Abstract
In this paper, we present a novel PCR method, termed SiteFinding-PCR, for gene or chromosome walking. The PCR was primed by a SiteFinder at a low temperature, and then the target molecules were amplified exponentially with gene-specific and SiteFinder primers, and screened out by another gene-specific primer and a vector primer. However, non-target molecules could not be amplified exponentially owing to the suppression effect of stem–loop structure and could not be screened out. This simple method proved to be efficient, reliable, inexpensive and time-saving, and may be suitable for the molecules for which gene-specific primers are available. More importantly, large DNA fragments can be obtained easily using this method. To demonstrate the feasibility and efficiency of SiteFinding-PCR, we employed this method to do chromosome walking and obtained 16 positive results from 17 samples.
INTRODUCTION
Several PCR methods have been developed for isolating an unknown segment adjacent to a known DNA sequence, including inverse PCR (1–6), ligation-mediated PCR (7–18) and randomly primed PCR (19–24). Each of these methods has particular advantages. For example, inverse PCR has high specificity. TAIL-PCR is especially suitable for large-scale manipulation because it can be easily manipulated and can be automated (19). However, most of these methods require complicated manipulations, including restriction cleavage, Southern blot analysis, ligation or tailing before PCR amplification. With some of these methods, arbitrary priming produces amplification of non-target molecules, which constitute the bulk of the final product (18). Furthermore, most products of them are limited to a length of <1 kb.
Here, we report a simple and efficient PCR method, i.e. SiteFinding-PCR, for gene or chromosome walking. The principle and the procedure of SiteFinding-PCR are outlined in Figure 1, and the detailed thermal cycler settings are listed in Table 1. We verified the feasibility of our protocol in two proofs of principle studies: (i) we amplified and cloned the sequence contiguous to the known sequence of a novel cyanophage, and (ii) we cloned and sequenced the insertion sites of agrobacterium T-DNA (25–27) inserted into the Arabidopsis genome.
Figure 1.
Schematic outline of SiteFinding-PCR method for chromosome walking. Known and unknown sequences are depicted with thick and thin lines, respectively. Blue segments show the expected SiteFinder targets. Gene-specific primers (GSPs) 1–3 can anneal with known sequences (white arrows). (1) Original genomic double-strand templates, showing target molecule and non-target molecules. (2) SiteFinding reaction: after low temperature priming by a SiteFinder, one strand of the target gene was replaced by long Taq DNA polymerase, which generated double-stranded target molecules of different lengths. (3) Nested PCR: the target DNA was exponentially amplified by nested PCR with GSPs and SiteFinder primers (SFPs) 1 and 2, while non-target gene amplification was suppressed by the stem–loop structure of the DNA. (4) Cloning target molecules: after being cleaved with NotI, the PCR products (generated by GSP2 and SFP2) were purified by agarose gel electrophoresis, and then the purified DNA was cloned into a pBluescript SK(+) vector linearized by NotI and EcoRV. (5) Screening and sequencing: the clones were screened by colony-PCR with the third gene-specific primer (GSP3) and a vector primer (M13 reverse primer or T3 primer), and the target molecules were screened out and sequenced subsequently.
Table 1.
Cycling conditions used for SiteFinding-PCR on PTC-200 Peltier Termal Cycler
| Reaction | Cycle(s) | Thermal condition |
|---|---|---|
| SiteFinding | 1 | 92°C (2 min), 95°C (1 min), 25°C (1 min), ramp to 68°C over 3 min, 68°C (10 min) |
| Primary | 1 | 94°C (1 min) |
| 30 | 95°C (10 s), 68°C (6 min) | |
| 1 | 72°C (5 min) | |
| Secondary | 1 | 94°C (1 min) |
| 30 | 95°C (10 s), 68°C (6 min) | |
| 1 | 72°C (5 min) |
MATERIALS AND METHODS
Template DNA and oligonucleotides
Cyanophage P4 was isolated from Kunming Lake at the Summer Palace in Beijing and its genomic DNA was extracted from the lysate of a cyanobacteria in accordance with the M13 phage DNA preparation protocol described by Sambrook and Russell (28). The genomic DNAs of Arabidopsis mutants were extracted according to the method described by Liu et al (19). The oligonucleotides and the corresponding primers of the SiteFinders are shown in Figure 2A. The gene-specific primers employed are shown in Figure 2B and C.
Figure 2.
(A) Sequences of two SiteFinders and their primers (SFP1 and SFP2). SiteFinder-1 and 2 differed only at their 3′ ends, and contained a rare restriction enzyme site for NotI, which facilitates cloning with commonly used vectors, such as pBluescript SK(+). The four specific nucleotides underlined with blue bar at the 3′ ends of the SiteFinders, with the help of NNNNNN, were used to anneal with the complimentary sites on genomic DNAs at low temperature and initiate SiteFinding-PCR. SFP1 and SFP2 were used in the primary and secondary reactions, respectively. (B) Three gene-specific primers (GSPs) for Cyanophage P4 (P4-1, P4-2 and P4-3) are indicated by black arrows. The distance from P4-2 to P4-3 was 31 bp. (C) Three GSPs for Arabidopsis T-DNA insertion mutants (DL1, DL2 and DL3) designed based on the T-DNA sequence of pSki015 are indicated by black arrows. The distance from DL2 to DL3 was 59 bp.
SiteFinding
The PCR mixture included 2 μl of 10× long Taq DNA polymerase buffer, 2 μl of mixed dNTP solution (2.5 mM each of dATP, dTTP, dCTP and dGTP), 0.5 U of long Taq DNA polymerase (Beijing TianWei Times Technology Co. Ltd, China), 10 pmol of SiteFinder and 10–200 ng of template DNA. The final volume was brought to 20 μl with Milli-Q water, and then a single cycle PCR cycle was run (Table 1).
Nested PCR
For the primary round of PCR, 5 μl of primer mixture (50 pmol of SFP1, 10 pmol of GSP1 and 1× Taq DNA polymerase buffer) was added to PCR tubes or the wells of a PCR plate (25 μl final volume) on ice, and then the PCR was run for 30 cycles (Table 1). For the secondary reaction, 1 μl of the primary PCR products was diluted into 100–1000 μl Milli-Q water, and then 1 μl of the diluted products was combined with 49 μl of the secondary PCR mixture, which contained 1× long Taq DNA polymerase buffer, 25 μM dNTPs, 0.8 U of long Taq polymerase, 0.2 μM each of internal specific primer (GSP2) and internal SiteFinder primer (SFP2), and then the PCR was run for 30 cycles (Table 1). Meanwhile, another PCR was run in which all of the components and their concentrations were the same as described above except that GSP3 was used instead of GSP2. Finally, the products were separated on 1.2% agarose gels. Specificity was judged by the difference in product size, which was consistent with the distance between the positions of GSP2 and GSP3.
Cloning the target molecules
(i) The PCR products were directly cleaved as follows: 43 μl of PCR products were mixed with 5 μl of 10× enzyme buffer and 2 μl (20 U) of NotI (New England Biolabs, USA), and then incubated overnight at 37°C. (ii) The digested PCR products were separated by electrophoresis on 1.2% agarose gels and purified with Wizard® SV Gel and PCR Clean-Up System (Promega, USA). (iii) The purified DNA was cloned into the pBluescript SK(+) vector between the restriction sites of NotI and EcoRV.
Screening and sequencing
Several clones were selected at random and transferred individually into PCR tubes, each of which contained 15 μl of the following PCR mixture: 1× Taq DNA polymerase buffer, 25 μM dNTPs, 0.5 U of Taq DNA polymerase (Beijing TianWei Times technology Co. Ltd), 0.2 μM of the M13 reverse primer (5′-CAG GAA ACA GCT ATG AC-3′) or T3 sequencing primer (5′-AAT TAA CCC TCA CTA AAG GG-3′), and the corresponding gene-specific primer 3 (GSP3). The clones were subjected to 30 cycles of PCR, and the reaction parameters were as follows: denaturation at 95°C for 10 s, annealing at 56°C for 30 s and extension at 72°C for 2.5 min, plus an initial denaturation step of 1 min at 94°C and a final extension of 5 min at 72°C. The PCR products were examined on a 1.2% agarose gel, and the positive clones were selected for sequencing (DNA sequencing was carried out by Dalian TaKaRa Biotechnology Co. Ltd, China). DNA sequence analyses were carried out using the BLAST program (http://ncbi.nlm.nih.gov).
RESULTS
Identification of an unknown region on cyanophage DNA by SiteFinding-PCR
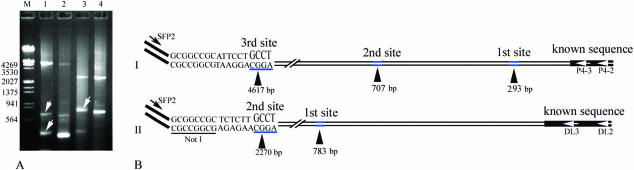
The Cyanophage P4 genomic DNA was extracted from the lysate of a cyanobacteria, and a 769 bp fragment was cloned into the pBluescript SK(+) vector and sequenced (M. Shi et al., unpublished data). Based on the known sequence, three primers, P4-1 (GSP1), P4-2 (GSP2) and P4-3 (GSP3), were designed (Figure 2B). After two rounds of PCR, the products were separated on a 1.2% agarose gel and examined. A DNA fragment of ∼4.5 kb was recovered and cloned into the pBluescript SK(+) vector. Eight clones were screened with the M13 reverse primer and P4-3, among which five positive clones were obtained. One clone with a ∼4.5 kb Cyanophage P4 DNA fragment was selected and sequenced. The sequencing results (Supplementary Material 1) showed that the cloned product was 4617 bp in length (Figure 3B, I) and contained another two GCCT sites, one of which was 293 bp away from the P4-2 locus, and the other was 707 bp away from the P4-2 locus (Figure 3B, I). The sequence was subsequently confirmed by Cyanophage P4 genome sequencing (M. Shi et al., unpublished data). To evaluate the feasibility of this approach for complicated genomes, DNA mixtures (0.1 ng of Cyanophage P4 DNA plus 300 ng of rice genome DNA) were utilized as original templates, and we obtained the same results as those using the P4 Cyanophage genome DNA as the original templates.
Figure 3.
Chromosome walking for the Cyanophage P4 and an Arabidopsis mutant using the SiteFinding-PCR method. SiteFinder-1 was used to initialize the SiteFinding reaction. (A) Products of the secondary round of PCR (lanes 1–2 for P4 Cyanophage and 3–4 for Arabidopsis mutant). Lanes 1–4 contained PCR products obtained with primer couples SFP2/P4-2, SFP2/P4-3, SFP2/DL2 and SFP2/DL3, respectively. (B) Cloned and sequenced PCR products. Lanes 1 and 2 both showed three specific products (A), and the largest one in lane 1 was cloned and sequenced as indicated in (I). There were another two GCCT sites on the 4617 bp fragment of the Cyanophage P4 sequence, which are indicated with the black arrowheads in (I) (first site and second site). These findings were consistent with the gel electrophoresis results (white arrows in lane 1). Lanes 3 and 4 both showed two specific products (A), and the larger one in lane 3 was cloned and sequenced as indicated in (II). There was another GCCT site on the 2270 bp fragment of the Arabidopsis DNA as indicated with the black arrowheads in (II) (first site), which was also consistent with the gel electrophoresis (white arrow in lane 3).
Identification of the T-DNA insertion sites of Arabidopsis mutants
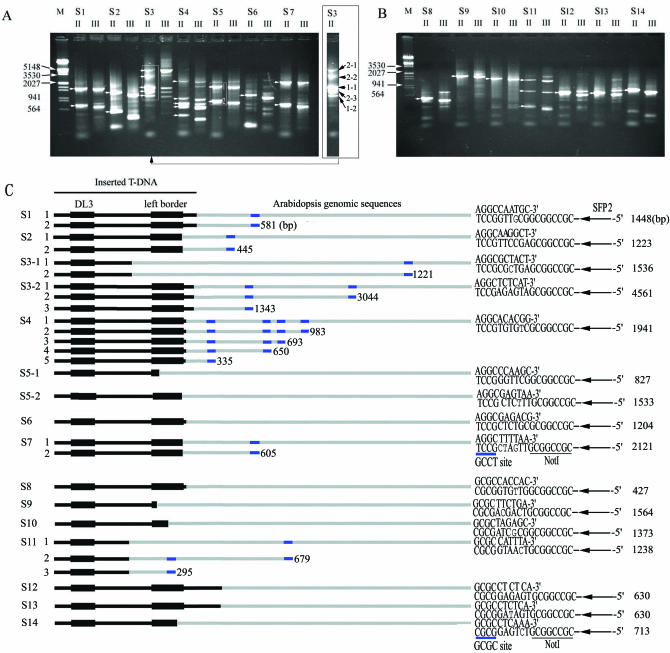
To verify the feasibility of this approach for a complicated genome, we used it to identify an Arabidopsis mutant site inserted by T-DNA (25–27) of the binary vector pSki015 (29,30), which had formerly been identified by TAIL-PCR (Supplementary Material 2). Three GSPs (DL1, DL2 and DL3), designed from known regions adjacent to the left border of the T-DNA in pSki015 (Figure 2C), were used together with two SFPs (SFP1 and SFP2). After two rounds of PCR, we obtained two specific DNA bands of ∼2.2 kb and ∼0.7 kb (Figure 3A, lane 3). The DNA was recovered from the ∼2.2 kb band, and then cloned and sequenced (Figure 3B, II). The sequencing results showed that the insertion site was consistent with that obtained by TAIL-PCR (Supplementary Material 2). To further demonstrate the feasibility, efficiency and universality of the SiteFinding-PCR method, the genomic DNAs from 15 other Arabidopsis mutants were used as templates to identify Arabidopsis genomic sequences flanking the T-DNA left borders. We obtained specific bands from 14 mutants (Figure 4A and B), among which 9 samples (samples 1–3 and 8–13) had formerly been identified by TAIL-PCR (Supplementary Materials 3 and 4). The others could not be identified successfully by TAIL-PCR. Here, only 1 out of the 15 samples did not show specific amplification (data not shown). We cloned and sequenced 28 specific bands indicated with white arrows in Figure 4A and B. Sequence analyses (Supplementary Material 4) revealed that we had identified the sites inserted by T-DNA for 14 samples, among which Samples 3 and 5 had two insertion sites (Figure 4A and C).
Figure 4.
Identification of T-DNA insertion sites of 14 Arabidopsis mutants with SiteFinding-PCR. Lanes II and III: the products generated by SFP2/DL2 and SFP2/DL3, respectively. Samples are labeled as S1–S14. (A) The products of S1–S7, initially primed by SiteFinder-1. White arrows in lane II indicated the cloned products. Digitals in boxed areas show different insertion sites. As for S3, 1-1 and 1-2 show two different products of the 1st insertion site; 2-1, 2-2 and 2-3 in S3 refer to three products of the 2nd insertion site. (B) The products of samples 8–14, initially primed by SiteFinder-2. White arrows in lane II indicated the cloned products. (C) Comparison of the 3′ end of the SiteFinder with the largest or larger fragment. Bold black line segments and grey line segments represent the T-DNA and Arabidopsis DNAs, respectively. The arrows represent SFP2. The sizes of the largest or larger specific fragment of the inserted site are indicated on the right, and the smaller ones are marked in the middle with blue bars. NNNNNN with 0–3 mismatch nt (courier) helped the 3′ ends (underlined with blue bars) of SiteFinder-1 and SiteFinder-2 to anneal accurately with 3′-CGGA-5′ (GCCT site) and 3′-CGCG-5′ (GCGC site), respectively.
DISCUSSION
Principle of SiteFinding-PCR
It is well known that the control of false priming is very important for PCR. However, at the 3′ end of a primer, there must be several nucleotides that complement the template accurately, even in the case of false priming. We utilized this characteristic to initialize SiteFinding-PCR in our protocol. The oligonucleotide 5′-NNNNNNGCCT-3′ at the 3′ end of SiteFinder-1 or 5′-NNNNNNGCGC-3′ at the 3′ end of SiteFinder-2 was utilized to find the GCCT or GCGC sites on the target molecules and non-target molecules. SiteFinding-PCR amplification could then be primed at low temperature. In non-target molecule amplification, the PCR products contained double-stranded SiteFinder sequences at both ends, and the ends of the individual DNA strands formed a stem–loop structure following every cycle due to the presence of inverted terminal repeats. Stem–loop structures are more stable than the primer-template hybrid, and therefore suppress exponential amplification (14,31,32). However, in the case of target molecule amplification, a distal gene-specific primer extends a DNA strand through the SiteFinder after the SiteFinding reaction, and the target molecule contains the SiteFinder sequence only at one end. As a result, the stem–loop structure does not form, and thus the PCR amplification can proceed smoothly, using a long distance thermostable DNA polymerase (long Taq DNA polymerase) with a two-temperature cycling protocol (Table 1).
Two key points pertaining to this method may be noted. First, the 4–6 nt oligonucleotide at the 3′ end of the SiteFinder initializes the reaction at low temperature; second, the stem–loop structure suppresses exponential amplification efficiently. In operation, we utilized two 4 nt oligonucleotides, GCCT and GCGC, to prime the PCR at 25°C. In fact, any 4, 5 or 6 nt oligonucleotides could be used, while the length and constitution of the oligonucleotides would influence the results. Importantly, the annealing temperature in the SiteFinding reaction should be adjusted appropriately for the specific oligonucleotides used. For example, we could use a 6 nt oligonucleotide at a higher temperature to obtain longer target molecules and a 4 nt oligonucleotide at a lower temperature to obtain shorter target molecules. In this paper, we obtained more specific DNA bands using GCCT (Figure 4A) than using GCGC (Figure 4B), because the latter exhibited a lower emergence frequency in the Arabidopsis genome (18). To help the SiteFinder anneal with complementary sites on genomic DNA, oligonucleotides with an appropriate number of random nucleotides or inosines triphosphate adjacent to the fixed 4–6 nt could be synthesized. In our experiment, sequencing results demonstrated that the 3′ ends of SiteFinder-1 and SiteFinder-2 accurately annealed with 3′-CGGA-5′ (GCCT site) and 3′-CGCG-5′ (GCGC site), respectively, under the help of NNNNNN with 0–3 mismatch nucleotides (courier in Figure 4C) owing to low stringency SiteFinding priming. Further analyses showed that the mismatch might be created during PCR cycles, because there were ∼0.1% error rates occurred by extending alignments with Arabidopsis genome sequence (Supplementary Material 4).
SiteFinders contain a rare restriction enzyme site for NotI, and PCR amplification was performed using a long Taq DNA polymerase, which could only produce the fragments with blunt ends. Therefore, after digesting the products (generated by SFP2 and GSP2) with NotI, we obtained target molecules with one staggered end and one blunt end (Figure 1, panel 4), which facilitated the cloning of target molecules into pBluescript SK(+) linearized by NotI and EcoRV (Figure 1, panel 4). In contrast, non-target molecules had either two adhesive terminals at both ends or a stem–loop structure with one adhesive terminal (Figure 1, panel 3), which prevented them from being cloned. Furthermore, an internal specific primer (GSP3) was used to screen the clones, which ensured that only those clones possessing the specific products would be screened out because only the target molecules possessed the complementary site for GSP3 (Figure 1, panel 5).
Advantages of SiteFinding-PCR
In general, there are three kinds of PCR methods available for chromosome walking: inverse PCR (1–6), LM–PCR (7–18) and RP–PCR (19–24). Inverse PCR and LM–PCR are dependent upon the use of restriction endonucleases before PCR and require additional steps, which may decrease the recovery efficiency of the product, increase the chance of contamination, and are time-consuming. In addition, it would be expensive because many restriction endonucleases had to be used due to unpredictable restriction endonuclease sites in unknown regions of the target molecule. RP–PCR, e.g. TAIL–PCR, is a popular method for chromosome walking, especially for the identification of T-DNA or transposon insertions, but its amplified products are usually small, and the optimum conditions have to be established empirically.
Compared with other PCR methods for chromosome walking, the SiteFinding-PCR method has several advantages. (i) Simplicity: the SiteFinding-PCR does not require additional manipulations (such as Southern blot analysis, restriction digestion, ligation or tailing) before PCR, and the products' specificity can be confirmed by a simple agarose gel electrophoresis analysis. (ii) Specificity: because of the stem–loop structure suppression effect, co-amplification of non-specific products is very weak. Furthermore, any non-specific product would not be cloned and screened out (Figure 1, panels 4 and 5). (iii) Sensitivity: we used 10 ng of Arabidopsis genomic DNA as the original template and obtained good results. (iv) Efficiency: we obtained 16 positive results from 17 samples, and only one reaction did not yield specific products, which may be due to the lack of corresponding sites adjacent to the known sequence. However, another SiteFinder could be used to complete the walking, i.e. SiteFinding-PCR may be potentially suitable for any DNA on which gene-specific primers can be designed. (v) Long specific product: we obtained 17 specific products that were >1 kb (Supplementary Material 4), 2 of which were >4.5 kb (Figures 3A and Figure 4A and B). (vi) Low cost and time-saving: the enzymes required for the procedure are only long Taq DNA polymerase, NotI and T4 DNA ligase; the PCR amplification can be completed consecutively within 6–8 h.
The potential applications of SiteFinding-PCR
The advantages of the SiteFinding-PCR method make it useful for the following applications: (i) rapidly obtaining a full gene sequence or finding promoters and regulatory elements from cloned cDNA fragments, (ii) determining the exon–intron boundaries within genes, (iii) walking upstream or downstream from sequence-tagged sites in known or unknown genomes for gene function studies, (iv) obtaining non-conserved regions of genes in uncharacterized species, according to the conserved sequences of reported genes and (v) filling in the gaps in genome sequencing.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Supplementary Material
Acknowledgments
We are grateful to Dr Xin Li and Dr Yanxia Zhang for providing Arabidopsis materials, and our colleagues Ye Wang, Shuangli Mi, Wanqiang Qian and Jiayu Gu for their helpful suggestions during the preparation of this manuscript. This work was supported by grants from the National Key Basic Science ‘973’ Program (No. G20000016204), the State ‘863’ High Technology R&D Program (No. 2004AA214072 and NO. 2002AA206631) and the Ministry of Education Doctor's Foundation Program (No. 20030001083) of the Chinese Government. Funding to pay the Open Access publication charges for this article was provided by Peking University, P.R. China.
Conflict of interest statement. None declared.
REFERENCES
- 1.Triglia T., Peterson M.G., Kemp D.J. A procedure for in vitro amplification of DNA segments that lie outside the boundaries of known sequences. Nucleic Acids Res. 1988;16:8186. doi: 10.1093/nar/16.16.8186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ochman H., Gerber A.S., Hartl D.L. Genetic applications of an inverse polymerase chain reaction. Genetics. 1988;120:621–623. doi: 10.1093/genetics/120.3.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Silverman G., A., Ye R.D., Pollock K.M., SadlerJ.E., Korsmeyer S.J. Use of yeast artificial chromosome clones for mapping and walking within human chromosome segment 18q21.3. Proc. Natl Acad. Sci. USA. 1989;86:7485–7489. doi: 10.1073/pnas.86.19.7485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huckaby C.S., Kouri R.E., Lane M.J., Peshick S.M., Carroll W.T., Henderson S.M., Faldasz B.D., Waterbury P.G., Vournakis J.N. An efficient technique for obtaining sequences flanking inserted retroviruses. Genet. Anal. Tech. Appl. 1991;8:151–158. doi: 10.1016/1050-3862(91)90024-l. [DOI] [PubMed] [Google Scholar]
- 5.Arveiler B., Porteous D.J. Amplification of end fragments of YAC recombinants by inverse polymerase chain reaction. Technique. 1991;3:24–28. [Google Scholar]
- 6.Keim M., Williams R.S., Harwood A.J. An inverse PCR technique to rapidly isolate the flanking DNA of dictyostelium insertion mutants. Mol. Biotechnol. 2004;26:221–224. doi: 10.1385/MB:26:3:221. [DOI] [PubMed] [Google Scholar]
- 7.Riley J., Butler R., Ogilvie D., Finniear R., Jenner D., Powell S., Anand R., Smith J.C., Markham A.F. A novel, rapid method for the isolation of terminal sequences from yeast artificial chromosome (YAC) clones. Nucleic Acids Res. 1990;18:2887–2890. doi: 10.1093/nar/18.10.2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rosenthal A., Jones D.S.C. Genomic walking and sequencing by oligo-cassette mediated polymerase chain reaction. Nucleic Acids Res. 1990;18:3095–3096. doi: 10.1093/nar/18.10.3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rosenthal A., MacKinnon R.N., Jones D.S.C. PCR waling from microdissection clone M54 identifies three exons from the human gene for the neural cell adhesion molecule L1 (CAM-L1) Nucleic Acids Res. 1991;19:5395–5401. doi: 10.1093/nar/19.19.5395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Espelund M., Jakobsen K.S. Cloning and direct sequencing of plant promoters using primer adapter mediated PCR on DNA coupled to a magnetic solid phase. BioTechniques. 1992;13:74–81. [PubMed] [Google Scholar]
- 11.Jones D.H., Winistorfer S.C. Sequence specific generation of a DNA panhandle permits PCR amplification of unknown flanking DNA. Nucleic Acids Res. 1992;20:595–600. doi: 10.1093/nar/20.3.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones D.H., Winistorfer S.C. Genome walking with 2-to 4-kb steps using panhandle PCR. PCR Methods Appl. 1993;2:197–203. doi: 10.1101/gr.2.3.197. [DOI] [PubMed] [Google Scholar]
- 13.Warshawsky D., Miller L. A rapid genomic walking technique based on ligation-mediated PCR and magnetic separation technology. BioTechniques. 1994;16:792–798. [PubMed] [Google Scholar]
- 14.Siebert P.D., Chenchik A., Kellogg D.E., Lukyanov K.A., Lukyanov S.A. An improved PCR method for walking in uncloned genomic DNA. Nucleic Acids Res. 1995;23:1087–1088. doi: 10.1093/nar/23.6.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sterky F., Holmberg A., Alexandersson G., Lundeberg J., Uhlen M. Direct sequenceing of bacterial artificial chromosone (BACs) and prokaryotic genomes by biotin-capture PCR. J. Biotechnol. 1998;246:810–813. doi: 10.1016/s0168-1656(97)00196-x. [DOI] [PubMed] [Google Scholar]
- 16.Dai S.M., Chen H.H., Chang C., Riggs A.D., Flanagan S.D. Ligation-mediated PCR for quantitative in vivo footprinting. Nat. Biotechnol. 2000;18:1108–1111. doi: 10.1038/80323. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Z., Gurr S.J. A ‘step down’ PCR-based technique for walking into and the subsequent direct-sequence analysis of flanking genomic DNA. Methods Mol. Biol. 2002;192:343–350. doi: 10.1385/1-59259-177-9:343. [DOI] [PubMed] [Google Scholar]
- 18.Yuanxin Y., Chengcai A., Li L., Jiayu G., Guihong T., Zhangliang C. T-linker-specific ligation PCR (T-linker PCR): an advanced PCR technique for chromosome walking or for isolation of tagged DNA ends. Nucleic Acids Res. 2003;31:e68. doi: 10.1093/nar/gng068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Y.G., Mitsukawa N., Oosumi T., Whittier R.F. Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions bu thermal asymmetric interlaced PCR. Plant J. 1995;8:457–463. doi: 10.1046/j.1365-313x.1995.08030457.x. [DOI] [PubMed] [Google Scholar]
- 20.Liu Y.G., Whittier R.F. Thermal asymmetric interlaced PCR: automatable amplification and sequencing of insert end fragments from P1 and YAC clones for chromosome walking. Genomics. 1995;25:674–681. doi: 10.1016/0888-7543(95)80010-j. [DOI] [PubMed] [Google Scholar]
- 21.Terauchi R., Kahl G. Rapid isolation of promoter sequences by TAIL-PCR: the 5-flanking regions of Pal and Pgi genes from yams (Dioscorea) Mol. Gen. Genet. 2000;263:554–560. doi: 10.1007/s004380051201. [DOI] [PubMed] [Google Scholar]
- 22.Weigel D., Ahn J.H., Blazquez M.A., Borevitz J.O., Christensen S.K., Fankhauser C., Ferrandiz C., Kardailsky I., Mlancharuvil E.J., Neff M.M., et al. Activation tagging in Arabidopsis. Plant Physiol. 2000;122:1003–1013. doi: 10.1104/pp.122.4.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Singer T., Burke E. High-throughput TAIL-PCR as a tool to identify DNA flanking insertions. Methods Mol. Biol. 2003;236:241–272. doi: 10.1385/1-59259-413-1:241. [DOI] [PubMed] [Google Scholar]
- 24.Antal Z., Rascle C., Fevre M., Bruel C. Single oligonucleotide nested PCR: a rapid method for the isolation of genes and their flanking regions from expressed sequence tags. Curr. Genet. 2004;46:240–246. doi: 10.1007/s00294-004-0524-6. [DOI] [PubMed] [Google Scholar]
- 25.Chang S.S., Park S.K., Kim B.C., Kang b.J., Kim D.U., Nam H.G. Stable genetic transformation of Arabidopsis thaliana by Agrobacterium inoculation in planta. Plant J. 1994;5:551–558. [Google Scholar]
- 26.Gelvin S.B. Agrobacterium-mediated plant transformation: the biology behind the ‘gene-jockeying’ tool. Microbiol Mol. Biol. Rev. 2003;67:16–37. doi: 10.1128/MMBR.67.1.16-37.2003. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tzfira T., Li J., Lacroix B., Citovsky V. Agrobacterium T-DNA integration: molecules and models. Trends Genet. 2004;20:375–383. doi: 10.1016/j.tig.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook J., Russell D.W. Molecular Cloning: A Laboratory Manual. 3rd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 29.Memelink J. T-DNA activation tagging. Methods Mol. Biol. 2003;236:345–362. doi: 10.1385/1-59259-413-1:345. [DOI] [PubMed] [Google Scholar]
- 30.Weigel D., Ahn J.H., Blázquez M.A., Borevitz J.O., Christensen S.K., Fankhauser C., Ferrándiz C., Kardailsky I., Malancharuvil E.J., Neff M.M., et al. Activation tagging in Arabidopsis. Plant Physiol. 2000;122:1003–1013. doi: 10.1104/pp.122.4.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matz M., Shagin D., Bogdanova E., Britanova O., Lukyanov S., Diatchenko L., Chenchik A. Amplification of cDNA ends based on template-switching effect and step-out PCR. Nucleic Acids Res. 1999;27:1558–1560. doi: 10.1093/nar/27.6.1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chenchik A., Diachenko L., Moqadam F., Tarabykin V., Lukyanov S., Siebert P.D. Full-length cDNA cloning and determination of mRNA 5′ and 3′ ends by amplification of adaptor-ligated cDNA. BioTechniques. 1996;21:526–534. doi: 10.2144/96213pf02. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.