Abstract
Microsatellite repeat and single nucleotide polymorphisms (SNPs) are abundant sources of genetic variation, but existing methodologies cannot simultaneously detect these variants in a facile or inexpensive way. We describe herein a thin-film biosensor chip based on an allele-discriminating oligonucleotide array that enables genotyping for both microsatellite repeats and SNPs in a single analysis. We validated this methodology for the functionally polymorphic −794 CATT5–8 repeat and −173 G/C SNP present in the promoter of the human gene for macrophage migration inhibitory factor (MIF). In a comparison of 30 samples collected at a rural hospital in Zambia, we observed a 100% concordance for both the CATT repeat and G/C SNP between the biosensor methodology and the conventional capillary electrophoresis. The biosensor chips are low in cost and once printed, they are robust and require no instrumentation for analysis. When combined with multiple displacement amplification, this methodology can be utilized in primitive settings for the genotyping of nanogram quantities of DNA present in blood, dried and stored on filter paper samples. We applied this methodology to a field study of MIF genotype in children with malaria, and provide first evidence for a potential association between MIF alleles and malaria infection. We also present data supporting significant population stratification of the low- versus high-expression forms of MIF that may bear on the role of this gene in infectious diseases.
INTRODUCTION
Emerging knowledge about the association of the variant alleles of different genes with the human diseases has focused attention on the development of technologies to rapidly discriminate between different genotypes (1). New methods are important, both to facilitate the acquisition of genotyping data and to better translate this knowledge into diagnostic and therapeutic applications. Studies of the genetics of host immunity, for example, are revealing novel pathogenic mechanisms that are important for understanding infectious diseases as they affect different human populations (2,3). Rapid and low-cost genotyping methods are especially important for studies in the developing world, where the facilities for DNA analysis are limited and where the timely, on-site utilization of genetic information could make an impact on morbidity and mortality.
Our studies of polymorphisms in the gene for the cytokine, macrophage migration inhibitory factor (MIF), and its role in malaria infection has prompted us to develop a robust genotyping method that can be performed on-site, in rural field settings, and at low-cost and turnaround time. MIF plays an important role in the innate and the adaptive immune response, and it is encoded by a functionally polymorphic gene that has been linked to the incidence or the severity of different inflammatory conditions (4,5). These MIF alleles differ in the structure of their promoter region, which varies by the presence of a tetranucleotide, CATT, that is repeated between 5 and 8 times in the promoter (−794 CATT5–8) (6). This repeat regulates basal and stimulus-induced transcriptional activity, which increases almost proportionally with repeat number in defined in vitro assay systems. A single nucleotide polymorphism (SNP) (−173 G/C) in the MIF promoter may also influence promoter activity, either by linkage disequilibrium or by an interaction with the tetranucleotide site (7–9).
The close interplay between the genes of the immune system and the host response to infection has led us to investigate the distribution of variant MIF alleles in populations, in which different infectious diseases are endemic, with the aim of better understanding the role of selective pressures on the evolution of the MIF locus. Toward this end, we have developed and implemented a thin-film optical biosensor chip for genotyping of both tetranucleotide CATT repeats and SNPs in a single analysis that greatly facilitates the examination of small quantities of DNA obtained in the field. These chips are based on an allele-discriminating oligonucleotide array that provides a direct visual readout of the different MIF promoter polymorphisms. We document, herein, the application of this technology to the analysis of MIF gene polymorphisms in dried blood spots collected in a rural hospital where malaria is endemic.
METHODS
Patient samples
DNA specimens from normal controls of known MIF genotype were used to develop the biosensor methodology. Validation was performed on a random selection of DNA specimens culled from ongoing studies of MIF genotype, and included Caucasians, African-Americans, North East Asians (Koreans) and the African sub-groups described in the text. The African sub-group specimens were collected either by K. K. Kidd and J. R. Kidd, or by one of their collaborators as part of their long-term studies on human genetic diversity. The malaria-infected samples were obtained as part of investigations at the Macha Mission Hospital in Choma, Zambia, which is holoendemic for Plasmodium falciparum malaria. These specimens consisted of blood samples that had been spotted on filter paper, dried and stored for several months. Parasitemia was assessed on hospital admission by enumeration of infected red cells in thick blood smears. All samples were obtained and utilized in accordance with institutional and local IRB protocols. The SAS statistical software package was used to perform χ2 analysis and logistic regression to determine the relationship between parasitemia and MIF genotype.
DNA extraction
Genomic DNA was isolated from patient blood or from the blood spots dried on filter papers using the QIAamp DNA Blood Mini kit (Qiagen, Valencia, CA) and the recommended protocols provided by the manufacturer.
DNA amplification by multiple displacement amplification
DNA extracted from blood spots dried on filter papers was amplified by multiple displacement amplification (MDA) (10). The MDA reactions were performed overnight at 31°C in 50 μl of reaction solution containing 37.5 mM Tris–HCl (pH 7.5), 50 mM KCl, 10 mM MgCl2, 5 mM (NH4)2SO4, 1 mM dNTP, 0.05 mM thiophosphate-modified random hexamer (5′-NNNNsNsN), 1–5 ng genomic DNA, 0.2 U yeast pyrophosphatase and 1 U Phi29 polymerase. The products of the MDA reaction were resolved on a 1% agarose gel stained with ethidium bromide. The yield of amplified genomic DNA was between 15 and 20 μg in a 50 μl reaction solution.
MIF DNA fragment amplification by PCR
Oligonucleotide primer sets were designed to amplify regions within the MIF promoter that corresponded to DNA product sizes of 119, 123, 127 and 131 bp for the 5-, 6-, 7- and 8-CATT repeat polymorphisms, respectively (Table 1). An additional set of primers was used for the −173 G/C SNP and produced a PCR product size of 129 bp. The PCRs were carried out in a 50 μl solution containing 1× PCR buffer (AmpliTaq Gold; Applied Biosystem), 2.5 mM MgCl2, 0.2 mM dNTP, 10 pmol of each forward and reverse primers, 100 ng of genomic DNA or MDA-amplified genomic DNA and 1 U AmpliTaq Gold DNA polymerase. For the CATT repeat, the PCR program was as follows: 95°C for 10 min, followed by 40 cycles at 94°C for 30 s, 56°C for 30 s, 72°C for 1 min and final extension at 72°C for 10 min, and for the −173 SNP, the PCR program was 95°C for 10 min, followed by 40 cycles of 94°C for 30 s, 62°C for 30 s, 72°C for 1 min and final extension at 72°C for 10 min.
Table 1.
Oligonucleotide sequences of capture probe P1, detection probe P2, synthetic targets and PCR primers
| CATT repeat | |
| Capture probe P1 | P1-CATT-1 5′-CHO-AAAAAAAAAACTGCTATGTCATGGCTTATCTTCTTTCACCCATT |
| P1-CATT-2 5′-CHO-AAAAAAAAAACTGCTATGTCATGGCTTATCTTCTTTCACCCATTCATT | |
| P1-CATT-3 5′-CHO-AAAAAAAAAACTGCTATGTCATGGCTTATCTTCTTTCACCCATTCATTCATT | |
| P1-CATT-4 5′-CHO-AAAAAAAAAACTGCTATGTCATGGCTTATCTTCTTTCACCCATTCATTCATTCATT | |
| Detection probe P2 | P2 biotin |
| 5′-phosphate-CATTCATTCATTCATTCAGCAGTATTAGTCAATGTC-biotin-3′ | |
| Synthetic target | Target AATG-5 |
| GACATTGACTAATACTGCTGAATGAATGAATGAATGAATGGGTGAAAGAAGATAAGCCATGACATAGCAG Target AATG-6 | |
| GACATTGACTAATACTGCTGAATGAATGAATGAATGAATGAATGGGTGAAAGAAGATAAGCCATGACATAGCAG Target AATG-7 | |
| GACATTGACTAATACTGCTGAATGAATGAATGAATGAATGAATGAATGGGTGAAAGAAGATAAGCCATGACATAGCAG Target AATG-8 | |
| GACATTGACTAATACTGCTGAATGAATGAATGAATGAATGAATGAATGAATGGGTGAAAGAAGATAAGCCATGACATAGCAG | |
| PCR primer and product | Forward primer: CTATGTCATGGCTTATCTTC |
| Reverse primer: TCCACTAATGGTAAACTCGG | |
| PCR products: 119, 123, 127 or 131 | |
| CTATGTCATGGCTTATCTTCTTTCACC(CATT)5–8CAGCAGTATTAGTCAATGTCTCTTGATATGCCTGGCACCTGCTAGATGGTCCCCGAGTTTACCATTAGTGGA | |
| SNP−173 G/C | |
| Capture probe P1 | P1-G 5′-CHO-AAAAAAAAAACCGGAACAGGCCGATTTCTAGCCGCCAAGTGGAGAACAGG |
| P1-C 5′-CHO-AAAAAAAAAACCGGAACAGGCCGATTTCTAGCCGCCAAGTGGAGAACAGC | |
| Detection probe P2 | P2 biotin |
| 5′-phosphate-TTGGAGCGGTGCGCCGGGCTTA-biotin-3′ | |
| Synthetic target | MIF−173 Target G AGCCCGGCGCACCGCTCCAACCTGTTCTCCACTTGGCGGCTAGAAATCGGCCTGTTCCGG |
| MIF−173 Target CAGCCCGGCGCACCGCTCCAAGCTGTTCTCCACTTGGCGGCTAGAAATCGGCCTGTTCCGG | |
| PCR primer and product | MIF−173 Forward ACTAAGAAAGACCCGAGGCG |
| MIF−173 Reverse GCAGGACCCTGGGCGACT | |
| PCR product 129 bp ACTAAGAAAGACCCGAGGCGAGGCCGGAACAGGCCGATTTCTAGCCGCCAAGTGGAGAACAGGTTGGAGCGGTGCGCCGGGCTTAGCGGCGGTTGCTGGAGGAACGGGCGGAGTCGCCCAGGGTCCTGC | |
The underlined sequences designate the unique repeated sequences (5–8 repeats) present in the synthetic oligonucleotide target. The bolded residue designates the –173 G/C SNP.
Oligonucleotide probe synthesis
The oligonucleotide probes were designed based on the general approach described previously (11), and with the detailed sequence information provided in Table 1. For the CATT repeat, the 4 P1 capture probes have 5′-aldehyde groups, 10 deoxyadenosine residues at their 5′ ends and 30 bases of upstream sequence, followed by a different number (from 1 to 4) of the CATT repeat, respectively. The P2 detection probe contains 4 copies of CATT and 20 bases of downstream sequence, with a biotin at its 3′ end for detection and a phosphate group at its 5′ end for ligation. The P1 and P2 probes were synthesized by the Yale Keck Biotechnology Facility at 40/50 nmol scale and were used without post-synthesis purification. Four complementary, single-stranded artificial targets with 5–8 copies of the AATG repeat flanked by 30 bases of the MIF promoter sequences both upstream and downstream of the repeat motif also were synthesized.
Detection of MIF CATT repeat polymorphism and −173 G/C SNP on thin-film biosensor chips
The MIF CATT repeat polymorphism and the −173 G/C SNP detection are performed by modifying solid-phase techniques described previously (11). The P1 capture probes are manually spotted in a format shown in Figure 1B on hydrazine-functionalized, thin-film biosensor chips (Thermo Electron, Louisville, CO) using 0.3 μl of 1 μM P1 in 0.1 M phosphate buffer, pH 7.8 and 10% glycerol. After incubation for 2 h at room temperature in a humid chamber, the chips are washed with 0.1% SDS at 60°C for 30 min, rinsed with water and dried. A ligation mixture containing 20 mM Tris–HCl, pH 8.3, 25 mM KCl, 10 mM MgCl2, 0.5 mM NAD, 0.01% Triton X-100, 5 mg/ml acid-treated casein, 10% formamide and 0.04 U/μl mutant Ampligase (Lys294Arg of Thermus thermophilus ligase) is applied to each chip and prewarmed to 60°C. The synthetic targets or MIF PCR amplicons from patient samples at a concentration of 100 fmol in 10 μl water are denatured at 95°C for 3 min in the presence of 100 fmol of the P2-biotin probe. After denaturation, 10 μl of this solution is immediately added to the pre-incubated ligation mixture and incubated at 60°C for 20 min. A stringent wash with 0.01 M NaOH at 60°C is applied three times, and this is followed by briefly rinsing three times with 0.1× standard saline citrate (SSC) at room temperature. The chips are then incubated at room temperature for 10 min with 100 μl of an anti-biotin–IgG–horseradish peroxidase conjugate (Jackson ImmunoResearch Lab; 1:1000 dilution from a 1 mg/ml stock in a buffer containing 5× SSC and 5 mg/ml acid-treated casein). After briefly washing three times with 0.1× SSC, 100 μl of a tetramethyl benzidine formulation from BioFX Lab is added to the chips and incubated for 5 min at room temperature, then rinsed with ddH2O and air dried. The CATT and SNP polymorphisms are scored visually and the results are recorded by imaging under a dissection microscope fitted with a digital camera.
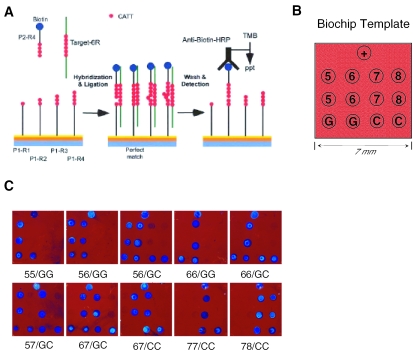
Figure 1.
(A) Schematic representation for the detection of the MIF CATT tetranucleotide repeat by ligation of biotinylated detection probe P2 to a set of capture probes P1 with different copies of CATT repeat immobilized on thin-film biosensor chip surface in the presence of certain CATT target (i.e. CATT 6). (B) Array template for the detection of the 5-, 6-, 7- and 8-CATT repeats, and the −173 G/C SNP. Oligonucleotides are arrayed in duplicates, as shown. +: positive control, an aldehyde modified dA20-biotin probe. (C) Visual appearance of the biochip arrays of the representative MIF genotypes.
RESULTS AND DISCUSSION
Figure 1 illustrates the hybridization and ligation-based detection strategy of the biosensor methodology used for detecting the MIF promoter CATT repeat, which is present in a copy number of 5–8, and the −173 G/C SNP. Of importance, prior ligation-based assays have been found to be efficient only for discriminating polymorphic alleles of short microsatellite repeats <30 bases (11,12). Ligation of longer repeats lacks specificity because of the looping out of repeat units in the probe/target hybrids. The loopout often occurs in an area outside of the ligase footprint that can span 26–30 bases (12). To avoid the formation of loopouts in the MIF CATT repeat sequence, we divided the repeat into the capture probes ‘P1’, with CATT 1, 2, 3 or 4, and ‘P2’, with CATT4 (Figure 1A), thereby ensuring that the ligase footprint covers 6–7-CATT repeat units. Four capture probes with 1, 2, 3 and 4 CATT repeat units, respectively, together with 30 bases of the upstream adjacent unique MIF sequence (see detailed sequences in Table 1) are covalently arrayed by the reaction between an aldehyde group at their 5′-termini and the hydrazine group on the chip surface. A detection probe, P2, with 4 copies of the CATT unit and 20 bases of the downstream adjacent and unique MIF sequence carries a biotin at the 3′ end for detection and a phosphate at its 5′ end for ligation. When target DNA with a variable number of the CATT unit (from 5 to 8, e.g. the 6-CATT shown in Figure 1A), is hybridized with the P1 and P2 probes on the chip surface in the presence of a thermostable DNA ligase, the enzyme selects only perfectly matched P1+P2/target for ligation. This reaction results in the immobilization of the biotinylated P2 probe at the matched P1 probe position on the chip surface. After a stringent wash to remove all non-ligated molecules, a biotin-associated detection procedure is applied to generate a color that can be visualized by the unaided human eye. The biochips can also be printed by high-throughput ‘spotting’ of the requisite capture probes onto hydrazine-functionalized thin-film biosensor chips. The entire methodology is low in cost and once printed, the chips are robust and require no instrumentation for analysis.
Using single-strand synthetic targets with tetranucleotide repeats of 5, 6, 7 or 8 AATG, we first tested the detection specificity for the different MIF CATT repeat sequences. A modified ligation condition with 10% formamide in the standard ligation mixture (11) gave unambiguous discrimination of the CATT repeat copy number. The allele-specific ligation strategy was also used for the simultaneous detection of the MIF SNP G/C −173.
To test for the accuracy of genotyping the CATT repeat polymorphism and the −173 SNP simultaneously by the biosensor chip, we analyzed an initial 30 genomic DNA samples that had been previously studied for MIF genotype. In our standard methodology, the CATT repeat polymorphism is detected by capillary electrophoresis of PCR products amplified to span the tetranucleotide region, and the −173 G/C SNP is determined by the ddNTP primer extension method and capillary electrophoresis, or by pyrosequencing (6). The typings of the CATT repeat and −173 SNP obtained by using the biosensor methodology were compared initially in a blinded fashion. Representative images are shown in Figure 1C and the optical biosensor genotypes are listed in Table 2. There was complete concordance of the −173 G/C SNP genotypes between the two methods. We observed one discrepancy for a previously analyzed DNA sample bearing the 8-CATT repeat. The initial chromatographic analysis for sample 1418 showed that one PCR product peak eluted in an intermediate position for that expected is either the 7-CATT or the 8-CATT repeat. The peak was scored as ‘8-CATT’ by the capillary electrophoresis (5/8), but 7-CATT by the biosensor chip (5/7). Subsequent reanalysis of this DNA sample by capillary electrophoresis verified the presence of 7-CATT and the scoring of this sample as ‘5/7’. The concordance with typing data for the −173 G/C SNP and the ability to identify errors in the CATT repeat obtained by capillary electrophoresis indicate that genotyping by optical biosensor chips is highly accurate.
Table 2.
Comparison of MIF genotypes of the CATT tetranucleotide repeat and the −173 G/C SNP in a selection of human DNA samples by optical biosensor chip and capillary electrophoresis
| Sample ID | CATT repeat | −173 G/C SNP | ||
|---|---|---|---|---|
| Capillary electrophoresis | Optical biosensor chip | Capillary electrophoresis | Optical biosensor chip | |
| 0009 | 5/6 | 5/6 | G/G | G/G |
| 0010 | 6/6 | 6/6 | G/G | G/G |
| 0013 | 6/6 | 6/6 | G/G | G/G |
| 0014 | 5/5 | 5/5 | G/G | G/G |
| 0015 | 6/7 | 6/7 | C/G | C/G |
| 0132 | 5/5 | 5/5 | G/G | G/G |
| 0134 | 5/5 | 5/5 | G/G | G/G |
| 0209 | 5/6 | 5/6 | G/G | G/G |
| 0306 | 5/6 | 5/6 | G/G | G/G |
| 0422 | 5/6 | 5/6 | G/G | G/G |
| 0425 | 5/7 | 5/7 | G/C | G/C |
| 0428 | 6/6 | 6/6 | G/G | G/G |
| 0429 | 5/7 | 5/7 | G/C | G/C |
| 0431 | 5/6 | 5/6 | G/G | G/G |
| 0506 | 6/6 | 6/6 | G/G | G/G |
| 0507 | 6/7 | 6/7 | C/C | C/C |
| 0602 | 7/7 | 7/7 | C/C | C/C |
| 0603 | 5/6 | 5/6 | G/C | G/C |
| 0807 | 6/7 | 6/7 | G/C | G/C |
| 1003 | 6/6 | 6/6 | G/G | G/G |
| 1203 | 6/6 | 6/6 | G/G | G/G |
| 1206 | 6/6 | 6/6 | G/C | G/C |
| 1207 | 6/6 | 6/6 | G/C | G/C |
| 1308 | 5/6 | 5/6 | G/G | G/G |
| 1418a | 5/8a | 5/7a | G/C | G/C |
| 1603 | 6/6 | 6/6 | G/G | G/G |
| 1606 | 5/6 | 5/6 | G/G | G/G |
| 0036 | 7/7 | 7/7 | C/C | C/C |
| 0914 | 7/8 | 7/8 | C/C | C/C |
| 1214 | 6/7 | 6/7 | C/C | C/C |
Different samples were first typed by biosensor chip methodology, and then verified by capillary electrophoresis and pyrosequencing.
aA discrepancy reading between optical biosensor chip and capillary electrophoresis.
As one phase of an ongoing project to study the association between MIF polymorphisms and severe malarial disease, we plan to genotype several hundred DNA specimens obtained in rural field settings. Finger stick blood samples are routinely collected and dried onto filter papers at local hospitals and stored on-site under ambient conditions. Limited amounts of genomic DNA are present in these blood-spotted filter papers, and a pre-amplification step, typically by MDA technology (10), allows these precious samples to be used for hundreds of PCRs. To evaluate concordance between MDA products and unamplified genomic DNA for the ligation-based optical biosensor methodology, the 30 samples genotyped in the previous experiment were re-evaluated after MDA. Complete fidelity was observed between the unamplified genomic DNA and the MDA products for both the MIF CATT polymorphism and the −173 G/C SNP genotype. It is important to point out that with respect to the applicability of this method to other genes, some sequences may not be amenable to genotyping if they reside in repeated regions or are closely homologous to sequences found elsewhere in the genome, and additional optimization studies may be necessary.
The first 40 samples from patients in Zambia with P.falciparum malaria were collected and dried onto filter papers, stored at ambient temperature and the genomic DNA extracted with the QIAamp DNA Blood Mini kit (Qiagen). DNA yields were reliably 10–20 ng per sample. Ten nanograms of the isolated, genomic DNA was used for whole genome amplification by MDA in a standard 50 μl reaction, resulting in 15–20 μg of the MDA product per reaction. Targets for the MIF polymorphic genotyping were prepared by PCR of 100 ng of MDA-amplified products. The genotyping was performed on optical biosensor chips, and the genotypes and allele frequencies were compared with that of a previously reported Caucasian control group (Table 3). The initial data indicated a possible difference in the allelic distribution of MIF genotypes in different populations. In particular among the Zambian samples, there was a relative increase in the proportion of the low-expression 5-CATT allele, and a corresponding decrease in the proportion of the high-expression 6-CATT and 7-CATT alleles when compared with the recent studies in Caucasian populations (6,13). Furthermore, the SNP analysis revealed a relative decrease in the −173G and a corresponding increase in the −173C in the Zambian samples when compared with previously studied Caucasian groups. We then analyzed small representative sets of DNA samples obtained from four different North African tribes and one African-American group. The allelic distribution, when summed over the different groups, appeared closer in distribution to the Zambian group than that described previously in Caucasian groups (included in Table 3) (6,13).
Table 3.
Allele and genotype frequencies of the MIF polymorphisms in Caucasian and African populations
| MIF polymorphism | Genotype, frequency no. (%) | Allele frequency (%) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| CATT(5–8) | 5/5 | 5/6 | 5/7 | 6/6 | 6/7 | 7/7 | 5 | 6 | 7 |
| Caucasiana (n = 159) | 8 (5.0) | 61 (38.3) | 10 (6.3) | 53 (33.3) | 25 (15.7) | 0 (0) | 27.3 | 60.3 | 11.0 |
| Zambia malaria (n = 40) | 14 (35.0) | 14 (35.0) | 3 (7.5) | 5 (12.5) | 1 (2.5) | 3 (7.5) | 56.3 | 31.2 | 12.5 |
| African control groups (n = 40) | 5 (12.5) | 19 (47.5) | 4 (10.0) | 5 (12.5) | 7 (17.5) | 0 (0) | 41.3 | 45.0 | 13.7 |
| −173 SNP G/C | G/G | G/C | C/C | G | C | ||||
| Caucasianb (n = 88) | 67 (76.1) | 21 (23.9) | 0 (0) | 88.1 | 11.9 | ||||
| Zambia malaria (n = 40) | 12 (30.0) | 12 (30.0) | 16 (40.0) | 45.0 | 55.0 | ||||
| African control groups (n = 40) | 6 (15.0) | 19 (47.5) | 15 (37.5) | 38.7 | 61.3 | ||||
With respect to clinical malaria infection, the small number of samples from Zambia did not permit us to make any meaningful determinations with respect to MIF alleles and disease complications or severity. Nevertheless, logistic regression analysis revealed that patients with the 5/X genotype (where X = 6-, 7- or 8-CATT allele) were significantly less likely to have parasitemia >10 000 than those with an X/X genotype (P = 0.04) (Table 4). Whether this finding indeed signifies a protective effect of low-expression MIF alleles is yet to be confirmed by larger studies. Such an association could provide prognostic information and influence treatment options, which frequently necessitates blood transfusions that are costly and may carry risk of HIV or hepatitis infection.
Table 4.
Association between parasitemia level on admission and MIF genotype for children evaluated at the Macha Mission Hospital in Zambia
| Parasitemia | MIF genotype (n, %) | ||
|---|---|---|---|
| 5/X | X/X | P-value | |
| ≤10 000 | 27 (84.4) | 5 (71.4) | 0.04 |
| >10 000 | 5 (15.6) | 2 (28.6) | |
5 = low-expression 5-CATT MIF allele; X = higher expression 6-, 7- or 8-CATT MIF alleles.
The data suggesting ethnic variation in the allelic frequency of the functional MIF CATT promoter polymorphism prompted us to study additional samples, including a set of 100 control (disease-free) specimens obtained from Korea. This analysis showed an allelic distribution that was intermediate in 5-CATT (62%, 5/5 or 5/X) and non-5-CATT (38%, X/X) frequency when compared with the African or previously analyzed Caucasian groups (6,13), and is in agreement with the proportions reported in a recent study of a Japanese population (14).
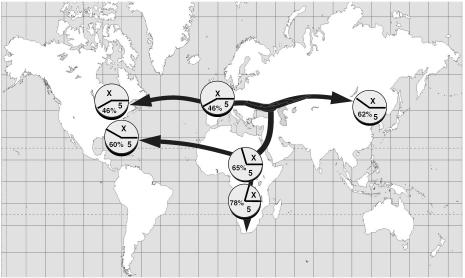
Table 5 summarizes the data that we obtained for the frequency of the MIF alleles in different populations around the world. By a conservative test, none of the populations with sample size >100 deviated significantly from Hardy–Weinberg ratios for the CATT polymorphism. Additional studies are necessary to determine if this distribution in CATT repeats reflect the relatedness between populations, as may be suggested by the representation in Figure 2. Ethnic variations in the distribution of different MIF alleles is an important finding, given the emerging association between these alleles and different inflammatory and infectious diseases (5–9,13,14). Racial or geographic variations in the population distribution of MIF alleles also suggest the possibility of different selective pressures acting on the MIF locus. Given the high prevalence of low-expression MIF alleles in Africa, these pressures could include historical exposure to tropical infections such as malaria, in which MIF is believed to play an important pathogenic role (15–17).
Table 5.
Compilation of MIF allele frequencies for the CATT repeat in different population groups determined by both biochip and standard methodologies
| 5/5 (%) | 5/6 (%) | 5/7 (%) | 5/8 (%) | 6/6 (%) | 6/7 (%) | 6/8 (%) | 7/7(%) | 5/5 +5/X(%) | X/X (%) | N | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Zambia | 52 (27.6) | 86 (45.7) | 6 (3.2) | 2 (1.1) | 18 (9.6) | 18 (9.6) | — | 6 (3.2) | 146 (77.7) | 42 (22.3) | 188 |
| N. Africa | 3 (8.8) | 17 (50) | 2 (5.9) | — | 4 (11.7) | 5 (14.7) | — | 3 (8.8) | 22 (64.7) | 12 (35.3) | 34 |
| NE Asia | 18 (18) | 31 (31) | 13 (13) | — | 25 (25) | 12 (12) | — | 1 (1) | 62 (62) | 38 (38.0) | 100 |
| Caucasian | 37 (6.2) | 185 (30.9) | 49 (8.2) | 1 (0.2) | 243 (40.6) | 69 (11.5) | — | 13 (2.1) | 272 (45.5) | 325 (54.5) | 597 |
| African Am. | 31 (14.5) | 73 (34.2) | 22 (10.3) | 1 (0.5) | 45 (21.4) | 35 (16.4) | 2 (0.9) | 4 (1.9) | 127 (59.6) | 86 (40.3) | 213 |
N. Africa, North African; NE Asia, Korean; African Am., African-American; X: 6-, 7- or 8-CATT MIF allele.
Figure 2.
Distribution of MIF CATT allele frequencies in different populations superimposed on human migration patterns.
In conclusion, the robust optical biosensor methodology described herein has high fidelity when compared with the currently used methods and instrumentation, and it can be readily combined with the MDA technique to analyze minute quantities of DNA extracted from dried, whole blood specimens. This method will greatly facilitate the genotyping and population studies in different field settings, and it makes possible the rapid translation of genotyping information in the clinic into medical intervention.
Acknowledgments
We thank Drs J. R. Kidd and K. K. Kidd for generously providing DNA samples from the Yoruba, Chagga, Hausa and Ibo African tribes, which were collected by their collaborators: Drs Sylvester L. J. Kajuna and Nganyirwa J. Karoma (Department of Biochemistry and Molecular Biology, Hubert Kairuki Memorial University, Dar es Salaam, Tanzania), Dr Selemani Kungulilo (Muhimbili University College of Health Sciences, Dar es Salaam Tanzania), Dr Adekunle Odunsi (Department of Gynecological Oncology, Roswell Park Cancer Institute, Buffalo, NY), Drs Friday Okonofua and F. Oronsaye (Department of Obstetrics and Gynecology, Faculty of Medicine, University of Benin, Benin City, Nigeria) and African-American DNA samples from the NIGMS Human Genetic Mutant Cell Repository, Coriell Institute for Medical Research. We thank Dr Efim Golub for preparing mutant Ampligase and Phi29 polymerase and Dr K. K. Kidd for thoughtful discussion. This work was supported by NIH grants 1RO1AI051306, 5RO1AI042310 and 1R01AR049610 (R.B.), 2P01GM57672 (D.C.W. and X.Z.), a Downs's Fellowship from the Yale School of Epidemiology and Public Health (A.B.), and grant R11-2002-098-0000-0 from KOSEF through the Rheumatism Research Center at Catholic University, Seoul, Korea (S.-H.P.). A portion of these studies comprised the MPH thesis of A.B., which was awarded the 2005 Wilbur G. Downs International Health Prize for the study of biochip genotyping in patients with malaria. Funding to pay the Open Access publication charges for this article was provided by NIH grant 1RO1AI051306 (R.B.).
Conflict of interest statement. Robert Jenison is an employee of Thermo Electron, Corp., which provided the thin-film biosensor chips gratis for this study. The other authors have declared no conflicts.
REFERENCES
- 1.Guttmacher A., Collins F.S. Genomic medicine—a primer. N. Engl. J. Med. 2002;347:1512–1520. doi: 10.1056/NEJMra012240. [DOI] [PubMed] [Google Scholar]
- 2.Cooke G.S., Hill A.V. Genetics of susceptibility to human infectious disease. Nature Rev. Genet. 2001;2:967–977. doi: 10.1038/35103577. [DOI] [PubMed] [Google Scholar]
- 3.Dronamraju K.R., editor. Infectious Disease and Host–Pathogen Evolution. Cambridge, UK: Cambridge University Press; 2004. [Google Scholar]
- 4.Calandra T., Roger T. Macrophage migration inhibitory factor: a regulator of innate immunity. Nature Rev. Immunol. 2003;3:791–800. doi: 10.1038/nri1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gregersen P., Bucala R. Macrophage migration inhibitory factor, MIF alleles, and the genetics of inflammatory disorders: incorporating disease outcome into the definition of phenotype. Arthritis Rheum. 2003;48:1171–1176. doi: 10.1002/art.10880. [DOI] [PubMed] [Google Scholar]
- 6.Baugh J.A., Chitnis S., Donnelly S.C., Monteiro J., Lin X., Plant B.J., Wolfe F., Gregersen P.K., Bucala R. A functional promoter polymorphism in the macrophage migration inhibitory factor (MIF) gene associated with disease severity in rheumatoid arthritis. Genes Immun. 2002;3:170–176. doi: 10.1038/sj.gene.6363867. [DOI] [PubMed] [Google Scholar]
- 7.De Benedetti F., Meazza C., Vivarelli M., Rossi F., Pistorio A., Lamb R., Lunt M., Thomson W., Ravelli A., Donn R., Martini A., British Paediatric Rheumatology Study Group Functional and prognostic relevance of the −173 polymorphism of the macrophage migration inhibitory factor gene in systemic-onset juvenile idiopathic arthritis. Arthritis Rheum. 2003;48:1398–1407. doi: 10.1002/art.10882. [DOI] [PubMed] [Google Scholar]
- 8.Donn R., Alourfi Z., De Benedetti F., Meazza C., Zeggini E., Lunt M., Stevens A., Shelley E., Lamb R., Ollier W.E., et al. Mutation screening of the macrophage migration inhibitory factor gene: positive association of a functional polymorphism of macrophage migration inhibitory factor with juvenile idiopathic arthritis. Arthritis Rheum. 2002;46:2402–2409. doi: 10.1002/art.10492. [DOI] [PubMed] [Google Scholar]
- 9.Donn R.P., Shelley E., Ollier W.E., Thomson W., British Paediatric Rheumatology Study Group A novel 5′-flanking region polymorphism of macrophage migration inhibitory factor is associated with systemic-onset juvenile idiopathic arthritis. Arthritis Rheum. 2001;44:1782–1785. doi: 10.1002/1529-0131(200108)44:8<1782::AID-ART314>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 10.Dean F.B., Hosono S., Fang L., Wu X., Faruqi A.F., Bray-Ward P., Sun Z., Zong Q., Du Y., Du J., et al. Comprehensive human genome amplification using multiple displacement amplification. Proc. Natl Acad. Sci. USA. 2002;99:5261–5266. doi: 10.1073/pnas.082089499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhong X.B., Reynolds R., Kidd, Kidd K.K., Jenison R., Marlar R.A., Ward D.C. Single-nucleotide polymorphism genotyping on optical thin-film biosensor chips. Proc. Natl Acad. Sci. USA. 2003;100:11559–11564. doi: 10.1073/pnas.1934783100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zirvi M., Bergstrom D.E., Saurage A.S., Hammer R.P., Barany F. Improved fidelity of thermostable ligases for detection of microsatellite repeat sequences using nucleoside analogs. Nucleic Acids Res. 1999;27:e41. doi: 10.1093/nar/27.24.e41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barton A., Lamb R., Symmons D., Silman A., Thomson W., Worthington J., Donn R. Macrophage migration inhibitory factor (MIF) gene polymorphism is associated with susceptibility to but not severity of inflammatory polyarthritis. Genes Immun. 2003;4:487–491. doi: 10.1038/sj.gene.6364014. [DOI] [PubMed] [Google Scholar]
- 14.Hizawa N., Yamaguchi E., Takahashi D., Nishihira J., Nishimura M. Functional polymorphisms in the promoter region of macrophage migration inhibitory factor and atopy. Am. J. Respir. Crit. Care Med. 2004;169:1014–1018. doi: 10.1164/rccm.200307-933OC. [DOI] [PubMed] [Google Scholar]
- 15.Martiney J.A., Sherry B., Metz C.N., Espinoza M., Ferrer A.S., Calandra T., Broxmeyer H.E., Bucala R. Macrophage migration inhibitory factor release by macrophages after ingestion of Plasmodium chabaudi-infected erythrocytes: possible role in the pathogenesis of malarial anemia. Infect. Immun. 2000;68:2259–2267. doi: 10.1128/iai.68.4.2259-2267.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chaisavaneeyakorn S., Moore J.M., Othoro C., Otieno J., Chaiyaroj S.C., Shi Y.P., Nahlen B.L., Lal A.A., Udhayakumar V. Immunity to placental malaria. IV. Placental malaria is associated with up-regulation of macrophage migration inhibitory factor in intervillous blood. J. Infect. Dis. 2002;186:1371–1375. doi: 10.1086/344322. [DOI] [PubMed] [Google Scholar]
- 17.Clark I., Awburn M. Migration inhibitory factor in the cerebral and systemic endothelium in sepsis and malaria. Crit. Care Med. 2002;30:S263–S267. doi: 10.1097/00003246-200205001-00015. [DOI] [PubMed] [Google Scholar]