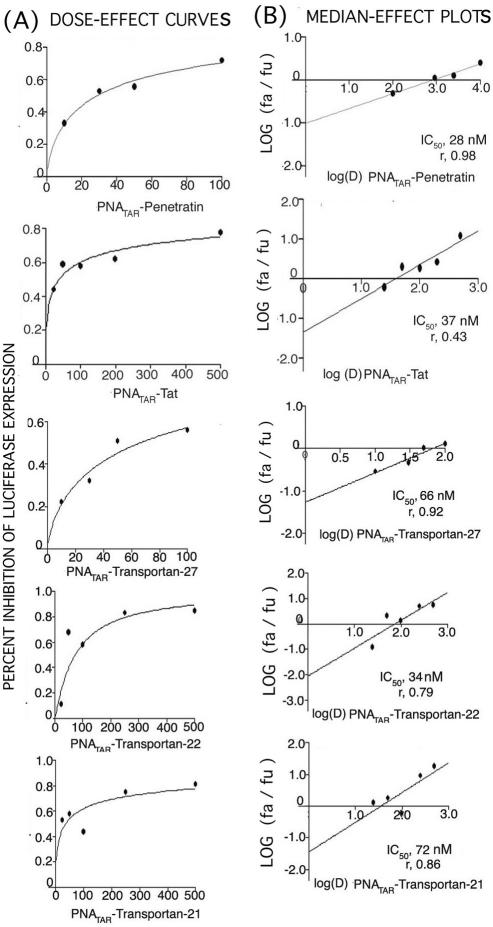
Table 1.
Comparative antiviral efficacy and virucidal activity of PNATAR in presence of different peptides
| PNATAR–peptide conjugate | Antiviral efficacy | Virucidal activity | ||||
|---|---|---|---|---|---|---|
| IC50 (nM) | m | r | IC50 (nM) | m | r | |
| PNATAR–penetratin | 800 | 2.8 | 0.93 | 28 | 0.69 | 0.98 |
| PNATAR–tat-peptide | 720 | 2.6 | 0.87 | 37 | 0.94 | 0.43 |
| PNATAR–transportan-27 | 400 | 1.4 | 0.98 | 66 | 0.69 | 0.92 |
| PNATAR–transportan-21 | 1000 | 0.6 | 0.99 | 34 | 0.46 | 0.79 |
| PNATAR–transportan-22 | 1100 | 1.78 | 0.98 | 72 | 0.86 | 0.86 |
For carrying out antiviral efficacy experiments CEM cells were infected with VSV-G pseudotyped S1 strain of HIV-1 and then grown in the presence of increasing concentrations of PNATAR–MTD peptides. Cells were harvested, lysed and assayed for luciferase expression 48 h post infection. The effects of PNATAR–MTD peptide concentrations on the luciferase expression were analyzed using the Calcusyn software (Biosoft) based on Median-effect equation of Chou (34,35). The IC50 values determined from the ratio of the log of luciferase expression in treated cells (fa) and untreated cells (fu) as a function of log concentration of PNATAR–MTD peptide. Sigmoidicity (m) of the dose–effect curve is derived from slope of median-effect plot and values of 1, >1, <1 indicate hyperbolic, sigmoidal, and negative sigmoidal shapes, respectively. Linear correlation coefficient of the median-effect plot is represented by r and a value of 1 indicates perfect conformity.
For carrying out virucidal activity experiments PNATAR conjugated with different peptides at varying concentrations (25, 50, 100, 250 and 500 nM) were incubated at 37°C with HIV-1 virions for 2 h. The virion particles were spun down by ultracentrifugation and then the virions were used for infection of CEM cells for 48 h at 37°C in a 5% CO2 incubator. The cells were harvested, lysed and assayed for luciferase activity. The median dose effect was calculated using Calcusyn software program (Biosoft). Penetratin displays the greatest virucidal efficacy among this group of MTD peptides.
The virucidal effects of different PNATAR–peptide conjugates
The virucidal effects of PNATAR–peptide conjugates were evaluated by treating the HIV-1 pseudovirus (14 ng, eq. p24) with varying concentrations of PNATAR–peptide conjugates (Figure 6). The treated virions were used to infect the CEM cells. The infected cells were then incubated for 48 h and lysed. The expression levels of a luciferase reporter gene were measured in the cell lysates to evaluate the effect of individual PNATAR–peptide conjugate on viral infectivity. Untreated virions were used as a control in this experiment. The IC50 for PNATAR–peptide conjugate on viral infectivity are tabulated in Table 1. As apparent from the median-effect plot and dose–response curve, PNATAR–penetratin was highly virucidal showing the dose median of 28.0 nM closely followed by PNATAR–transportan-21 with a value of 34.0 nM and by PNATAR–tat-peptide with a value of 37.0 nM. The virucidal activity of PNATAR–transportan-21 is not congruent with its pattern of reduced uptake compared to parent transportan, as shown by FACScan studies, suggesting that the mode of interaction with virus envelope is different from its interaction with the cell membrane. The deletion of the cationic domain in the mastoparan unit shows that there is no alteration in uptake pattern as suggested by FACScan studies and the virucidal activity remains comparable to the parent transportan. Although PNATAR–transportan-27 and PNATAR–transportan-22 show dose median values of 66.0 and 72.0 nM, respectively, these are still in the <100 nM range. The IC50 values for virucidal and antiviral activities are in a narrow range not showing correlation with the uptake efficiency of each of the peptides. The most plausible explanation is that the peptides are used here as a cell-transducing vector and once inside the cell the disulfide bond will be cleaved, leaving the PNA free to reach its target. As the experiments were done for 48 h, the effect of rapid entry is likely to be minimized after 48 h.
Figure 6.
(A) Dose–effect curves for virucidal activity of PNATAR–peptides. The VSV-G pseudotyped HIV-1 virions were first incubated with indicated concentrations of individual PNATAR–peptide conjugates as described in the Materials and Methods. The treated virions were used to infect CEM cells (0.5 × 106 cells). The infected cells were grown for 48 h, harvested, lysed and assayed for luciferase activity. The level of luciferase expression as a function of virucidal effect of individual PNATAR–peptide concentration on HIV-1 virions in CEM cells infected with treated and untreated virions, was analyzed using the Calcusyn software (Biosoft). (B) Median-effect Plots for virucidal activity: The IC50 for virucidal effect of individual PNATAR–peptide conjugates was determined from the ratio of the log of luciferase expression in CEM cells infected with treated (fa) and untreated HIV-1 virions (fu) as a function of the log concentration of PNATAR–peptide (Table 1).
DISCUSSION
AIDS is a sexually transmitted disease that has become a global pandemic. The numbers of patients infected with HIV are increasing at an alarming rate mainly due to transmission during sexual contacts. The reasons for the huge number of new infections due to sexual transmission (2) include: ignorance of the HIV status of the sexual partners, unavailability of condoms and anti-HIV spermicides, and fear of confronting male partners regarding safe sex prior to intercourse. Despite massive efforts to develop an effective vaccine against HIV-1 (39), a vaccine that will confer long-term protective immunity has yet to be developed. For these reasons, it is imperative to develop a strategy that will minimize new infections. Along these lines, efforts are underway to develop topical microbicides (4–6) and some agents, such as (+)-calanolide A, are advancing in clinical trials (5). This strategy will be helpful in decreasing the number of new infections, a critical step towards effective control of the AIDS epidemic. Antisense technology has great potential as part of this prophylaxis strategy and PNAs are proven antisense agents (40,41). Although PNAs are very potent antisense molecules in vitro, they are poorly internalized by cells (42). A number of methods for their intracellular delivery have been developed which include microinjection (43), electroporation (44), co-transfection with DNA (45,46) and conjugation with transporter peptides (23,24).
In the present study, we have used various MTD peptides as delivery vectors for anti-HIV-1 PNA and have evaluated their influence on the cellular uptake of PNA, as each transporter peptide will alter both the biodelivery and bioavailability profiles of the PNA. Gel shift assays clearly indicated that the conjugation with different MTD peptides does not decrease the affinity of the PNA for its target RNA sequence. We have earlier shown that uptake of PNATAR–transportan conjugate is temperature independent and not affected by phenylarsine treatment of the cells suggesting non-involvement of endocytosis or receptor-mediated internalization of the conjugate (24). Similar uptake patterns were also noted for PNATAR–penetratin and PNATAR–tat conjugates. Our observations are consistent with the reported study on the uptake of penetratin and transportan peptides (37) while in disagreement with the nature of uptake of the tat-peptide (36,47,48). Initially, Vivès et al. (36), have shown that endocytosis pathway is not involved in the uptake of tat-peptide as no inhibition of uptake was noted at 4°C. Later, they contradicted their own observations and suggested that entry of tat-peptide is ATP and temperature dependent, likely involving an endocytotic mechanism (47,48). Recently, Wadia et al. (49) have demonstrated that the uptake of a large Tat–Cre fusion protein (38 kDa) is mediated by lipid-raft dependent micropinocytosis which is also inhibited at 4°C. However, in our hands, the uptake of PNATAR–tat conjugate as determined by FACScan analysis was found to be temperature independent indicating that endocytosis pathways may not be operative for the conjugate. It is possible that the mode of uptake for tat-peptide alone versus tat–PNA conjugates may differ significantly.
The uptake kinetics have revealed a sigmoidal curve for all the five PNATAR–MTD conjugates indicating a common uptake mechanism, likely involving cooperative interaction between the cellular membrane and the conjugate. The observed cooperativity of uptake may be due to micelle formation, which requires a concentration threshold of components. A cooperativity index of 2.5 observed with PNATAR–tat conjugate suggests that in order to increase the uptake velocity from 0.1 to 0.9 Vmax, only a 2.5-fold increase in the concentration of conjugate is required. The cooperativity indices with other PNATAR–peptide conjugates were in the range of 4.0–10.6. This pattern of sigmoidal uptake is expected to be more effective than a hyperbolic curve which would require a much higher concentration of the substrate (90-fold increase) to achieve similar velocity from 0.1 to 0.9 Vmax. The anti-HIV-1 PNA in this study was designed to target the TAR region of the HIV-1 genome. Conjugation of this PNATAR–MTD peptides facilitated intracellular delivery with the goal of inhibiting HIV-1 production by down regulation of tat-mediated transactivation of HIV-1 transcription. All the conjugates were indeed inhibitory to HIV-1 production when supplemented in cell culture. We have shown that infection of CEM cells by HIV-1 virions was also drastically reduced in the presence of PNATAR–transportan-27 conjugate (23,24). Similar results were also obtained with PNA conjugated with penetratin, tat, and two truncated derivatives of transportan. However, PNATAR–penetratin conjugate was the most effective virucidal agent displaying an IC50 of 28 nM. The rendering of HIV-1 virions as non-infectious indicates that these conjugates may either interfere with viral entry by altering or disrupting the viral envelope or may permeate the virus envelope and bind to the target sequence on the viral genome. We have earlier shown that endogenous reverse transcription of HIV-1 virions pretreated with PNATAR–transportan conjugate is completely aborted in the TAR region since the PNA conjugate bound to the TAR region cannot be displaced by reverse transcriptase (24).
The potent virucidal activity displayed by this class of PNA–MTD conjugates may not be unique to HIV-1 but may also be relevant for other enveloped viruses. Most currently available anti-HIV drugs do not inactivate circulating HIV-1 virions and their mode of action is limited to inhibiting HIV-1 replication after the infection (HIV entry) has taken place. The only exceptions to this are entry inhibitors that block HIV-1 entry into the cells without inactivating the virion particles (50–52) and enfuvirtide is the only entry inhibitor currently in clinical use. In this context, anti-HIV-1 PNAs conjugated with MTD peptides are distinct in that these are not only inhibitory to intracellular HIV-1 replication but may also have the ability to block HIV-1 infection by rendering HIV-1 virions non-infectious.
Acknowledgments
This research was supported by grants from the NIAID/NIH (AI42520). We would like to thank Dr. Beverly E. Barton, Department of Surgery, UMDNJ, for FACScan analysis and helpful discussions. Funding to pay the Open Access publication charges for this article was provided by National Institute of Allergy and Infectious Diseases, National Institute of Health.
Conflict of interest statement. None declared.
REFERENCES
- 1.Kress K.D. HIV update: emerging clinical evidence and a review of recommendations for the use of highly active antiretroviral therapy. Am. J. Health Syst. Pharm. 2004;61:S3–S14. doi: 10.1093/ajhp/61.suppl_3.S3. [DOI] [PubMed] [Google Scholar]
- 2.Merson M.H. Slowing the spread of HIV: agenda for the 1990s. Science. 1993;260:1266–1268. doi: 10.1126/science.8493570. [DOI] [PubMed] [Google Scholar]
- 3.Ketas T.J., Klasse P.J., Spenlehauer C., Nesin M., Frank I., Pope M., Strizki J.M., Reyes G.R., Baroudy B.M., Moore J.P. Entry inhibitors SCH-C, RANTES, and T-20 Block HIV Type 1 replication in multiple cell types. AIDS Res. Hum. Retroviruses. 2003;19:177–186. doi: 10.1089/088922203763315678. [DOI] [PubMed] [Google Scholar]
- 4.Borkow G., Salomon H., Wainberg M.A., Parniak M.A. Attenuated infectivity of HIV type 1 from epithelial cells pretreated with a tight-binding non-nucleoside reverse transcriptase inhibitor. AIDS Res. Hum. Retroviruses. 2002;18:711–714. doi: 10.1089/088922202760072339. [DOI] [PubMed] [Google Scholar]
- 5.Creagh T., Ruckle J.L., Tolbert D.T., Giltner J., Eiznhamer D.A., Dutta B., Flavin M.T., Xu Z.Q. Safety and pharmacokinetics of single doses of (+)-calanolide A, a novel, naturally occurring non-nucleoside reverse transcriptase inhibitor, in healthy, human immunodeficiency virus-negative human subjects. Antimicrob Agents Chemother. 2001;45:1379–1386. doi: 10.1128/AAC.45.5.1379-1386.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Piret J., Lamontagne J., Bestman-Smith J., Roy S., Gourde P., Désormeaux A., Omar R.F., Juhász J., Bergeron M.G. In vitro and in vivo evaluations of sodium lauryl sulfate and dextran sulfate as microbicides against herpes simplex and human immunodeficiency viruses. J. Clin. Microbiol. 2000;38:110–119. doi: 10.1128/jcm.38.1.110-119.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Muesing M.A., Smith D.H., Capon D.J. Regulation of mRNA accumulation by a human immunodeficiency virus trans-activator protein. Cell. 1987;48:691–701. doi: 10.1016/0092-8674(87)90247-9. [DOI] [PubMed] [Google Scholar]
- 8.Ratner L., Haseltine W., Patarca R., Livak K.J., Starcich B., Josephs S.F., Doran E.R., Rafalski J.A., Whitehorn E.A., Baumeister K., et al. Complete nucleotide sequence of the AIDS virus, HTLV-III. Nature. 1985;313:277–284. doi: 10.1038/313277a0. [DOI] [PubMed] [Google Scholar]
- 9.Sherman P.A., Dickson M.L., Fyfe J.A. Human-immunodeficiency virus type 1 integration protein: DNA sequence requirements for cleaving and joining reactions. J. Virol. 1992;66:3593–3601. doi: 10.1128/jvi.66.6.3593-3601.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wakefield J.K., Kang S., Morrow C.D. Construction of a type 1 human immunodeficiency virus that maintains a primer-binding site complementary to tRNA (His) J. Virol. 1996;70:966–975. doi: 10.1128/jvi.70.2.966-975.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vink C., VanGent D.C., Elgersma Y., Plasterk R.H.A. Human immunodeficiency virus integrase protein requires a sub-terminal position of its viral DNA recognition sequence for efficient cleavage. J. Virol. 1991;65:4636–4644. doi: 10.1128/jvi.65.9.4636-4644.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cullen B.R., Greene W.C. Functions of the auxiliary gene products of the human immunodeficiency virus type 1. Virology. 1990;178:1–5. doi: 10.1016/0042-6822(90)90373-y. [DOI] [PubMed] [Google Scholar]
- 13.Feng S., Holland E.C. HIV-1 Tat trans-activation requires the loop sequence within TAR. Nature. 1988;334:165–167. doi: 10.1038/334165a0. [DOI] [PubMed] [Google Scholar]
- 14.Harrich D., Ulich C., Garcia-Martinez L.F., Gaynor R.B. Tat is required for efficient HIV-1 reverse transcription. EMBO J. 1997;16:1224–1235. doi: 10.1093/emboj/16.6.1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kurreck J. Antisense technologies. Eur. J. Biochem. 2003;270:1628–1644. doi: 10.1046/j.1432-1033.2003.03555.x. [DOI] [PubMed] [Google Scholar]
- 16.Crooke S.T. Progress in antisense technology. Ann. Rev. Med. 2004;55:61–95. doi: 10.1146/annurev.med.55.091902.104408. [DOI] [PubMed] [Google Scholar]
- 17.Neilsen P.E., Egholm M., Berg R.H., Buchardt O. Sequence-selective recognition of DNA by strand displacement with a thymidine-substituted polyamide. Science. 1991;254:1497–1500. doi: 10.1126/science.1962210. [DOI] [PubMed] [Google Scholar]
- 18.Lee R., Kaushik N., Modak M.J., Vinayak R., Pandey V.N. Polyamide nucleic acid targeted to the primer binding site of the HIV-1 RNA genome blocks in vitro HIV-1 reverse transcription. Biochemistry. 1998;37:900–910. doi: 10.1021/bi972197m. [DOI] [PubMed] [Google Scholar]
- 19.Kaushik N., Pandey V.N. PNA targeting the PBS and A-loop sequences of HIV-1 genome destabilizes packaged in the virions and inhibits HIV-1 replication. Virology. 2002;303:297–308. doi: 10.1006/viro.2002.1630. [DOI] [PubMed] [Google Scholar]
- 20.Kaushik N., Talele T.T., Monel R., Palumbo P., Pandey V.N. Destabilization of from the primer-binding site of HIV-1 genome by anti-A loop polyamide nucleotide analog. Nucleic Acids Res. 2001;29:5099–5106. doi: 10.1093/nar/29.24.5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mayhood T., Kaushik N., Pandey P.K., Kashanchi F., Deng L., Pandey V.N. Inhibition of Tat-mediated transactivation of HIV-1 LTR transcription by polyamide nucleic acid targeted to TAR hairpin element. Biochemistry. 2000;39:11532–11539. doi: 10.1021/bi000708q. [DOI] [PubMed] [Google Scholar]
- 22.Kaushik N., Basu A., Pandey V.N. Inhibition of HIV-1 replication by anti-trans-activation responsive polyamide nucleotide analog. Antiviral Res. 2002;56:13–27. doi: 10.1016/s0166-3542(02)00024-4. [DOI] [PubMed] [Google Scholar]
- 23.Kaushik N., Basu A., Palumbo P., Myers R.L., Pandey V.N. Anti-TAR polyamide nucleotide analog conjugated with a membrane-permeating peptide inhibits human immunodeficiency virus type 1 production. J. Virol. 2002;76:3881–3891. doi: 10.1128/JVI.76.8.3881-3891.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chaubey B., Tripathi S., Ganguly S., Harris D., Casale R.A., Pandey V.N. A PNA-transportan conjugate targeted to the TAR region of the HIV-1 genome exhibits both antiviral and virucidal properties. Virology. 2005;331:418–428. doi: 10.1016/j.virol.2004.10.032. [DOI] [PubMed] [Google Scholar]
- 25.Derossi D., Calvet S., Trembleau A., Brunissen A., Chassaing G., Prochiantz A. Cell internalization of the third helix of the antennapedia homeodomain is receptor-independent. J. Biol. Chem. 1996;271:18188–18193. doi: 10.1074/jbc.271.30.18188. [DOI] [PubMed] [Google Scholar]
- 26.Derossi D., Chassaing G., Prochiantz A. Trojan peptides: the penetratin system for intracellular delivery. Trends Cell Biol. 1998;8:84–87. [PubMed] [Google Scholar]
- 27.Schwarze S.R., Ho A., Vocero-Akbani A., Dowdy S.F. In vivo protein transduction: delivery of a biologically active protein into the mouse. Science. 1999;285:1569–1572. doi: 10.1126/science.285.5433.1569. [DOI] [PubMed] [Google Scholar]
- 28.Soomets U., Lindgren M., Gallet X., Hällbrink M., Elmquist A., Balaspiri L., Zorko M., Pooga M., Brasseur R., Langel Ü. Deletion analogues of transportan. Biochim. Biophys. Acta. 2000;1467:165–176. doi: 10.1016/s0005-2736(00)00216-9. [DOI] [PubMed] [Google Scholar]
- 29.Bernatowicz M.S., Matsueda R., Matsueda G.R. Preparation of Boc- [S- (3-nitro-2-pyridinesulfenyl)]-cysteine and its use for unsymmetrical disulfide bond formation. Int. J. Pept. Protein Res. 1986;28:107–112. doi: 10.1111/j.1399-3011.1986.tb03236.x. [DOI] [PubMed] [Google Scholar]
- 30.Gunnery S., Green S.R., Mathews M.B. Tat-responsive region RNA of human immunodeficiency virus type 1 stimulates protein synthesis in vivo and in vitro: relationship between structure and function. Proc. Natl Acad. Sci. USA. 1992;89:11557–11561. doi: 10.1073/pnas.89.23.11557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sanger F., Nicklen S., Coulson A.R. DNA sequencing with chain terminating inhibitors. Proc. Natl Acad. Sci. USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kashanchi F., Shibata R., Ross E.K., Brady J.N., Martin M.A. Second-site long terminal repeat (LTR) revertants of replication-defective human immunodeficiency virus: effects of revertant TATA box motifs on virus infectivity, LTR-directed expression, in vitro RNA synthesis and binding of basal transcription factors TFIID and TFIIA. J. Virol. 1994;68:3298–3307. doi: 10.1128/jvi.68.5.3298-3307.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Planelles V., Bachelerie F., Jowett J.B., Haislip A., Xie Y., Banooni P., Masuda T., Chen I.S. Fate of the human immunodeficiency virus type 1 provirus in infected cells: a role for vpr. J. Virol. 1995;69:5883–5889. doi: 10.1128/jvi.69.9.5883-5889.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chou T.-C. Relationships between inhibition constants and fractional inhibitions in enzyme-catalysed reations with different numbers of reactants, different reaction mechanisms, and different types of mechanisms of inhibition. Mol. Pharmacol. 1974;39:235–247. [PubMed] [Google Scholar]
- 35.Chou T.-C. Derivation and properties of Michaelis–Menten type and Hill equations for reference ligands. J. Theoret. Biol. 1977;39:345–356. doi: 10.1016/0022-5193(76)90169-7. [DOI] [PubMed] [Google Scholar]
- 36.Vivès E., Brodin P., Lebleu B. A truncated HIV-1 Tat protein basic domain rapidly translocates through the plasma membrane and accumulates in the cell nucleus. J. Biol. Chem. 1997;272:16010–16017. doi: 10.1074/jbc.272.25.16010. [DOI] [PubMed] [Google Scholar]
- 37.Pooga M., Hällbrink M., Zorko M., Langel Ü. Cell penetration by transportan. FASEB. 1998;12:67–77. doi: 10.1096/fasebj.12.1.67. [DOI] [PubMed] [Google Scholar]
- 38.Dom G., Shaw-Jackson C., Matis C., Bouffioux O., Picard J.J., Prochiantz A., Mingeot-Leclercq M., Brasseur R., Rezsohazy R. Cellular uptake of Antennapedia penetratin peptides is a two-step process in which phase transfer precedes a tryptophan-dependent translocation. Nucleic Acids Res. 2003;31:556–561. doi: 10.1093/nar/gkg160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Estcourt M.J., McMichael A.J., Hanke T. DNA vaccine against human immunodeficiency virus type 1. Immunol. rev. 2004;199:144–155. doi: 10.1111/j.0105-2896.2004.00151.x. [DOI] [PubMed] [Google Scholar]
- 40.Buchardt O., Egholm M., Berg R.H., Nielsen P.E. Peptide nucleic acids and their potential applications in biotechnology. Trends Biotechnol. 1993;11:384–386. doi: 10.1016/0167-7799(93)90097-S. [DOI] [PubMed] [Google Scholar]
- 41.Marin V.L., Roy S., Armitage B.A. Recent advances in the development of peptide nucleic acid as a gene-targeted drug. Expert Opin. Biol. Ther. 2004;4:337–348. doi: 10.1517/14712598.4.3.337. [DOI] [PubMed] [Google Scholar]
- 42.Koppelhus U., Nielsen P.E. Cellular delivery of peptide nucleic acid (PNA) Adv. Drug Delivery Rev. 2003;55:267–280. doi: 10.1016/s0169-409x(02)00182-5. [DOI] [PubMed] [Google Scholar]
- 43.Taylor R.W., Chinnery P.F., Turnbull D.M., Lightowlers R.N. Selective inhibition of mutant human mitochondrial DNA replication in vitro by peptide nucleic acids. Nature Genet. 1997;15:212–215. doi: 10.1038/ng0297-212. [DOI] [PubMed] [Google Scholar]
- 44.Shammas M.A., Simmons C.G., Corey D.R., ShmooklerReis R.J. Telomerase inhibition by PNA reverses ‘immortality’ of transformed human cells. Oncogene. 1999;18:6191–6200. doi: 10.1038/sj.onc.1203069. [DOI] [PubMed] [Google Scholar]
- 45.Herbert B.-S., Pitts A.E., Baker S.I., Hamilton S.E., Wright W.E., Shay J.W., Corey D.R. Inhibition of human telomerase in immortal human cells leads to progressive telomere shortening and cell death. Proc. Natl Acad. Sci. USA. 1999;96:14276–14281. doi: 10.1073/pnas.96.25.14276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nulf C.J., Corey D. Intracellular inhibition of hepatitis C virus (HCV) internal ribosomal entry site (IRES)-dependent translation by peptide nucleic acids (PNAs) and locked nucleic acids (LNAs) Nucleic Acids Res. 2004;32:3792–379847. doi: 10.1093/nar/gkh706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Richard J.P., Melikov K., Vives E., Ramos C., Verbeure B., Gait M.J., Chernomordik L.V., Lebleu B. Cell-penetrating peptides. J. Biol. Chem. 2003;278:585–590. doi: 10.1074/jbc.M209548200. [DOI] [PubMed] [Google Scholar]
- 48.Richard J.P., Melikov K., Brooks H., Prevot P., Lebleu B., Chernomordik L.V. Cellular uptake of unconjugated tat-peptide involves clathrin dependent endocytosis and heparan sulfate receptors. J. Biol. Chem. 2005;280:15300–15306. doi: 10.1074/jbc.M401604200. [DOI] [PubMed] [Google Scholar]
- 49.Wadia J.S., Stan R.V., Dowdy S.F. Transducible TAT-HA fusogenic peptide enhances escape of TAT-fusion proteins after lipid raft macropinocytosis. Nature Medicine. 2004;10:310–315. doi: 10.1038/nm996. [DOI] [PubMed] [Google Scholar]
- 50.Wild C., Greenwell T., Matthews T. A synthetic peptide from HIV-1 gp41 is a potent inhibitor of virus-mediated cell-cell fusion. AIDS Res. Hum. Retroviruses. 1993;9:1051–1053. doi: 10.1089/aid.1993.9.1051. [DOI] [PubMed] [Google Scholar]
- 51.Derdeyn C.A., Decker J.M., Sfakianos J.N., Wu X., O'Brien W.A., Ratner L., Kappes J.C., Shaw G.M., Hunter E. Sensitivity of human immunodeficiency virus type 1 to the fusion inhibitor T-20 is modulated by coreceptor specificity defined by the V3 loop of gp120. J. Virol. 2000;74:8358–8367. doi: 10.1128/jvi.74.18.8358-8367.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nagashima K.A., Thompson D.A., Rosenfield S.I., Maddon P.J., Dragic T., Olson W.C. Human immunodeficiency virus type 1 entry inhibitors PRO 542 and T-20 are potently synergistic in blocking virus–cell and cell–cell fusion. J. Infect. Dis. 2001;183:1121–1125. doi: 10.1086/319284. [DOI] [PubMed] [Google Scholar]