Abstract
The gene density of eukaryotic nuclear genomes is generally low relative to prokaryotes, but several eukaryotic lineages (many parasites or endosymbionts) have independently evolved highly compacted, gene-dense genomes. The best studied of these are the microsporidia, highly adapted fungal parasites, and the nucleomorphs, relict nuclei of endosymbiotic algae found in cryptomonads and chlorarachniophytes. These systems are now models for the effects of compaction on the form and dynamics of the nuclear genome. Here we report a large-scale investigation of gene expression from compacted eukaryotic genomes. We have conducted EST surveys of the microsporidian Antonospora locustae and nucleomorphs of the cryptomonad Guillardia theta and the chlorarachniophyte Bigelowiella natans. In all three systems we find a high frequency of mRNA molecules that encode sequence from more than one gene. There is no bias for these genes to be on the same strand, so it is unlikely that these mRNAs represent operons. Instead, compaction appears to have reduced the intergenic regions to such an extent that control elements like promoters and terminators have been forced into or beyond adjacent genes, resulting in long untranslated regions that encode other genes. Normally, transcriptional overlap can interfere with expression of a gene, but these genomes cope with high frequencies of overlap and with termination signals within expressed genes. These findings also point to serious practical difficulties in studying expression in compacted genomes, because many techniques, such as arrays or serial analysis of gene expression will be misleading.
Keywords: genome compaction, microsporidia, nucleomorph, overlapping transcription
Eukaryotic genomes are generally considered to be relatively spacious compared to those of prokaryotes; however, there is a great deal of variability in both size and density of nuclear genomes. At one end of both of these spectra lie the genomes of microsporidian parasites and nucleomorphs. Microsporidia are obligate intracellular parasites related to fungi with genomes as small as 2.3 Mbp (1–3). The 2.9-Mbp genome of the microsporidian Encephalitozoon cuniculi has been fully sequenced, and it has a gene density of ≈0.97 genes per kilobase (1–3). Nucleomorphs, on the other hand, are relict nuclei of red and green algal endosymbionts that are found in cryptomonad and chlorarachniophyte algae, respectively (4). These are not free-living organisms, but hyperreduced organelles with genomes smaller than those of microsporidia. The completely sequenced nucleomorph genome of the cryptomonad Guillardia theta is 551 kbp and has a gene density of 1.02 genes per kilobase (5), and the nucleomorph of the chlorarachniophyte Bigelowiella natans is 380 kbp with a gene density of 0.88 genes per kilobase (4).
The reduction of these genomes is the result of the combined effect of several processes: a reduction in the total number of genes and the compaction of the remaining genes into a smaller space. The first process is relatively easy to understand because microsporidia are parasites and nucleomorphs are organelles, so both are highly dependent on their host. Compaction is harder to explain in general, but we can identify several distinct aspects of this process that have been found to operate in various combinations in both microsporidia and nucleomorphs. “Non-essential” elements like mobile elements or introns may be lost or reduced in number or size, the average length of the remaining genes may decrease in length, and the noncoding intergenic regions may shrink substantially (4–9). This last process is probably the single greatest contributor to the increase in gene density in these genomes, because most eukaryotic genomes have large buffer regions that insulate individual genes from one another. Normally, intergenic regions encode essential regulatory elements, such as promoters and terminators, which direct the accurate initiation and termination of transcription and prevent the expression of one gene from interfering with that of neighboring genes, and in eukaryotes these regions can be large (10). In contrast, the mean intergenic distances in the genomes of the microsporidia E. cuniculi and Antonospora locustae are only 129 bp and 211 bp, respectively (6, 8), whereas nucleomorphs intergenic regions are further reduced (4, 5).
When genomes reach this level of compaction, it is likely that fundamental processes like transcription are substantially affected. Indeed, the sequences of two transcripts from the B. natans nucleomorph have been shown to encode more than one gene, suggesting either that termination control is substantially altered or that nucleomorphs use polycistronic messages (7), like prokaryotes and, in some rare cases, eukaryotes (11, 12). Determining whether these are exceptional cases requires more data, but no systematic analysis of gene expression has been carried out in any highly compacted nuclear genome. Here we report EST surveys of three independently evolved compact genomes: the microsporidian A. locustae and the nucleomorphs of B. natans and G. theta. A large proportion of mRNAs from all three genomes encode multiple genes or gene fragments, sometimes as many as three additional genes apart from the one assumed to be the target of expression. Overall, transcript structure in these organisms suggests that promoter elements and termination signals may have been squeezed from shrinking intergenic regions and embedded in adjacent genes. Genome reduction may therefore result in paradoxically wasteful transcription systems that must at the same time cope with levels of transcriptional overlap that would probably not be tolerated in most genomes.
Materials and Methods
cDNA Library Construction and Sequencing. A total of 1 × 108 purified spores of A. locustae (ATCC 3086) from M&R Durango Biocontrol (Bayfield, CO) were ground under liquid nitrogen and RNA-purified by using TRIzol reagent (Invitrogen). mRNA was extracted from total RNA by using oligo dT cellulose powder. This mRNA was reverse-transcribed by using oligo dT primers, and second-strand synthesis was carried out by using E. coli DNA polymerase and RNase H. cDNA was directionally cloned into pcDNA3.1. G. theta (CCMP 327) was grown in f/2-Si medium at 16°C (12:12-h light:dark cycle). Cells were harvested by centrifugation and ground under liquid nitrogen. Ground cells were resuspended in TRIzol without allowing them to thaw, and total RNA was isolated according to the manufacturer's instructions. mRNA was purified from 0.2 mg of total RNA with an Oligotex mRNA kit (Qiagen, Hilden, Germany). A cDNA library was constructed in the pDNR-LIB vector by using the long-distance PCR method of the Creator SMART cDNA Library Construction Kit as described by the manufacturer (Clontech). A B. natans (CCMP 621) Lambda Zap cDNA library was a generous gift from G. I. McFadden and P. R. Gilson (University of Melbourne).
From the A. locustae library, 726 clones were sequenced from both ends and 466 additional clones were sequenced from the 5′ end, resulting in a total of 1,146 clones with usable sequence. From G. theta and B. natans the 5′ ends of 2,125 and 3,448 clones were sequenced, respectively. Most of these correspond to host genes, so nucleomorph transcripts were identified by similarity to nucleomorph genomic sequence. All EST sequences were clustered and compared with public databases for gene identification by using PEPdb (http://megasun.bch.umontreal.ca/pepdb/pepdb.html). Multigene sequences were identified in microsporidia by blastx search against National Center for Biotechnology Information databases, and all clones matching two genes or where sequence from opposite directions matched different genes were completely sequenced. All nucleomorph transcripts were completely sequenced, and multigene transcripts were identified by comparison with genomic sequence. All EST sequences are available from PEPdbPUB and have been submitted to dbEST and GenBank (accession numbers DQ057484–DQ057579 and DQ071178–DQ071262).
5′ Cap-Dependent and 3′ RACE Cloning. Total RNA was extracted from A. locustae as described above and used directly for 3′ RACE or polyA-purified by using a PolyA pure kit (Ambion, Austin, TX) and used for 5′ RLM-RACE (Ambion) according to the manufacturer's protocol. Two gene-specific primers (TCCACTTGCGCTTGAATGCCTTGAA and GACCTGCAGAAGCTGAACGCTTTGC) were designed to the second downstream ORF of the multigene transcript encoding a DNA polymerase α complex and rpl24. Nested PCR resulted in two distinct products, which were cloned by using TOPO TA vector and sequenced. The A. locustae frataxin was amplified by 3′ RACE by using nested PCR with two gene-specific primers (GTTCTGTCATGGAAAGTAGACGGTGT and GAGTACGTGTTCAATAAGCAGACAC), and products were cloned and sequenced as above.
Results and Discussion
Expressed mRNAs from the Microsporidian A. locustae. Microsporidia are obligate intracellular parasites, and the only stage that may be isolated from the host in substantial quantities is the largely dormant but infectious spore. To construct a cDNA library free of host contamination we isolated mRNA from purified spores of the microsporidian A. locustae. Because microsporidian genomes encode few genes and the spore is only one stage of its life cycle, a relatively small number of ESTs from this library provided a good representation of the spore transcriptome. Of 1,146 cDNA clones sequenced, 871 (76%) had recognizable homologues in public databases. This proportion is high relative to many genome surveys, but expectedly so, because the complete genome of the microsporidian E. cuniculi encodes homologues of most known A. locustae genes (6, 8). In fact, this number of unrecognized sequences in the EST sampling is higher than would be expected based on the number of genes shared between these two organisms.
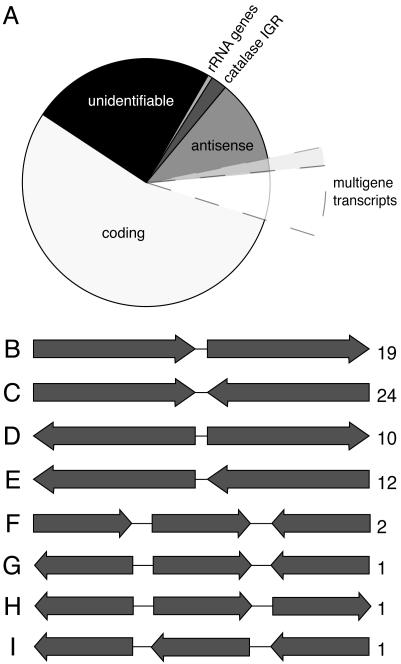
The breakdown of unique spore transcripts by functional categories closely matches that of the E. cuniculi genome as a whole, suggesting that there is not a strong bias in the kinds of mRNAs present in spores (data not shown). Unexpectedly, however, a large proportion of transcripts with similarity to known sequences are non-protein-coding, which fall into several classes (Fig. 1A). Two classes were unremarkable: 7 transcripts encoded rRNA fragments, and another 24 were highly similar (but not identical) to intergenic regions of the A. locustae catalase. This region is repeated in the genome (13), and these mRNA are inferred to be from expressed repetitive elements. More interesting is that 144 transcripts (17% of total matches) encoded the antisense of identifiable protein-coding genes. This large proportion of antisense transcripts may represent a high level of posttranscriptional control as found in other eukaryotes (14, 15). However, among the coding and antisense transcripts, several mRNA encode fragments of more than one gene (see below), suggesting another explanation.
Fig. 1.
Distribution of cDNA types from A. locustae ESTs. (A) Classification of all 1,146 A. locustae mRNA clones according to what they encode. Twenty-four percent have no identifiable similarity to known sequences, 1% are fragments of the A. locustae rRNA operon, and 2% are an expressed repeated element known in the catalase intergenic region from A. locustae. Thirteen percent encode only the antisense of protein-coding genes, and 15% of these (extended wedge) encode antisense of more than one gene. Sixty-one percent encode sense of identifiable genes, and, of these, 11% (extended wedge) encode fragments of two or more genes in sense or antisense strands. (B–I) Schematic diagrams of multigene transcript types, with the numbers of instances of each type indicated by numbers on the right. All cDNAs are oriented with the polyA to the right. Arrows are not to scale, because each type may represent several different loci.
Of the 871 clones found to encode recognizable genes, 97 transcripts (11%) from 70 distinct loci encoded sequence from more than one gene (Fig. 1 A; see also Table 1, which is published as supporting information on the PNAS web site). The polyA sites of these clones do not correspond to polyA tracts in the genome, so they are unlikely to derive from DNA contamination (see also below), but instead come from polyA RNA. In prokaryotes, polycistronic mRNAs commonly code for multiple proteins (11), but with few exceptions (12) eukaryotic mRNAs encode a single gene. A. locustae multigene transcripts encode two or three genes or gene fragments in various orientations (Fig. 1 B–I), but they cannot all be polycistronic messages because there is no bias for genes being on the same strand. Indeed, in the majority of multigene transcripts coding regions appear on both strands (and rarely also include tRNAs). Antisense sequences cannot be translated, so, although we cannot rule out polycistronic mRNAs for certain individual cases, multigene transcripts probably direct the translation of a single protein in general and reflect the movement of transcription control elements beyond their canonical intergenic location.
Examining the structure of various multigene transcripts shows that transcription termination and probably also transcription initiation are affected. The first coding region is most often in the sense strand (Fig. 1 B, C, and F). These mRNAs can be interpreted as run-on transcripts where termination signals are absent from the downstream intergenic regions and are instead found within or between downstream genes. Transcriptional termination and promotion within adjacent genes have been observed in other eukaryotes, including fungi, but these situations appear to be relatively rare and in some cases are known to have a deleterious effect on the transcription of adjacent genes (16–19). In A. locustae they are abundant. In other A. locustae cDNAs, the first coding region is in the antisense strand (Fig. 1 D, E, and G–I), presenting two possible explanations. On one hand, these cDNAs may represent 3′ run-on transcripts that are substantially 5′ truncated (many cDNAs in this library are 5′ truncated, which is common in cDNA library construction) but in some cases would suggest that transcripts are very large (in Fig. 1 G–I, for example, the transcripts would contain at least four genes). On the other hand, transcription initiation may also have been moved from the upstream intergenic region into the adjacent gene. The latter possibility has been shown to be the case for the A. locustae photolyase gene, where transcription initiates 430 bp into the upstream gene (20). The EST data suggest that such initiation within upstream genes is not restricted to one or two genes in A. locustae but could be found throughout the genome.
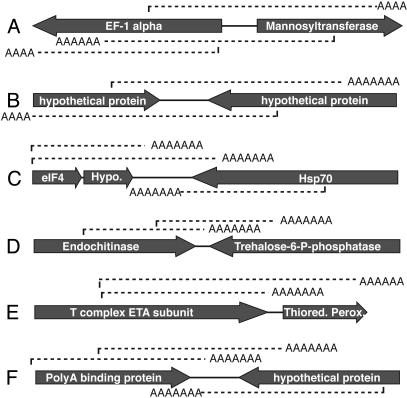
An interesting implication of both of these possibilities is a significant uncertainty as to which gene is actually being expressed in microsporidian transcripts. One could assume that the first sense-strand coding region is the target of expression, but, in many cases where transcripts include two sense genes, transcripts terminate downstream of the stop codon of the following gene (15 of 70 cases). Moreover, when single gene transcripts are also considered, many genes are represented in more than one transcript type (Fig. 2). It is sometimes possible to put forward a credible hypothesis for which gene is actually expressed. In many other cases, however, apparently different transcripts overlap substantially. In Fig. 2E, for example, T-complex protein transcripts might read into thioredoxin peroxidase, or thioredoxin peroxidase transcription might initiate within the T-complex protein. In other cases, transcripts from two adjacent genes appear to terminate within or beyond one another (Fig. 2B).
Fig. 2.
Patterns of cDNAs at A. locustae loci with multiple ESTs. Gray arrows represent the position and direction of the genes in genomic DNA (with gene names given above). Beneath the arrows, mRNA transcripts detected in ESTs are dashed lines oriented by their polyA tail.
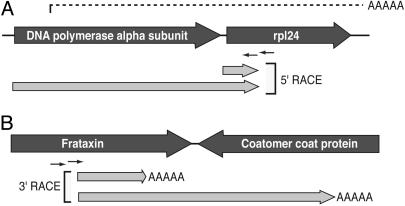
To provide additional evidence that multigene transcripts are derived from mRNA and to examine the nature of the transcripts, we used cap-dependent 5′ RACE and polyA dependent 3′ RACE. The most abundant multigene transcript encodes DNA polymerase α and ribosomal protein L24 (rpl24). These genes are separated by an intergenic region of only 23 bp, and in the 10 ESTs representing this fragment both genes are at least partially represented (Fig. 3A). Using primers within rpl24, cap-dependent 5′ RACE revealed two distinct transcripts: one starting one base upstream of the ATG of rpl24 and a second starting at the ATG of the DNA polymerase α gene. Overall, it appears that both genes have unique promoters, but DNA polymerase transcripts terminate downstream of rpl24, possibly using the same transcription termination signals. 3′ RACE similarly suggests overlap due to transcription termination points within genes. Using primers specific to the frataxin gene, we amplified transcripts terminating before the end of frataxin and within the downstream gene coding for a coatomer complex protein (Fig. 3B). These transcripts suggest that the frataxin mRNA terminates within the coatomer complex protein gene, but also that frataxin sequence forms part of the 3′ UTR of the upstream gene transcript. Results from neither method are consistent with contaminating DNA. Although these examples illustrate that A. locustae transcription patterns are unusual, transcripts of the most common single gene ESTs have a strong tendency to end at a single particular polyadenylation site (Table 2, which is published as supporting information on the PNAS web site), indicating that transcription termination/polyadenylation can be consistent.
Fig. 3.
Transcript ends for adjacent genes in A. locustae.(A) Cap-dependent 5′ RACE at the DNA polymerase α/rpl24 locus, the most abundant multigene transcript. Genomic and EST data are represented as in Fig. 2, and light gray arrows illustrate fragments amplified by 5′ RACE using a specific primer 76 and 109 bp into the rpl24 gene. (B)3′ RACE products at the frataxin/coatomer coat protein locus. Here 3′ RACE products were amplified by using primers 108 and 134 Bp into the frataxin gene. In both cases dual overlapping products can be inferred to come from transcripts for two adjacent genes in the same strand.
The diversity and complexity in structure of A. locustae multigene transcripts raise questions about the large proportion of cDNAs that encode only the antisense of identifiable genes (Fig. 1 A). Given the structure of many multigene transcripts and the fact that some cDNAs encode fragments of more than one antisense gene, it is possible that antisense transcripts may represent truncated cDNAs of multigene transcripts. These data suggest that the overall proportion of multigene transcripts may be higher than currently recognized: multigene plus antisense transcripts together represent approximately a quarter of A. locustae cDNAs with homologues in National Center for Biotechnology Information databases.
Microsporidian genomes are atypical, conspicuously in their highly reduced and compacted nature. The content and characteristics of these genomes have been studied in some detail (6, 8, 9), but whether this has any effects on genome function is not clear. The unusual nature of transcription in A. locustae indicates that compaction may indeed have an impact on expression. One of the obvious characteristics of compacted microsporidian genomes is the short intergenic regions: in A. locustae intergenic spaces average only 211 bp (8). If the pressure to shrink intergenic regions is strong enough, they could be reduced beyond the minimal size needed to encode the essential control regions for expression. The severe reduction of a 3′ intergenic space could, for example, eliminate existing transcription termination signals and force transcription termination fortuitously within the next gene or, if it is in the same strand, using the downstream gene's existing termination signals. Consistent with this idea, the mean intergenic space between genes encoded on multigene transcript mRNAs is only 119 bp, just over half the average for this genome.
Transcript Structure in Nucleomorphs. If multigene transcripts in A. locustae are the result of its compacted genome, then we might expect transcription in other gene-dense nuclear genomes to share similar characteristics; therefore, we carried out EST surveys of G. theta and B. natans to examine transcription of nucleomorph genes. Not only are these nucleomorph genomes the smallest and most compact of any nuclear genomes, but the G. theta and B. natans nucleomorphs have evolved in parallel from a red and green alga, respectively (4), so they give us two independent points of comparison. Two multigene transcripts from the B. natans nucleomorph have been characterized previously (7); however, with only two examples it is not clear whether these messages are representative or not. Because in one case the coding regions characterized were in the sense strand, they could be interpreted as polycistronic or processed messages (7). A larger sample from both nucleomorphs would discern these possibilities from multigene transcripts as observed in microsporidia.
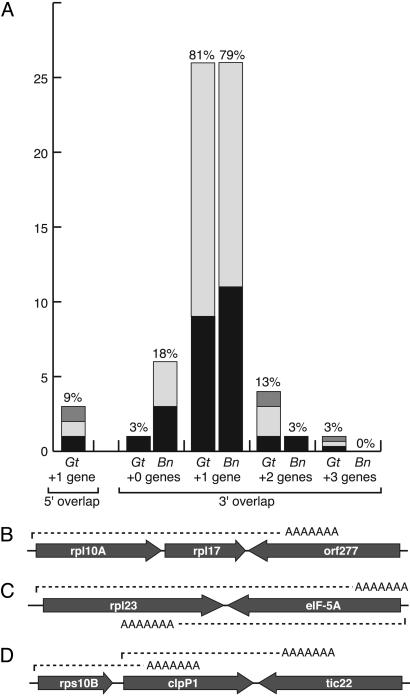
A total of 2,125 and 3,448 ESTs were sequenced from G. theta and B. natans, respectively, and transcripts derived from nucleomorph genes were identified by comparison with genomic sequence resulting in 52 and 38 nucleomorph loci, respectively (the vast majority of transcripts being from host nuclear genes because nucleomorph transcription does not form a large proportion of expression in the cell). Once completely sequenced, 19 and 3 of these loci, respectively, appeared to represent spurious products that do not clearly correspond to mRNAs of any particular gene or are truncated at A-tracks in the genome, and these were not analyzed further. The structure of the cDNAs for the remaining loci were examined for all genes and gene fragments encoded within them (Tables 3 and 4, which are published as supporting information on the PNAS web site). The overall picture from these data are that multigene transcripts are more common in nucleomorphs than in microsporidia (Fig. 4A), but they are simpler in some respects. Identifying the likely expressed gene was not difficult, because virtually every cDNA began with a sense-strand gene. Three G. theta cDNAs and 28 B. natans cDNAs corresponded to intron-containing genes, one G. theta gene a formerly unannotated rpl30. All three G. theta introns were spliced in the cDNA, and all three were part of multigene transcripts. In B. natans not all introns were spliced, but all cDNAs contained at least one spliced intron, confirming beyond doubt that they correspond to mRNA. Only three G. theta transcripts appear to initiate within an upstream coding sequence, two in protein-coding genes and one within a tRNA gene (nearly all B. natans cDNAs were truncated near the 5′ end of the first gene, suggesting that 5′ leaders are likely not long). However, at the 3′ end, many cDNAs overlap with at least one additional gene, and sometimes several (Fig. 4A). In B. natans, cDNAs from 27 of 35 loci included sequence of more than one gene, and one of these included sequence from three genes. In G. theta, the proportions were higher, with cDNAs from 31 of 32 loci including sequence from at least one additional gene, and five cases included two or three additional genes or gene fragments. In G. theta, tRNA genes were found on five multigene transcripts, whereas none were found in B. natans, although one ended at a tRNA gene. In all cases in both nucleomorphs, multigene transcripts ended within a downstream coding region rather than beyond it, as was sometimes found in microsporidia, which suggests that processing signals are encoded within many genes. Indeed, in multiple cases from both nucleomorph genomes, transcripts from two adjacent genes were identified that show that mRNA processing can occur within otherwise actively transcribed genes. In some cases these genes were convergent (Fig. 4C), and in others they are transcribed from the same strand (Fig. 4D).
Fig. 4.
Summary of nucleomorph transcripts from G. theta and B. natans. (A) Graph showing frequency of overlap with adjacent genes. The y axis shows (left to right) overlap with another gene at the 5′ end (nearly all B. natans cDNAs were 5′ truncated, so only G. theta data are shown), no overlap at the 3′ end (i.e., a single gene transcript), or 3′ overlap of one, two, or three additional genes. The x axis represents the total occurrence of each class of cDNA, and the bars are further subdivided to indicate whether the adjacent genes are protein-coding genes in the same strand as the target gene (black), in the opposite strand (light gray), or tRNA genes (gray). The percentage of the total number of cDNAs is shown above each bar. (B–D) Examples of G. theta multigene transcripts including cases where two genes are encoded in the same strand (B), where transcripts for two convergent adjacent genes overlap (C), and where transcripts from two parallel adjacent genes overlap (D). The last case is of particular interest because it shows that transcripts can read through termination signals for upstream transcripts.
Compaction and Multigene Transcripts. Considering microsporidian and nucleomorph genomes together, there is a correlation between compaction and the presence of multigene transcripts, particularly in light of the further correlation between multigene transcription and short intergenic size within the A. locustae genome. This finding has several interesting implications for the process of genome compaction and for the process of transcription.
Why genomes compact is not obvious, but several reasons have been proposed. One of the underlying themes to explain compaction is the economy of reducing the amount of DNA to replicate (21, 22). This may be true in part, but this economy also appears to spawn waste at a different level, because many transcripts have exceedingly long untranslated regions, perhaps because the short intergenic regions have lost the capacity to encode control elements and these are replaced fortuitously upstream or downstream with whatever happens to work. Whatever the cause of compaction, another effect appears to be the stagnation of genomic rearrangements. This stagnation has been attributed to various constraints (23), including the reduction in intergenic spaces making it difficult to reorganize genes without introducing deleterious breakpoints (8, 23). The presence of multigenic transcripts adds another possible cause. The movement of promoter and terminator elements into adjacent genes will make it more difficult to reorder the genome, because removing a gene from one region of a genome might also remove control elements important for the expression of adjacent genes. It is likely that complex traits like this are caused by the sum of a variety of forces.
Multigene transcripts also raise some interesting questions about the process of transcription in nuclear genomes in general. Instances of overlapping transcription are certainly known from bacteria (24) and other eukaryotes, including well studied examples in other fungi, some of which have no detectable effect on gene expression (17, 18). However, on balance, overlapping transcription appears to be rare in systems where it has been examined globally (16). One of the reasons for this rarity is that transcriptional overlap can interfere with the expression of genes where it is normally prevented by control elements. This transcriptional interference can take several forms. Transcriptional collision between two convergent genes has been shown in an artificially convergent construct in Saccharomyces cerevisiae (25) or when the promoter of one yeast gene is enhanced to the point that transcripts read through the normal termination sites into a downstream gene (19). In both cases, the expression of one or both genes was disrupted. Similarly, enhancing the yeast ARO4 promoter so that transcription reads into the downstream HIS7 gene, which is on the same strand, blocks expression of HIS7 (26). Interestingly, this effect has also been used by yeast to control expression at the SER3 locus. Here, a small upstream noncoding transcript that extends into the SER3 promoter acts as an inhibitor of SER3 expression (27). Examples of both of these situations can be seen in compacted genomes (e.g., Fig. 4 B and C). As the SER3 locus shows, overlapping expression has to be considered in time as well as space, because temporal separation of expression could mitigate any deleterious effects of overlap. Indeed, it has been shown that coexpressed genes tend to be divergent when they are adjacent, perhaps to limit the possibility of promoter occlusion (28). However, it seems unlikely that this temporal separation of expression could explain the massive amounts of overlap observed in microsporidia and particularly nucleomorphs. These systems have more likely adapted in different ways to cope with high levels of overlapping expression.
One of the interesting aspects of this coping mechanism must be found in adjacent genes in the same strand where the upstream gene transcript terminates within the downstream gene (e.g., Fig. 4D). If there are termination signals within the downstream gene, how are they recognized by one transcription complex and not another? It is possible that transcription of the downstream gene is terminated at some frequency at these positions, but we have not observed this. It is also possible that relatively weak termination signals may be strong enough to terminate transcripts that have proceeded some distance from the promoter but not others that are still relatively short. Unfortunately, the process of transcription termination is complex and still only partially understood. It is clearly not as simple as a set of signals recognized by a complex but instead is tightly coupled to other processes, such as elongation (25, 29).
Implications for Other Systems. Transcription in nuclear genomes is not as simple as once thought. Numerous studies have found high levels of noncoding transcripts (sometimes called “orphan” or “sterile”), which sometimes overlap with protein-coding genes but apparently do not direct the translation of those genes (30–32). These important complexities are potentially universal to nuclear genomes but different from that described here. The major distinction is that multigene transcripts in microsporidian and nucleomorph genomes are not a class of transcripts apart from those that direct the expression of protein-coding genes. Instead, multigene transcripts are canonical mRNAs altered by extreme conditions in the genome. This effect is most obvious in the nucleomorphs, where multigene transcripts represent nearly 100% of mRNAs. In the microsporidia the situation is harder to interpret, but some of the very highly represented cDNAs (e.g., DNA polymerase α) are multigene transcripts. It is possible that many microsporidian cDNAs are noncoding transcripts like those found in other nuclear systems, but there is also another layer of complexity apparently introduced by the density of the genome, and this complexity is specific to such genomes.
Although microsporidia and nucleomorphs represent the most compact nuclear genomes presently known, there are also other genomes to which these observations may be extended. The marine picoalga Ostreococcus has a small genome (33), and it is reasonable to assume that it is likely compact as well. Similarly, complete genome sequence of the apicomplexan parasite Cryptosporidium has shown it to be reduced and somewhat compacted (34). We examined a small number of short Cryptosporidium EST sequences from public databases (www.cryptodb.org) and found at least 10 of 576 instances where cDNAs encoded fragments of more than one gene. Whether these all correspond to mRNA is not verifiable, and, because the sequences are short, these data are not appropriate to estimate the frequency of multigene transcripts in this organism. Nevertheless, observations from microsporidia and nucleomorphs suggest that this process should be sought in Cryptosporidium. Last, there is no reason to expect that multigene transcripts are restricted to cases where whole genomes are compacted. Transcription of genes within compacted regions of otherwise normal genomes may follow similar patterns, as indeed has been shown in one region of the yeast genome (35).
A high frequency of multigene transcripts also has important practical implications. Several of the eukaryotes with compact genomes (such as microsporidia or Cryptosporidium) are parasites of medical or commercial importance. Because of their requirement for a host, studying these parasites and their interactions with their hosts can be challenging. One method to circumvent these problems when a complete genome is available is to examine expression profiles at various stages of infection using methods such as arrays or serial analysis of gene expression. However, these methods will be misleading in organisms with high levels of multigene transcripts, because “gene” sequences will be encoded in untranslated regions of other genes (some methods distinguish strands, and others do not). Indeed, in A. locustae we have many instances of completely sequenced cDNAs, and it is still not possible to conclusively state which gene is being expressed. By using less direct methods, the chance for error is clear. It will be important to determine the frequency of multigene transcripts in other microsporidia where transcription profiles are desirable (e.g., the human parasite E. cuniculi) and to confirm whether they are also abundant in Cryptosporidium and other organisms with genomic characteristics to suggest that they may be prevalent.
Supplementary Material
Acknowledgments
We thank G. I. McFadden and P. R. Gilson for providing the B. natans cDNA library and for use of the nucleomorph sequence. This work was supported by a grant from the Canadian Institutes for Health Research (CIHR) and a New Investigator Award from the Burroughs–Wellcome Fund. A. locustae and G. theta EST sequencing was supported by the Protist EST Program of Genome Canada/Genome Atlantic, and B. natans EST sequencing was supported by a grant from the Natural Sciences and Engineering Research Council. P.J.K. is a scholar of the Canadian Institute for Advanced Research and a New Investigator of the CIHR and Michael Smith Foundation for Health Research.
Author contributions: B.A.W., C.H.S., N.J.P., N.M.F., and P.J.K. designed research, performed research, analyzed data, and wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviation: rpl24, ribosomal protein L24.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. DQ057484–DQ057579 and DQ071178–DQ071262).
References
- 1.Peyretaillade, E., Biderre, C., Peyret, P., Duffieux, F., Méténier, G., Gouy, M., Michot, B. & Vivarès, C. P. (1998) Nucleic Acids Res. 26, 3513-3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Keeling, P. J. & Fast, N. M. (2002) Annu. Rev. Microbiol. 56, 93-116. [DOI] [PubMed] [Google Scholar]
- 3.Biderre, C., Pagès, M., Méténier, G., Canning, E. U. & Vivarès, C. P. (1995) Mol. Biochem. Parasitol. 74, 229-231. [DOI] [PubMed] [Google Scholar]
- 4.Gilson, P. R. & McFadden, G. I. (2002) Genetica 115, 13-28. [DOI] [PubMed] [Google Scholar]
- 5.Douglas, S., Zauner, S., Fraunholz, M., Beaton, M., Penny, S., Deng, L. T., Wu, X., Reith, M., Cavalier-Smith, T. & Maier, U. G. (2001) Nature 410, 1091-1096. [DOI] [PubMed] [Google Scholar]
- 6.Katinka, M. D., Duprat, S., Cornillot, E., Méténier, G., Thomarat, F., Prenier, G., Barbe, V., Peyretaillade, E., Brottier, P., Wincker, P., et al. (2001) Nature 414, 450-453. [DOI] [PubMed] [Google Scholar]
- 7.Gilson, P. R. & McFadden, G. I. (1996) Proc. Natl. Acad. Sci. USA 93, 7737-7742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Slamovits, C. H., Fast, N. M., Law, J. S. & Keeling, P. J. (2004) Curr. Biol. 14, 891-896. [DOI] [PubMed] [Google Scholar]
- 9.Vivarès, C. P., Gouy, M., Thomarat, F. & Méténier, G. (2002) Curr. Opin. Microbiol. 5, 499-505. [DOI] [PubMed] [Google Scholar]
- 10.Nelson, C. E., Hersh, B. M. & Carroll, S. B. (2004) Genome Biol. 5, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jacob, F. & Monod, J. (1961) J. Mol. Biol. 3, 318-356. [DOI] [PubMed] [Google Scholar]
- 12.Blumenthal, T. (1998) BioEssays 20, 480-487. [DOI] [PubMed] [Google Scholar]
- 13.Fast, N. M., Law, J. S., Williams, B. A. & Keeling, P. J. (2003) Eukaryot. Cell 2, 1069-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kiyosawa, H., Yamanaka, I., Osato, N., Kondo, S. & Hayashizaki, Y. (2003) Genome Res. 13, 1324-1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vanhee-Brossollet, C. & Vaquero, C. (1998) Gene 211, 1-9. [DOI] [PubMed] [Google Scholar]
- 16.Hurowitz, E. H. & Brown, P. O. (2003) Genome Biol. 5, R2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hansen, K., Birse, C. E. & Proudfoot, N. J. (1998) EMBO J. 17, 3066-3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gerads, M. & Ernst, J. F. (1998) Nucleic Acids Res. 26, 5061-5066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peterson, J. A. & Myers, A. M. (1993) Nucleic Acids Res. 21, 5500-5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Slamovits, C. H. & Keeling, P. J. (2004) J. Mol. Biol. 341, 713-721. [DOI] [PubMed] [Google Scholar]
- 21.Cavalier-Smith, T. (2005) Ann. Bot. (London) 95, 147-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van'T Hof, J. & Sparrow, A. H. (1963) Proc. Natl. Acad. Sci. USA 49, 897-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hurst, L. D., Williams, E. J. & Pal, C. (2002) Trends Genet. 18, 604-606. [DOI] [PubMed] [Google Scholar]
- 24.Sameshima, J. H., Wek, R. C. & Hatfield, G. W. (1989) J. Biol. Chem. 264, 1224-1231. [PubMed] [Google Scholar]
- 25.Prescott, E. M., Proudfoot, N. J., Furger, A., Dye, M. J. & Greger, I. H. (2002) Proc. Natl. Acad. Sci. USA 99, 8796-8801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Springer, C., Valerius, O., Strittmatter, A. & Braus, G. H. (1997) J. Biol. Chem. 272, 26318-26324. [DOI] [PubMed] [Google Scholar]
- 27.Martens, J. A., Laprade, L. & Winston, F. (2004) Nature 429, 571-574. [DOI] [PubMed] [Google Scholar]
- 28.Kruglyak, S. & Tang, H. (2000) Trends Genet. 16, 109-111. [DOI] [PubMed] [Google Scholar]
- 29.Proudfoot, N. (2004) Curr. Opin. Cell Biol. 16, 272-278. [DOI] [PubMed] [Google Scholar]
- 30.Elmendorf, H. G., Singer, S. M. & Nash, T. E. (2001) Nucleic Acids Res. 29, 4674-4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen, J., Sun, M., Kent, W. J., Huang, X., Xie, H., Wang, W., Zhou, G., Shi, R. Z. & Rowley, J. D. (2004) Nucleic Acids Res. 32, 4812-4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dahary, D., Elroy-Stein, O. & Sorek, R. (2005) Genome Res. 15, 364-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Courties, C., Perasso, R., Chretiennot-Dinet, M. J., Gouy, M., Guillou, L. & Troussellier, M. (1998) J. Phycol. 34, 844-849. [Google Scholar]
- 34.Abrahamsen, M. S., Templeton, T. J., Enomoto, S., Abrahante, J. E., Zhu, G., Lancto, C. A., Deng, M., Liu, C., Widmer, G., Tzipori, S., et al. (2004) Science 304, 441-445. [DOI] [PubMed] [Google Scholar]
- 35.Puig, S., Perez-Ortin, J. E. & Matallana, E. (1999) Curr. Microbiol. 39, 369-373. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.