Abstract
Huntington's disease is a progressive neurodegenerative disorder caused by a polyglutamine repeat expansion in the first exon of the huntingtin (Htt) protein. N-terminal Htt peptides with polyglutamine tracts in the pathological range (51–122 glutamines) form high-molecular-weight protein aggregates with fibrillar morphology in vitro, and they form discrete inclusion bodies in a cell-culture model. However, in some studies, formation of discrete Htt inclusions does not correlate well with cell death. We coexpressed N-terminal Htt fragments containing 91 glutamines fused to different affinity tags in HEK293 cells, and we isolated small aggregates by double sequential-affinity chromatography to assure the isolation of multimeric molecules. Transmission electron microscopy and atomic force microscopy revealed the isolated aggregates as globules or clusters of globules 4–50 nm in diameter without any detectable fibrillar species. Because small nonfibrillar oligomers, not mature fibrils, recently have been suggested to be the principal cytotoxic species in neurodegenerative disease, these Htt globular aggregates formed in cells may represent the pathogenic form of mutant Htt.
Keywords: Huntington's disease, protein purification
Huntington's disease (HD) is a progressive autosomal dominant neurodegenerative disorder characterized by psychiatric manifestations, motor impairment, and dementia (1). It is associated with selective neuronal cell death occurring primarily in the cortex and striatum (2). The genetic lesion that underlies HD is the expansion of CAG triplet repeats coding for polyglutamine stretches in huntingtin (Htt), a 350-kDa protein of unknown function (3). Mutant Htt containing expanded polyglutamine tracts forms aggregates in vitro and in cultured cells, whereas the wild-type protein, with <35–40 glutamines, does not (4–7). The length of polyglutamine tracts in Htt exhibits a strong inverse correlation with the age of onset of HD and with the frequency of microscopically visible inclusion bodies in neocortex and in cell-culture models (6–8). Increased vulnerability of affected neurons in HD is mimicked by increased susceptibility to apoptotic stress of cultured cells expressing mutant Htt (4, 8). Thus, the cell-culture model is useful to examine the molecular mechanism by which aggregation induces cell damage. However, a direct toxic role for polyglutamine aggregation has been questioned, because of the imperfect correlation between microscopically detectable large inclusion bodies and pathology (9–18).
Examination of HD postmortem brain reveals intranuclear and cytoplasmic neuronal inclusions and other deposits consisting of fibrillar material. In vitro, expanded Htt also forms fibrillar aggregates with morphological and biophysical properties similar to those formed by amyloid (Aβ) peptide (19). However, mounting evidence suggests a role for nonfibrillar Aβ aggregates in the pathogenesis of Alzheimer's disease (20–26). Nonfibrillar peptide species are also implicated in Lewy body-associated disorders, such as Parkinson's disease (27, 28). Poirier et al. recently observed that globular and protofibrillar intermediates of Htt participate in the genesis of mature fibrils in vitro (29). It is possible that the pathway from soluble Htt to fibrillar aggregates is also a multistep process in vivo, with the toxic species formed before the mature fiber. However, small nonfibrillar aggregates formed in vivo have not been characterized.
In this study, we isolated Htt aggregates from cultured cells by using a variant of tandem-affinity chromatography (30). Transmission electron microscopy (TEM) and atomic force microscopy (AFM) reveal that aggregates isolated from cultured cells appear as small globular structures or clusters of globules of remarkably consistent dimensions that lack detectable filamentous material. We used the same method to isolate small aggregates of mutant cystic fibrosis transmembrane conductance regulator (CFTR), a highly aggregation-prone polytopic membrane protein that also formed globular aggregates indistinguishable from the Htt aggregates. These data indicate that tandem-affinity chromatography is a powerful approach to isolate oligomeric protein complexes such as protein aggregates and reveal that aggregates isolated from cells share consistent physical shape and dimensions.
Materials and Methods
Constructs. pQ91/GFP (3) was kindly provided by G. M. Lawless (University of California, Los Angeles). All constructs were cloned into pcDNA 3.1 (Invitrogen) or pEGFP (Clontech).
Cell Culture and Transfection. HEK293 cells were maintained in DMEM supplemented with 10% FBS, penicillin G, and streptomycin. Transfection was performed by using TransIT-LT1 (Mirus, Madison, WI) according to the manufacturer's instructions.
Isolation of Aggregates. For isolation of aggregates under nondenaturing conditions, the modified tandem-affinity purification (TAP) method (30) was applied. An outline of the procedure is shown in Fig. 1b, and all manipulations were done at 4°C unless indicated otherwise. HEK293 cells (6 × 107) cotransfected with pQ91/GFP, pQ91/ProtA, and pQ91/CBP were incubated for 48 h. The harvested cell pellets were homogenized in ice-cold buffer A (50 mM Tris·HCl, pH 7.5/1 μg/ml leupeptin/1 mM PMSF) by 30 strokes with Dounce homogenizer and centrifuged at 100,000 × g for 30 min. The pellet was sonicated for 20 s in 1.6 ml of buffer B (10 mM Tris·HCl, pH 8.0/150 mM NaCl/0.1% Nonidet P-40/1 μg/ml leupeptin). The suspension was centrifuged at 700 × g for 5 min, and the resulting supernatant was incubated with 200 μl of IgG Sepharose 6 Fast Flow (Amersham Biosciences) for 2 h. The resin was washed with buffer B extensively and then incubated with 100 units of TEV protease (Invitrogen) in buffer B containing 1 mM DTT and 0.5 mM EDTA at 16°C for 2 h. The eluate obtained by collecting the column flow-through was incubated with 3 ml of buffer C (10 mM Tris·HCl, pH 8.0/150 mM NaCl/1 μg/ml leupeptin/1 mM Mg acetate/1 mM imidazole/2 mM CaCl2/0.1% Nonidet P-40/1 mM DTT) and 200 μl of calmodulin beads (Stratagene) for 2 h. After the resin was washed sequentially with buffer C and buffer D (10 mM Tris·HCl, pH 8.0/1 mM Mg acetate/1 mM imidazole/2 mM CaCl2/1 mM DTT), 200-μl fractions were eluted with buffer E (10 mM Tris·HCl, pH 8.0/1 mM Mg acetate/1 mM imidazole/2 mM EGTA/1 mM DTT).
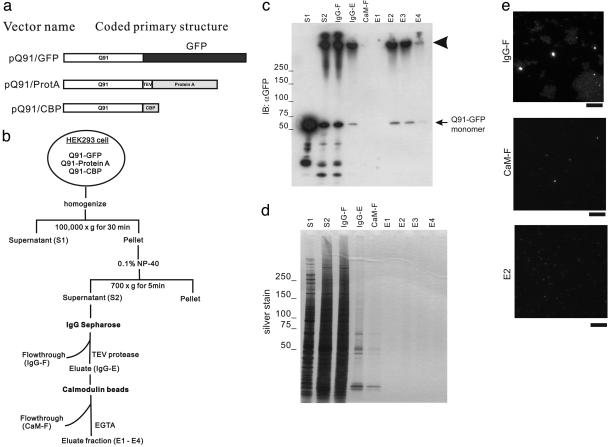
Fig. 1.
Isolation of HDQ91 aggregates under nondenaturing conditions. (a) Constructs. The GFP tag is indicated by a black box, and affinity tags (protein A and CBP) for isolation of HDQ91 aggregates are indicated by gray boxes. Q91, Htt exon 1 (3) protein with 91 glutamine repeats; TEV, TEV protease recognition sequence. (b) Diagram showing the isolation procedure for HDQ91 aggregates under nondenaturing conditions. (c) Immunoblotting (IB) of column fractions was performed using anti-GFP antibody (JL-8). Molecular mass markers are indicated in kDa at the left. The arrowhead indicates the bottom of the loading wells. (d) Silver staining of column fractions. The same amount of each indicated column fraction from c was applied to SDS/PAGE, and silver stained. (e) Fluorescence microscopy of column fractions. The individual fractions are as indicated in b. (Scale bar, 10 μm.)
An outline of the isolation procedure in the presence of urea is shown in Fig. 4b. HEK293 cells (6 × 107) cotransfected with pQ91/His and either pFLAG/S/Q91 or pHA/S/Q91 were incubated for 48 h. The harvested cell pellets were homogenized in ice-cold buffer A by Dounce homogenizer for 30 strokes and centrifuged at 100,000 × g for 30 min. The pellet was sonicated for 20 s in buffer F (20 mM Tris·HCl, pH 8.0/100 mM NaCl) containing 2 M urea. The suspension was centrifuged at 700 × g for 5 min, and the supernatant was incubated with 400 μl of S protein agarose (Novagen) for 1 h. After washing the resin with buffer F, the sample was eluted with buffer F containing 8 M urea. The 2.4-ml eluate was incubated with 200 μl of Ni-NTA agarose (Qiagen) with 20 mM imidazole for 1 h. After the resin was washed with buffer F containing 8 M urea and 50 mM imidazole, the 200-μl fractions were eluted with buffer F containing 8 M urea and 250 mM imidazole.
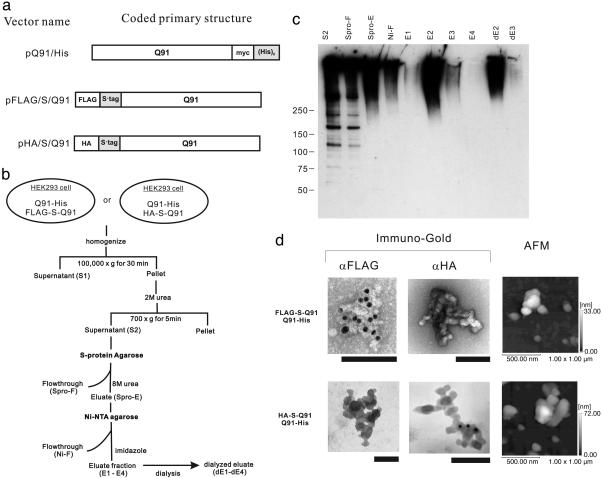
Fig. 4.
Isolation of HDQ91 aggregates in the presence of urea. (a) Constructs. Affinity tags [(His)6 tag and S·tag] for isolation of HDQ91 aggregates are indicated by gray boxes. Q91, Htt exon 1 (11) protein with 91 glutamine repeats; myc, myc tag; HA, HA tag; FLAG, FLAG tag. (b) Diagram showing the isolation step of HDQ91 aggregates in the presence of urea. (c) Immunoblotting of column fractions was performed by using anti-FLAG (M2) antibody. The individual fractions are as indicated in b. Molecular mass markers (in kDa) are indicated on the left. (d) TEM and AFM of HDQ91 aggregates isolated in the presence of urea. The size of the gold particles was 10 nm. (Scale bar, 100 nm.) The composition of the aggregates is indicated on the left.
Antibodies. The monoclonal antibody JL-8 and polyclonal antibody against GFP were purchased from Clontech. Anti-HA monoclonal antibody 12CA5 was purchased from Roche, and anti-FLAG monoclonal antibody M2 was purchased from Sigma. Goat F(ab′)2 anti-rabbit or anti-mouse IgG conjugated with 10-nm gold (British Biocell International, Cardiff).
Electrophoresis and Immunoblotting. Aliquots of the fractions were mixed with SDS sample buffer (31) and applied to 5–15% gradient precast polyacrylamide gels (Bio-Rad). Silver staining of the gel was performed by using the silver-staining kit II (WAKO Pure Chemical, Osaka). Immunoblotting was performed by using poly(vinylidene difluoride) (PVDF) membrane according to standard procedures. Membranes were blocked with 5% normal goat serum in PBS containing 0.05% Triton X-100 (PBS-T) and incubated with each primary antibody, as indicated in the figure legends. The secondary antibody was a peroxidase-conjugated anti-mouse antibody, and immunoreactive protein was detected by using the enhanced chemiluminescence method (Amersham Biosciences).
Electron Microscopy of the Isolated Proteins. Three μl of the isolated aggregates was placed on a formvar and carbon-coated grid (Ted Pella, Redding, CA) and blotted after 3 min. For immunogold labeling, the grid was incubated for 15 min with purified 0.1% normal goat IgG (Sigma) and then incubated for 15 min in PBS-T with purified 0.1% normal goat IgG containing the polyclonal antibody against GFP (1:100), anti-12CA5 (1:100), or M2 (1:100), as indicated. After three 1-min washes in PBS-T, the secondary antibody conjugated with 10-nm gold (1:50) was applied in PBS-T with purified 0.1% normal goat IgG for 15 min at room temperature. After three 1-min washes in PBS-T, followed by washing in distilled water twice, the sample was stained for 3 min with 1% uranyl acetate, washed with distilled water briefly, and viewed with an H-7100 (Hitachi) electron microscope.
AFM of the Isolated Aggregates. Aliquots (3 μl) were deposited on freshly cleaved mica (Nilaco, Tokyo). After 60 s, the substrate was gently rinsed twice with 50 μl of water to remove salt and loosely bound proteins. The samples were then dried in the atmosphere at room temperature. Each sample was imaged immediately with an SPM-9500 J2 (Shimadzu) in the noncontact mode. Etched silicon probes, attached to triangular microcantilevers 160 μm in length (Olympus), were operated at resonances of 220–320 kHz. The images were taken in air, ambient conditions, at a scan frequency of 0.8 Hz.
Results
Isolation of HDQ91 Aggregates by Tandem-Affinity Chromatography Under Nondenaturing Conditions. To isolate Htt aggregates, HEK293 cells cotransfected with three plasmids [HDQ91 tagged with CBP, protein A, and GFP (Fig. 1a)] were subjected to double-affinity chromatography using IgG Sepharose and calmodulin beads sequentially. The sequential application of protein A–IgG Sepharose and CBP calmodulin beads, known as tandem-affinity purification (TAP), allows protein complex purification under native conditions (30). In the original TAP procedure, the two affinity tags were all fused to the same protein, which could be isolated by sequential rounds of affinity purification. Our method theoretically allows isolation of at least heteromultimeric aggregates that have both the protein A tag and CBP tag without contamination of monomeric soluble HDQ91 protein. Immunoblotting using anti-GFP antibody showed that the high-molecular-weight complex that remained at the top of the gel was recovered in the eluate fractions from calmodulin beads (Fig. 1c, arrowhead). A faint band at ≈70 kDa in the eluate fraction probably corresponds to the small amount of monomeric form of HDQ91-GFP liberated by dissolution of HDQ91 aggregates in the SDS-sample buffer (see below). Silver staining revealed that these eluate fractions contained barely detectable levels of protein (Fig. 1d). The column fractions contained fine GFP-labeled particles that could be observed by microscopy (Fig. 1e) and flow cytometry (data not shown).
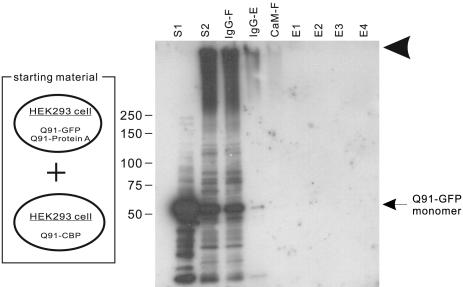
To rule out the possibility that SDS-resistant HDQ91 aggregates were formed in vitro during the isolation step, we applied the same isolation procedure to a mixture of cells expressing either HDQ91-CBP or HDQ91-protein A together with HDQ91-GFP (Fig. 2). No GFP immunoreactive bands were detectable in the eluate fractions from calmodulin beads, suggesting that HDQ91 aggregates were not produced during lysis or sample preparation. Thus, the oligomers isolated by the tandem-affinity procedure from cotransfected cells were produced in vivo.
Fig. 2.
Specificity of protein aggregation isolated by double-affinity method. The column fractions from a mixture of cells transfected with pQ91/GFP and pQ91/ProtA and cells transfected with only pQ91/CBP was immunoblotted by using anti-GFP antibody (JL-8). Molecular mass markers (in kDa) are indicated on the left. The naming of the individual fractions is the same as in Fig. 1b. The arrowhead indicates the bottom of loading wells.
Morphological Analysis of HDQ91 Aggregates Isolated Under Nondenaturing Conditions. HDQ91 aggregates isolated by tandem-affinity chromatography were analyzed by TEM (Fig. 3 a and b). These aggregates appeared as clusters of globules of <50 nm in diameter that occasionally appeared to be composed of smaller structures. No fibrillar species were observed. We did not observe these structures in control samples obtained by the same procedures from untransfected cells or proteasome inhibitor (N-acetyl-leucinyl-leucinyl-norleucinal, ALLN)-treated untransfected cells (data not shown). The anti-GFP antibody JL-8 was found to decorate the globules by using the secondary antibody conjugated with 10 nm of gold (Fig. 3 a and b).
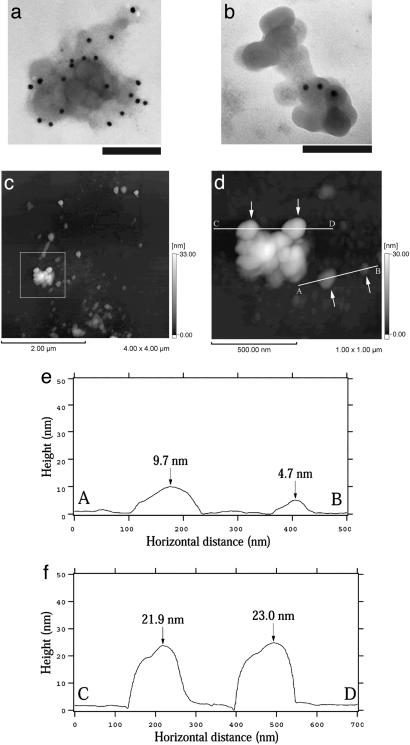
Fig. 3.
Structure of isolated HDQ91 aggregates. (a and b) TEM of uranyl acetate-stained immunogold-labeled HDQ91 aggregates. The size of the gold particles was 10 nm. (Scale bar, 100 nm.) (c) AFM height image of HDQ91 aggregates. (d) Magnified image of the boxed area in c. (e and f) The AFM surface profile along A–B and C–D axes in d, respectively. The arrows in d indicate the globular aggregates, corresponding to the arrows in e and f.
To independently confirm these data, the column eluates were assessed by AFM. By this technique, the HDQ91 aggregates also appeared as globules or clusters of globules (Fig. 3 c–f). Height analysis indicated that the size of a prominent small globule was 4.7 ± 1.0 nm (n = 93), and larger globule seemed to be a cluster of these smaller globules. These structures are consistent with the data observed by TEM.
Isolation and Characterization of HDQ91 Aggregates by Tandem-Affinity Chromatography Under Denaturing Conditions. It is possible that the morphology of the HDQ91 aggregates described in the preceding section could be influenced by the presence of noncovalently attached proteins. Moreover, the sizes of the GFP and protein A tags (27 and 15 kDa, respectively) are relatively large compared with that of the HDQ91 monomer and could also influence its aggregation behavior. To circumvent these potential problems, we prepared a set of HDQ91 constructs containing tags that could be used for purification under strongly denaturing conditions (Fig. 4a). These constructs were coexpressed in HEK293 cells and purified by sequential-affinity chromatography and purified in the presence of 8 M urea (Fig. 4b). Immunoblotting with anti-FLAG antibody showed that these urea- and SDS-resistant aggregates migrated at the at the top of the gel indicating that they were in complexes with very high molecular weight (Fig. 4c). Similar results were obtained when fractions from cells coexpressing HA-S·tag-HDQ91 with HDQ91-(His)6 were immunoblotted with anti-HA antibody (data not shown). FLAG-S·tag-HDQ91/HDQ91-(His)6 and HA-S·tag-HDQ91/HDQ91-(His)6 complexes appeared as globular structures indistinguishable from those obtained from the HDQ91-GFP complex isolated in the absence of urea (Fig. 4d). These structures were immunoreactive with only the specific antibody for the particular tag. The shape of these structures, assessed by AFM, were consistent with the TEM images. The height of small globules was 4.2 ± 1.1 nm (n = 70) for FLAG-S·tag-HDQ91/HDQ91-(His)6 complex, and 3.7 ± 0.9 nm (n = 36) for HA-S·tag-HDQ91/HDQ91-(His)6 complex, respectively, in close agreement with the results obtained for the aggregates isolated under nondenaturing conditions. These results suggest that HDQ91 forms globular aggregates in HEK293 cells of similar dimensions irrespective of the types of the tag or the presence of denaturing solvent.
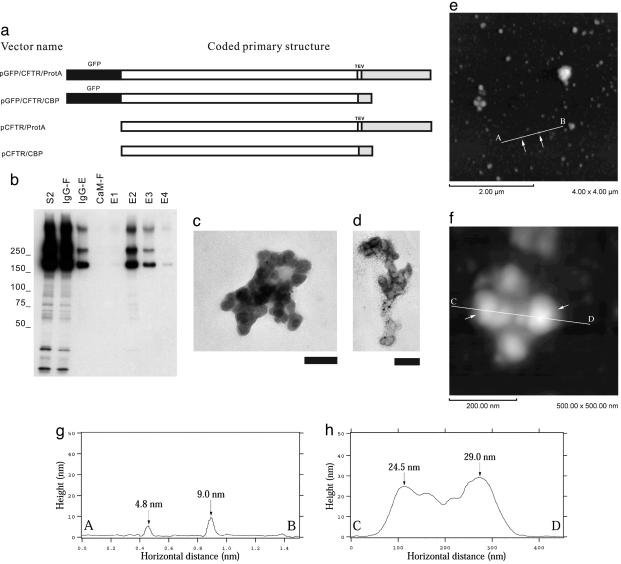
Isolation and Characterization of Mutant CFTR Aggregates by Tandem-Affinity Chromatography Under Nondenaturing Conditions. To examine whether the formation of the small globular aggregates in cells is unique to the polyglutamine-aggregation process, we used a similar approach to express and purify an unrelated aggregation-prone protein in cells. CFTR is a hydrophobic polytopic membrane protein, and the ΔF508 mutant of CFTR and GFP-CFTR have been shown to quantitatively misfold (32) and accumulate in cytoplasmic inclusion bodies upon overexpression (33). Expression constructs for GFP-CFTRΔF508 fused to protein A and CBP (Fig. 5a) were cotransfected into HEK293 cells, and GFP-CFTRΔF508 aggregates were isolated by sequential double-affinity chromatography using IgG Sepharose and calmodulin beads under nondenaturing conditions, as described for HDQ91. Immunoblotting revealed that the eluate fractions were enriched in high-molecular-weight oligomers (Fig. 5b). TEM analysis showed that GFP-CFTRΔF508 aggregates also appeared as clusters of globules and lacked any filamentous structures (Fig. 5 c and d). The anti-GFP antibody JL-8 was found to decorate the globules by using a secondary antibody conjugated with 10-nm gold. CFTR aggregates isolated from HEK293 cells cotransfected with tagged GFP-free constructs (Fig. 5a) revealed similar structures by TEM (data not shown). AFM analysis of these aggregates revealed globular structures with unit height 3.8 ± 1.0 nm (n = 107). These data suggest that small globular aggregates of similar dimensions are widely formed irrespective of the primary structure and the molecular size of the original protein in mammalian cells.
Fig. 5.
Isolation of CFTRΔF508 aggregates under nondenaturing conditions. (a) Constructs. GFP tag is indicated by black box, and affinity tags (protein A and CBP) for isolation of CFTRΔF508 aggregates are indicated by gray boxes. TEV, TEV protease recognition sequence. (b) Immunoblotting of column fractions was performed by using anti-GFP antibody (JL-8). The individual fractions are designated by the convention introduced in Fig. 1b. Molecular mass markers (in kDa) are indicated on the left. (c and d) TEM of uranyl acetate-stained immunogold-labeled CFTRΔF508 aggregates. The size of the gold particles was 10 nm. (Scale bar, 100 nm.) (e–h) The AFM height images of CFTRΔF508 aggregates (e and f) and the surface profiles (g and h) along the A–B (e and g) and C–D (f and h) axes, respectively. The arrows in e indicate the globular aggregates, corresponding to the arrows in g. The arrows in f indicate the globular aggregates, corresponding to the arrows in h.
Discussion
In this article, we describe the isolation of globular aggregates of HDQ91 and CFTRΔF508 by using double-affinity chromatography. The isolation of recombinant polyglutamine aggregates as fibrillar structures formed in cultured cells has been reported (34–36). Those studies used methods focused on the isolation of inclusion bodies by using detergent insolubility and the strong fluorescence of GFP tags. It is probable that the supernatants from detergent and salt-treated samples, which contain the smaller aggregates, were discarded because most of the aggregates isolated by our method could not be pelleted efficiently by centrifugation at 100,000 × g for 1 h in the presence of detergent or urea (data not shown). The double-affinity isolation method described here assures the isolation of dimerized or multimerized Htt, which is the smallest unit of aggregate without contamination of monomer. Furthermore, as shown in Fig. 1e, massive aggregates were eluted in the column flow-through. Our double-affinity method is rapid, which is important for minimizing the formation of aggregates ex vivo, and it can be used under high- or low-stringency conditions. The finding that HDQ91 and CFTRΔF508 aggregates can be isolated by this method suggests that it may be generally applicable to the isolation of small aggregates from different disease models.
Protein aggregation has been widely considered to be a nonspecific coalescence of misfolded proteins, driven by interactions between solvent-exposed hydrophobic surfaces that are normally buried with the interior of a protein. However, we have found that protein aggregation exhibits exquisite specificity even among extremely hydrophobic substrates expressed at very high levels in mammalian cells (37). Although the present data indicate that HDQ91 fused to various different tags can coaggregate in culture cells, it is likely that aggregation in these preparations is driven by common structural features of the misfolded protein and is not strongly influenced by the nature of the affinity tag.
Recently, Bucciantini et al. (38) reported that the normally harmless SH3 domain of phosphatidylinosotol-3′-kinase can form granular aggregates 4–200 nm in diameter in vitro in a low-pH acetate buffer containing trifluoroethanol and that these aggregates are toxic to cells, whereas the fibrillar aggregates formed from the same protein are nontoxic. Together with the report demonstrating disease-related toxicity of globular Aβ oligomers (26), these data indicate that it is possible that relatively uniform small globular aggregates can be formed from structurally unrelated proteins and could have a common toxic character irrespective of the structure or function of the original protein. Despite the similarities between small globular aggregates of Htt and CFTR isolated in this study and the small nonfibrillar aggregates from Aβ and SH3 described above, our data do not address the issue of cytotoxicity. Nonetheless, the similarities noted among these different aggregates lead us to speculate that small globular aggregates might reflect another “default” state of protein structure, analogous to what has been suggested for the cross-β structure of amyloid (39). Also note that a mixture of globular and filamentous structures are observed during aggregation of the amyloidogenic prion Sup35 (40).
We have no direct evidence as to whether these globules of Htt aggregates are identical to the intermediate form before mature amyloid fibrillogenesis reported in in vitro experiments (29), because inclusion bodies formed in mammalian cells are thought to contain many proteins such as molecular chaperones and components of the ubiquitin-proteasome system (41). It is tempting to hypothesize that there is a homogeneous globular unit of aggregation ≈4 nm in diameter. Perhaps, in some cases, relatively larger globules could not be clearly resolved into clusters of smaller units because of the resolution limit of the triangular cantilever of our AFM. If this hypothesis is true, the diameter of the unit seems to be close to the size of the HDQ44 oligomers (4–5 nm in diameter) formed in vitro reported by Poirier et al. (29). It is possible that these units and small assemblies thereof are the preferred substrates for microtubule-dependent retrograde transport to aggresomes. Biochemical and biophysical examination of these small globular aggregate would help to clarify the precise pathological cascade underlying polyglutamine and other protein-deposition diseases.
Acknowledgments
We thank Jon Mulholland for help with initial TEM experiments. This work was supported in part by grants from the National Institute of Diabetes and Digestive and Kidney Diseases and the Coalition for the Cure of the Huntington's Disease Society of America (to R.R.K.).
Author contributions: H.M., N.F.B., Y.O., and R.R.K. designed research; H.M., T.I., E.G., S.T., and A.T. performed research; H.M. and R.R.K. analyzed data; N.F.B. contributed new reagents/analytic tools; and H.M. and R.R.K. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: HD, Huntington's disease; Htt, huntingtin; TEM, transmission electron microscopy; AFM, atomic force microscopy; CBP, calmodulin-binding peptide; HA, hemagglutinin; PBS-T, PBS containing 0.05% Triton X-100; CFTR, cystic fibrosis transmembrane conductance regulator; TEV, tobacco etch virus.
References
- 1.Harper, P. (1996) Huntington's Disease (Saunders, Philadelphia).
- 2.Vonsattel, J. P., Myers, R. H., Stevens, T. J., Ferrante, R. J., Bird, E. D. & Richardson, E. P., Jr., (1985) J. Neuropathol. Exp. Neurol. 44, 559-577. [DOI] [PubMed] [Google Scholar]
- 3.The Huntington's Disease Collaborative Research Group (1993) Cell 72, 971-983. [DOI] [PubMed] [Google Scholar]
- 4.Cooper, J. K., Schilling, G., Peters, M. F., Herring, W. J., Sharp, A. H., Kaminsky, Z., Masone, J., Khan, F. A., Delanoy, M., Borchelt, D. R., et al. (1998) Hum. Mol. Genet. 7, 783-790. [DOI] [PubMed] [Google Scholar]
- 5.Hackam, A. S., Singaraja, R., Wellington, C. L., Metzler, M., McCutcheon, K., Zhang, T., Kalchman, M. & Hayden, M. R. (1998) J. Cell Biol. 141, 1097-1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li, S. H. & Li, X. J. (1998) Hum. Mol. Genet. 7, 777-782. [DOI] [PubMed] [Google Scholar]
- 7.Lunkes, A. & Mandel, J. L. (1998) Hum. Mol. Genet. 7, 1355-1361. [DOI] [PubMed] [Google Scholar]
- 8.Martindale, D., Hackam, A., Wieczorek, A., Ellerby, L., Wellington, C., McCutcheon, K., Singaraja, R., Kazemi-Esfarjani, P., Devon, R., Kim, S. U., et al. (1998) Nat. Genet. 18, 150-154. [DOI] [PubMed] [Google Scholar]
- 9.Cummings, C. J. & Zoghbi, H. Y. (2000) Hum. Mol. Genet. 9, 909-916. [DOI] [PubMed] [Google Scholar]
- 10.Gutekunst, C. A., Li, S. H., Yi, H., Mulroy, J. S., Kuemmerle, S., Jones, R., Rye, D., Ferrante, R. J., Hersch, S. M. & Li, X. J. (1999) J. Neurosci. 19, 2522-2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khoshnan, A., Ko, J. & Patterson, P. H. (2002) Proc. Natl. Acad. Sci. USA 99, 1002-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuemmerle, S., Gutekunst, C. A., Klein, A. M., Li, X. J., Li, S. H., Beal, M. F., Hersch, S. M. & Ferrante, R. J. (1999) Ann. Neurol. 46, 842-849. [PubMed] [Google Scholar]
- 13.Reddy, P. H., Williams, M. & Tagle, D. A. (1999) Trends Neurosci. 22, 248-255. [DOI] [PubMed] [Google Scholar]
- 14.Saudou, F., Finkbeiner, S., Devys, D. & Greenberg, M. E. (1998) Cell 95, 55-66. [DOI] [PubMed] [Google Scholar]
- 15.Sisodia, S. S. (1998) Cell 95, 1-4. [DOI] [PubMed] [Google Scholar]
- 16.Tobin, A. J. & Signer, E. R. (2000) Trends Cell Biol. 10, 531-536. [DOI] [PubMed] [Google Scholar]
- 17.Zoghbi, H. Y. & Orr, H. T. (1999) Curr. Opin. Neurobiol. 9, 566-570. [DOI] [PubMed] [Google Scholar]
- 18.Arrasate, M., Mitra, S., Schweitzer, E. S., Segal, M. R. & Finkbeiner, S. (2004) Nature 431, 805-810. [DOI] [PubMed] [Google Scholar]
- 19.Scherzinger, E., Lurz, R., Turmaine, M., Mangiarini, L., Hollenbach, B., Hasenbank, R., Bates, G. P., Davies, S. W., Lehrach, H. & Wanker, E. E. (1997) Cell 90, 549-558. [DOI] [PubMed] [Google Scholar]
- 20.Goldberg, M. S. & Lansbury, P. T., Jr. (2000) Nat. Cell Biol. 2, E115-E119. [DOI] [PubMed] [Google Scholar]
- 21.Hartley, D. M., Walsh, D. M., Ye, C. P., Diehl, T., Vasquez, S., Vassilev, P. M., Teplow, D. B. & Selkoe, D. J. (1999) J. Neurosci. 19, 8876-8884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim, H. J., Chae, S. C., Lee, D. K., Chomy, B., Lee, S. C., Park, Y. C., Klein, W. L., Krafft, G. A. & Hong, S. T. (2003) FASEB J. 17, 118-120. [DOI] [PubMed] [Google Scholar]
- 23.Klein, W. L., Krafft, G. A. & Finch, C. E. (2001) Trends Neurosci. 24, 219-224. [DOI] [PubMed] [Google Scholar]
- 24.Lambert, M. P., Barlow, A. K., Chromy, B. A., Edwards, C., Freed, R., Liosatos, M., Morgan, T. E., Rozovsky, I., Trommer, B., Viola, K. L., et al. (1998) Proc. Natl. Acad. Sci. USA 95, 6448-6453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walsh, D. M., Klyubin, I., Fadeeva, J. V., Cullen, W. K., Anwyl, R., Wolfe, M. S., Rowan, M. J. & Selkoe, D. J. (2002) Nature 416, 535-539. [DOI] [PubMed] [Google Scholar]
- 26.Zhu, Y. J., Lin, H. & Lal, R. (2000) FASEB J. 14, 1244-1254. [DOI] [PubMed] [Google Scholar]
- 27.Conway, K. A., Lee, S. J., Rochet, J. C., Ding, T. T., Williamson, R. E. & Lansbury, P. T., Jr. (2000) Proc. Natl. Acad. Sci. USA 97, 571-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tompkins, M. M. & Hill, W. D. (1997) Brain Res. 775, 24-29. [DOI] [PubMed] [Google Scholar]
- 29.Poirier, M. A., Li, H., Macosko, J., Cai, S., Amzel, M. & Ross, C. A. (2002) J. Biol. Chem. 277, 41032-41037. [DOI] [PubMed] [Google Scholar]
- 30.Rigaut, G., Shevchenko, A., Rutz, B., Wilm, M., Mann, M. & Seraphin, B. (1999) Nat. Biotechnol. 17, 1030-1032. [DOI] [PubMed] [Google Scholar]
- 31.Laemmli, U. K. (1970) Nature 227, 680-685. [DOI] [PubMed] [Google Scholar]
- 32.Ward, C. L. & Kopito, R. R. (1994) J. Biol. Chem. 269, 25710-25718. [PubMed] [Google Scholar]
- 33.Johnston, J. A., Ward, C. L. & Kopito, R. R. (1998) J. Cell Biol. 143, 1883-1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suhr, S. T., Senut, M. C., Whitelegge, J. P., Faull, K. F., Cuizon, D. B. & Gage, F. H. (2001) J. Cell Biol. 153, 283-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hazeki, N., Tsukamoto, T., Yazawa, I., Koyama, M., Hattori, S., Someki, I., Iwatsubo, T., Nakamura, K., Goto, J. & Kanazawa, I. (2002) Biochem. Biophys. Res. Commun. 294, 429-440. [DOI] [PubMed] [Google Scholar]
- 36.Mitsui, K., Nakayama, H., Akagi, T., Nekooki, M., Ohtawa, K., Takio, K., Hashikawa, T. & Nukina, N. (2002) J. Neurosci. 22, 9267-9277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rajan, R. S., Illing, M. E., Bence, N. F. & Kopito, R. R. (2001) Proc. Natl. Acad. Sci. USA 98, 13060-13065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bucciantini, M., Giannoni, E., Chiti, F., Baroni, F., Formigli, L., Zurdo, J., Taddei, N., Ramponi, G., Dobson, C. M. & Stefani, M. (2002) Nature 416, 507-511. [DOI] [PubMed] [Google Scholar]
- 39.Dobson, C. M. (2003) Nature 426, 884-890. [DOI] [PubMed] [Google Scholar]
- 40.Serio, T. R., Cashikar, A. G., Kowal, A. S., Sawicki, G. J., Moslehi, J. J., Serpell, L., Arnsdorf, M. F. & Lindquist, S. L. (2000) Science 289, 1317-1321. [DOI] [PubMed] [Google Scholar]
- 41.Kopito, R. R. (2000) Trends Cell Biol. 10, 524-530. [DOI] [PubMed] [Google Scholar]