Abstract
Expression of the Arabidopsis CGS1 gene that codes for cystathionine γ-synthase is feedback regulated at the step of mRNA stability in response to S-adenosyl-L-methionine (AdoMet). A short stretch of amino acid sequence, called the MTO1 region, encoded by the first exon of CGS1 itself is involved in this regulation. Here, we demonstrate, using a cell-free system, that AdoMet induces temporal translation elongation arrest at the Ser-94 codon located immediately downstream of the MTO1 region, by analyzing a translation intermediate and performing primer extension inhibition (toeprint) analysis. This translation arrest precedes the formation of a degradation intermediate of CGS1 mRNA, which has its 5′ end points near the 5′ edge of the stalled ribosome. The position of ribosome stalling also suggests that the MTO1 region in nascent peptide resides in the ribosomal exit tunnel when translation elongation is temporarily arrested. In addition to the MTO1 region amino acid sequence, downstream Trp-93 is also important for the AdoMet-induced translation arrest. This is the first example of nascent peptide-mediated translation elongation arrest coupled with mRNA degradation in eukaryotes. Furthermore, our data suggest that the ribosome stalls at the step of translocation rather than at the step of peptidyl transfer.
Keywords: S-adenosyl-L-methionine, translation arrest, mRNA stability, MTO1 region, cystathionine γ-synthase, feedback regulation
Cystathionine γ-synthase (CGS; EC 2.5.1.48) catalyzes the first committed step of methionine biosynthesis in higher plants (Matthews 1999) and is encoded by the CGS1 gene in Arabidopsis thaliana (gene ID At3g01120; Kim and Leustek 1996). Expression of the CGS1 gene is regulated by negative feedback at the step of mRNA stability in response to methionine application in Arabidopsis. When wild-type calli were treated with methionine, the amount of full-length CGS1 mRNA was decreased, and a short CGS1 RNA species formed that is truncated at its 5′ region. This 5′-truncated RNA species is likely an intermediate of CGS1 mRNA degradation. The mto1 mutants of Arabidopsis, which carry single amino acid sequence alterations within the first exon of CGS1, are deficient in this regulation and overaccumulate soluble methionine (Chiba et al. 1999). A short stretch of amino acid sequence, termed the MTO1 region, which is encoded within CGS1 exon 1 and covers the mto1 mutation sites, is involved in this process (Ominato et al. 2002). Since CGS1 exon 1 acts in cis, we proposed that this regulation occurs during translation, when the nascent polypeptide and its mRNA are in close proximity (Chiba et al. 1999; Suzuki et al. 2001). In support of this idea, the regulation is abolished by application of a translation inhibitor (Lambein et al. 2003).
CGS1 exon 1-mediated post-transcriptional regulation was reproduced in an in vitro translation system using wheat germ extract, and S-adenosyl-L-methionine (AdoMet), a direct metabolite of methionine, was identified as the natural effector of this regulation (Chiba et al. 2003). When an in vitro-transcribed capped RNA carrying the CGS1 exon 1 fused to a reporter gene was translated in vitro, reporter activity was down-regulated in response to AdoMet. In addition, 5′-truncated RNA species were detected that had the same 5′ end points as those detected in vivo (Chiba et al. 2003). Although this suggested that the same mRNA degradation event was occurring both in vivo and in vitro, the amount of full-length RNA after in vitro translation in the presence of AdoMet did not correlate with the reporter activity. This discrepancy raises the possibility that CGS1 exon 1-mediated post-transcriptional regulation is primarily a translational regulation.
In some eukaryotic genes, a cis determinant in the coding region induces translational pausing and destabilizes its mRNA (Caponigro and Parker 1996; Lemm and Ross 2002). We hypothesized that MTO1-dependent translation arrest may occur during translation of CGS1 exon 1, causing mRNA degradation. In this study, we tested this possibility using an in vitro translation system of wheat germ extract (Chiba et al. 2003). Identification of a translation intermediate and primer extension inhibition (toeprint) analysis revealed that, prior to CGS1-specific mRNA degradation, translation elongation is arrested temporarily downstream of the MTO1 region in response to AdoMet. We also present evidence suggesting that translation is arrested at the step of ribosome translocation.
Results
Identification of a translation arrest product
If translation elongation arrest is induced in CGS1 exon 1, an incomplete translation product may accumulate when CGS1 exon 1-containing RNA is translated in vitro in the presence of AdoMet. To detect an incomplete translation product, a glutathione S-transferase (GST) tag sequence was fused in-frame to the 5′ end of the CGS1 exon 1 coding sequence and linked to a firefly luciferase (LUC) reporter. This DNA was then transcribed in vitro to prepare capped GST:Ex1:Luc RNA with an A30 tail (Fig. 1A). Addition of the GST tag did not affect the AdoMet-induced down-regulation of reporter activity (Supplementary Fig. S1).
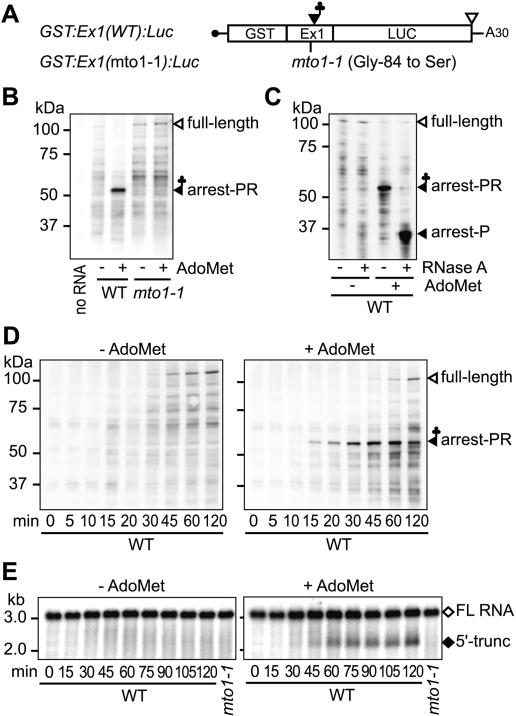
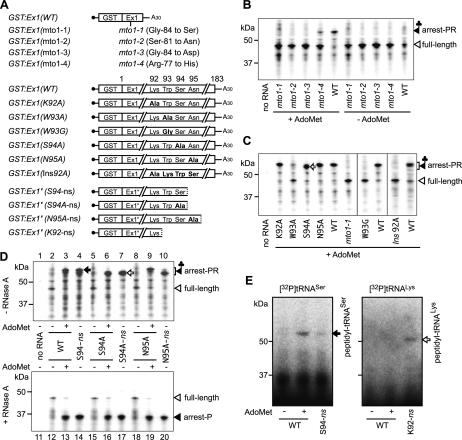
Figure 1.
Translation elongation arrest and RNA degradation. (A) Schematic representation of GST:Ex1:Luc RNA. (B,C) Immunoblot analysis. GST:Ex1(WT):Luc RNA or GST:Ex1(mto1-1): Luc RNA was translated for 30 min in the presence (+) or absence (-) of 1 mM AdoMet. The translation products were separated by SDS-PAGE and subjected to immunoblot analysis with anti-GST antibody. In C, the WT samples were treated with RNase A as indicated before separation by SDS-PAGE. The 106-kDa full-length translation product (full-length), the 55-kDa AdoMet-dependent peptidyl-RNA (arrest-PR), and the AdoMet-dependent band shifted by the RNase A treatment (arrest-P) are marked. (D,E) Time course analyses. GST:Ex1(WT):Luc RNA was translated in the presence (right) or absence (left) of AdoMet. Aliquots were withdrawn at the indicated time points and subjected to immunoblot analysis with anti-GST antibody (D) or Northern blot analysis with LUC 3′ probe (E). The full-length RNA (FL RNA) and the 5′-truncated RNA (5′-trunc) are marked in E. Representative results of duplicate to triplicate experiments are shown.
After translating GST:Ex1:Luc RNA in wheat germ extract for 30 min, the translation products were analyzed by immunoblot analysis with anti-GST antibody (Fig. 1B). When GST:Ex1(WT):Luc RNA was translated in the presence of 1 mM AdoMet, a band with an apparent molecular weight of ∼55 kDa was observed in addition to the full-length product (106 kDa). The 55-kDa band was also detectable at 0.1 mM AdoMet (Supplementary Fig. S2); however, for the maximum detection of the 55-kDa band, an AdoMet concentration of 1 mM was used in this study. When GST:Ex1(mto1-1):Luc RNA carrying the mto1-1 mutation (Chiba et al. 1999, 2003) was translated, no such band was observed irrespective of AdoMet application.
If accumulation of this 55-kDa band is due to translation arrest, tRNA may remain attached to the C terminus of the polypeptide. As shown in Figure 1C, RNase A treatment shifted the AdoMet-dependent 55-kDa band to ∼35 kDa. The result indicates that the 55-kDa translation product contains an RNA moiety of ∼20 kDa, which is most likely a tRNA. Further evidence that this RNA moiety is a tRNA will be presented in later sections. The 35-kDa size corresponds to that of the partial polypeptide from the N terminus down to the MTO1 region, suggesting that accumulation of the 55-kDa peptidyl-RNA is due to translation elongation arrest that occurred at around the MTO1 region.
Translation elongation arrest precedes formation of the 5′-truncated RNA
In order to compare the timing of translation arrest and mRNA degradation, time course analyses were carried out. As shown in Figure 1D, when GST:Ex1(WT):Luc RNA was translated in the presence of AdoMet, the 55-kDa peptidyl-RNA was first detectable after a 10-15-min incubation and increased until 45 min. In contrast, the AdoMet-dependent 5′-truncated RNA was first detectable at 30-45 min and increased until 60 min (Fig. 1E). Therefore, accumulation of the peptidyl-RNA precedes that of the RNA degradation intermediate.
To test whether translation elongation resumes after the arrest, we performed a pulse-chase analysis using [35S]methionine (Fig. 2). For this experiment, GST:Ex1 RNA without the LUC reporter was used. The GST ORF contains nine methionine codons, while the CGS1 exon 1 region contains only the initial methionine codon (Fig. 2A). Edeine, an inhibitor of translation initiation, was used to inhibit new rounds of translation (Fang et al. 2004). The 55-kDa arrested product was detected after a 15-min incubation in the presence of AdoMet. The intensity of this band increased until 30 min and then declined. As the 55-kDa band decreased, the intensity of the full-length band (45 kDa) increased until 60 min in the presence of AdoMet, and the increase was slower than that in the absence of AdoMet (Fig. 2B,C). These results suggest that AdoMet induces a temporal translation elongation arrest at around the MTO1 region.
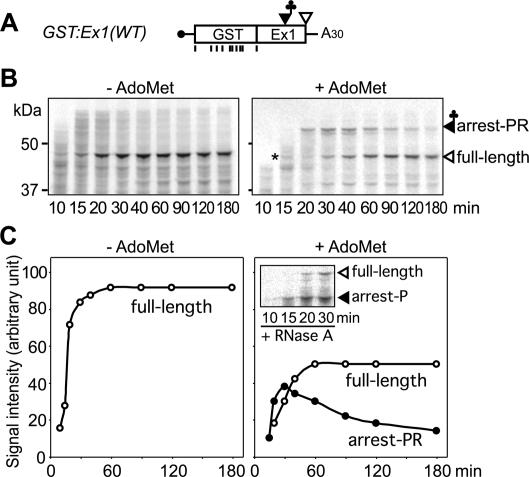
Figure 2.
Pulse-chase analysis of the translation arrest products. (A) Schematic representation of GST:Ex1(WT) RNA. Positions of methionine codons are marked with vertical bars. (B) GST:Ex1(WT) RNA was translated in the presence of [35S]methionine with (right) or without (left) AdoMet. Edeine was added 5 min after the start of translation. Aliquots were withdrawn at the indicated time points, and the radioactive signals were detected after separation by SDS-PAGE. The 45-kDa full-length translation product (full-length) and the 55-kDa AdoMet-dependent peptidyl RNA band (arrest-PR) are marked. The asterisk indicates a 45-kDa background signal detected at 15 min. (C) The radioactive signals in B were quantified by NIH Image software. Open circle indicates 45-kDa full-length product; solid circle indicates 55-kDa AdoMet-dependent peptidyl RNA. (Inset) The 45-kDa background signal in B was ignored as this signal was no longer detectable after RNase A treatment. Representative results of triplicate experiments are shown.
Association of the peptidyl-RNA with a ribosome
If the translation arrest results in the accumulation of the peptidyl-RNA, the peptidyl-RNA should be associated with an arrested ribosome. To test this possibility, translation products of GST:Ex1(WT) RNA were fractionated using sucrose density gradient centrifugation. The AdoMet-induced 55-kDa peptidyl-RNA was detected in the 80S ribosome fractions and polysome fractions, but not in fractions without translating ribosomes (Fig. 3A, right panel). In contrast, the full-length translation product (45 kDa) was abundant in the top fractions, as well as in the translating ribosome fractions, and this pattern was similar to that of the full-length polypeptide produced in the absence of AdoMet (Fig. 3A, left panel). Incubation of the in vitro translation mixture with puromycin, which releases nascent polypeptide as a peptidyl-puromycin (Carrasco et al. 1976), caused most of the 55-kDa-band signals to shift to 35 kDa and to fractions without translating ribosomes (Fig. 3B, right panel). These results indicate that the peptidyl-RNA is associated with a translating ribosome.
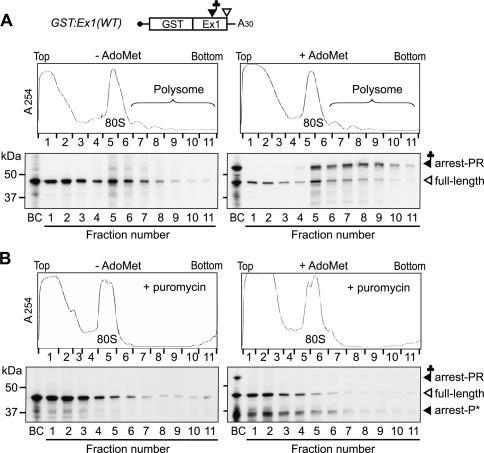
Figure 3.
Association of the peptidyl-RNA with ribosomes. (A) GST:Ex1(WT) RNA was translated for 60 min in the presence (right panels) or absence (left panels) of AdoMet. The samples were fractionated by 10%-30% (w/v) sucrose density gradient centrifugation. UV absorbance profile at 254 nm (upper panels) and immunoblot analysis of each fraction with anti-GST antibody (lower panels) are shown. (B) The same as in A except that the samples were incubated for 30 min with 1 mM puromycin before centrifugation. The 45-kDa full-length translation product (full-length), the 55-kDa AdoMet-dependent peptidyl-RNA band (arrest-PR), and a 35-kDa band, possibly a peptidyl-puromycin (arrest-P★), are marked. Lanes BC represent samples before centrifugation. Representative results of quadruplicate (A) or duplicate (B) experiments are shown.
Determination of the position of the translation arrest
The data in Figure 1C suggested that translation is arrested near the MTO1 region. To determine the position of the translation arrest, we introduced a stop codon around the MTO1 region and tested how far translation needs to progress for the translation arrest to occur. Although a GST tag is attached at the N terminus, the positions of the amino acid residues of CGS1 exon 1 are numbered from the first methionine of CGS1.
Initial experiments, in which a stop codon was introduced into Arg-78, Ile-88, Lys-92, Pro-97, and Ser-111, indicated that the critical region resides between Lys-92 and Pro-97 (data not shown). Therefore, we substituted the 93rd-96th codons of CGS1 exon 1 with a UAG stop codon. As shown in Figure 4A, when UAG was substituted for Lys-92 (K92UAG), Trp-93 (W93UAG), or Ser-94 (S94UAG), the AdoMet-induced 55-kDa band was not detected. In contrast, when UAG was substituted for Asn-95 (N95UAG), Asn-96 (N96UAG), or Pro-97 (P97UAG), the AdoMet-induced 55-kDa band was detected as seen in the wild-type construct.
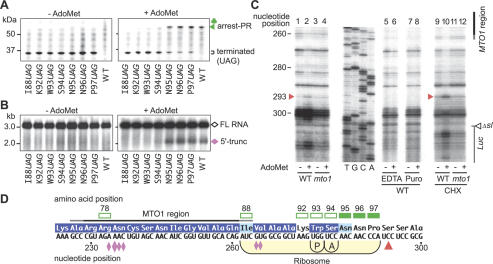
Figure 4.
Determination of the position of the translation elongation arrest. (A,B) Effects of stop codon substitutions. GST:Ex1:Luc RNAs carrying the stop codon substitution constructs were translated for 30 min (A) or 90 min (B) in the presence (right) or absence (left) of AdoMet. Aliquots were subjected to immunoblot analysis with anti-GST antibody (A) or Northern blot analysis with LUC 3′ probe (B). The full-length polypeptide (106 kDa) is not seen in A. Brackets in A mark the complete translation products terminated at the introduced stop codon. (C) Toeprint analysis. GST:Ex1Δsl(WT):Luc RNA (lanes 1,2,5-10) or GST:Ex1Δsl(mto1-1):Luc RNA (lanes 3,4,11,12) was translated for 30 min in the presence (+) or absence (-) of AdoMet. In lanes 5 and 6, EDTA was added to a final concentration of 5 mM at 30 min, and the samples were incubated for another 5 min. MgCl2 was added to 10 mM prior to the primer extension reaction. In lanes 7 and 8, puromycin (Puro) was added to 1 mM at 30 min, and the reaction mixtures were incubated for an additional 10 min. In lanes 9-12, 0.5 mg/mL CHX was added at 30 min. The red arrowheads mark the most 5′-proximal AdoMet-dependent toeprint signals at the 293rd nucleotide position. The sequence ladder (shown in the sense strand sequence) was synthesized using the same primer as used for toeprinting. The MTO1 region and the Δsl:Luc junction are indicated. (D) Schematic representation of the translation elongation arrest position. The red arrowhead indicates the position of the most 5′-proximal AdoMet-dependent toeprint signal. The positions of the P and A sites of the stalled ribosome deduced from the toeprint signal are shown. Open squares indicate codons whose substitution to a stop codon abolished production of the AdoMet-dependent peptidyl-tRNA and 5′-truncated RNA, while solid squares indicate codons without any effect. Amino acid residues that are conserved among plant CGSs are reversed in purple and those with conservative changes are shaded in light blue (Ominato et al. 2002). The pink diamonds indicate the positions of the 5′ ends of the 5′-truncated RNA species with the large one pointing to the most prominent one (Chiba et al. 2003). Representative results of triplicate to quadruplicate experiments are shown.
Next, we tested the effect of these stop codon substitutions on the production of the 5′-truncated RNA (Fig. 4B). In N95UAG, N96UAG, and P97UAG, the 5′-truncated RNA was detected, whereas in I88UAG, K92UAG, W93UAG, and S94UAG, the 5′-truncated RNA was no longer detected. Therefore, translation needs to proceed as far as the Ser-94 codon to produce the 5′-truncated RNA species, as well as the peptidyl-RNA, suggesting a close linkage between the translation arrest and RNA degradation.
As an alternative approach to determining the position of the translation arrest, a toeprint analysis was performed to determine the position of ribosome stalling. For this analysis, GST:Ex1Δsl:Luc RNA was used. This RNA is devoid of a large stem-loop structure located downstream of the MTO1 region, which inhibited elongation of the reverse transcription reaction (data not shown). As shown in Figure 4C, AdoMet-dependent toeprint signals were observed downstream of the MTO1 region when GST:Ex1Δsl(WT):Luc RNA was translated in the presence of AdoMet, while no such signals were observed for the GST:Ex1Δsl(mto1-1):Luc RNA (Fig. 4C, lanes 1-4). When samples were incubated in the presence of EDTA or puromycin prior to the reverse transcription reaction, the AdoMet-dependent toeprint signals disappeared, suggesting that translating ribosomes are responsible for the toeprint signals (Fig. 4C, lanes 5-8). The most 5′-proximal AdoMet-dependent toeprint signal was detected at the 293rd nucleotide from the first AUG of the CGS1 exon 1 (Fig. 4C, arrowhead). This toeprint signal became more prominent when cycloheximide (CHX) was added after in vitro translation to inhibit the progression of translating ribosome during the reverse transcription reaction (Fig. 4C, lanes 9-12).
Based on the relationship between the positions of the toeprint signal and the P and A sites of eukaryotic ribosomes (Anthony and Merrick 1992; Law et al. 2001; Sachs et al. 2002), the toeprint signal at the 293rd nucleotide represents a ribosome that stalled when the Trp-93 and Ser-94 codons are located at the P and A sites, respectively (Fig. 4D). Taken together with the results of the stop codon substitution experiments, these results suggest that translation is arrested in response to AdoMet at the position where the A site of a ribosome resides at the Ser-94 codon.
Importance of the MTO1 region
The MTO1 region was identified by mto1 mutations and has been defined as the region whose amino acid sequence is important for the post-transcriptional regulation of CGS1 (Chiba et al. 1999; Ominato et al. 2002). To assess the importance of the MTO1 region in translation arrest, the effects of different alleles of mto1 mutation were tested (Fig. 5A,B). When GST:Ex1(mto1-2) or GST:Ex1(mto1-3) RNA was translated, the AdoMet-dependent peptidyl-RNA was not detected as in the case of GST:Ex1(mto1-1). In contrast, when GST:Ex1(mto1-4) RNA was used, the AdoMet-dependent peptidyl-RNA was observed, although less abundantly than in GST:Ex1(WT) RNA. In vivo, mto1-4 is a leaky allele, while mto1-1, mto1-2, and mto1-3 are stringent ones (Ominato et al. 2002). The data are consistent with the idea that the MTO1 region amino acid sequence is important for the translation arrest to occur. As for the RNA degradation, the 5′-truncated RNA was barely detectable with GST:Ex1 RNAs carrying mto1-1, mto1-2, and mto1-3 mutations, while a small amount of the 5′-truncated RNA was detected with GST:Ex1(mto1-4) RNA (data not shown).
Figure 5.
Amino acid changes affecting translation elongation arrest and determination of tRNA species in the arrest product. (A) Schematic representation of GST:Ex1 RNAs carrying mutations and GST:Ex1′ nonstop RNAs. (B) GST:Ex1 RNAs carrying different alleles of the mto1 mutation were translated for 30 min in the presence or absence of AdoMet, and the translation products were analyzed by immunoblot analysis with anti-GST antibody. (C) GST:Ex1 RNAs carrying amino acid substitutions or insertion were translated and analyzed as in B. The open arrow marks the AdoMet-dependent band shifted by substitution of Ser-94 to alanine (S94A). (D) Comparison of the mobility in SDS-PAGE. GST:Ex1 RNAs carrying alanine substitutions and GST:Ex1′ nonstop RNAs were translated and analyzed as in B. The samples were separated by SDS-PAGE before (upper) or after (lower) RNase A treatment. The solid and open arrows mark the peptidyl-tRNASer and peptidyl-tRNAAla, respectively. (E) In vitro translation in the presence of labeled tRNAs. 32P-labeled tRNASer(AGA) (left) or tRNALys(CUU) (right) (2 × 105 cpm) was preincubated for 10 min at 25°C in wheat germ extract without an RNA template. Then, GST:Ex1(WT) RNA was translated in a 20-μL reaction mixture for 30 min with (+) or without (-) AdoMet, and 5 μL of each sample was separated by SDS-PAGE. The radioactive signals were visualized by an image analyzer. Nonstop RNAs GST:Ex1′(S94-ns) (left) or GST:Ex1′(K92-ns) (right) was used as a positive control. Representative results of duplicate to triplicate experiments are shown.
The importance of the amino acid sequence of the MTO1 region in translation arrest was also shown by testing the effects of frameshift mutations (Supplementary Fig. S3).
Trp-93 is also important for translation arrest
Among the amino acid residues at around the translation arrest site, Trp-93 and Ser-94 are conserved among angiosperms, including both dicot and monocot plant species (Fig. 4D; Ominato et al. 2002). To test the importance of these residues, 92nd-95th codons of the CGS1 exon 1 were individually substituted with alanine (Fig. 5A), and their effects on the accumulation of the peptidyl-RNA were tested (Fig. 5C). Of these substitutions, only the substitution of alanine for Trp-93 (W93A) showed a prominent reduction in the accumulation of the AdoMet-induced peptidyl-RNA, although a faint band of the peptidyl-RNA was still observed. Substitution of glycine for Trp-93 (W93G) showed even stronger effect with virtually no detectable AdoMet-specific band (Fig. 5C). To test the importance of Trp-93 in vivo, we performed transfection experiments (Supplementary Fig. S4). The effects of these substitutions were intermediate of wild-type and mto1-1 mutation, with W93G exhibiting a stronger effect than W93A. The results suggest that, both in vivo and in vitro, Trp-93 is important for the translation arrest, although it may not be absolutely necessary.
To test the importance of distance between the MTO1 region and Trp-93, we inserted an alanine codon before Lys-92 (Ins92A). As shown in Figure 5C, Ins92A abolished accumulation of the AdoMet-dependent peptidyl-RNA, suggesting that the distance between the MTO1 region and the arrest site is also important for the translation arrest.
Determination of the RNA moiety of the peptidyl-RNA as tRNASer
Changing Ser-94 into alanine (S94A) did not substantially reduce the accumulation of the AdoMet-induced peptidyl-RNA; however, it did affect migration of the peptidyl-RNA in SDS-PAGE (Fig. 5C, open arrow). This difference in mobility is likely due to a difference in the tRNA species linked to the polypeptide, and implies that the RNA moiety of peptidyl-RNA is a tRNA decoding the Ser-94 codon. If our interpretation is correct, we may determine the tRNA species in peptidyl-tRNA by using this mobility difference.
We synthesized nonstop RNAs GST:Ex1′(S94-ns) and GST:Ex1′(S94A-ns), which are 3′-truncated forms of GST:Ex1 RNAs without a stop codon and terminated at the 94th codon of GST:Ex1(WT) and GST:Ex1(S94A), respectively (Fig. 5A). In vitro translation of GST:Ex1′(S94-ns) and GST:Ex1′(S94A-ns) RNAs should produce peptidyl-tRNASer and peptidyl-tRNAAla, respectively, and their production will be independent of the presence of AdoMet. We compared the mobility of the AdoMet-dependent peptidyl-RNA produced by GST:Ex1(WT) and GST:Ex1(S94A) RNAs with that of the nonstop RNAs. As shown in Figure 5D, upper panel, the AdoMet-dependent band produced by GST:Ex1(WT) RNA comigrated with peptidyl-tRNASer (Fig. 5D, solid arrow) produced by the GST:Ex1′(S94-ns) RNA (Fig. 5D, upper panel, lanes 3,4), and the one from GST:Ex1(S94A) RNA comigrated with peptidyl-tRNAAla (Fig. 5D, open arrow) produced by the GST:Ex1′(S94A-ns) RNA (Fig. 5D, upper panel, lanes 6,7). As a control, we synthesized GST:Ex1′(N95A-ns) RNA and compared the mobility of peptidyl-tRNAAla produced from this nonstop RNA with that from GST:Ex1(N95A) RNA. As expected, the AdoMet-dependent band produced from GST:Ex1(N95A) RNA did not comigrate with a product from the GST:Ex1′(N95A-ns) RNA (Fig. 5D, upper panel, lanes 9,10).
After RNase A treatment, the difference in mobility of the AdoMet-dependent band between the wild-type and S94A constructs was no longer discernible, indicating that the difference in mobility was indeed due to a difference in the RNA moieties of the peptidyl-RNA (Fig. 5D, lower panel). These results suggest that the AdoMet-dependent peptidyl-RNA consists of a tRNASer and a polypeptide produced by translation as far as the Ser-94 codon of the CGS1 exon 1. This result is consistent with the position of translation arrest determined by the stop codon substitution experiments and toeprint analysis.
Further evidence that the AdoMet-dependent peptidyl-RNA contains tRNASer was obtained from an in vitro translation analysis using labeled tRNAs. After preincubation of 32P-labeled tRNASer in wheat germ extract, in vitro translation was carried out for 30 min, and the radioactive signals were detected following SDS-PAGE (Fig. 5E). As a control, GST:Ex1′(K92-ns) RNA, a nonstop RNA terminated at Lys-92, was constructed (Fig. 5A) and used with 32P-labeled tRNALys. Translation of GST:Ex1′(K92-ns) RNA produces a 50-kDa band, instead of the 55-kDa band in GST:Ex1′(S94-ns) RNA (data not shown). When 32P-labeled tRNASer and tRNALys were used with GST:Ex1′(S94-ns) and GST:Ex1′(K92-ns) RNA, respectively, the labeled tRNA produced bands of the expected sizes, indicating that the in vitro-transcribed tRNAs are functional in the wheat germ extract system (Akama and Beier 2003). When 32P-labeled tRNASer was used with GST:Ex1(WT) RNA, a labeled AdoMet-dependent 55-kDa band was detected, while no labeled AdoMet-dependent band was detected when labeled tRNALys was used. These results indicate that the AdoMet-dependent peptidyl-RNA contains tRNASer.
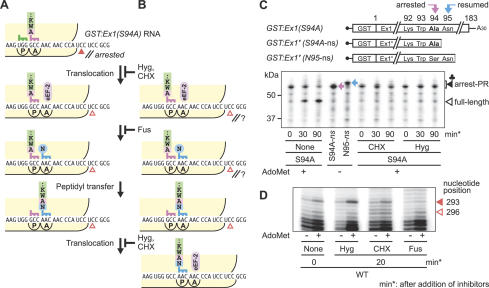
Translation elongation is arrested temporarily at the step of ribosome translocation
The results of the toeprint analyses suggested that the Ser-94 codon is located at the A site of the stalled ribosome, which implies that the peptidyl-tRNASer resides at the A site of the ribosome (Fig. 4D). If this is the case, translation elongation should be arrested at the step of ribosome translocation (Fig. 6A). To test this possibility, the effects of translocation inhibitors, hygromycin B (Hyg) and CHX, were examined. Hyg fixes peptidyl-tRNA in the A site (Bashan et al. 2003) and CHX inhibits eukaryotic elongation factor-2 (eEF-2)-dependent translocation (Fig. 6A,B; McKeehan and Hardesty 1969; Obrig et al. 1971). If translation elongation is arrested temporarily at the step of ribosome translocation, resumption of translation will be blocked by Hyg and CHX without proceeding to the next peptidyl transfer reaction (Fig. 6A). In contrast, if translation elongation is arrested temporarily at one of the steps following translocation, including the next peptidyl transfer step, one round of peptidyl transfer will still occur in the presence of Hyg or CHX upon resumption of translation (Fig. 6B).
Figure 6.
Determination of the arrested step in translation elongation. (A,B) Schematic representations of the ribosome stalled at the translocation step at Ser-94 (A) or at one of the steps after the translocation (B), and the reactions that follow the resumption of translation. The steps inhibited by CHX, Hyg, and Fus are indicated. The solid and open arrowheads represent the toeprint signals at the 293rd and 296th nucleotides, respectively. (C) Effects of translation elongation inhibitors on the first round of peptidyl transfer after the resumption of translation. GST:Ex1(S94A) RNA was translated in the presence of AdoMet. Edeine was added at 5 min. Hyg (1 mM) or CHX (1 mg/mL) was added at 30 min, and the reaction mixtures were further incubated as indicated. Positions of the peptidyl-tRNAAla (pink arrow) and peptidyl-tRNAAsn (blue arrow) are marked. (D) Effects of translation elongation inhibitors on toeprinting. GST:Ex1Δsl(WT):Luc RNA was translated in the presence (+) or absence (-) of AdoMet. Hyg (1 mM), CHX (1 mg/mL), or Fus (1 mM) was added at 30 min, and the reaction mixtures were incubated for additional 20 min prior to the primer extension reaction. The solid and open arrowheads mark the AdoMet-dependent toeprint signals at the 293rd and 296th nucleotides, respectively. Representative results of triplicate (C) or duplicate (D) experiments are shown.
In this experiment, GST:Ex1(S94A) RNA was used, and peptidyl transfer from peptidyl-tRNAAla to Asn-tRNAAsn was monitored using the difference in the mobility of peptidyl-tRNAAla (Fig. 6C, pink arrow) and peptidyl-tRNAAsn (blue arrow). Edeine was used to prevent new rounds of translation initiation. Hyg or CHX was added at 30 min and the mixture was incubated for 30 or 90 min more. As shown in Figure 6C, the amount of peptidyl-tRNA declined in the absence of a translocation inhibitor, indicating that translation resumes during the incubation, whereas after the addition of CHX or Hyg, the mobility and amount of the AdoMet-dependent peptidyl-tRNA did not change. The results indicate that the peptidyl transfer from peptidyl-tRNAAla to Asn-tRNA-Asn does not occur in the presence of translocation inhibitors, even when translation would normally resume.
Next, we examined the position of the stalled ribosome after incubation in the presence of translocation inhibitors by toeprint analysis. After GST:Ex1Δsl(WT): Luc RNA was translated for 30 min, the reaction mixture was incubated with a translocation inhibitor for 20 min before proceeding to the primer extension reaction. Incubation with CHX or Hyg did not affect the toeprint signal at the 293rd nucleotide (Fig. 6D, solid arrowhead). This indicates that Hyg and CHX immobilized the position of the stalled ribosomes, which is consistent with the result of the above immunoblot analysis. In contrast, after incubation with fusidic acid (Fus), which blocks dissociation of eEF-2 after translocation (Joseph 2003), the toeprint signal at the 293rd nucleotide was diminished, indicating that a large portion of the stalled ribosomes proceeded for at least one round of translocation during the 20-min incubation period (Fig. 6D). These results are consistent with the notion that translation elongation is arrested at the step of ribosome translocation (Fig. 6A).
Discussion
Translation elongation arrest is the primary event induced by AdoMet in CGS1 exon 1-mediated post-transcriptional regulation
In the present study, we demonstrated that peptidyl-tRNASer accumulates in response to AdoMet during in vitro translation of mRNA carrying CGS1 exon 1 and presented evidence suggesting that this peptidyl-tRNA is a translation intermediate produced by temporal translation arrest that occurred at the Ser-94 codon.
We have previously shown that CGS1 mRNA levels are regulated in a novel feedback mechanism that involves degradation of the mRNA in response to AdoMet (Chiba et al. 1999, 2003). A strong correlation between the translation arrest and mRNA degradation was observed in this study. In the stop codon substitution analyses, changing Ser-94 to a stop codon abolished the accumulation of both the peptidyl-tRNA and 5′-truncated RNA, while changing Asn-95 to a stop codon did not affect the accumulation of either. Substitution of Trp-93 with alanine or glycine also markedly reduced both the accumulation of the peptidyl-tRNA (Fig. 5C) and the 5′-truncated RNA in response to AdoMet in vitro (Y. Haraguchi, Y. Nagami, H. Onouchi, S. Naito, unpubl.).
Time course analyses showed that the peptidyl-tRNA accumulation precedes mRNA degradation. In addition, in vitro translation experiments using rabbit reticulocyte lysate reproduced the AdoMet-induced translation arrest, but not the 5′-truncated RNA production (H. Onouchi, M. Nakamoto, Y. Haraguchi, Y. Nagami, S. Naito, in prep.). These observations suggest that the translation arrest is the primary event induced by AdoMet in the CGS1 exon 1-mediated regulation.
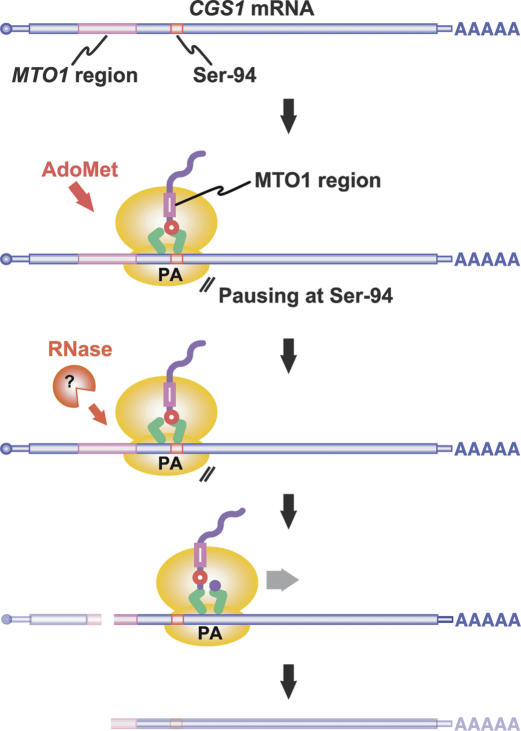
Based on these findings, we propose the following model for CGS1 exon 1-mediated post-transcriptional regulation (Fig. 7). The translation of CGS1 mRNA is arrested temporarily at the Ser-94 codon in response to AdoMet after the MTO1 region is translated. Following the translation arrest, mRNA degradation occurs near the 5′ edge of the stalled ribosome (Fig. 4D). The 5′-truncated RNA thus formed is detected as a degradation intermediate of CGS1 mRNA (Chiba et al. 1999, 2003; Lambein et al. 2003).
Figure 7.
Model for CGS1 exon 1-mediated post-transcriptional regulation. When the cellular concentration of AdoMet is high, translation of CGS1 mRNA is temporarily arrested when the A site of the ribosome resides at the Ser-94 codon of CGS1. The MTO1 region in the nascent polypeptide acts within the ribosomal exit tunnel to cause translation elongation arrest. Following the translation arrest, mRNA degradation occurs upstream of the stalled ribosome, resulting in production of the 5′-truncated RNA species. The ribonuclease (Pacman) responsible for the mRNA degradation remains to be identified.
Although we do not have direct in vivo evidence of translation arrest in CGS1 regulation, translational control was suggested by our previous data obtained from analysis of transgenic Arabidopsis carrying the CGS1 exon 1 fused to a reporter gene (Suzuki et al. 2001). In these transgenic plants, down-regulation of reporter activity was more prominent than the reduction of the amount of the full-length mRNA. This result implies that the CGS1 exon 1-mediated post-transcriptional regulation in vivo is also primarily a translational control. In support of this notion, the effects of strong and weak alleles of mto1, as well as W93A and W93G substitutions, on the translation arrest in vitro (Fig. 5B,C) paralleled those on the down-regulation of reporter activity in vivo (Supplementary Fig. S4; Chiba et al. 1999).
The MTO1 region in the nascent polypeptide acts within the ribosome to cause translation elongation arrest
The MTO1 region is the cis element in the post-transcriptional regulation of CGS1, and the amino acid changes in the MTO1 region abolish the regulation (Chiba et al. 1999; Ominato et al. 2002). RNAs carrying the mto1 mutations diminished accumulation of the translation arrest product. In addition, frameshift mutations covering the MTO1 region abolished accumulation of the translation arrest product (Supplementary Fig. S3). These results suggest that the amino acid sequence of the MTO1 region is responsible for the translation arrest in vitro. Together with our previous observations that the MTO1 region acts in cis (Chiba et al. 1999; Suzuki et al. 2001) and that ongoing translation is required for the CGS1 regulation (Lambein et al. 2003), we may conclude that the MTO1 region in the nascent polypeptide acts to inhibit translation elongation in the presence of AdoMet (Fig. 7).
In mammalian c-myc (Lemm and Ross 2002) and Saccharomyces cerevisiae MATα1 (Caponigro et al. 1993), degradation of mRNA coupled with translation elongation arrest has been reported. These mRNAs carry the cis determinant of mRNA instability within the coding region, and rare codons are responsible for the translational pausing. In CGS1, involvement of rare codons is unlikely, because, in addition to the fact that Asn-95 is not a rare codon in Arabidopsis, the in vitro translation system used in this study contains an ample amount of added tRNA. Rather, the nascent peptide of the MTO1 region would have a role in inducing the translation arrest in CGS1.
In both prokaryotes and eukaryotes, nascent peptide-mediated translation arrest has been reported (Gong et al. 2001; Nakatogawa and Ito 2001; for reviews, see Lovett and Rogers 1996; Morris and Geballe 2000). Among these systems, the nascent polypeptide encoded by the secM gene of Escherichia coli has been suggested to interact with components of the ribosomal exit tunnel, L22 protein and 23S rRNA, to cause translation elongation arrest (Nakatogawa and Ito 2002). A similar interaction between the nascent polypeptide and the ribosomal exit tunnel has also been suggested for the E. coli tnaC gene (Gong and Yanofsky 2002). Since a ribosome covers ∼40 amino acid residues of the nascent peptide (Matlack and Walter 1995), a simple interpretation of our results is that the MTO1 region resides in the ribosomal exit tunnel when translation is arrested at Ser-94 (Fig. 7). An intriguing implication of this notion is that the MTO1 region acts within the exit tunnel to cause translation arrest. Our observation that the distance between the MTO1 region and the translation arrest site is important (Fig. 5C) argues for this possibility.
CGS1 is unique among the regulatory systems involving nascent peptide-mediated translation arrest
In some nascent peptide-mediated regulatory systems in bacteria, such as the catA86, cmlA, and secM genes, a ribosome stalls in the middle of the ORF as observed in CGS1 (Lovett and Rogers 1996; Nakatogawa and Ito 2001). In contrast, in all the known natural examples in eukaryotes, the ribosome stalls at the stop codon of the upstream ORF (uORF), i.e., translation is arrested at the step of translation termination (Cao and Geballe 1996; Wang and Sachs 1997; Law et al. 2001). In fact, in the uORFs of the cytomegalovirus gpUL4 and mammalian AdoMet decarboxylase genes, the ribosome stalling has been shown to be dependent on the stop codon of the uORF (Cao and Geballe 1995; Mize et al. 1998; Janzen et al. 2002).
Unlike the uORF systems of gpUL4 and AdoMet decarboxylase genes, the nascent peptide encoded by the uORF of Neurospora crassa arg-2 can still cause ribosome stalling when the uORF is fused in-frame to a reporter gene (Wang et al. 1998; Fang et al. 2004). This artificial arg-2 and the CGS1 exon 1 systems are the only examples of nascent peptide-mediated translation elongation arrest in eukaryotes. In the arg-2 system, however, no link to mRNA degradation has been reported.
Nascent peptide-mediated mRNA cleavage has been reported in E. coli daaP (Loomis et al. 2001) and Bacillus subtilis ermC (Drider et al. 2002) genes. In addition, examples of ribosome stalling coupled with mRNA cleavage have been reported recently in E. coli, although mRNA cleavage occurs at the A site of the stalled ribosome (Hayes and Sauer 2003; Sunohara et al. 2004a,b).
The only nascent peptide-mediated mRNA degradation reported in eukaryotes other than CGS1 is the mammalian β-tubulin gene. The N-terminal tetrapeptide of the nascent β-tubulin acts in cis to feedback regulate its own mRNA instability in response to unassembled tubulin subunits. Unlike CGS1 regulation, the regulatory domain of β-tubulin must be located at the N terminus, and acts after emerging from the ribosome, as the β-tubulin mRNA needs to be translated for >40 codons from the N terminus (Theodorakis and Cleveland 1993).
Translation elongation of CGS1 mRNA is arrested at the step of ribosomal translocation
The relative positions of the stalled ribosome and the peptidyl-tRNA showed that at least the anticodon end of the peptidyl-tRNA resides at the A site of the ribosome (Fig. 4D), which suggests that translation elongation is arrested at the step of ribosome translocation. Our analyses using translocation inhibitors also supported the translation arrest at this step (Fig. 6A). To the best of our knowledge, this is the first report of nascent peptide-mediated inhibition of translation elongation at the step of ribosomal translocation.
During translocation, large conformational changes occur in the ribosome (for review, see Wilson and Noller 1998). We speculate that the MTO1 region in the nascent polypeptide inhibits such conformational changes in the presence of AdoMet by acting within a ribosome. Alternatively, the nascent polypeptide could directly inhibit movement of the peptidyl-tRNA from the A site to the P site of a ribosome.
Although the mechanism of how the MTO1 region causes translation elongation arrest remains to be elucidated, the data presented in this study revealed a novel mechanism of nascent peptide-mediated post-transcriptional regulation in eukaryotes. This includes an AdoMet-induced translation elongation arrest at the step of ribosome translocation, which is followed by CGS1-specific mRNA degradation.
Materials and methods
Chemicals
AdoMet and Fus were purchased from Sigma-Aldrich. CHX and Hyg were obtained from Wako Pure Chemicals. Edeine was a gift from the National Cancer Institute. The other chemicals used are listed in Chiba et al. (2003).
Plasmid construction
pMI21(WT) and pMI21(mto1-1), which carry the Ex1(WT):Luc and Ex1(mto1-1):Luc DNA in the pSP64 Poly(A) vector (Promega), respectively, were described previously (Chiba et al. 2003).
Plasmid pYN10(WT) carries the GST:Ex1(WT):Luc DNA in the pSP64 Poly(A) vector. To construct pYN10(WT), the GST coding region was amplified by PCR from pGEX-6P-1 (Amersham) using primers GSTf and GSTr (Supplementary Table S1), and the amplified fragment was digested with XbaI and inserted into the XbaI site of pMI21(WT). pYN10(mto1-1) was constructed by inserting the XbaI-digested GST fragment of pYN10(WT) into pMI21(mto1-1).
Plasmid pMN1(WT) harbors the GST:Ex1(WT) DNA in the pSP64 Poly(A) vector. To generate this plasmid, complementary oligonucleotides Ex1stop1 and Ex1stop2 (Supplementary Table S1) and the SphI-BamHI fragment containing GST:Ex1(WT) from pYN10(WT) or Ex1(WT) from pMI21(WT) were ligated and inserted between the SphI and SacI sites of a pSP64 Poly(A) vector. mto1-1, mto1-2, mto1-3, and 4 mutant versions of pMN1 were constructed by replacing a 0.37 kbp AflII-BamHI fragment of pMN1(WT) with a corresponding AflII-BamHI fragment of pMI21(mto1-1), pMI21(mto1-2), pMI21(mto1-3), and pMI21(mto1-4), respectively.
pOH31(WT) and pOH31(mto1-1) carry the GST:Ex1Δsl(WT): Luc and GST:Ex1Δsl(mto1-1):Luc DNA, respectively, which lack the large stem-loop structure located downstream of the MTO1 region. To construct pOH31(WT) and pOH31(mto1-1), the 0.5-kbp BglII-NcoI fragment of pYN10(WT) was replaced with BglII- and NcoI-digested PCR products amplified from pYN10(WT) and pYN10(mto1-1), respectively, using primers Ex1P1 (Chiba et al. 1999) and CD3Xr (Supplementary Table S1).
Stop codon (TAG) and alanine substitution mutagenesis was accomplished by the overlap extension PCR method (Ho et al. 1989; Pogulis et al. 1996). In the K92A and W93A constructs, the Lys-92 and Trp-93 codons were changed to GCG (Ala). In the S94A and N95A constructs, the Ser-94 and Asn-95 codons were changed to GCC (Ala). In the W93G construct, the Trp-93 codon was changed to GGG (Gly). In the Ins92A construct, a GCA (Ala) codon was inserted between Ala-91 and Lys-92. The exon 1 fragments were recloned into pMN1(WT) (alanine and glycine substitutions and Ins92A) or pYN10(WT) (stop codon substitutions). Sequences of the primers used will be provided upon request.
Templates for 32P-labeled tRNA were prepared as follows. Plasmids carrying Arabidopsis tRNALys(CUU) and tRNASer(AGA) genes (Beier et al. 1991; Chen et al. 1998) in pBluescript II KS(+) vector were kindly provided by K. Akama (Shimane University, Matsue, Japan). Fragments harboring a T7 promoter and each tRNA gene were amplified by PCR from these plasmids using oligonucleotides Ser(AGA)f and Ser(AGA)r, and Lys(CUU)f and Lys(CUU)r, respectively, digested with EcoRI, and inserted into the EcoRI site of pUC19. The resulting plasmids pMN3 and pMN4 carry tRNALys(CUU) and tRNASer(AGA) genes, respectively.
In all of the constructs, the integrity of the PCR-amplified regions was confirmed by sequence analysis.
In vitro transcription
DNA templates in pSP64 poly(A) vector (Promega) were linearized with EcoRI and purified as described (Chiba et al. 2003). Templates for nonstop RNAs were prepared by amplifying the corresponding region from pMN1(WT), pMN1(S94A), and pMN1(N95A) by PCR with KOD-plus DNA polymerase (Toyobo). For PCR amplification, forward primer SP65′fP and reverse primers S94r, S94Ar, N95r, N95Ar, and K92r (Supplementary Table S1) were used to synthesize GST:Ex1′(S94-ns), GST:Ex1′(S94A-ns), GST:Ex1′(N95-ns), GST:Ex1′(N95A-ns), and GST:Ex1′(K92-ns) RNAs, respectively. The amplified fragments were purified using a QIAquick PCR Purification kit (Qiagen). In vitro transcription in the presence of a cap analog m7G[5′]ppp[5′]GTP was accomplished as described (Chiba et al. 2003).
In vitro translation and immunoblot analysis
In vitro translation in wheat germ extract (Promega) was carried out at 25°C as described (Chiba et al. 2003). AdoMet was added at 1 mM. For immunoblotting, 1 pmol GST:Ex1 or GST:Ex1:Luc RNA, or 2 pmol GST:Ex1′ nonstop RNAs were translated in a 20 μL reaction mixture. The products were separated on a NuPAGE 4%-12% Bis-Tris Gel (Invitrogen), transferred to a Hybond-ECL nitrocellulose membrane (Amersham), probed with a polyclonal anti-GST antibody (Sigma-Aldrich or Zymed), and visualized using ECL plus the Immunoblotting Detection System (Amersham). For RNase A treatment, RNase A was added at 0.5 mg/mL and incubated for 15 min at 37°C.
Pulse-chase experiment
[35S]methionine (37 TBq/mmol; Amersham) was added instead of unlabeled methionine at a final concentration of 3.7 kBq/μL (0.1 μM). In vitro translation was carried out with 50 fmol/μL RNA. Edeine was used to inhibit new rounds of translation.
Sucrose density gradient centrifugation
A sucrose density gradient (10%-30% [w/v], 1.8 mL) in buffer B (50 mM Tris-HCl at pH 6.5, 25 mM KCl, 10 mM MgCl2; modified from Davies and Abe 1995) was layered over 0.2 mL of 50% (w/v) sucrose cushion in buffer B. After in vitro translation of 25 pmol GST:Ex1 RNA in a 50-μL reaction mixture, the sample was diluted with 150 μL buffer B, layered over the sucrose gradient, and centrifuged in a Beckman TLS-55 rotor at 30,000 rpm (∼60,000 × g) for 65 min at 4°C. Four-drop fractions were collected using the ISCO 520C density gradient system.
Northern analysis
GST:Ex1:Luc RNA (1.2 pmol) was translated in a 600-μL reaction mixture. RNA extraction and Northern blot analysis were as described (Suzuki et al. 2001; Chiba et al. 2003) except that 1.0% (w/v) agarose-formaldehyde gels were used. The LUC 3′ probe was described previously (Chiba et al. 2003).
Toeprint analysis
After in vitro translation of 200 fmol GST:Ex1Δsl:Luc RNA in a 20-μL reaction mixture, toeprint analysis was performed as described by Wang and Sachs (1997). The oligonucleotide LucTP3 (Supplementary Table S1) was labeled at its 5′ terminus with T4 polynucleotide kinase (Takara) and [γ-32P]ATP (110 TBq/mmol; Amersham) and was purified using a QIAquick Nucleotide Removal kit (Qiagen). Reverse transcription samples were separated on an 8% polyacrylamide/7 M urea gel. DNA sequence ladders were prepared using the 5′-32P-labeled LucTP3 primer, the pOH31 plasmid as a template, and a Takara Taq Cycle Sequencing kit. The gel image was analyzed using a Fuji BAS 1000 Bio Image Analyzer.
Preparation of labeled tRNA
The plasmids pMN3 and pMN4 carrying the Arabidopsis tRNALys(CUU) and tRNASer(AGA) genes, respectively, were digested with MvaI to generate a CCA trinucleotides end at their 3′ termini, and used as a template for in vitro transcription. 32P-labeled tRNAs were prepared by in vitro transcription in the presence of [α-32P]CTP (110 TBq/mmol; Amersham) using a T7 MEGA Shortscript High Yield Transcription kit (Ambion). Intact tRNA bands were excised from the gel by electroelution after separating on an 8% polyacrylamide/8 M urea gel.
Acknowledgments
We are grateful to Dr. Kazuhito Akama for valuable discussion and clones of the Arabidopsis tRNA genes, Drs. Toshiyuki Fukuhara and Hans Bohnert for ice plant seeds, Dr. Derek B. Goto for critical reading of the manuscript, Dr. Yukako Chiba for her contribution during the initial stages of this study as well as for valuable discussions, Ms. Saeko Yasokawa for skillful technical assistance, and Ms. Kumi Fujiwara for general assistance. We used the Radioisotope Laboratory of the Graduate School of Agriculture, Hokkaido University. This work was supported in part by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan (12138201, 13440233, and 17026001 to S.N., and 14035208 to H.O.). R.S. is supported by the Japan Society for the Promotion of Science.
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1317105.
References
- Akama K. and Beier, H. 2003. Translational nonsense codon suppression as indicator for functional pre-tRNA splicing in transformed Arabidopsis hypocotyl-derived calli. Nucleic Acids Res. 31: 1197-1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony D.D. and Merrick, W.C. 1992. Analysis of 40 S and 80 S complexes with mRNA as measured by sucrose density gradients and primer extension inhibition. J. Biol. Chem. 267: 1554-1562. [PubMed] [Google Scholar]
- Bashan A., Zarivach, R., Schluenzen, F., Agmon, I., Harms, J., Auerbach, T., Baram, D., Berisio, R., Bartels, H., Hansen, H.A., et al. 2003. Ribosomal crystallography: Peptide bond formation and its inhibition. Biopolymers 70: 19-41. [DOI] [PubMed] [Google Scholar]
- Beier D., Stange, N., Gross, H.J., and Beier, H. 1991. Nuclear tRNA-Tyr genes are highly amplified at a single chromosomal site in the genome of Arabidopsis thaliana. Mol. Gen. Genet. 225: 72-80. [DOI] [PubMed] [Google Scholar]
- Cao J. and Geballe, A.P. 1995. Translational inhibition by a human cytomegalovirus upstream open reading frame despite inefficient utilization of its AUG codon. J. Virol. 69: 1030-1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 1996. Coding sequence-dependent ribosomal arrest at termination of translation. Mol. Cell. Biol. 16: 603-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caponigro G. and Parker, R. 1996. mRNA turnover in yeast promoted by the MATα1 instability element. Nucleic Acids Res. 24: 4304-4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caponigro G., Muhlrad, D., and Parker, R. 1993. A small segment of the MATα1 transcript promotes mRNA decay in Saccharomyces cerevisiae: A stimulatory role for rare codons. Mol. Cell. Biol. 13: 5141-5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrasco L., Fernandez-Puentes, C., and Vazquez, D. 1976. Antibiotics and compounds affecting translation by eukaryotic ribosomes. Mol. Cell. Biochem. 16: 97-122. [DOI] [PubMed] [Google Scholar]
- Chen Z., Ulmasov, B., and Folk, W.R. 1998. Nonsense and missense translational suppression in plant cells mediated by tRNALys. Plant Mol. Biol. 36: 163-170. [DOI] [PubMed] [Google Scholar]
- Chiba Y., Ishikawa, M., Kijima, F., Tyson, R.H., Kim, J., Yamamoto, A., Nambara, E., Leustek, T., Wallsgrove, R.M., and Naito, S. 1999. Evidence for autoregulation of cystathionine γ-synthase mRNA stability in Arabidopsis. Science 286: 1371-1374. [DOI] [PubMed] [Google Scholar]
- Chiba Y., Sakurai, R., Yoshino, M., Ominato, K., Ishikawa, M., Onouchi, H., and Naito, S. 2003. S-adenosyl-L-methionine is an effector in the posttranscriptional autoregulation of the cystathionine γ-synthase gene in Arabidopsis. Proc. Natl. Acad. Sci. 100: 10225-10230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies E. and Abe, S. 1995. Methods for isolation and analysis of polyribosomes. In Methods in cell biology (eds., D.W. Galbraith et al.), pp. 209-222. Academic Press, San Diego, CA. [DOI] [PubMed]
- Drider D., DiChiara, J.M., Wei, J., Sharp, J.S., and Bechhofer, D.H. 2002. Endonuclease cleavage of messenger RNA in Bacillus subtilis. Mol. Microbiol. 43: 1319-1329. [DOI] [PubMed] [Google Scholar]
- Fang P., Spevak, C.C., Wu, C., and Sachs, M.S. 2004. A nascent polypeptide domain that can regulate translation elongation. Proc. Natl. Acad. Sci. 101: 4059-4064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong F. and Yanofsky, C. 2002. Instruction of translating ribosome by nascent peptide. Science 297: 1864-1867. [DOI] [PubMed] [Google Scholar]
- Gong F., Ito, K., Nakamura, Y., and Yanofsky, C. 2001. The mechanism of tryptophan induction of tryptophanase operon expression: Tryptophan inhibits release factor-mediated cleavage of TnaC-peptidyl-tRNAPro. Proc. Natl. Acad. Sci. 98: 8997-9001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes C.S. and Sauer, R.T. 2003. Cleavage of the A site mRNA codon during ribosome pausing provides a mechanism for translational quality control. Mol. Cell 12: 903-911. [DOI] [PubMed] [Google Scholar]
- Ho S.N., Hunt, H.D., Horton, R.M., Pullen, J.K., and Pease, L.R. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene (Amst.) 77: 51-59. [DOI] [PubMed] [Google Scholar]
- Janzen D.M., Frolova, L., and Geballe, A.P. 2002. Inhibition of translation termination mediated by an interaction of eukaryotic release factor 1 with a nascent peptidyl-tRNA. Mol. Cell. Biol. 22: 8562-8570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph S. 2003. After the ribosome structure: How does translocation work? RNA 9: 160-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. and Leustek, T. 1996. Cloning and analysis of the gene for cystathionine γ-synthase from Arabidopsis thaliana. Plant Mol. Biol. 32: 1117-1124. [DOI] [PubMed] [Google Scholar]
- Lambein I., Chiba, Y., Onouchi, H., and Naito, S. 2003. Decay kinetics of autogenously regulated CGS1 mRNA that codes for cystathionine γ-synthase in Arabidopsis thaliana. Plant Cell Physiol. 44: 893-900. [DOI] [PubMed] [Google Scholar]
- Law G.L., Raney, A., Heusner, C., and Morris, D.R. 2001. Polyamine regulation of ribosome pausing at the upstream open reading frame of S-adenosylmethionine decarboxylase. J. Biol. Chem. 276: 38036-38043. [DOI] [PubMed] [Google Scholar]
- Lemm I. and Ross, J. 2002. Regulation of c-myc mRNA decay by translational pausing in a coding region instability determinant. Mol. Cell. Biol. 22: 3959-3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loomis W.P., Koo, J.T., Cheung, T.P., and Moseley, S.L. 2001. A tripeptide sequence within the nascent DaaP protein is required for mRNA processing of a fimbrial operon in Escherichia coli. Mol. Microbiol. 39: 693-707. [DOI] [PubMed] [Google Scholar]
- Lovett P.S. and Rogers, E.J. 1996. Ribosome regulation by the nascent peptide. Microbiol. Rev. 60: 366-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matlack K.E. and Walter, P. 1995. The 70 carboxyl-terminal amino acids of nascent secretory proteins are protected from proteolysis by the ribosome and the protein translocation apparatus of the endoplasmic reticulum membrane. J. Biol. Chem. 270: 6170-6180. [DOI] [PubMed] [Google Scholar]
- Matthews B.F. 1999. Lysine, thereonine, and methionine biosynthesis. In Plant amino acids: Biochemistry and biotechnology (ed., B.K. Singh), pp. 205-225. Marcel Dekker, New York.
- McKeehan W. and Hardesty, B. 1969. The mechanism of cycloheximide inhibition of protein synthesis in rabbit reticulocytes. Biochem. Biophys. Res. Commun. 36: 625-630. [DOI] [PubMed] [Google Scholar]
- Mize G.J., Ruan, H.J., Low, J.J., and Morris, D.R. 1998. The inhibitory upstream open reading frame from mammalian S-adenosylmethionine decarboxylase mRNA has a strict sequence specificity in critical positions. J. Biol. Chem. 273: 32500-32505. [DOI] [PubMed] [Google Scholar]
- Morris D.R. and Geballe, A.P. 2000. Upstream open reading frames as regulators of mRNA translation. Mol. Cell. Biol. 20: 8635-8642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatogawa H. and Ito, K. 2001. Secretion monitor, SecM, undergoes self-translation arrest in the cytosol. Mol. Cell 7: 185-192. [DOI] [PubMed] [Google Scholar]
- ____. 2002. The ribosomal exit tunnel functions as a discriminating gate. Cell 108: 629-636. [DOI] [PubMed] [Google Scholar]
- Obrig T.G., Culp, W.J., McKeehan, W.L., and Hardesty, B. 1971. The mechanism by which cycloheximide and related glutarimide antibiotics inhibit peptide synthesis on reticulocyte ribosomes. J. Biol. Chem. 246: 174-181. [PubMed] [Google Scholar]
- Ominato K., Akita, H., Suzuki, A., Kijima, F., Yoshino, T., Yoshino, M., Chiba, Y., Onouchi, H., and Naito, S. 2002. Identification of a short highly conserved amino acid sequence as the functional region required for posttranscriptional autoregulation of the cystathionine γ-synthase gene in Arabidopsis. J. Biol. Chem. 277: 36380-36386. [DOI] [PubMed] [Google Scholar]
- Pogulis R.J., Vallejo, A.N., and Pease, L.R. 1996. In vitro recombination and mutagenesis by overlap extension PCR. Methods Mol. Biol. 57: 167-191. [DOI] [PubMed] [Google Scholar]
- Sachs M.S., Wang, Z., Gaba, A., Fang, P., Belk, J., Ganesan, R., Amrani, N., and Jacobson, A. 2002. Toeprint analysis of the positioning of translation apparatus components at initiation and termination codons of fungal mRNAs. Methods 26: 105-114. [DOI] [PubMed] [Google Scholar]
- Sunohara T., Jojima, K., Tagami, H., Inada, T., and Aiba, H. 2004a. Ribosome stalling during translation elongation induces cleavage of mRNA being translated in Escherichia coli. J. Biol. Chem. 279: 15368-15375. [DOI] [PubMed] [Google Scholar]
- Sunohara T., Jojima, K., Yamamoto, Y., Inada, T., and Aiba, H. 2004b. Nascent-peptide-mediated ribosome stalling at a stop codon induces mRNA cleavage resulting in nonstop mRNA that is recognized by tmRNA. RNA 10: 378-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki A., Shirata, Y., Ishida, H., Chiba, Y., Onouchi, H., and Naito, S. 2001. The first exon coding region of cystathionine γ-synthase gene is necessary and sufficient for downregulation of its own mRNA accumulation in transgenic Arabidopsis thaliana. Plant Cell Physiol. 42: 1174-1180. [DOI] [PubMed] [Google Scholar]
- Theodorakis N.G. and Cleveland, D.W. 1993. Translationally coupled degradation of tubulin mRNA. In Control of messenger RNA stability (ed., J.G. Belasco and G. Brawerman), pp. 219-238. Academic Press, New York.
- Wang Z. and Sachs, M.S. 1997. Ribosome stalling is responsible for arginine-specific translational attenuation in Neurospora crassa. Mol. Cell. Biol. 17: 4904-4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Fang, P., and Sachs, M.S. 1998. The evolutionarily conserved eukaryotic arginine attenuator peptide regulates the movement of ribosomes that have translated it. Mol. Cell. Biol. 18: 7528-7536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson K.S. and Noller, H.F. 1998. Molecular movement inside the translational engine. Cell 92: 337-349. [DOI] [PubMed] [Google Scholar]