Abstract
A comprehensive differential gene expression screen on a panel of 54 breast tumors and >200 normal tissue samples using DNA microarrays revealed 15 genes specifically overexpressed in breast cancer. One of the most prevalent genes found was trichorhinophalangeal syndrome type 1 (TRPS-1), a gene previously shown to be associated with three rare autosomal dominant genetic disorders known as the trichorhinophalangeal syndromes. A number of corroborating methodologies, including in situ hybridization, e-Northern analysis using ORF EST (ORESTES) and Unigene EST abundance analysis, immunoblot and immunofluorescence analysis of breast tumor cell lines, and immunohistochemistry, confirmed the microarray findings. Immunohistochemistry analysis found TRPS-1 protein expressed in >90% of early- and late-stage breast cancer, including ductal carcinoma in situ and invasive ductal, lobular, and papillary carcinomas. The TRPS-1 gene is also immunogenic with processed and presented peptides activating T cells found after vaccination of HLA-A2.1 transgenic mouse. Human T cell lines from HLA-A*0201+ female donors exhibiting TRPS-1-specific cytotoxic T lymphocyte activity could also be generated.
Keywords: gene expression profiling, immunohistochemistry
Breast cancer is a major worldwide health problem that affects >10% of women in the western world (1, 2). Despite the introduction of several new drugs for breast cancer (3, 4), there has been continued interest in finding new therapeutic targets. New intracellular genes, especially signaling molecules and transcriptional factors, are of special interest because they can form new targets for drug therapies and immunotherapies, such as antigen-specific cancer vaccines.
We used a multidisciplinary scheme to identify intracellular gene products overexpressed in breast cancer involving gene profiling with DNA microarrays, validation of gene expression at the protein level, and elucidation of T cell reactivity against predicted peptide epitopes. This screening approach led to the detection of a number of overexpressed, breast tumor-specific genes. One of the most highly overexpressed genes was found to be a new GATA family transcriptional regulator called trichorhinophalangeal (TRP) syndrome type 1 (TRPS-1) previously found associated with three rare autosomal dominant genetic disorders called the TRP syndromes (5-8). Gene expression at the protein level was also confirmed in breast tumor cell lines and human biopsy samples. Initial immunological characterization of the gene found that it is also capable of generating Abs upon immunization and CD8+ T cell responses to defined nonamer peptides.
Materials and Methods
Tumors and Nonpathogenic Tissues. Primary breast cancer tumors and lymph node metastases were obtained from the archives of the Peter MacCallum Cancer Institute. Institutional ethics approval was obtained for all tumor material used in this study. Total RNA from nonpathogenic human tissues and organs was obtained commercially (Clontech and Invitrogen) or was isolated from fresh-frozen cadaveric samples (Zoion Diagnostics, Shrewsbury, MA) from trauma victims.
RNA Preparation and DNA Microarrays. A custom-designed oligonucleotide microarray using Affymetrix GeneChip technology was used (PDL-Hu03) containing 400,000 perfect-match probes (≈59,000 probe sets) for interrogating essentially all expressed human genes in the public domain at the time of array design (September, 2001). Consensus sequences for interrogation were selected from the expressed mRNA and EST databases in GenBank (9) and compiled with the clustering and alignment tool software (DoubleTwist, Oakland, CA); additional mRNAs were predicted from the Human Genome Project by using the fgenesh algorithm of ab initio exon prediction (10). The custom GeneChip expression probe arrays were hybridized with biotinylated cRNA (made from 10 μg of total RNA) according to standard protocols (11, 12). The fluorescent intensity data from the arrays were normalized with a gamma distribution model (12); an average intensity for each probe set was calculated by using a tri-mean of the intensities of the constituent probes (12, 13). For expression ratio calculations, a minimum value of 10 average intensity units was used for both the numerator and the denominator.
Cloning into Expression Vectors. The TRPS-1 gene was isolated by RT-PCR from the BT-474 breast cancer cell line by using the forward primer 5′-CGGGATCCACCA TGGTCCGGAAAAAGAACCCC-3′ (designated as TRPS1-BamH1/F1), and the reverse primer 5′-CGGGATCCCTCTTTAGGTTTTCCATTTTTTTCCAC-3′ (designated TRPS-1-BamH1/R1). A 3.8-kb PCR fragment with a CACC Kozak sequence was cloned into the pcDNA3.1/Zeo(+) vector (Invitrogen) and into NYVAC, an attenuated Vaccinia vector.
Polyclonal Ab (pAb) and mAb Generation. pAbs were generated against 22-mer or 23-mer peptides conjugated to keyhole limpet hemocyanin (KLH) in New Zealand rabbits (Harlan Laboratories, Haslett, MI). The peptide sequences were KLHMVRKKNPPLRNVASEG EGQILE (CLP2589), KLH-SPKATEETGQAQSGQANCQGLS (CLP2590), KLHVA KPSEKNSNKSIPALQSSDSG (CLP25691), KLH-NHLQGSDGQQSVKESKEHSCTK (CLP2592), KLH-NGEQIIRRRTRKRLNPEALQAE (CLP2593), and KLH-ANGASKEK TKAPPNVKNEGPLNV (CLP2594). mAbs were generated against a 54-kDa His-tagged, N-terminal recombinant protein of TRPS-1 (TRPS1-N54) in BALB/c mice. Two hybridoma clones (designated 8D11 and 8A1) exhibited optimal reactivity and were purified by using Protein G chromatography.
Immunofluorescence Microscopy. Breast cancer cell lines (BT-474, MDA-MB 453, and MDA-MB-231) and control cell lines (COS and HeLa) were fixed with 3% paraformaldehyde in PBS and permeabilized in PBS/0.1% saponin for 10 min. The cells were stained with primary Abs followed by Alexa Fluor 488-coupled secondary Abs with 0.05% Evans blue counterstaining. The slides were analyzed with a Zeiss LSM confocal fluorescence microscope.
Immunohistochemical Studies. Formalin-fixed paraffin-embedded breast cancer biopsy specimens were obtained from the tissue bank in the Department of Anatomic Pathology at Sunnybrook and Women's College Health Sciences Center. Fifty tumor sections consisting of ductal carcinoma in situ (DCIS), invasive ductal and lobular carcinoma, and mixed DCIS and invasive carcinoma were stained with the 8D11 and 8A1 mAb. Immunohistochemistry (IHC) staining was performed as described in ref. 14. Arrays of normal tissues (MaxArray, Zymed Laboratories) were also stained by using the same protocol. Slides were scored according to the percentage of cells and the percentage of tumor specimens staining positive. The intensity of staining in the sections was scored as low, intermediate, or high. A parallel set of sections from the same tumor specimens were also stained for estrogen receptor 1 (ER-1), progesterone receptor, and human epidermal growth factor receptor 2 (HER-2)/neu (data not shown).
Immunogenicity Studies in Human T Cell Cultures. Nonamer peptides predicted to bind to HLA-A*0201 were chosen by using a combination of three computer algorithms: bimas (15), syfpeithi (16), and an in-house neural network program. Peptides were synthesized by using the fluorenylmethoxycarbonyl methodology with the 396 Multiple Biomolecular Synthesizer (Advanced ChemTech). Female donors were prescreened for HLA-A*0201 expression by using DNA sequencing and leukapherisis products collected with consent according to the institutional guidelines of Sunnybrook and Women's College Health Sciences Centre. Activated human T cell lines were generated in peripheral blood mononuclear cell cultures against pools of peptides predicted to bind to HLA-A*0201 using sequential stimulation with peptide-pulsed mature dendritic cells and autologous B cells activated with CD40-ligand (10 μg/ml peptide pulse). Enzyme-linked immunospot (ELISPOT) analysis for IFN-γ production was performed after the third, fourth, and fifth stimulation with the peptides. ELISPOT plates were coated with 2 μg/ml of anti-human IFN-γ blocked with 1% BSA/PBS.
Vaccination Studies in Mice. HLA-A2.1/Kb transgenic mice (A2/Kb Tg) expressing chimeric human HLA-A*0201 α1-α2 domain fused to murine H-2Kb α3 were bred and housed under specific-pathogen-free conditions (17). Female mice (age, 6-15 weeks) were primed and boosted after 28 days with pcDNA3.1-TRPS-1 intramuscularly (tibial muscle). Spleen cells were isolated 3-4 weeks after boosting and reactivated in vitro with peptide-pulsed syngeneic naïve A2/Kb Tg spleen cells. After 5 days, T cell responses were determined with IFN-γ ELISPOT analysis.
Results
Discovery of the TRPS-1 Gene in Breast Cancer by Gene Profiling. We conducted a differential gene expression screen designed to find intracellular genes specifically overexpressed in breast cancer that have potential as targets for immunotherapy. Our screen was designed to detect high- and low-abundance targets, with the latter being of special interest because of the lower likelihood of being subject to immune tolerance. Gene expression was evaluated by hybridizing mRNA from 54 tumor-enriched breast cancer specimens and from 289 nonpathogenic specimens representing ≈75 unique adult tissues or organs on a set of custom-designed Affymetrix GeneChip expression arrays containing probes from >50,000 known genes, EST, and predicted exons. The different tumor pathologies represented in this breast cancer collection were 10 DCIS, 38 invasive ductal carcinomas, and 6 lymph node metastases. Associated pathology data, including tissue location, lymph node involvement, tumor size, grade, and surgical description is given in Table 2, which is published as supporting information on the PNAS web site. To select candidate breast cancer-specific mRNAs, we derived a three-stage data analysis process that used criteria of statistical significance, effect size, and protein localization.
To identify genes with significant overexpression in breast cancer, we converted the average intensity scores into interquartile range (IQR) units by subtracting the median value of the 289 nonpathogenic specimens from the average intensity, then dividing by the IQR of the same 289 normal tissues. This conversion is similar to that for standard deviations over the mean (Z scores) but is less susceptible to outliers. We initially selected genes that were at least 5 IQR units above the median in at least five of the 54 breast tumors. Only 147 genes passed this initial criterion, including many genes previously found overexpressed in primary breast cancer, such as HER-2/neu (18), MUC-1 (19), mammaglobin (20), NY-BR-1 (21), ER-1 (22), pS2 (23), progesterone receptor (24), and SBEM (25).
We then selected genes expressed at minimal levels in all normal tissues and abundantly expressed in breast cancer. The initial 147 genes were further filtered so that the tumor-to-normal tissue expression ratio was ≥2.5, where the average tumor expression was calculated as the 90th percentile value of the 54 breast tumors and the average normal tissue expression was the 99th percentile value of the 289-specimen body atlas. This method positively selected genes with at least 10% prevalence in the breast cancer cohort and negatively selected genes with >1% prevalence in the normal body atlas. Of the 21 candidate TAA genes selected by this step, many “tumor-specific” genes were excluded because of abundant expression in normal tissues (e.g., mammaglobin is expressed in skin; SBEM is expressed in skin and salivary glands).
Finally, we excluded overexpressed genes predicted to encode cell-surface-accessible or extracellular proteins and concentrated on sequences predicted to encode intracellular proteins. A total of 15 sequences fulfilled these criteria. Table 1 lists the GenBank accession and Unigene identification numbers for these genes along with their putative identities, chromosomal location, overall degree of overexpression, and proportion of tumors having at least a 2.5-fold increased expression over the normal tissue panel. Of the 15 genes, a significant number are known putative transcription factors (NY-BR-1, forkhead box A1, TRPS-1, and AP-A2). Another group of genes code for detoxification enzymes involved in chemotherapy resistance (CYP4Z1, and NAT-1). Another potentially important gene was assigned to the PEM-3 family of KH-domain RNA binding proteins that includes MEX-3 from Caenorhabditis elegans and a recently cloned mammalian RNA-binding protein called TINO (26, 27). The top-scoring gene in terms of fold overexpression was NY-BR-1 (overexpressed on average by a factor of 52.5-fold over normal tissues). This gene was previously discovered in breast cancer through serological analysis of recombinant cDNA expression libraries (28). The NY-BR-1 gene encodes a 150- to 160-kDa protein containing a nuclear localization and bZIP motifs, suggesting that it is a transcription factor.
Table 1. List of genes and their putative assignments found specifically overexpressed in a panel of 54 breast cancer specimens by using microarray screening in relation to a panel of 289 normal tissue RNA samples.
| GenBank accession no. | Unigene ID no. | Cytoband | Gene title | Tumor/normal ratio | Prevalence in 54 tumors |
|---|---|---|---|---|---|
| NM_052997 | Hs.326736 | 10p11.21 | Breast cancer antigen NY-BR-1 (NY-BR-1) (ANKRD30A) | 52.4 | 0.76 |
| AW138959 | Hs.437551 | 8q24.11 | EST | 17.6 | 0.56 |
| NM_152597 | Hs.129598 | 15q14 | Fibrous sheath interacting protein 1 (FSIP-1) | 7.0 | 0.46 |
| AA464510 | Hs.152812 | 2q35 | EST | 5.8 | 0.76 |
| AI267700 | Hs.529095 | 14q11.1 | Hypothetical gene XM_498570 (LOC440156) | 5.6 | 0.57 |
| AW341473 | Hs.469965 | 2q21.1 | Hypothetical gene XM_498913 (LOC440916) | 5.4 | 0.43 |
| NM_004496 | Hs.163484 | 14q21.1 | Forkhead box A1 (FOXA1) (HNF-3α) | 5.1 | 0.81 |
| AW248508 | Hs.370809 | 1q22 | Hypothetical gene XM_044166 (LOC92312); similar to PEM-3 (Ciona savignyi) | 4.7 | 0.46 |
| NM_178134 | Hs.176588 | 1p33 | Cytochrome P450, family 4, subfamily Z, polypeptide 1 (CYP4Z-1) | 4.4 | 0.72 |
| NM_014112 | Hs.253594 | 8q24 | TRPS-1 | 3.8 | 0.83 |
| NM_003221 | Hs.33102 | 6p12.3 | AP-2β transcription factor | 3.5 | 0.44 |
| NM_000662 | Hs.155956 | 8p22 | N-acetyltransferase 1 (arylamine N-acetyltransferase) | 3.2 | 0.41 |
| AI793124 | Hs.144479 | 5p12 | EST | 3.2 | 0.44 |
| NM_005853 | Hs.435730 | 16q12.2 | Iroquois homeobox protein 2A (IRX-2A) (IRX-5) | 3.2 | 0.63 |
| U79293 | Hs.159264 | 15q23 | EST | 3.0 | 0.37 |
A microarray analysis was performed by using a custom Affymetrix GeneChip (PDL-Hu03) containing 400,000 perfect-match probes (≈59,000 probesets) interrogating all human genes in the public domain at the time of array design (September, 2001). The 54 breast tumor RNAs and six normal breast RNAs were run independently against 289 normal tissue RNA samples. Analysis of the microarray (see Materials and Methods) revealed that 147 genes had at least 5 interquartile range units above the mean relative gene expression in the 289 normal tissue RNA panel in at least five of the 54 breast cancers studied. This list was further filtered to include only genes encoding intracellular proteins (potential CTL-based vaccine targets) having a tumor-to-normal-tissue expression ratio of at least 2.5 and at least 10% prevalence in the breast cancer panel and <1% prevalence in the normal tissue panel. This process resulted in a list of the 15 genes shown above. The TRPS-1 gene (shown in bold) was expressed in the highest proportion of tumors in the panel according to our cut-off of a minimum 2.5-fold increased expression over the average of the normal tissue panel.
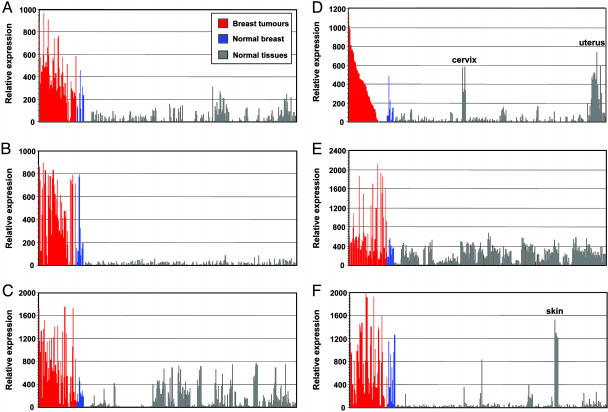
In addition to the total average fold overexpression of the genes, we were also interested in the overall percentage of tumors exhibiting at least a 2.5-fold increased mRNA expression over the normal tissue panel. As shown in Table 1, this analysis revealed that the TRPS-1 gene (shown in bold) had the highest degree of prevalence (83%), despite having a lower fold increase in expression over normal tissues than NY-BR-1. This gene had not been previously found in primary human breast cancer at the mRNA level. Fig. 1 shows the expression profiles of TRPS-1 relative to its expression in mRNA from 289 normal tissues (including normal breast tissue) in comparison to DNA microarray results for NY-BR-1, ER-1, HER-2/neu, MUC-1, and mammaglobin. The results of the microarray analysis for these five genes were near predicted values based on the published expression data.
Fig. 1.
TRPS-1 is overexpressed at the mRNA level in breast cancer relative to normal tissues, as found by using DNA microarray analysis. (A) The relative expression of TRPS-1 in 54 breast tumor biopsy specimens in comparison to a body atlas consisting of samples from 289 normal tissue RNAs and nine normal breast specimens. Relative expression was determined after subtraction of the background hybridization intensities from the GeneChip arrays. (B-F) Expression profiles for other previously known breast cancer-associated genes, NY-BR-1 (B), MUC-1 (C), ER-1 (D), HER-2/neu (E), and mammaglobin (F), are shown as comparison, each with its expression in the 54 breast tumors, 9 normal breast tissues, and the 289 other normal tissue samples. As shown in the ER-1 expression profiles, the 54 breast tumors were arranged according to ER-1 expression (highest to lowest).
Further testing using in situ hybridization in tissue microarrays found medium to strong expression of TRPS-1 in 93% DCIS (28 of 30 samples in the array), 75% of invasive carcinoma (93 of 124), and 90% (18 of 20) of the metastases in the panel (data not shown). Only minimal patchy staining in ≈16% (7 of 44) of normal breast tissue was found with essentially no staining in normal colon and prostate (data not shown). TRPS-1 was found expressed in a number of tumor cell lines, including MDA-MB-453, BT474, U118 MG, and aPRWE-2-1, with the highest expression found in the BT474 cell line (data not shown). Further bioinformatics assessments with e-Northern analysis using a number of EST libraries, including the Unigene cluster, ORF EST (ORESTES), and Incyte LifeSeq, revealed that TRPS-1 has a high degree of prevalence in mammary gland libraries, especially those containing a high number of breast cancer-derived ESTs (Fig. 5, which is published as supporting information on the PNAS web site).
By using the TRPS-1 3′ and 5′ sequences as a template, a full-length clone of TRPS-1 was isolated by RT-PCR from the BT474 cell line that turned out to be identical to the TRPS-1 sequence (GenBank accession no. AF183810) originally isolated from a human fetal brain cDNA library (5). TRPS-1 is an unusual GATA-like transcriptional regulator consisting of only one C2H2 GATA domain in the C terminus instead of three domains found in GATA-1 to GATA-6 (29). We further subcloned TRPS-1 into a number of expression vectors, including pcDNA3.1 and New York Vaccinia (NYVAC).
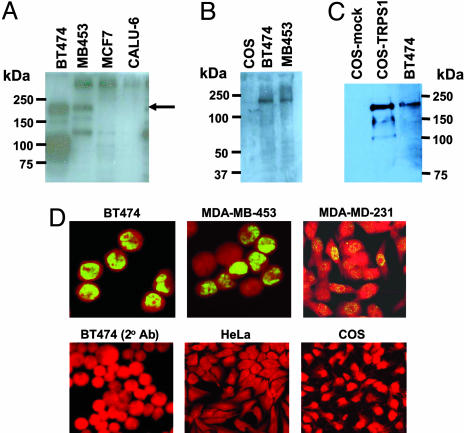
Detection of TRPS-1 Protein in Human Breast Cancer. pAbs against a series of 22-mer peptides in the TRPS-1 sequence recognized a specific band in lysates from BT474 and MDA-MB-453 breast tumor cells migrating ≈180 kDa after SDS/PAGE (Fig. 2A). This result was confirmed with a mAb (8D11) (Fig. 2 B and C) that detected the same size protein in BT474 and MDA-MB-453 breast cancer cells (Fig. 2B). A same size band was also specifically detected in TRPS-1-transfected COS cells (Fig. 2C), confirming protein size and mAb specificity. Immunofluorescence microscopy found that TRPS-1 is expressed in the nucleus of BT474, MDA-MB-453, and MDA-MB-231 breast tumor lines (Fig. 2D).
Fig. 2.
TRPS-1 expression in human breast cancer cell lines, as detected by immunoblot and immunofluorescence analysis. (A) TRPS-1 protein expression was detected in BT474 and MDA-MB-453 breast tumor lines by using a rabbit pAb as a band near 180 kDa (arrow). No expression was found in MCF-7 and CALU-6, cell lines that were negative for TRPS-1 mRNA. (B and C) The expression and apparent molecular weight of TRPS-1 in breast cancer lines was confirmed by using anti-TRPS-1 mAb 8D11. (B) Detection of TRPS-1 protein in BT474 and MDA-MB-453 in immunoblots by using 8D11 mAb. (C) Detection of TRPS-1 protein by using mAb 8D11 in TRPS-1-transfected COS cells but not in mock-transfected COS cells. (D) TRPS-1 protein specifically expressed in the nucleus of breast cancer cell lines, such as BT-474, MDA-MB-453, and MDA-MB-231, as detected by staining with fluorescence microscopy with 8D11 mAb. No expression was found in HeLa cells or COS cells, used as negative controls.
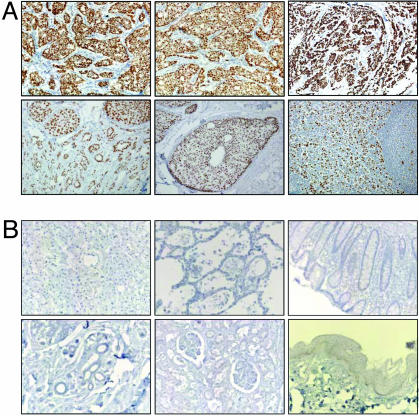
The 8D11 and 8A1 mAbs were used to perform IHC screening of a panel of 50 primary breast tumor biopsies collected from at the Department of Anatomic Pathology in Sunnybrook and Women's College Health Sciences Center. The samples were chosen in an unbiased fashion from the repository and consisted of 40 cases of invasive carcinomas from patients with or without regional lymph node involvement and 10 cases of DCIS. Strong nuclear staining was evident in invasive ductal carcinoma, DCIS, and mixed invasive ductal carcinoma and DCIS (Fig. 3A; see also Fig. 6, which is published as supporting information on the PNAS web site). A high percentage of nuclei were stained in most of the tumor sections analyzed (for distribution of nuclear staining, see Tables 3 and 4, which are published as supporting information on the PNAS web site). Expression of TRPS-1 in a RLN metastasis sample was also found (Fig. 3A). Overall, substantial expression in all stages of breast cancer was found with nine of 10 DCIS samples staining positive (see Table 4 for staining summary). In addition, we acquired two cases of atypical ductal hyperplasia and found positive staining in >25% of the hyperplastic cells (data not shown), again indicating the presence of TRPS-1 in early-stage breast cancer. A striking result of the IHC analysis was the absence of TRPS-1 in a variety of normal adult tissues, such as liver, lung, colon, stomach, kidney, and skin (Fig. 3B). Similar results were obtained in 30 different adult tissues on a commercial normal tissue array (Zymed Laboratories) stained with 8D11 or 8A1 (see Fig. 7, which is published as supporting information on the PNAS web site). Staining in normal breast epithelium in the tumor samples was also evaluated. Most tumors exhibited no staining or weak to moderate staining of the nuclei in the normal epithelium, whereas a small percentage had strong staining in the normal epithelium in 90-100% of the cells. Fig. 8A, which is published as supporting information on the PNAS web site, summarizes the extent of TRPS-1 staining in the tumor panel. Looking specifically at the invasive ductal carcinomas (n = 40), 90% of the samples had 75% or more cells stained with the 8D11 mAb (Fig. 8B). Similar results were obtained with clone 8A1, with 84% of the tumors having at least 75% of the cells stained positive.
Fig. 3.
Immunohistochemistry analysis demonstrates that the TRPS-1 protein is intensely and homogeneously expressed in invasive ductal carcinomas and in DCIS of the breast, with no staining found in normal tissues. (A) (Upper) Strong nuclear staining found in nearly all of the cells in three invasive ductal carcinomas shown stained with mAb clones 8D11 or 8A1. (Lower) Strong and homogeneous nuclear staining for TRPS-1 in most tumor cells is also seen in specimens with mixed invasive and DCIS components (Left), DCIS only (Center), and in a lymph node (LN) metastases (Right). TRPS-1 protein is not detected in normal tissues or breast tissue from noncancerous individuals. (B) Absence of TRPS-1 in normal tissue after staining with the 8D11 mAb: liver (Upper Left), lung (Upper Center), colonic crypts (Upper Right), breast (Lower Left), kidney and glomeruli (Lower Center), and skin (Lower Right).
We also looked at the relationship between ER-1 and HER-2/neu status and TRPS-1 expression for cases of invasive carcinoma and DCIS and found expression in ER-1+ and ER1- (Fig. 8C) and in HER2/neu+ and HER2/neu- tumors (Fig. 8D). These results indicate that TRPS-1 protein is highly tumor-specific and has a comprehensive expression throughout the patient population (>90% positive) in the early and late stages of breast cancer.
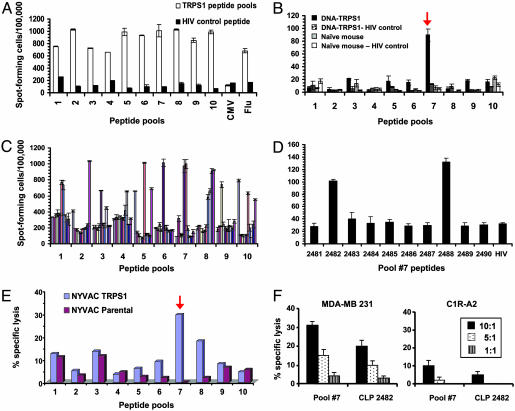
Human T Cell Reactivity Against TRPS-1 and Identification of Processed Epitopes. We next determined whether TRPS-1 was immunogenic and could elicit the expansion of human CD8+ T cells from female HLA-A*0201+ donors. A library of 100 nonamer TRPS-1 peptides were designed based on their potential to bind to HLA-A*0201 with a variety of binding prediction algorithms, including the syfpeithi and bimas programs (15, 16) and an in-house neural net program (for a complete listing of peptides, see Table 5, which is published as supporting information on the PNAS web site). Binding of peptides to HLA-A*0201 was confirmed with plasmon resonance spectroscopy (data not shown). TRPS-1 peptide-induced T cell lines were tested for antigen-specific IFN-γ secretion in ELISPOT assays and in cytotoxic T lymphocyte (CTL) assays by using 51Cr-labeled target cells. We first tested the different T cell cultures against their respective pool of 10 TRPS-1 peptides and found a significant number of lines that reacted positively (Fig. 4A). The reactive peptide pools were then deconvoluted in another series of IFN-γ ELISPOT assays to isolate single reactive peptides from each pool. This process identified a number of individual immunogenic peptides (Fig. 4C).
Fig. 4.
TRPS-1 is immunogenic and generates CTL responses against specific peptides in human T cell lines and after vaccination in mice. (A) IFN-γ secretion by human CD8+ T cells stimulated with pools of TRPS-1 peptides (10 peptides per pool). T cells from HLA-A*0201+ female donors were stimulated with pools of TRPS-1 peptides (pools 1-10) or with HLA-A*0201-binding CMV pp65 or influenza matrix peptides. (B) TRPS-1 plasmid vaccination in HLA-A2/Kb-transgenic mice identified pool 7 peptides as major processed and presented epitopes. Pool 7 TRPS-1 peptides, including the CLP 2482 and CLP 2488 peptides, were recognized by human T cells. (C) Single reactive peptides in each group were identified by using another round of IFN-γ ELISPOT analysis using T cells activated with the different TRPS-1 peptide pools. (D) Deconvolution of TRPS-1 peptide pool 7 found that peptides CLP 2482 and CLP 2488 also had dominant activity in IFN-γ ELISPOT assays after ex vivo restimulation of spleen cells. (E and F) Human HLA-A*0201-restricted T cell lines specific for pool 7 peptides or peptide CLP 2482 exhibit CTL activity in 51Cr-release assays, with C1R-A2 targets infected with the NYVAC-TRPS-1 vector (E) and against an HLA-A*0201+ breast cancer cell line (MDA-MB-231) expressing TRPS-1 protein (F).
DNA vaccination experiments in HLA-A2.1/Kb transgenic mice identified peptide pool 7 of TRPS-1 to be processed and presented on HLA-A*0201 during vaccination (Fig. 4B). One peptide in particular in this pool, ASLGLLTPV (TRPS-1 809-817; designated as CLP 2482) reproducibly exhibited high activity in a series of vaccination experiments with pcDNA3.1-TRPS-1 (Fig. 4D). An HLA-A*0201-expressing C1R lymphoma line infected with a NYVAC vector expressing TRPS-1 was also specifically killed by CD8+ T cell lines specific for the TRPS-1 pool 7 peptides containing the CLP 2482 ASLGLLTPV peptide (Fig. 4E, red arrow). In addition, the HLA-A*0201+ MDA-MB-231 breast tumor cell line expressing TRPS-1 was specifically killed by both TRPS-1 pool 7 peptide-specific and CLP 2482-specific T cells (Fig. 4F). Thus, TRPS-1-expressing cells process and present epitopes recognized by TRPS-1-specific CTL.
Discussion
A comprehensive DNA microarray-based approach designed to detect high- and low-abundance targets specific for breast cancer revealed that the most prevalent overexpressed gene was TRPS-1, originally associated with a series of autosomal-dominant genetic disorders called the TRP syndromes. Subsequent IHC analysis found that the gene was highly expressed at the protein level in >90% of invasive breast tumors and DCIS, with little or no protein expression found in a large array of normal tissues. The lack of TRPS-1 expression at the protein level in many normal tissues suggests that TRPS-1 may have potential as a diagnostic marker for breast cancer. Although previous work (30) has suggested that TRPS-1 mRNA is expressed in other adult tissues, such as the ovary, liver, and aorta, our microarray screen and IHC analysis with two independent mAbs did not detect appreciable expression. It must be noted, however, that the original RNA blotting studies were performed with total RNA dot blots under low-stringency hybridization conditions (30). Another interesting finding was the expression of TRPS-1 at the protein level in two cases of preneoplastic atypical ductal hyperplasia. These results together with the high expression in DCIS suggest that TRPS-1 overexpression may be an early event during breast cancer development.
We also developed a strategy to validate whether the new gene products were immunogenic and capable of activating naïve human CD8+ T cells from female donors. This strategy involved a peptide-screening process using IFN-γ ELISPOT analysis and genetic vaccination of HLA transgenic mice that was capable of identifying HLA-A*0201-binding epitopes processed and presented during vaccination. Our results suggest that the TRPS-1 protein is immunogenic, suggesting that tolerance to the protein can be broken through immunologic intervention (e.g., vaccination). Based on these initial findings on the immunogenicity of TRPS-1, its potential as a cancer vaccine target may be a fruitful area of further research.
The high degree of TRPS-1 expression in human breast cancer of different histological grades and stages indicates that the gene may play an important function in the proliferation and/or survival of tumor cells. TRPS-1 was discovered by means of loss of heterozygosity analysis at a breakpoint on the long arm of chromosome 8 (region q24) in individuals suffering from a rare autosomal dominant genetic disorder called TRPS-I (5). This haplo-insufficiency induces a malformation syndrome involving craniofacial and skeletal abnormalities due to the overgrowth and incorrect patterning of skeletal and muscular tissues. Individuals with TRPS-1 have sparse scalp hair, a bulbous tip of the nose, and protruding forehead and ears (5, 6). Truncations and missense and nonsense mutations leading to a loss of TRPS-1 function have also been detected (8, 31). The long arm of chromosome 8q24, where TRPS-1 is located, is a region of amplification in breast cancer and contains genes, such as MYC and EIF3S3, in addition to TRPS-1 (32). Recently, mRNA expression analysis using RT-PCR has also found TRPS-1 to be amplified along with the MYC and EIF3S3 in prostate cancer biopsies and cell lines (32-35).
At present, it is not known whether overexpression of the TRPS-1 protein alters growth regulation in normal or cancer cells. One possible function of TRPS-1 may be in the repression of GATA-dependent gene transcription involved in the differentiation of epithelial cells (36, 37). Another possible function for the gene product is in the regulation of steroid receptor signaling. Recently, TRPS-1 has been shown to interact with the RING finger protein RNF4 (38). The region in RNF4 (amino acids 6-65) associating with TRPS-1 has also been found to bind to steroid receptors (37). These and other recent results (38) suggest a link between TRPS-1, GATA family regulation, and steroid receptor signaling in breast cancer.
Supplementary Material
Acknowledgments
We thank Drs. Brian Barber, Patrick Halloran, Thomas Fuerst, Olivier Armant, Peter Emtage, and Natalia Martin-Orozco for intellectual contributions. This work was partially funded by a Technology Partnerships Canada loan to Sanofi Pasteur.
Author contributions: L.R., C.L., K.G., W.H., P.D., R.O., J.T., and N.L.B. designed research; L.R., D.S.-S., S.G., C.L., A.P., G.M., K.G., K.K., W.H., J.H., J.S., and M.D. performed research; L.R., D.S.-S., S.G., A.P., K.G., J.A., D.V., and M.P. contributed new reagents/analytic tools; L.R., D.S.-S., S.G., C.L., A.P., K.G., W.H., J.Z., R.O., and N.L.B. analyzed data; and L.R. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: TRP, trichorhinophalangeal; TRPS-1, TRP syndrome type 1; IHC, immunohistochemistry; DCIS, ductal carcinoma in situ; ER-1, estrogen receptor 1; ELISPOT, enzyme-linked immunospot; pAb, polyclonal antibody; HER-2, human epidermal growth factor receptor 2; CTL, cytotoxic T lymphocyte.
Data deposition: The microarray data in this paper have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo [accession nos. GSE1477 (series) and GSM25045-GSM25107 (samples)].
References
- 1.Levin, M. (2003) Drug News Perspect. 16, 395-398. [PubMed] [Google Scholar]
- 2.Jemal, A., Tiwari, R. C., Murray, T., Ghafoor, A., Samuels, A., Ward, E., Feuer, E. J. & Thun, M. J. (2004) CA Cancer J. Clin. 54, 8-29. [DOI] [PubMed] [Google Scholar]
- 3.Coombes, R. C., Hall, E., Gibson, L. J., Paridaens, R., Jassem, J., Delozier, T., Jones, S. E., Alvarez, I., Bertelli, G., Ortmann, O., et al. (2004) N. Engl. J. Med. 350, 1081-1092. [DOI] [PubMed] [Google Scholar]
- 4.Rivera, E., Valero, V., Francis, D., Asnis, A. G., Schaaf, L. J., Duncan, B. & Hortobagyi, G. N. (2004) Clin. Cancer Res. 10, 1943-1948. [DOI] [PubMed] [Google Scholar]
- 5.Momeni, P., Glockner, G., Schmidt, O., von Holtum, D., Albrecht, B., Gillessen-Kaesbach, G., Hennekam, R., Meinecke, P., Zabel, B., et al. (2000) Nat. Genet. 24, 71-74. [DOI] [PubMed] [Google Scholar]
- 6.Malik, T. H., Von Stechow, D., Bronson, R. T. & Shivdasani, R. A. (2002) Mol. Cell. Biol. 22, 8592-8600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ludecke, H. J., Schaper, J., Meinecke, P., Momeni, P., Gross, S., von Holtum, D., Hirche, H., Abramowicz, M. J., Albrecht, B., Apacik, C., et al. (2001) Am. J. Hum. Genet. 68, 81-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaiser, F. J., Brega, P., Raff, M. L., Byers, P. H., Gallati, S., Kay, T. T., de Almeida, S., Horsthemke, B. & Ludecke, H. J. (2004) Eur. J. Hum. Genet. 12, 121-126. [DOI] [PubMed] [Google Scholar]
- 9.Benson, D. A., Karsch-Mizrachi, I., Lipman, D. J., Ostell, J., Rapp, B. A. & Wheeler, D. L. (2002) Nucleic Acids Res. 30, 17-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Salamov, A. A. & Solovyev, V. V. (2000) Genome Res. 10, 516-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lockhart, D. J., Dong, H., Byrne, M. C., Follettie, M. T., Gallo, M. V., Chee, M. S., Mittmann, M., Wang, C., Kobayashi, M., Horton, H. & Brown, E. L. (1996) Nat. Biotechnol. 14, 1675-1680. [DOI] [PubMed] [Google Scholar]
- 12.Platzer, P., Upender, M. B., Wilson, K., Willis, J., Lutterbaugh, J., Nosrati, A., Willson, J. K., Mack, D., Ried, T. & Markowitz, S. (2002) Cancer Res. 62, 1134-1138. [PubMed] [Google Scholar]
- 13.Tukey, J. W. (1977) Exploratory Data Analysis (Addison-Wesley, Reading, MA).
- 14.Hanna, W., Alowami, S. & Malik, A. (2003) Breast J. 9, 485-490. [DOI] [PubMed] [Google Scholar]
- 15.Rotzschke, O., Falk, K., Stevanovic, S., Jung, G. & Rammensee, H. G. (1992) Eur. J. Immunol. 22, 2453-2456. [DOI] [PubMed] [Google Scholar]
- 16.Parker, K. C., Bednarek, M. A. & Coligan, J. E. (1994) J. Immunol. 152, 163-175. [PubMed] [Google Scholar]
- 17.Borenstein, S. H., Graham, J., Zhang, X. L. & Chamberlain, J. W. (2000) J. Immunol. 165, 2341-2353. [DOI] [PubMed] [Google Scholar]
- 18.Slamon, D. J., Clark, G. M., Wong, S. G., Levin, W. J., Ullrich, A. & McGuire, W. L. (1987) Science 235, 177-182. [DOI] [PubMed] [Google Scholar]
- 19.Ginestier, C., Charafe-Jauffret, E., Bertucci, F., Eisinger, F., Geneix, J., Bechlian, D., Conte, N., Adelaide, J., Toiron, Y., Nguyen, C., et al. (2002) Am. J. Pathol. 161, 1223-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Watson, M. A. & Fleming, T. P. (1996) Cancer Res. 56, 860-865. [PubMed] [Google Scholar]
- 21.Jager, D., Stockert, E., Gure, A. O., Scanlan, M. J., Karbach, J., Jager, E., Knuth, A., Old, L. J. & Chen, Y. T. (2001) Cancer Res. 61, 2055-2061. [PubMed] [Google Scholar]
- 22.Inoue, A., Yoshida, N., Omoto, Y., Oguchi, S., Yamori, T., Kiyama, R. & Hayashi, S. (2002) J. Mol. Endocrinol. 29, 175-192. [DOI] [PubMed] [Google Scholar]
- 23.Rio, M. C., Bellocq, J. P., Gairard, B., Rasmussen, U. B., Krust, A., Koehl, C., Calderoli, H., Schiff, V., Renaud, R. & Chambon, P. (1987) Proc. Natl. Acad. Sci. USA 84, 9243-9247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horwitz, K. B. & McGuire, W. L. (1975) Steroids 25, 497-505. [DOI] [PubMed] [Google Scholar]
- 25.Miksicek, R. J., Myal, Y., Watson, P. H., Walker, C., Murphy, L. C. & Leygue, E. (2002) Cancer Res. 62, 2736-2740. [PubMed] [Google Scholar]
- 26.Huang, N. N., Mootz, D. E., Walhout, A. J., Vidal, M. & Hunter, C. P. (2002) Development (Cambridge, U.K.) 129, 747-759. [DOI] [PubMed] [Google Scholar]
- 27.Donnini, M., Lapucci, A., Papucci, L., Witort, E., Jacquier, A., Brewer, G., Nicolin, A., Capaccioli, S. & Schiavone, N. (2004) J. Biol. Chem. 279, 20154-20166. [DOI] [PubMed] [Google Scholar]
- 28.Jager, D., Unkelbach, M., Frei, C., Bert, F., Scanlan, M. J., Jager, E., Old, L. J., Chen, Y. T. & Knuth, A. (2002) Cancer Immun. 2, 5-17. [PubMed] [Google Scholar]
- 29.Molkentin, J. D. (2000) J. Biol. Chem. 275, 38949-38952. [DOI] [PubMed] [Google Scholar]
- 30.Chang, G. T., Steenbeek, M., Schippers, E., Blok, L. J., van Weerden, W. M., van Alewijk, D. C., Eussen, B. H., van Steenbrugge, G. J. & Brinkmann, A. O. (2000) J. Natl. Cancer Inst. 92, 1414-1421. [DOI] [PubMed] [Google Scholar]
- 31.Kobayashi, H., Hino, M., Shimodahira, M., Iwakura, T., Ishihara, T., Ikekubo, K., Ogawa, Y., Nakao, K. & Kurahachi, H. (2002) Am. J. Med. Genet. 107, 26-29. [DOI] [PubMed] [Google Scholar]
- 32.Savinainen, K. J., Linja, M. J., Saramaki, O. R., Tammela, T. L., Chang, G. T., Brinkmann, A. O. & Visakorpi, T. (2004) Br. J. Cancer 90, 1041-1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chang, G. T., Blok, L. J., Steenbeek, M., Veldscholte, J., van Weerden, W. M., van Steenbrugge, G. J. & Brinkmann, A. O. (1997) Cancer Res. 57, 4075-4081. [PubMed] [Google Scholar]
- 34.Chang, G. T., van den Bemd, G. J., Jhamai, M. & Brinkmann, A. O. (2002) Apoptosis 7, 13-21. [DOI] [PubMed] [Google Scholar]
- 35.Gao, X., Sedgwick, T., Shi, Y. B. & Evans, T. (1998) Mol. Cell. Biol. 18, 2901-2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang, H., Lu, M. M., Zhang, L., Whitsett, J. A. & Morrisey, E. E. (2002) Development (Cambridge, U.K.) 129, 2233-2246. [DOI] [PubMed] [Google Scholar]
- 37.Kaiser, F. J., Moroy, T., Chang, G. T., Horsthemke, B. & Ludecke, H. J. (2003) J. Biol. Chem. 278, 38780-38785. [DOI] [PubMed] [Google Scholar]
- 38.Hoch, R. V., Thompson, D. A., Baker, R. J. & Weigel, R. J. (1999) Int. J. Cancer 84, 122-128. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.