Abstract
Phosphatidylinositol 3-kinase [PI (3)K]/Akt signaling is a critical pathway in cell survival. Here, we demonstrate a mechanism where membrane alteration by the n-3 fatty acid status affects Akt signaling, impacting neuronal survival. Docosahexaenoic acid (DHA), an n-3 polyunsaturated fatty acid highly enriched in neuronal membranes, promotes neuronal survival by facilitating membrane translocation/activation of Akt through its capacity to increase phosphatidylserine (PS), the major acidic phospholipid in cell membranes. The activation of PI (3)K and phosphatidylsinositol triphosphate formation were not affected by DHA, indicating that membrane interaction of Akt is the event responsible for the DHA effect. Docosapentaenoic acid, which replaces DHA during n-3 fatty acid deficiency, was less effective in accumulating PS and translocating Akt and thus less effective in preventing apoptosis. Consistently, in vivo reduction of DHA by dietary depletion of n-3 fatty acids decreased hippocampal PS and increased neuronal susceptibility to apoptosis in cultures. This mechanism may contribute to neurological deficits associated with n-3 fatty acid deficiency and support protective effects of DHA in pathological models such as brain ischemia or Alzheimer's disease.
Keywords: apoptosis, phosphatidylserine, hippocampus, docosapentaenoic acid, n-3 fatty acid deficiency
The phosphatidylinositol 3-kinase [PI (3)K]/Akt signaling is a critical mechanism in neuronal survival (1-3), and its regulation by external stimuli has been well documented (4). Docosahexaenoic acid (DHA, 22:6, n-3), a long-chain n-3 polyunsaturated fatty acid, is highly enriched in the nervous system (5). Inadequate supply of n-3 fatty acids during prenatal and postnatal development decreases the levels of DHA in neuronal tissues with a reciprocal increase of docosapentaenoic acid (DPA, 22:5 n-6) (6), leading to a variety of visual, cognitive, and/or behavioral deficits in animal models (7-9). Conversely, dietary supply of DHA during infancy has been shown to improve mental development in humans (10, 11). In addition, it has been indicated that low levels of DHA in brain are associated with neurodegenerative diseases, such as generalized peroxisomal disorders (12) and Alzheimer's disease (13). Furthermore, DHA supplementation has been shown to alleviate symptoms in some patients with peroxisomal disorders (14), prevent dendritic pathology observed in Alzheimer's disease mouse models (15), and reduce neuronal injury in experimental brain ischemia (16, 17). Although all this evidence indicates the beneficial effect of DHA and suggest a requirement of DHA accretion in neuronal cells, underlying mechanisms are not well understood.
In neuronal membranes, DHA particularly accumulates in aminophospholipids, phosphatidylethanolamine, and phosphatidylserine (PS) (5). The phospholipid composition of eukaryotic biomembranes is stable and difficult to be perturbed (18). Nevertheless, evidence shows that DHA can modulate the PS levels in vivo and in vitro. For example, intraamniotic injection of DHA during pregnancy can increase PS in a newborn rat brain (19). Similarly, DHA supplementation in cultured glia or neuronal cells has been shown to increase PS content (20, 21). In contrast, depletion of DHA under n-3 fatty acid deficiency led to a reduced PS level in neuronal tissues, including rat brain cortex, hippocampus, pineal, and olfactory system (22-24), despite the compensatory replacement of DHA with DPA. Consistently, DHA containing phospholipids were found to be the preferred substrate to DPA species for PS biosynthesis (25).
It has been reported that DHA promotes survival of rat retinal photoreceptors during development in vitro (26). We also have shown that neuronal apoptosis induced by trophic factor removal or staurosporine treatment is inhibited by DHA, and its ability to promote PS accumulation in cell membranes is important for this effect (21, 27). All of this evidence suggests a unique role of DHA in neuronal membranes by modulating PS levels and subsequent signaling events involved in cell survival. In this regard, studying the role of DHA in survival signaling processes may provide a mechanistic basis supporting the beneficial effects of DHA observed in neurodegenerative diseases. An insight for the essential role of DHA and the adverse implication of n-3 fatty acid deficiency may be gained by investigating the differential effect of DHA and DPA on survival signaling affected by PS.
Materials and Methods
Cell Culture, Transfection, and Fatty Acid Supplementation. Neuro 2A (mouse neuroblastoma) cells (American Type Culture Collection) were cultured and supplemented with DHA (Nu Chek Prep, Elysian, MN) and/or DPA with the final fatty acid concentration of 25 μM in the presence of 40 μM vitamin E and 1-2% FBS as reported in ref. 21. Nonenriched controls received the similar treatment, but fatty acids were omitted. Vitamin E alone influenced neither the phospholipid content nor cell survival under our experimental condition. In some experiments, serine was depleted from the cell culture medium for ≈1 week before supplementation. Cells were seeded on six-well plates or Delta T4 dishes (Bioptechs, Butler, PA) for time-lapse studies. Neuro 2A cells were transfected with GFP-AktPH (a kind gift from Tobias Meyer, Stanford University, Stanford, CA) or its mutants (R15A, K20A, R67A, and R69A, mutations made at Veritas Laboratory, Rockville, MD) by using Lipofectamine 2000 (Invitrogen) and supplemented with fatty acids after 12 h of recovery. After 48 h of supplementation, cells were used for microscopy, harvested for phospholipid analysis, or serum starved for 2 days to induce apoptosis in the presence or absence of inhibitors. Caspase-3 activity was measured as described in ref. 21. Before stimulation with insulin-like growth factor (IGF)-1, cells were serum starved for 14 h.
PS Molecular Species Analysis by HPLC-Electrospray Ionization (ESI)-MS. PS molecular species were separated and determined by using reverse-phase HPLC-ESI-MS with a C18 column (150 × 2.0 mm, 5 mm) as described in ref. 21.
TUNEL Assay. The apoptotic nuclei containing free 3′OH termini in DNA were detected by using TUNEL assay kit (In Situ Cell Death Detection, Roche Applied Science Indianapolis). Hippocampal neurons were counterstained with hematoxylin (Sigma) before mounting.
Animals, Diet, and Hippocampal Cultures. Pregnant females (250-300 g), Sprague-Dawley rats from Charles River Laboratories, Portage, MI) were fed for 16 days with two different diets from the second day of pregnancy with AIN-93 based diets, containing 0.04% or 3.1% α-linolenic acid (18:3n-3) for the n-3 fatty acid deficient or adequate diet, respectively (28). Embryonic hippocampal cells were obtained from embryonic day 18 rat hippocampi, and cultured in neurobasal medium with N2 supplements as described in ref. 28, with an exception that the cell density used in this study was 60,000 cells per cm2. After 4 days, in vitro cells were deprived of trophic factors for 15 h to induce apoptosis.
EGFP-AktPH Translocation and Image Analysis. Translocation of EGFP-AktPH was monitored by fluorescence microscopy by using an inverted semimotorized IX81 Olympus (Melville, NY) microscope with a GFP filter and a ×60 oil objective. Cells were stimulated with IGF (10 ng/ml) at 35°C, maintained with a Delta T4 culture dish controller (Bioptechs). After recording the initial image, medium was exchanged with IGF-containing buffer, and time series of images were recorded every 30 seconds from 1.5 min after the IGF addition with a Roper coolsnap FX 12-bit 20-MHz camera (Roper Scientific, Trenton, NJ). The time series were analyzed by using microsuite (Soft Imaging System, Lakewood, CO). The ratio [R = (plasma membrane fluorescence - cytosolic fluorescence)/cytosolic fluorescence] representing the relative increase of fluorescence intensity in the plasma membrane over the cytosol (29) was calculated in three to five cells per condition. The R values obtained were normalized to the plateau value and then averaged. The R values presented in Fig. 7, which is published as supporting information on the PNAS web site, were not normalized to clearly indicate the lesser degree of EGFP-AktPH translocation in the mutant cells.
In Vitro PI (3)K Assay and Endogenous PIP3 Formation. In vitro PI (3)K assay was performed as reported by Varticovski et al. (30). Cell lysate (0.8-1 mg of protein) was incubated with anti-PI (3)K p85 antibody conjugated to agarose (Upstate Biotechnology, Lake Placid, NY) overnight at 4°C. Immunoprecipitated PI (3)K was incubated with 4 μg of phospholipids (PI, PIP, PIP2, and PS) in the presence of 25 μCi (1 Ci = 37 GBq) [32P]ATP (PerkinElmer) for 20 min at 37°C. Endogenous phospholipids were metabolically labeled according to Traynor-Kaplan et al. (31). Briefly, ≈1.2 × 106 cells per sample were labeled with 12.5 μCi H332PO4 (PerkinElmer) in Hepes buffer for 1 h, washed, and stimulated with 10 ng/ml IGF for the indicated time periods. Total lipids were isolated, separated on TLC as reported in refs. 30 and 31, and PIP3 formation was identified by exposing the TLC plate to x-ray film.
RT-PCR Analysis. Total RNA was isolated by using TRIzol reagent (Life Technologies, Rockville, MD) for the synthesis of first-strand cDNA by using Superscript III RT (M-MLV Reverse Transcriptase, Life Technologies) according to manufacturer's protocol. For PCR (PCR master mix, Roche Molecular Biochemicals), the following primers were used: PSS1 forward primer (5′-GGC CAT GAA GGC CTT GTT GAT CCG TAG T-3′, a 28-mer positioned at 1207-1234) and reverse primer (5′-TAT GAA TGT CCT TGA AGC TTG CCC A-3′, a 25-mer positioned at 1442-1418); PSS2 forward primer (5′-GGC TGG GCA TCT ACT GTG G-3′, a 19-mer positioned at 815-833) and reverse primer (5′-GTA GAA GGG CAG TGA CAG G-3′, 1284-1266). G3PDH primers (Clontech) were used as the positive control by providing a 0.45-kb band. A band of 235 and 469 base pairs of PCR products were visualized on 2% agarose gel stained with ethidium bromide (Sigma) for PSS1 and PSS2, respectively.
Results
Inhibition of Apoptosis by Polyunsaturates Is PS-Dependent. It is well known that most cells in culture contain very low levels of long-chain highly unsaturated fatty acids such as DHA because of the lack of δ 4-desaturation activity (32). Neuro 2A cells contained DHA and DPA at a minor level, each <1% of the total fatty acid. By enriching the cells with 25 μM DHA for 48 h, the DHA level was raised up to 15-16%, the approximate level normally found in neuronal membranes. Because DPA replaces DHA in n-3 fatty acid deficiency, the cells were supplemented with DHA and DPA at various ratios to model the varying extent of n-3 fatty acid deficiency. DHA and DPA differentially affected the PS accumulation and neuronal survival under a serum starvation condition (Fig. 1). The cells enriched with DHA or DPA alone showed significantly higher levels of the total PS in comparison to the nonenriched control (Fig. 1A). However, the level of PS attained by DPA was <80% of that observed with DHA enrichment. As the proportion of DHA with respect to DPA increased, a gradual enhancement in the total PS content was observed, in agreement with the previous report that DPA phospholipids are less effective than DHA species as substrates in PS biosynthesis (25). The protective effect against apoptotic cell death evaluated by caspase-3 activity gradually increased with increasing DHA proportion (Fig. 1B), indicating that the PS content and antiapoptotic effect correlated well. The apoptotic cell death evaluated by in situ DNA fragmentation showed consistent results (Fig. 1 C and D). DHA enrichment significantly decreased the serum starvation-induced TUNEL positive cells in comparison with the nonenriched control. Cells enriched with DPA also contained a reduced number of TUNEL positive cells, however, to a lesser extent, again confirming the correlation between the PS content and the antiapoptotic effect. Depleting serine from the enrichment media significantly reduced the PS increase caused by DHA or DPA (Fig. 1E), in agreement with the serine requirement for the mammalian PS synthesis by using the serine-base exchange reaction (33). The protective effect of these polyunsaturated fatty acids against cell death was accordingly diminished in these cells, indicating that PS accumulation is primarily responsible for the protective effect of both DHA and DPA. Silencing the pss1 and pss2 genes, which are the responsible enzymes in PS biosynthesis, decreased their mRNA levels considerably (Fig. 1F). However, the gene silencing affected neither the PSS protein level nor the PS accumulation increased by DHA. Only the cells enriched with DHA in a serine-depleted condition showed inhibited PS accumulation. The data suggest that these very long-chain polyunsaturated fatty acids are uniquely capable of expanding the PS pool in living cells. This observation is consistent with the fact that neuronal tissues, which are abundant with DHA, contain higher levels of PS in comparison with nonneuronal tissues (34, 35). It is also in agreement with the PS reduction previously observed selectively in neuronal tissues with n-3 fatty acid deficiency
Fig. 1.
Effect of DHA on PS accumulation and apoptotic cell death. (A and B) Differential effects of DHA and DPA on PS accumulation (A) and caspase-3 activity induced by serum starvation (B). (C-E) PS-dependent inhibition of apoptosis evidenced by TUNEL positive cells (C) and representative micrographs (D), with respect to PS accumulation altered by polyunsaturated fatty acids and serine depletion (E). (F) Effect of pss1 and pss2 gene silencing on DHA-induced PS accumulation. Caspase-3 activity was expressed as the percentage to the basal value from the nonenriched control. Statistical significance was tested against nonenriched (A and F) or nonenriched serum-free control (B and C). ***, P < 0.001; **, P < 0.01; *, P < 0.05; NS, not significant.
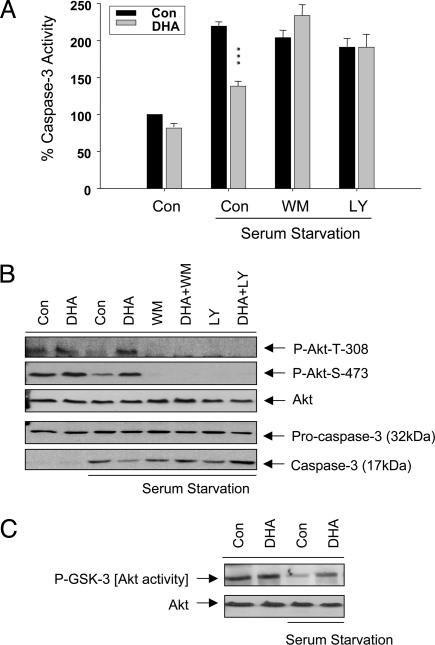
Protection by DHA Is PI (3)K-Dependent. When cells were incubated with PI (3)K inhibitors such as wortmanin (WM) and LY-294002 (LY), the protective effect of DHA enrichment shown by the suppressed caspase-3 activation was completely abolished, indicating that PI (3)K pathway is required for the protection by DHA (Fig. 2 A and B). Phosphorylation of Akt at Thr-308 and Ser-473, the consequence of PI (3)K activation (36), was significantly reduced by serum starvation and enrichment of cells with DHA before serum starvation, partially prevented the reduction of Akt phosphorylation at Thr-308 and Ser-473 (Fig. 2B). In vitro kinase assay by using immunoprecipitated Akt consistently revealed the reduction of Akt activity under serum starvation, which was partially rescued by DHA enrichment (Fig. 2C). These data indicate that DHA's antiapoptotic effect was mediated through the PI (3)K/Akt pathway.
Fig. 2.
Role of PI (3)/Akt kinase pathway in DHA's antiapototic effect. (A) The presence of wortmanin (WM) (10 nM) or LY 294002 (LY) (50 μM) during serum starvation abolished DHA's inhibition of caspase-3 activity in serum-starved cells. Results are percentages of the nonenriched control value, and statistical significance was tested against serum-free nonenriched control. ***, P < 0.001. (B) Western blot data showing the reduction of Akt phosphorylation upon serum starvation and the prevention of this reduction by DHA and abolition of DHA's protection by WM and LY. (C) Akt activity maintained by DHA during serum deprivation. Results are representative of at least two independent experiments.
DHA Facilitates Akt Translocation. To determine the target of the PS- and PI (3)K/Akt pathway-dependent protection by DHA, we examined the translocation of Akt, the prerequisite step for the phosphorylation and activation of Akt (37). Akt translocation, resulting from the interaction between the PH domain of Akt and the plasma membrane PIP3 generated by PI (3)K activation, was evaluated by using the PH domain of Akt tagged with the enhanced green fluorescence protein (EGFP-AktPH) (38). Upon stimulation with IGF, Neuro 2A cells enriched with DHA and DPA showed considerably faster translocation in comparison with the non-supplemented control (Fig. 3 A and B). The ratio [R = (plasma membrane fluorescence - cytosolic fluorescence)/cytosolic fluorescence] was used to represent the relative increase in the fluorescence intensity at the plasma membrane over the cytosol as reported elsewhere (29). Whereas translocation of EGFP-AktPH to the plasma membrane reached a plateau at ≈5.5 min in control cells, cells enriched with DHA or DPA GFP-AktPH showed such translocation within 2.5 or 3.5 min, respectively. The faster translocation observed in DHA and DPA enriched cells was due to the PS concentration, because the inhibition of PS accumulation by depleting serine from the enrichment media prevented the facilitated translocation observed in these cells (Fig. 3B). Therefore, the differential ability to raise the PS concentration by DPA with respect to DHA is likely responsible for the slower translocation observed in DPA-enriched cells in comparison with DHA-enriched cells. Movies 1 and 2, which are published as supporting information on the PNAS web site, show faster translocation of EGFP-AktPH in DHA-enriched Neuro 2A cells upon IGF stimulation in comparison with nonenriched control cells.
Fig. 3.
Effect of DHA and mutation on Akt translocation and phosphorylation. (A) Representative micrographs showing translocation of GFP-AktPH at indicated time points after IGF-1 stimulation with 10 ng/ml in cells supplemented with fatty acids with or without serine. Insets show the fluorescence intensity profile across a transverse section in one cell. (B) Time course of translocation by using the averaged relative R values normalized to the plateau R value for each group (see Materials and Methods). (C) Representative micrographs of IGF-induced translocation of GFP-AktPH mutants (R67A, R69A, R15A, and K20A). (D) Time course of Akt phosphorylation detected with antibodies specific to phospho-Akt Thr-308. The representative data from at least four independent experiments, performed in duplicate, are shown.
The high resolution crystal structure has indicated that besides the binding pocket for PIP3, the PH domain of Akt contains basic residues, Arg-15, Lys-20, Arg-67, and Arg-69, which form a flat surface for a possible interaction with acidic membrane phospholipids (39). We observed that the mutation of Arg-15 or Lys-20 to Ala resulted in the complete inhibition of EGFP-AktPH translocation in response to IGF at concentrations of up to 1 μg/ml (Fig. 3C). Because these basic residues are located near the phosphatidylinositol triphosphate (PIP3) binding pocket, the lack of translocation is most likely due to structural changes at the PIP3 binding site caused by the mutation. When Arg-67 or Arg-69, which are located further away from the PIP3 binding pocket, were mutated to Ala, translocation did occur, but to a far lesser degree in comparison to the wild type (Figs. 3C and 7). DHA supplementation did not produce any significant effect in these mutants, supporting the suggested interaction of these basic residues with acidic membrane components, in this case PS, and its importance for the observed acceleration of Akt translocation by DHA.
The observed faster translocation of EGFP-AktPH should be translated into faster Akt phosphorylation (37), because the consequence of Akt translocation from cytosol to plasma membrane is its phosphorylation. As indicated by the Western blot data in Fig. 3D, both nonenriched control and DHA-enriched cells showed time-dependent phosphorylation of Akt at Thr-308 upon IGF stimulation. In agreement with the observed faster translocation of EGFP-AktPH, Akt phosphorylation was considerably faster in DHA-enriched cells in comparison with nonenriched control (Fig. 3D).
DHA Has No Effect on PI (3)K Activation. The faster translocation and phosphorylation of Akt observed in DHA-enriched cells could reflect the possible effect of DHA enrichment on the signaling events, which are upstream of Akt translocation. However, the data from in vitro kinase activity of immunoprecipitated PI (3)K shown in Fig. 4A indicated that PI (3)K was similarly activated in nonenriched control or DHA-enriched cells in response to IGF stimulation. In addition, endogenous PIP3 formation monitored by metabolic 32P labeling (31) followed virtually the same time course for both control and DHA-enriched cells (Fig. 4B). These data clearly demonstrate that it is not the upstream events preceding Akt translocation but the Akt translocation to plasma membrane itself that is affected by DHA enrichment in survival signaling.
Fig. 4.
Effect of DHA on the time course of PI (3)K activation or PIP3 formation. (A) In vitro PI phophorylation by immunoprecipitated PI (3)K from control and DHA-enriched cells stimulated with IGF (10 ng/ml) for indicated time periods. (B) PI phosphorylation in control and DHA-enriched cells that were metabolically prelabeled with H332PO4 for 1 h and stimulated for indicated time periods. In both experiments, total lipids were extracted, separated on TLC plates, and exposed to x-ray film. Representative radiographs from two independent experiments are shown.
In Vivo DHA Depletion Promotes Neuronal Cell Death. Based on the PS-dependent survival signaling observed in transformed cells, the implication in neuronal survival of the in vivo PS reduction, resulting from the depletion of DHA under n-3 fatty acid-deficient condition, was examined by using embryonic day (E)18 hippocampal primary cultures. When pregnant female rats were fed with an n-3 fatty acid-deficient diet from the day 2 of pregnancy, reciprocal replacement of DHA by DPA was observed in E18 fetal hippocampi although the DHA accretion was low (3.3%) at this stage of development (Fig. 5A). Consistently, the PS content in the fetal hippocampi from the n-3 fatty acid-deficient group was lower by 15% in comparison with the n-3 fatty acid adequate group (Fig. 5A). The spontaneous apoptosis under basal condition was slightly but significantly higher in hippocampal primary culture from n-3 fatty acid-deficient animals in comparison with the n-3 fatty acid adequate culture (32% vs. 25%) (Fig. 5B). Under trophic factor withdrawal condition, a dramatic increase in cell death (42%) was observed in n-3 fatty acid-deficient cultures, whereas n-3 fatty acid adequate cultures did not show significant increase from the basal level, as indicated by the representative micrographs in Fig. 5C. The increased susceptibility to cell death in DHA-deficient neurons where PS was reduced is consistent with the PS-dependent protection by DHA against neuronal apoptosis observed in Neuro2A cells.
Fig. 5.
Effect of in vivo depletion of n-3 fatty acids on apoptotic cell death and the fatty acid and PS profile in embryonic day 18 hippocampal cultures. (A) Reciprocal replacement of DHA with DPA and significant decrease in PS content by n-3 fatty acid depletion. (B and C) TUNEL-positive cells increased significantly in n-3 fatty acid-deficient culture in basal and overnight trophic factor withdrawal (TFW) conditions (B) as shown in representative micrographs (C). Results represent three independent experiments performed in quadruplicate with ≈1,500 cells evaluated per treatment. The statistical significance was tested against adequate (A) and adequate basal (B) values. *, P < 0.05; ***, P < 0.001.
Discussion
Our present data demonstrate that PS levels were in direct correlation with the DHA content in neuronal cells. The protection against apoptotic cell death induced by serum starvation was sensitive to the PS level altered by the DHA status. The increase of PS by DHA resulted in faster translocation and phosphorylation of Akt without altering PI (3)K activation, indicating that translocation of Akt is the target for the PS involvement in survival signaling pathways. DPA is less effective than DHA in increasing PS levels, resulting in less-efficient translocation of Akt and less protection from cell death. Our current data represent a mechanistic demonstration that membrane modification introduced by DHA influences a targeted event in survival signaling.
The PS level in mammalian cells is difficult to perturb, as evidenced by the little changes in PS levels, even after inhibiting proteins or knocking out the genes responsible for PS biosynthesis (40, 41). Considering the well established notion that changes in the phospholipid proportion are not easily introduced (18), unlike fatty acyl composition within phospholipids, the modulation of the PS pool in neuronal membranes by the DHA status appears to be a unique mechanism, allowing the disruption of membrane homeostasis in living animal cells. Previous demonstration that DHA is readily released from astroglia and supplied to a neuron (42, 43) suggested a trophic action of DHA. In agreement with this suggestion, DHA enrichment supports the neuronal survival by preventing apoptotic cell death under adverse conditions (21). The antiapoptotic effect of DHA depends on its ability to accumulate PS in neuronal membranes, because depletion of serine during the enrichment or decreasing the DHA proportion by replacing it with DPA, a less favorable substrate for PS synthesis (25), significantly diminished the antiapoptotic capability. These data strongly suggest that PS accumulation by DHA in neuronal membranes is an important aspect in transmitting the survival signal.
The role of Akt, a downstream effector of PI (3)K pathway, in cell survival has been extensively studied (1-4). The abolition of DHA's antiapoptotic effect by PI (3)K inhibitors observed in this study indicated the role of PI (3)K/Akt in DHA-mediated protection in neuronal survival under a serum-free condition. Moreover, DHA enrichment partially prevented the reduction of Akt phosphorylation and activity caused by serum starvation, suggesting that DHA promoted cell survival by assisting in the maintenance of basal Akt activity under an adverse condition.
Membrane translocation is an event that is prerequisite for the full activation of Akt (33) by enabling successive phosphorylation at Thr-308 and Ser-473 with kinases such as PDK (44, 45). The faster translocation of EGFP-AktPH and Akt-phosphorylation observed in DHA-enriched cells are likely due to the interaction of accumulated PS with the PH domain of Akt, because such effect was PS-dependent and observed only for the Akt-translocation and phosphorylation, but not at the PI (3)K activation level. Similar unspecific ionic interaction of the PH domain of ARNO (46) with membrane PS has been reported. Also, the involvement of PS in other signaling pathways such as Raf-1 and PKC activation (21, 47, 48) has been documented, and the importance of the PS as the potential switch for the GTPase substrate preference for GTPase activating protein has been recently demonstrated (49). The interaction between PS and Akt was supported by the abolition or significant reduction of translocation after the mutation of the basic residues, which have been suggested to interact with negatively charged membrane surfaces (39). It appears that translocation of Akt to the plasma membrane is neither efficient nor stable upon interaction with PIP3 only but also requires the interaction with acidic phospholipids in membrane apart from the PIP3 binding pocket (39). The PS-dependent acceleration of Akt translocation/activation may be particularly important for cell survival under suboptimal conditions, such as in trophic factor depleted condition where PIP3 generation is limited. Fig. 6 summarizes the effect of DHA in survival signaling. DHA enrichment in neuronal membranes increases PS. Increasing the membrane PS concentration promotes the interaction of the PH domain of Akt with the plasma membrane, facilitating translocation and phosphorylation of Akt. The phosphorylation and activation of Akt facilitated by the membrane PS-Akt interaction suppress caspase-3 activation and cell death, thus promoting cell survival, especially under conditions where PIP3 generation is diminished. Under those adverse conditions, cell death due to the reduction of PIP3 signaling may be rescued by the high content of PS in neuronal membranes. In this regard, the capacity to concentrate membrane PS by DHA is an important determinant for modulating survival signaling. DPA that replaces DHA under n-3 fatty acid deficiency is as effective as DHA neither in accumulating membrane PS nor in facilitating Akt activation, consequently compromising survival signaling. The cell survival inadequately supported, especially under stressed conditions, may have a significant implication in neurological deficit often observed in n-3 fatty acid deficiency. This mechanism may also provide an explanation to the previously reported protective effect of DHA in brain ischemia and Alzheimer's disease, although other mechanisms such as neuroprotectin formation (50) and antioxidant defense (16) also might contribute to neuronal survival.
Fig. 6.
Positive effect of DHA on neuronal survival: a schematic model. DHA is provided from astroglia, incorporated into neurons, and promotes PS accumulation by serving as the preferred substrate for PS biosynthesis. Membrane concentration of PS facilitates Akt translocation through interaction with basic residues in the PH domain, resulting in efficient phosphorylation and activation of Akt and suppressing caspase-3 activation and cell death, especially under adverse conditions where PIP3 generation is limited. DPA, replacing DHA in n-3 fatty acid deficiency, is not as effective as DHA in promoting PS accumulation and Akt translocation and, consequently, less effective in supporting cell survival.
Supplementary Material
Acknowledgments
We thank Dr. Osamu Kuge (Kyushu University, Fukuoka, Japan) for providing the phosphatidylserine synthase-1 (PSS1) antibody.
Author contributions: H.-Y.K. designed research; M.A., F.C., and Z.W. performed research; M.A., F.C., Z.W., and H.-Y.K. analyzed data; and M.A., F.C., and H.-Y.K. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: DHA, docosahexaenoic acid; DPA, docosapentaenoic acid; PS, phosphatidylserine; PI, phosphatidylinositol; PIP, phosphatidylsinositol phosphate; PIP3, phosphatidylinositol triphosphate; PI (3)K, phosphatidylinositol 3-kinase; IGF, insulin growth factor; PH domain, pleckstrin homology domain.
References
- 1.Dudek, H., Datta, S. R., Franke, T. F., Birnbaum, M. J., Yaou, R., Cooper, G. M., Segal, R. A., Kaplan, D. R. & Greenberg, M. E. (1997) Science 275, 661-665. [DOI] [PubMed] [Google Scholar]
- 2.Crowder, J. & Freeman, R. S. (1998) J. Neurosci. 18, 2933-2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Philpott, K. L., McCarthy, M. J., Klippel, A. & Rubin, L. L. (1997) J. Cell Biol. 139, 809-815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Segal, R. A. (2003) Annu. Rev. Neurosci. 26, 299-330. [DOI] [PubMed] [Google Scholar]
- 5.Salem, N., Jr., Kim, H.-Y. & Yergey, J. A. (1986) in Health Effects of Polyunsaturated Fatty Acids in Seafoods, eds. Simopoulos, A. P. & Kifer, R. R. (Academic, New York), pp. 263-317.
- 6.Schiefermeier, M. & Yavin, E. (2002) J. Lipid Res. 43, 124-131. [PubMed] [Google Scholar]
- 7.Wheeler, T. G., Benolken, R. M. & Anderson, R. E. (1975) Science 188, 1312-1314. [DOI] [PubMed] [Google Scholar]
- 8.Niu, S.-L., Mitchell, D. C., Lim, S. Y., Wen, Z. M., Kim, H.-Y., Salem, N., Jr., & Litman, B. J. (2004) J. Biol. Chem. 279, 31098-31104. [DOI] [PubMed] [Google Scholar]
- 9.Moriguchi, T., Greiner, R. S. & Salem, N., Jr. (2000) J. Neurochem. 75, 2563-2573. [DOI] [PubMed] [Google Scholar]
- 10.Birch, E. E., Garfield, S., Hoffman, D. R., Uauy, R. & Birch, D. G. (2000) Develop. Med. Child Neurol. 42, 174-181. [DOI] [PubMed] [Google Scholar]
- 11.Willatts, P., Forsyth, J. S., Di Modugno, M. K., Varma, S. & Colvin, M. (1998) Lancet 352, 688-691. [DOI] [PubMed] [Google Scholar]
- 12.Martinez, M. (1990) Neurology 40, 1292-1298. [DOI] [PubMed] [Google Scholar]
- 13.Soderberg, M., Edlund, C., Kristensson, K. & Dallner, G. (1991) Lipids 26, 421-425. [DOI] [PubMed] [Google Scholar]
- 14.Martinez, M., Vazquez, E., Garcia-Silva, M. T., Manzanares, J., Bertran, J. M., Caltello, F. & Mougan, I. (2000) Am. J. Clin. Nutr. 71, S376-S385. [DOI] [PubMed] [Google Scholar]
- 15.Calon, F., Lim, G. P., Yang, F., Morihara, T., Teter, B., Obeda, O., Rostaing, P., Triller, A., Salem, N., Jr., Ashe, K. H., et al. (2004) Neuron 43, 633-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choi-Kwon, S., Park, K. A., Lee, H. J., Park, M. S., Lee, J. H., Jeon, S. E., Choe, M, A. & Park, K. C. (2004) Brain Res. Dev. Brain Res. 152, 11-18. [DOI] [PubMed] [Google Scholar]
- 17.Okada, M., Amamoto, T., Tomonaga, M., Kawachi, A., Yazawa, K., Mine, K. & Fujiwara, M. (1996) Neuroscience 71, 17-25. [DOI] [PubMed] [Google Scholar]
- 18.Wolf, C. & Quinn, P. J. (2004) Subcell. Biochem. 37, 317-357. [DOI] [PubMed] [Google Scholar]
- 19.Green, P. & Yavin, E. (1998) J. Neurosci. Res. 52, 129-136. [DOI] [PubMed] [Google Scholar]
- 20.Garcia, M., Ward, G., Ma, Y.-C., Salem, N., Jr., & Kim, H.-Y. (1998) J. Neurochem 10, 24-30. [DOI] [PubMed] [Google Scholar]
- 21.Kim, H.-Y., Akbar, M., Lau, A. & Edsall, L. (2000) J. Biol. Chem. 275, 35215-35223. [DOI] [PubMed] [Google Scholar]
- 22.Hamilton, J., Greiner, R., Salem, Jr., N. & Kim, H.-Y. (2000) Lipids 8, 863-869. [DOI] [PubMed] [Google Scholar]
- 23.Murthy, M., Hamilton, J., Greiner, R. S., Moriguchi, M. & Salem, N., Jr. (2002) J. Lipid Res. 43, 611-617. [PubMed] [Google Scholar]
- 24.Zhang, H., Hamilton, J. H., Salem, N., Jr., & Kim, H.-Y. (1998) J. Lipid Res. 39, 1397-1403. [PubMed] [Google Scholar]
- 25.Kim, H.-Y., Bigelow, J. & Kevala, J. H. (2004) Biochemistry 43, 1030-1036. [DOI] [PubMed] [Google Scholar]
- 26.Rotstein, N. P., Aveldano, M. I., Barantes, F. J. & Politi, L. E. (1996) J. Neurochem. 66, 1851-1859. [DOI] [PubMed] [Google Scholar]
- 27.Akbar, M. & Kim, H.-Y. (2002) J. Neurochem. 82, 655-666. [DOI] [PubMed] [Google Scholar]
- 28.Calderon, F. & Kim, H.-Y. (2004) J. Neurochem. 90, 979-988. [DOI] [PubMed] [Google Scholar]
- 29.Oancea, E. & Meyer, T. (1998) Cell 95, 307-318. [DOI] [PubMed] [Google Scholar]
- 30.Varticovski, L., Harrison-Findik, D., Keeler, M. L. & Susa, M. (1994) Biochim. Biophys. Acta 1226, 1-11. [DOI] [PubMed] [Google Scholar]
- 31.Traynor-Kaplan, A. E., Thompson, B. L., Harris, A. L., Taylor, P., Omann, G. M. & Sklar, L. A. (1989) J. Biol. Chem. 264, 15668-15673. [PubMed] [Google Scholar]
- 32.Spector, A. A. & Yorek, M. A. (1985) J. Lipid Res. 9, 1015-1035. [PubMed] [Google Scholar]
- 33.Kanfer, J. N. (1980) Can. J. Biochem. 58, 1370-1380. [DOI] [PubMed] [Google Scholar]
- 34.Rouser, G., Nelson, G. J., Fleischer, S. & Simon, G. (1968) Biological Membranes: Physical Fact and Function, ed. Chapman, D. (Academic, New York), pp. 5-69.
- 35.Ansell, G. B. & Spanner, S. (1982) in Phospholipids, eds. Hawthorne, J. N. & Ansell, G. B. (Elsevier, New York), pp. 1-49.
- 36.Alessi, D. R., James, S. R., Downes, C. P., Holmes, A. B., Gaffney, P. R. J., Reese, C. I. & Cohen, P. (1997) Curr. Biol. 7, 261-269. [DOI] [PubMed] [Google Scholar]
- 37.Andjelkovic, M., Alessi, D. R., Meier, R., Fernandez, A., Lamb, N. J., Frech, M., Cron, P., Cohen, P., Lucocq, J. M. & Hemmings, B. A. (1997) J. Biol. Chem. 272, 31515-31524. [DOI] [PubMed] [Google Scholar]
- 38.Haugh, J. M., Codazzi, F., Teruel, M. & Meyer, T. (2000) J. Cell. Biol. 151, 1269-1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thomas, C. C., Deak, M., Alessi, D. R. & van Aalten, D. M. (2002) Curr. Biol. 12, 1256-1262. [DOI] [PubMed] [Google Scholar]
- 40.Kuge, O., Hasegawa, K., Saito, K. & Nishijima, M. (1998) Proc. Natl. Acad. Sci. USA 95, 4199-4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bergo, M. O., Gavino, B. J., Steenbergen, R., Sturbois, B., Parlow, A. F., Sanan, D. A., Skarnes, W. C., Vance, J. E. & Young, S. G. (2002) J. Biol. Chem. 277, 47701-47708. [DOI] [PubMed] [Google Scholar]
- 42.Garcia, M. C. & Kim, H.-Y. (1997) Brain Res. 768, 43-48. [DOI] [PubMed] [Google Scholar]
- 43.Moore, S. A. (1993) Adv. Exp. Med. Biol. 331, 229-233. [DOI] [PubMed] [Google Scholar]
- 44.Anderson, K. E., Coadwell, J., Stephens, L. R. & Hawkins, P. T. (1998) Curr. Biol. 8, 684-691. [DOI] [PubMed] [Google Scholar]
- 45.Balendran, A., Casamayer, A., Deak, M., Patterson, E., Gaffney, P., Currie, R., Down, C. P. & Alessi, D. R. (1999) Curr. Biol. 9, 393-404. [DOI] [PubMed] [Google Scholar]
- 46.Macia, E., Paris, S. & Chabre, M. (2000) Biochemistry 39, 5893-5901. [DOI] [PubMed] [Google Scholar]
- 47.Ghosh, S., Strumm, J. C., Siorra, V. A., Daniel, L. & Bell, R. M. (1996) J. Biol. Chem. 271, 8472-8480. [DOI] [PubMed] [Google Scholar]
- 48.Newton, A. C. & Keranen, L. M. (1994) Biochemistry 33, 6651-6658. [DOI] [PubMed] [Google Scholar]
- 49.Ligeti, E., Dagher, M. C., Hernandez, S. E., Koleske, A. G. & Settleman, J. (2004) J. Biol. Chem. 279, 5055-5058. [DOI] [PubMed] [Google Scholar]
- 50.Mukherjee, P. K., Marcheselli, V. L., Serhan, C. N. & Bazan, N. G. (2004) Proc. Natl. Acad. Sci. USA 101, 8491-8496. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.