Abstract
In this work, we examine how the reported dual decay processes of rhodopsin and binding site stereospecificity can be accounted for by the recently available crystal structure of rhodopsin. Arguments are presented for possible presence of two rhodopsin “rotamers” that fit within the binding cavity. Directed pathways of decay could account for the observed excited decay processes. We summarize evidence for the possible existence of two different ground-state configurations that give rise to two different excited species.
Keywords: molecular modeling, retinal conformation, retinal binding site
The crystal structure of rhodopsin was first reported in late 2000 (1). Although it confirmed many of the postulated structural features of the seven helical transmembrane structure of the visual protein, there are also surprising features not fully appreciated before (2). For example, the retinyl chromophore is not centrally located amidst the seven helices. Rather, it is situated near the edge of the helical bundle facing the extracellular domain, with the longest, but rigid, 4,5-transmembrane (4,5-TM) loop “serving as a plug” to hold the chromophore in place (2). This unique location recently was suggested as a key feature that leads to the enhanced rates and quantum yield of isomerization of rhodopsin (3). In this work, we point out other unique features of the binding site that contribute to the chemical characteristics of the visual pigment.
The Dual Decay Processes of Excited Rhodopsin
The report on real time fluorescence decay of excited rhodopsin (4) was a very thought-provoking study on ultrafast kinetics. By following decay at two wavelengths, Chosrowjan et al. (4) were able to resolve the complex decay kinetics of rhodopsin fluorescence decay. Neither the observed decay at 578 nm nor at 635 nm was single exponential. Instead, they were shown to consist of two major but different fast components: an 80% 146-fsec component at 578 nm and a 70% 330-fsec component at 635 nm. Both of these processes are considerably faster than decay of the relaxed excited rhodopsin (1–2 psec) (4). The faster component (146 fsec) correlated well with the 200-fsec appearance time of the primary photoproduct, photorhodopsin, reported in a separate time-resolved absorption study (5). Therefore, logically this ultrafast process was assigned to that of the isomerization channel (4).
In a subsequent work (6), the same group proposed a “branch model” (see discussion below) for decay of excited rhodopsin. In it, the major (≈70%) decay pathway was assigned to the isomerization channel and the minor (≈30%) decay pathway to the photophysical channel by means of a separate fluorescent intermediate. They pointed out that these numbers were consistent with the quantum yield of rhodopsin (recently redetermined to be 0.65) (7).
A puzzling question that followed was if these two major decay processes originated from the same excited species (say, the Franck–Condon species), why one could determine two separate decay constants instead of a single constant equal to the sum of the two. The situation is understandable if the method allows detection of two different products rather than disappearance of a common reactant. It is not clear that the fluorescent experiment corresponds to this situation. Another possibility is the involvement of primarily two different forms of rhodopsin starting from the ground state. The latter possibility will be examined in some detail in this work.
Conformational Properties of the 11-cis-Retinyl Chromophore
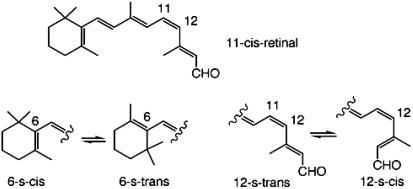
First, we examine the conformational properties of the related 11-cis-retinal (see, e.g., refs. 8 and 9). It is known to exist in the 6-s-cis-12-s-trans conformation (10). In solution the 6-s-cis form is likely to undergo rapid equilibration with the higher energy 6-s-trans form (10, 11). However, when fitted into the hydrophobic pocket of opsin, evidence suggests that the retinyl chromophore exists exclusively in the 6-s-cis form (12). [Interestingly, for the corresponding hydrophobic pocket in bacterioopsin, the ring-chain conformation of the all-trans-retinyl chromophore is exclusively 6-s-trans (13).] Around the hindered 11-cis geometry, the more stable conformation is the twisted 12-s-trans, which in solution is in rapid equilibration with the higher energy, twisted 12-s-cis (11) (see Scheme 1).
Scheme. 1.
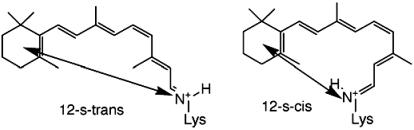
Because of reversal of bond order in the excited state, upon excitation of these rapidly equilibrating conformers, a set of nonequilibrating isomeric excited species would be expected by virtue of the nonequilibrating excited rotamers (NEER) phenomenon (14) [analogous to the isomeric diene triplets believed to be involved in the triplet-sensitized dimerization of dienes (15)]. Selective decay processes could lead to detection of different rates of the excited species. However, when 11-cis-retinal is bound to opsin, the chromophore exists exclusively in the 12-s-trans form (1). The 12-s-cis form is not likely to exist even in small equilibrium concentration, because the resultant pigment (a protonated Schiff base) will have a much altered longitudinal distance (see arrows in Scheme 2) between the hydrophobic pocket and the iminium nitrogen (or the α-carbon of Lys-296) (16). Such an equilibration is clearly out of the question for a chromophore fixed within the binding pocket. Therefore, if different rotamers exist in rhodopsin, they must be different from the traditional s-cis/s-trans conformational equilibration nearby the twisted 6,7 or 12,13 bond. We suspect their existence is, instead, a result of the interaction of the chromophore with nearby protein residue(s) (a supramolecular effect).
Scheme. 2.
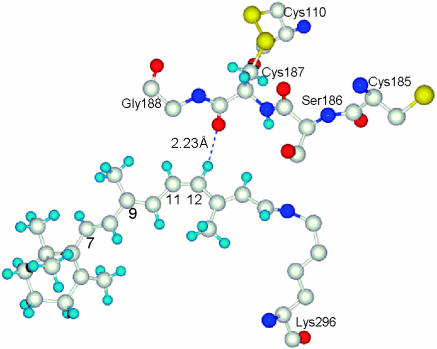
A specific form of the interaction is suggested by recent F-NMR data of rhodopsin analogs (17, 18) and x-ray crystal structure of rhodopsin (1, 19). From a set of fluorine opsin shift values (F chemical shift of a pigment analog minus that of the corresponding protonated Schiff base in solution), it was concluded that H-12 of rhodopsin is likely in close contact with a protein residue (3). The x-ray crystal structure of rhodopsin (partially shown in Fig. 1) pinpointed that residue to be Cys-187, the second closest unit to the retinyl chromophore (the first being the counteranion Glu-113). The distance between the carbonyl oxygen of Cys-187 and C12 is 3.25 Å, which gives an estimated distance between the oxygen and H-12 of ≈2.23 Å.
Fig. 1.
Partial crystal structure of rhodopsin showing close proximity of C=O of Cys-187 (Upper) and the 11,12-unit of the retinyl chromophore (Lower). The 2.23 value is a hypothetical distance (see text).
In making these estimates, one must keep in mind the relatively low resolution [2.8 (1) and 2.6 Å] (19) of the two available x-ray crystal structures of rhodopsin. The positions of the atoms of the chromophore are even less exact as indicated by the fact that a convergent solution for the retinyl chromophore was possible only after assuming a structure for the 6-s-cis,12-s-trans polyene chain (1). Needless to say, the exact orientation of any C–H and C–C bonds (hence the polyene conformation) is far from certain. The discussion below is unavoidably somewhat speculative in nature.
The estimated distance of ≈2.23 Å between the carbonyl oxygen and H-12 is shorter than the sum of van der Waals radii of an oxygen (1.4 Å) and a hydrogen (1.2 Å) (20) atom. Therefore, the C=O bond and the C12-H bond are probably not coplanar. Because Cys-187 is part of a rigid β-sheet, the position of the carbonyl is likely to be unyielding to any perturbation by the chromophore. Hence, the adjustment to avoid excessive carbonyl oxygen and H-12 crowding must take place on the polyene side. An obvious solution is to move the H-atom to either side of the carbonyl group. It should be clear that conformational interconversion of such two forms in the ground state (forced to go through the colinear alignment) will be difficult. Because of expected increased steric crowding in the excited state (3), such an interconversion upon light excitation will be even more unlikely. This finding gives rise to possible formation of two “isomeric” excited species, a feature consistent with the observed two major decay processes for excited rhodopsin. We have, therefore, initiated a search for supporting evidence for possible presence of two forms of rhodopsin. The tool used in this search is the molecular modeling approach used by two of us before (21), but now we are using the crystal structure of the binding cavity of rhodopsin that has since become available (1).
Binding-Site Specificity of Rhodopsin
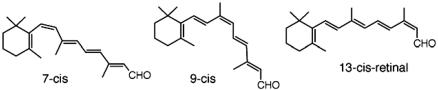
Rhodopsin pigment regeneration studies using all available stable retinal isomers showed stable isomeric pigments can be formed from the following isomers: 11-cis, 9-cis, 7-cis, 9,13-dicis, 7,13-dicis, 9,11-dicis, 7,11-dicis, 7,9-dicis, 7,9,11-tricis, and 7,9,13-tricis (22, 23) with varying rates of pigment formation (22) (see Scheme 3). With the exception of 9-cis-retinal (8), all isomers require much longer times to give isomeric pigments at reduced yields (22).
Scheme. 3.
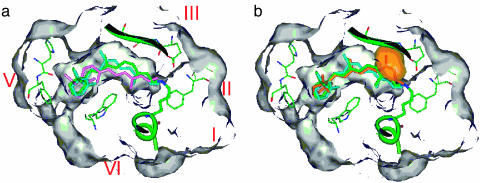
In a molecular modeling study to map the maximum possible perimeter of the binding site of rhodopsin (21), all carbon atoms of the binding isomeric rhodopsin analogs were superimposed after retaining maximum overlap of the trimethylcyclohexenyl rings and the α-carbons of the lysine side chains of isomeric pigments. Now, by using the crystal structures of rhodopsin (1, 19), we can test the idea of a common binding site in a more exact manner. In Fig. 2, we have reproduced the binding cavity of rhodopsin (reflecting walls consisting of van der Waals radii of atoms of all close lying protein residues) containing the 11-cis chromophore of the reported coordinates (Fig. 2a). Superimposed are structures of two binding mono-cis isomers of retinal (9-cis and 7-cis). It is clear that all atoms of the latter two isomeric pigment analogs fall within the binding cavity. In Fig. 2b, we replaced the 7-cis pigment with atoms of the 13-cis protonated Schiff base. It is immediately clear that the 13-methyl group (and partly C13 and C14) of the 13-cis chromophore projects far beyond the binding pocket (the orange area). More specifically, it overlaps with atoms in the β-sheet of the 4,5-TM loop. Hence, the negative result of the 13-cis is not surprising (and for the same reason rejecting the nonbinding all-trans isomer). This result confirms the presence of a “forbidden zone” in the binding cavity as concluded from the earlier molecular modeling study (21). The much reduced rate for pigment formation for the 7-cis isomer is likely due to the altered ring conformation (increased 5,6,7,8-dihedral angle), the relocated 9-methyl group, and a slight shift of the somewhat shortened polyene chain.
Fig. 2.
Binding site of rhodopsin. (a) The gray areas are the empty space within rhodopsin with the middle area (below the β-sheet in green) being the binding cavity as revealed by the crystal structure of rhodopsin (1). The 11-cis-retinyl chromophore (green) is that reported. The modeled 9-cis-retinyl (aqua) and 7-cis-retinyl (pink) chromophores are superimposed with the 11-cis, allowing maximum possible overlap of the cyclohexenyl rings and the iminium nitrogens. (b) The nonbinding 13-cis isomer is superimposed with 11-cis and 9-cis. The orange area marks the zone of conflict, thus, negating possible binding interaction of 13-cis-retinal with opsin.
It is also clear that the binding site (Fig. 2a) contains an empty space of sufficient size to allow binding interaction of other isomers of retinal that have doubly or triply bent polyene side chains (as shown in the previous modeling study) (21). In this sense, the opsin recognition for 11-cis-retinal should not be considered a snug lock-and-key interaction. Rather, its substrate recognition mechanism involves only a few key points of contact that any binding isomer must be able to meet. First, there is a longitudinal distance requirement [a concept first introduced by Matsumoto and Yoshizawa (16)] defined at one end by the hydrophobic pocket for the trimethylcyclohexenyl ring and the other end by the iminium nitrogen atom (24). The latter exists as an ion pair with the carboxylate ion of Glu-113. Tyr-268, Ala-292, and Ala-117 are part of the hydrophobic pocket that also recognizes a specific twisted ring-chain conformation necessary for positive binding interaction. Some movement of the iminium nitrogen is likely but not to the extent to disrupt the ion pair interaction (i.e., permitted by the minor conformation reorganization of the side chain of Glu-113 within the binding pocket). The second major protein recognition site is the wall consisting of part of the β-sheet (Ala-171 to Cys-185) of the 4,5-TM loop. H-12 of the 11-cis linkage rests snugly against this wall (in close contact with Cys-187), making a cis bend at this site absolutely necessary for successful binding interactions (hence the negative result of all-trans and 13-cis). Between 5-methyl and 13-methyl, the rhodopsin-binding pocket is relatively open [allowing pigment formation for dicis, tricis isomers (21) and other retinal analogs] (25). However, the location of Glu-181, Tyr-191, and a water molecule (19) are strategically important to ensure the twisted conformation around the 12,13 bond. These units along with Tyr-268, Ala-292, and Ala-117 ensure the well known helical twist of the bound 11-cis-retinyl chromophore (26, 27).
Possible Presence of Two Rhodopsin Pigments
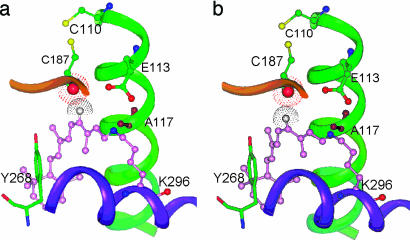
The available empty space in the rhodopsin-binding cavity suggests possible existence of more than one form of rhodopsin, of which the chromophore conformational difference could be minor for detection by spectroscopic techniques directed at the polyene side-chain. However, relative to a reference point on the protein pocket, the difference between the two forms could be significant. Because of the close lying of Cys-187 to the chromophore, its role as a key reference point deserves special attention. In Fig. 3, we show one possible pair of rhodopsin chromophores (see also Fig. 4).
Fig. 3.
Two possible rotamers of rhodopsin showing nearby amino acid residues protruding from helix-3 (green), helix-5 (purple), and part of a β-sheet (brown) of 4,5-TM loop. (a) From the reported crystal structure of rhodopsin showing the carbonyl oxygen of C187 (red) above H-12 (gray). (b) A rearranged 11-cis-retinyl chromophore in which the carbonyl oxygen (red) of C187 is now slightly below H-12 (gray).
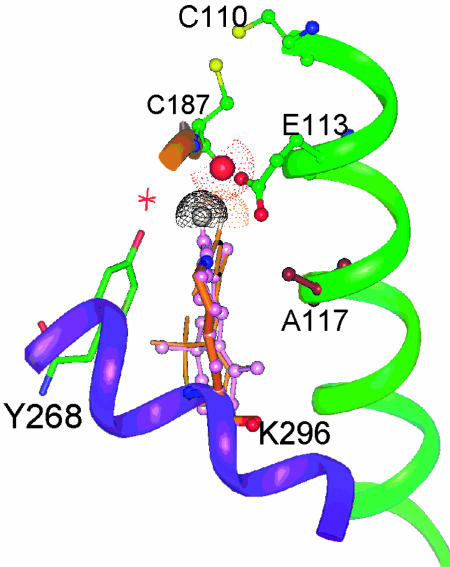
Fig. 4.
A stacked plot of two possible forms of rhodopsin (from Fig. 3) with the carbonyl oxygen sandwiched between the two H-12s. The figure has been rotated slightly from those in Fig. 3 to emphasize the similarity between the two structures and to show clearly the location of the water molecule (red asterisk) that is also the general direction of the empty space for isomerization.
Fig. 3 shows the 11-cis-retinyl chromophore along with the surrounding amino acid units projecting from helix-3 and -5 and part of a β-sheet of the 4,5-TM loop. This partial structure is reproduced from the reported crystal structure of rhodopsin (1). In this projection, the carbonyl oxygen (shown in red) of Cys-187 is above that of H-12 (in gray). In Fig. 3b is a rearranged structure of the 11-cis-retinyl chromophore in which the middle section of the polyene chain (primarily the 11,12-cis linkage) has been tilted slightly toward the left so that H-12 is now above the carbonyl oxygen of Cys-187. With the C=O and C12–H bonds now not aligned in a colinear manner, excessive van der Waals repulsion is avoided. Yet, H-12 remains in close contact with the carbonyl group. Importantly, we found that all atoms in this rearranged structure still fall within the binding pocket as defined in Fig. 2a.
Separately, we have carried out a molecular mechanics calculation for the relative energies of these two conformers. It should be clear that the energy difference is likely to be too small for accurate assessment if we include the energy of the entire protein. Therefore, we have approximated the situation by calculating the energies of the frozen conformers of the isolated 11-cis-retinal. We found that the constructed second structure is, on the average (depending on the method of energy calculation), within 1 kcal/mol of that reported in the crystal structure. There are other conformers similarly close in energy. The results suggest a possible lack of conformational homogeneity of the retinyl chromophore, and without a protein crystal structure of high precision, it will not be possible to determine the preferred conformation of the chromophore by calculations. Therefore, we emphasize that the current work should be considered a suggestion of a concept and not a proof for the existence of the proposed second conformer, especially regarding its exact structure.
Experimental Evidence Suggesting the Presence of Two Forms of Rhodopsin
The present suggestion of two forms of rhodopsin diverges from the traditional thought process of a single form of rhodopsin in consideration of rhodopsin photochemistry (28) or in construction of theoretical models for the isomerization process (29, 30). For reasons stated above, in the low-resolution crystal structures of rhodopsin (1, 19) it is not possible to distinguish between the two structures or to prove or disprove possible simultaneous existence of these two forms in rhodopsin. However, it will be of interest to examine some of the relevant experimental data in the literature.
There have been some discussions concerning the possible existence of two forms of rhodopsin because there is more than one set of decay rates of the later dark intermediates in the photobleaching processes of rhodopsin (31). However, the Kyoto group concluded that there were no spectrally different forms of rhodopsin (32). More recently, Mathies and coworkers (33) reported a small variation of quantum yield of photoisomerization of rhodopsin depending on the excitation wavelength (0.65 for light between 450–480 nm, dropping steadily from 500 to 570 nm by a maximum amount of 5%) (7). The variation was explained by: “... [a] dynamic internal conversion model... that a change in the excess energy of the wavepacket alters its torsional velocity and hence the quantum yield“ (7). However, it occurred to us that the observed quantum yield data are also consistent with possible involvement of two forms of rhodopsin of slightly different absorption characteristics. The longer-wavelength absorbing species would be the photochemically inert one. The 65% quantum efficiency could, in fact, reflect a statistical distribution of two forms of rhodopsin (65% for the reactive form and 35% for the unreactive form, the relative amounts probably determined kinetically when they are formed). Note that this interpretation of the wavelength-dependent quantum yield data is not necessarily in disagreement with the Kyoto group's conclusion of the absence of spectrally different forms of rhodopsin. The difference could simply be too small for detection.
We might add that the fluorescence spectra of rhodopsin recorded by a stationary method also showed variation depending on the excitation wavelength (34).
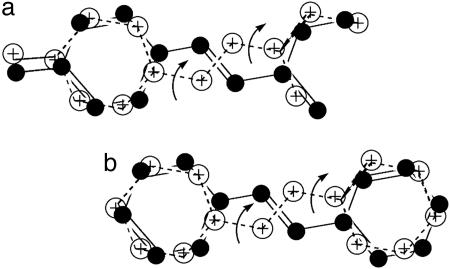
It is worth noting that in a recent reanalysis of the low temperature crystal structure of photoactive yellow protein (PYP), the trans-thiocinnamate chromophore was shown to consist of two forms (35), which presumably undergo ready interconversion at more elevated temperatures by means of what appears to be a bicycle-pedal process (Fig. 5a). Similarly, temperature-dependent x-ray crystallographic studies of trans-stilbene (Fig. 5b) and the closely related trans-azobenzene (with nitrogen connecting atoms) revealed conformational equilibration also in the form of bicycle-pedal processes (36).
Fig. 5.
Equilibrating rotamers (filled circles vs. open circles) in crystals of the trans-cinnamoyl chromophore of PYP (a) and trans-stilbene (b), with structures in both cases connected by bicycle-pedal motions. The structures are essentially those in refs. 35 and 36.
These results reinforce the notion that scaffolding within crystals can provide sufficiently large empty space for conformational or other type of molecular reorganization. The current proposed idea of the presence of two rhodopsin “rotamers” (although not interconverting) may be viewed as a simple extension of the established cases of crystals of PYP, stilbene, and azobenzene rather than a radically new one.
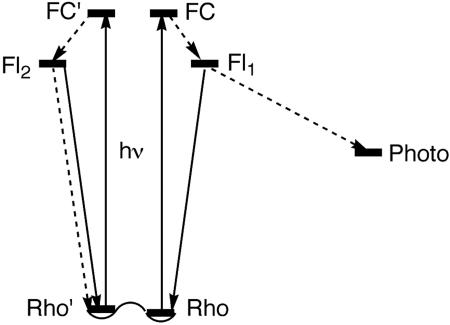
In conclusion, we have summarized evidence from molecular modeling studies and experimental results in the literature that suggest that rhodopsin might exist, primarily, in two different forms. The different excited-state structures derived from the two postulated forms of rhodopsin chromophore readily account for the two major decay processes reported for excited rhodopsin. Thus, decay of excited rhodopsin can be described in a modified “branch model” shown in Fig. 6. We must emphasize that such a model does not involve radically new ideas; rather, it is one that has retained many of the essential features in the “branch model” proposed by Kandori et al. (6), including the involvement of two different fluorescent species. The only change is the presence of two distinct FC-species formed from different conformers of rhodopsin. In carrying out the molecular modeling studies reported in this work, it was also clear to us that the current work could not exclude other minor structures of the rhodopsin chromophore. It is premature to speculate whether such minor structures could account for some of the reported minor decay pathways of the excited rhodopsin (4, 6).
Fig. 6.
A schematic diagram modified from the “branch model” in ref. 6 for the primary processes of rhodopsin. Solid and dashed lines represent, respectively, radiative and nonradiative processes to and from the Franck–Condon species (FC and FC′) of rhodopsin conformers. Fl1 and Fl2 are the same fluorescent states discussed before with relative abundance of ≈70% to ≈30% (6). Photo is the isomerized primary product photorhodopsin.
Lastly, we would like to add that the more recently available crystal structure at 2.2 Å resolution (37) showed little change in the conformation of the 11-cis-retinyl chromophore. Therefore, it does not change the conclusions in this work.
Acknowledgments
The work was supported by National Science Foundation Grant CHE-01-32250.
Author contributions: R.S.H.L. designed research; R.S.H.L. and T.M. performed research; R.S.H.L. and G.S.H. analyzed data; and R.S.H.L. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviation: 4,5-TM, 4,5-transmembrane.
References
- 1.Palczewski, K., Kumasaha, T., Hori, T., Benhke, C. A., Motoshima, H., Fox, B. A., Le Trong, I., Teller, D. C., Okada, T., Stemkamp, R. E., et al. (2000) Science 289, 739-745. [DOI] [PubMed] [Google Scholar]
- 2.Teller, D. C., Okada, T., Behnke, C. A., Palczewski, K. & Stenkamp, R. E. (2001) Biochemistry 40, 7761-7772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu, R. S. H. & Colmenares, L. U. (2003) Proc. Natl. Acad. Sci. USA 100, 14639-14644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chosrowjan, H., Mataga, N., Shibata, Y., Tachibanaki, S., Kandori, H., Shichida, Y., Okada, T. & Kouyama, T. (1998) J. Am. Chem. Soc. 120, 9706-9707. [Google Scholar]
- 5.Wang, Q., Schoenlein, R. W., Peteanu, L. A., Mathies, R. A. & Shank, C. V. (1994) Science 266, 422-424. [DOI] [PubMed] [Google Scholar]
- 6.Kandori, H., Furutani, Y., Nishimura, S., Shichida, Y., Chosrowjan, H., Shibata, Y. & Mataga, N. (2001) Chem. Phys. Lett. 334, 271-276. [Google Scholar]
- 7.Kim, J. E., Tauber, M. J. & Mathies, R. A. (2001) Biochemistry 40, 13774-13778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hubbard, R., Brown, P. K. & Bownds, D. (1971) Methods Enzymol. C 18, 615-653. [Google Scholar]
- 9.Wald, G. (1968) Science 162, 232-239. [Google Scholar]
- 10.Simmons, C. J., Liu, R. S. H., Denny, M. & Seff, K. (1981) Acta Crystallogr. B 37, 2197-2205. [Google Scholar]
- 11.Rowan, R., III, Warshel, A., Sykes, B. & Karplus, M. (1974) Biochemistry 13, 970-981. [DOI] [PubMed] [Google Scholar]
- 12.Fujimoto, Y., Ishihara, J., Maki, S., Fujioka, N., Wang, T., Furuta, T., Fishkin, T., Borhan, S., Berova, N. & Nakanishi, K. (2001) Chem. Eur. J. 7, 4198-4204. [DOI] [PubMed] [Google Scholar]
- 13.Lueke, H., Schobert, B., Richter, H. T., Cartailler, J.-P. & Lanyi, J. K. (1999) J. Mol. Biol. 291, 899-911. [DOI] [PubMed] [Google Scholar]
- 14.Havinga, E. (1962) Chimia 16, 145-172. [Google Scholar]
- 15.Hammond, G. S. & Liu, R. S. H. (1963) J. Am. Chem. Soc. 85, 477-478. [Google Scholar]
- 16.Matsumoto, H. & Yoshizawa, T. (1978) Vision Res. 18, 607-609. [DOI] [PubMed] [Google Scholar]
- 17.Colmenares, L. U., Niemczura, W. P., Asato, A. E. & Liu, R. S. H. (1996) J. Phys. Chem. 100, 9175-9180. [Google Scholar]
- 18.Colmenares, L. U. & Liu, R. S. H. (2003) J. Photosci. 10, 81-87. [Google Scholar]
- 19.Okada, T., Fujiyoshi, Y., Silow, M., Navarro, J., Landau, E. M. & Shichida, Y. (2002) Proc. Natl. Acad. Sci. USA 99, 5982-5987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pauling, L. (1960) The Nature of the Chemical Bond (Cornell Univ. Press, Ithaca, NY).
- 21.Liu, R. S. H. & Mirzadegan, T. (1989) J. Am. Chem. Soc. 110, 8617-8623. [Google Scholar]
- 22.Liu, R. S. H., Matsumoto, H., Kini, A., Asato, A. E., Denny, M., Kropf, A. & DeGrip, W. J. (1984) Tetrahedron 40, 473-482. [Google Scholar]
- 23.Liu, R. S. H. & Asato, A. E. (1990) in Chemistry and Biology of Synthetic Retinoids, eds. Dawson, M. & Okamura, W. H. (CRC, Boca Raton, FL), pp. 147-176.
- 24.Liu, R. S. H., Asato, A. E., Denny, M. & Mead, D. (1984) J. Am. Chem. Soc. 106, 8298-8300. [Google Scholar]
- 25.Nakanishi, K. & Crouch, R. (1995) Isr. J. Chem. 35, 253-272. [Google Scholar]
- 26.Buss, V., Kolster, K., Terstegen, F. & Vekrenhorst, R. (1998) Angew. Chem. Int. Ed. 33, 1893-1895. [Google Scholar]
- 27.Fujimoto, Y., Fishkin, N., Pescitelli, G., Decatur, J., Berova, N. & Nakanishi, K. (2002) J. Am. Chem. Soc. 124, 7294-7302. [DOI] [PubMed] [Google Scholar]
- 28.Yoshizawa, T. & Kandori, H. (1991) Prog. Retinal Res., 11, 33-55. [Google Scholar]
- 29.Ferre, N. & Olivucci, M. (2003) J. Am. Chem. Soc. 125, 6868-6869. [DOI] [PubMed] [Google Scholar]
- 30.Ishiguro, M., Hirano, Y. & Oyama, Y. (2003) ChemBioChem. 3, 228-231. [DOI] [PubMed] [Google Scholar]
- 31.Sasaki, N., Tokunaga, F. & Yoshizawa, T. (1980) FEBS Lett. 114, 1-3. [DOI] [PubMed] [Google Scholar]
- 32.Shichida, Y., Matuoka, S. & Yoshizawa, T. (1984) Photobiochem. Photobiophys. 7, 221-228. [Google Scholar]
- 33.Mathies, R. A. & Lugtenburg, J. (2000) in Handbook of Biological Physics, eds. Stavenga, D. G., deGrip, W. J. & Pugh, E. N., Jr. (Elsevier, Amsterdam), Vol. 3, pp. 55-90. [Google Scholar]
- 34.Kochendoerfer, G. G. & Mathies, R. A. (1996) J. Phys. Chem. 100, 14526-14532. [Google Scholar]
- 35.Anderson, S., Srajer, V. & Moffat, K. (2004) Photochem. Photobiol. 80, 7-14. [DOI] [PubMed] [Google Scholar]
- 36.Harada, J. & Ogawa, K. (2004) J. Am. Chem. Soc. 126, 3539-3544. [DOI] [PubMed] [Google Scholar]
- 37.Okada, T., Sugihara, M., Bondar, Ana-Nicoleta, Elstner, M., Entel, P. & Buss, V. (2004) J. Mol. Biol. 342, 571-583. [DOI] [PubMed] [Google Scholar]