Abstract
Cyclobutane pyrimidine dimers are the major DNA photoproducts produced upon exposure to UV radiation. If left unrepaired, these lesions can lead to replication errors, mutation, and cell death. Photolyase is a light-activated flavoenzyme that binds to pyrimidine dimers in DNA and repairs them in a reaction triggered by electron transfer from the photoexcited flavin cofactor to the dimer. Using gold electrodes modified with DNA duplexes containing a cyclobutane thymine dimer (T<>T), here we probe the electrochemistry of the flavin cofactor in Escherichia coli photolyase. Cyclic and square-wave voltammograms of photolyase deposited on these electrodes show a redox signal at 40 mV versus normal hydrogen electrode, consistent with electron transfer to and from the flavin in the DNA-bound protein. This signal is dramatically attenuated on surfaces where the π-stacking of the DNA bases is perturbed by the presence of an abasic site below the T<>T, an indication that the redox pathway is DNA-mediated. DNA repair can, moreover, be monitored electrically. Exposure of photolyase on T<>T-damaged DNA films to near-UV/blue light leads to changes in the flavin signal consistent with repair, as confirmed by parallel HPLC experiments. These results demonstrate the exquisite sensitivity of DNA electrochemistry to perturbations in base pair stacking and the applicability of this chemistry to probe reactions of proteins with DNA.
Keywords: DNA charge transport, DNA electrochemistry, thymine dimers
DNA-modified gold electrodes have proven to be invaluable tools in both the study and the application of DNA-mediated charge transport (CT) chemistry (1-10). In these systems, the supramolecular self-assembly of thiol-modified DNA duplexes on a gold surface yields well defined monolayers that can serve as platforms for electrochemical assays of DNA CT. In these assays, the base pair stack is interrogated through the reduction of an intercalated reporter such as methylene blue (5, 7) or daunomycin (2, 8); the yield of reduced probe provides a measure of the efficiency of DNA CT. A wealth of experimental data has established that DNA CT is exquisitely sensitive to even subtle perturbations in the base pair π-stack, including single base mismatches (3, 8, 9). Exploiting this dramatic effect, DNA-modified surfaces have been successfully used in the electrochemical detection of mismatches and base lesions (3, 9). Protein-DNA interactions can also be investigated by using these surfaces. Perturbations of the DNA base pair stack caused by protein binding through base flipping or kinking are clearly detected by using this methodology (10). Indeed, this technique can provide a sensitive probe of protein-DNA binding and reaction.
The remarkable sensitivity of CT efficiency to DNA lesions and mismatches, as well as the observation that DNA-binding proteins can modulate long-range CT (10, 11), has provided an impetus for the evaluation of a physiological role for DNA CT. DNA-modified gold surfaces have recently been used in the study of DNA repair proteins possessing [4Fe-4S] clusters, such as MutY and EndoIII (12, 13). Our investigations indicate that DNA binding is requisite for the redox activity of these proteins and that CT from the electrode surface to the [4Fe-4S] cluster occurs in a DNA-mediated fashion. Based on these observations, we have recently proposed a model for how base excision repair enzymes may use DNA CT as a scanning mechanism to redistribute themselves onto damaged regions of the genome (12-14). Notably, these studies of DNA repair proteins on modified gold surfaces demonstrate that a redox-active cofactor within a protein can serve as the reporter moiety in our electrochemical assays; we are not limited to redox-active intercalators in this regard. Furthermore, these results suggest that DNA CT could potentially be harnessed for monitoring DNA repair.
Here, we employ DNA-modified electrodes to probe the repair protein Escherichia coli photolyase. Photolyase is a flavoenzyme that repairs cyclobutane pyrimidine dimer lesions in a reductive catalytic cycle upon irradiation with blue light (15). These lesions form as a result of a photoinduced [2 + 2] cycloaddition between two adjacent pyrimidines, typically thymines, on the same DNA strand. Biochemical (16) and structural (17, 18) evidence indicates that, upon photolyase binding, the pyrimidine dimer is flipped out of the DNA helix into a pocket accessible to the flavin cofactor. Initially, this displaced thymine dimer (T<>T) lesion should disrupt DNA CT. After repair, however, the monomer thymine bases are returned to the base pair stack, and DNA CT should be restored. Thus, given that base flipping leads to a dramatic perturbation of DNA CT, photolyase binding and subsequent T<>T repair could potentially be visualized electrochemically.
The redox state of the flavin cofactor is also of critical importance to the photolyase repair photocycle; when photolyase is isolated and purified, the flavin is typically in the semiquinone oxidized form, whereas the catalytically active form of photolyase requires a fully reduced flavin (FADH-). Flavin cofactors typically range in redox potential from -450 to 150 mV versus normal hydrogen electrode, well within the window of potentials accessible by using DNA-modified electrodes (19). In view of its redox activity and its intimate role in photorepair, the flavin cofactor in photolyase represents an ideal probe for real-time monitoring of DNA repair on our modified electrodes. Here, we report the electrochemistry of the flavin cofactor in photolyase by using gold electrodes modified with DNA duplexes containing T<>T. We find that changes in the flavin signal upon photoreactivation can be used to track DNA repair.
Materials and Methods
Protein and T<>T DNA. Oligonucleotides were prepared on an Applied Biosystems 394 DNA synthesizer using standard phosphoramidite chemistry and were purified by reverse-phase HPLC. T<>T-containing oligonucleotides were prepared photochemically with acetophenone as a triplet photosensitizer and purified by reverse-phase HPLC as described in refs. 20 and 21. Preparation and purification of E. coli photolyase and the W306F mutant were adapted from established protocols (22) with the exception that DTT was excluded from all buffers. Proteins were kept in storage buffer (50 mM Tris·HCl, pH 7.5/1 mM EDTA/50 mM KCl/50% glycerol) at -20°C. Apoprotein was prepared as described in ref. 23 by dialyzing native photolyase against 50 mM Tris·HCl, pH 7.5/1 mM EDTA/100 mM KCl/2 M KBr at 4°C using Spectra/Por dialysis tubing (molecular weight cutoff of 12-14 kDa). The dialysis buffer was changed daily, and the UV-visible absorbance of the protein solution was monitored every second day. Once no absorption was detected in the region of 340-700 nm (5 days), the apoprotein was dialyzed against storage buffer for 8 h at 4°C and then stored at -20°C.
Electrochemical Measurements. Cyclic and square-wave voltammetry experiments were carried out at ambient temperature by using an electrochemical analyzer equipped with a picoamp booster and Faraday cage (CH Instruments, Austin, TX). A standard three-electrode configuration was used, consisting of a 0.2-cm2 gold-on-mica working electrode, a modified AgCl/Ag 1 M KCl reference electrode, and a platinum wire auxiliary electrode within a low-volume (50-200 μl) electrochemical cell designed and built in-house. Because of volume constraints, the reference electrode is separated from the solution by a tip filled with 4% agarose in 3 M NaCl.
DNA-Modified Electrodes. Single-stranded oligonucleotides modified with a thiol-terminated, σ-bonded tether (3′-DNA-5′-O2CNH(CH2)6NHCO(CH2)2SH) were prepared as described in ref. 24. After HPLC purification, the thiol-modified single strand was hybridized with its complement, either containing or lacking a T<>T, by heating a mixture of equimolar amounts of each strand (0.1 mM) in 5 mM sodium phosphate/50 mM NaCl, pH 7, to 90°C followed by slow cooling to room temperature. This solution of duplex DNA was deposited on the gold surface for 12 h, thoroughly rinsed with buffer, backfilled with 1 mM mercaptohexanol, and used for electrochemical experiments.
Photorepair Experiments. Precautions were taken to exclude all extraneous light before and during the experiments. Before irradiation, samples were prepared under “gold” light (using General Electric bulbs) to avoid exposure to near-UV/blue light. Photolyase samples (20-300 μM) in 50 mM Tris·HCl, pH 7.5/50 mM KCl/1 mM EDTA/10% glycerol were incubated on modified gold surfaces for 2 min before irradiation. DNA-modified gold electrodes were exposed to black light by using a F15T8/BLB lamp (λmax = 366 nm, 0.4 mW cm-2 at 354 nm) or blue LED (λmax = 470 nm, 0.25 mW cm-2 at 354 nm) for up to 130 min. Examination of solvent levels preirradiation and postirradiation ensured that no noticeable evaporation was occurring on the time scale of the experiments.
HPLC Assay for T<>T Repair. After photoreactivation, the unthiolated DNA oligonucleotides were removed from the electrode surface by washing with 500 μl of buffer (5 mM sodium phosphate, pH 7/50 mM NaCl) at 75°C in 100-μl aliquots. The washings were collected and analyzed by reverse-phase HPLC on a Microsorb-MV C18 reversed phase column (4.6-mm i.d., 30-cm length), eluting with a gradient of 50 mM NH4OAc/CH3CN (95:5 to 70:30 over 30 min). These samples were compared with oligonucleotide standards of identical sequence, either with or without a T<>T.
Results and Discussion
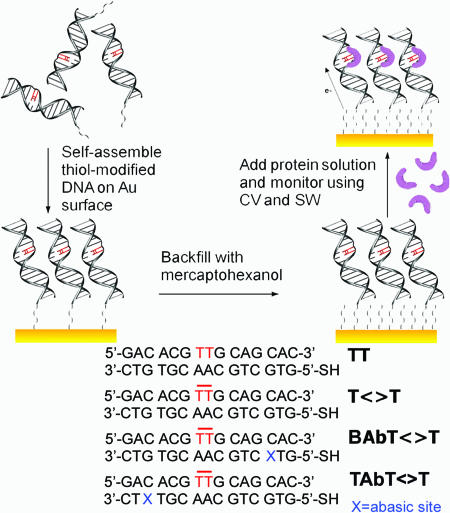
Electrochemistry of Photolyase on DNA-Modified Electrodes. DNA duplexes for electrochemical experiments are prepared by hybridizing either undamaged or T<>T-damaged ssDNA with its thiolated complement to yield duplexes TT or T<>T, respectively. As illustrated in Fig. 1, the duplexes are then assembled on the Au electrode surface in a loosely packed monolayer, allowing sufficient space for access of the protein to the DNA, and the surface is passivated with mercaptohexanol to preclude direct interaction of the photolyase with the electrode surface. The electrode is then incubated with protein, and electrochemical experiments are performed. Note that DTT is excluded from all protein samples and buffers to avoid degradation of the electrode surface.
Fig. 1.
Schematic of the experimental plan. Thiol-modified T<>T DNA duplexes are assembled on a Au electrode surface in a loosely packed monolayer. The electrode surface is then backfilled with mercaptohexanol. A solution of photolyase (purple) is incubated on the electrode, and electrochemical experiments are performed. The sequences used in the electrochemical experiments also are shown.
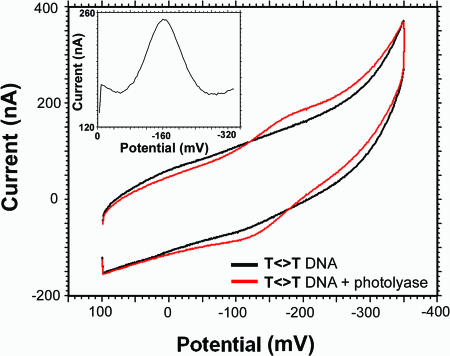
Fig. 2 shows the results of cyclic voltammetry and square-wave voltammetry experiments on the electrode surface modified with T<>T. A weak but reversible redox signal associated with photolyase is seen at -160 mV versus AgCl/Ag 1 M KCl. Based on the linear dependence of the peak current with the scan rate, the redox species is surface-bound (25). When apoprotein, which can bind DNA nonspecifically (23), is instead incubated on the T<>T-modified gold surface, no redox signal is observed. We therefore assign this redox couple of 40 mV versus normal hydrogen electrode to the DNA-bound flavin cofactor.
Fig. 2.
Cyclic voltammetry of photolyase (60 μM) on a gold electrode. The electrode was modified with T<>T in 50 mM Tris·HCl, pH 7.5/50 mM KCl/1 mM EDTA/10% glycerol. Potentials are measured versus a AgCl/Ag 1 M KCl reference electrode. (Inset) Square-wave voltammogram of photolyase (60 μM) on T<>T.
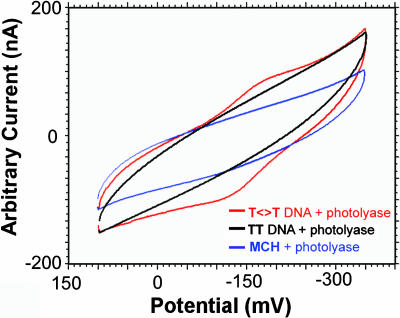
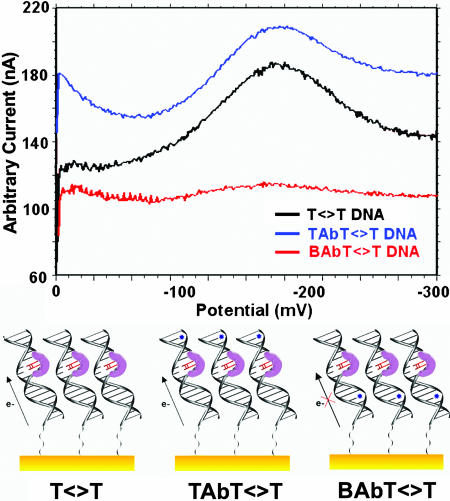
At low protein concentrations (20-100 μM), this flavin signal is seen only on electrode modified with T<>T-damaged DNA, not on mercaptohexanol lacking DNA nor on surfaces modified with undamaged DNA (Fig. 3). At higher photolyase concentrations (>150 μM), a weak signal at the same potential can be observed electrochemically; we associate this signal with protein binding to undamaged duplex. Experiments were also performed to establish the nature of the electron transfer pathway from the electrode to the flavin cofactor. An abasic site represents a significant perturbation in the DNA base pair stack that can also be detected electrochemically as an attenuation in signal from redox probes bound to DNA (3). We wanted to explore whether flavin reduction occurs through a DNA-mediated pathway or whether the T<>T-modified electrode serves to concentrate the protein, permitting direct flavin reduction at the surface. We therefore compared the photolyase redox couple on electrode surfaces modified with T<>T and BAbT<>T, the T<>T-containing oligomer with an intervening abasic site inserted in the sequence below the T<>T. As is evident in Fig. 4 with square-wave voltammetry, the presence of the intervening abasic site leads to a dramatic attenuation in the flavin redox signal compared with T<>T; in contrast, no attenuation in redox signal is observed compared with T<>T when an abasic site is positioned above the T<>T site (TAbT<>T). Thus, photolyase is not reduced simply as a result of being concentrated near the electrode surface. Rather, the charge transfer is DNA-mediated.
Fig. 3.
Cyclic voltammetry of photolyase (50 μM) on gold electrodes. The electrodes were modified with TT (black), mercaptohexanol (MCH) (blue), and T<>T (red).
Fig. 4.
Square-wave voltammetry of photolyase (50 μM) on T<>T-, TAbT<>T-, and BAbT<>T-modified gold electrodes. The abasic site below the dimer disrupts the pathway between the electrode and the photolyase, resulting in a greatly attenuated electrochemical signal.
It is noteworthy that the signal observed for the T<>T-bound photolyase is particularly weak and broad, even without an intervening abasic site. The intensity of the redox signal corresponds to a surface coverage of ≈1 pmol/cm2, markedly lower than the expected density of DNA on the surface, 12 pmol/cm2, as measured previously by radioactive tagging (10). The presence ofaT<>T within a DNA duplex leads to a kink in the DNA (26, 27), and such disruptions in base stacking are also expected to be associated with an attenuation in signal from DNA-bound redox probes. Indeed, using methylene blue in an assay for CT efficiency (9), we observe a decrease in the methylene blue redox signal on electrodes modified with T<>T DNA relative to those modified with undamaged DNA. Although this assay provides an underestimate of the loss of CT efficiency in the case of bulky lesions that prohibit close packing of the DNA duplexes, it provides clear evidence that DNA CT is disrupted by the T<>T. Hence, the weak flavin redox signal seen in our experiments suggests that the T<>T intervenes between the flavin cofactor and the DNA stack. This idea is supported by the fact that the signal intensity grows dramatically upon repair of the damaged DNA (see below).
These results establish that the DNA-bound photolyase can be accessed electrochemically. The DNA-bound flavin cofactor has a redox couple of 40 mV versus normal hydrogen electrode. The pathway for flavin reduction is DNA-mediated so that the flavin is electronically coupled with the base pair stack, albeit weakly, given the intervening T<>T.
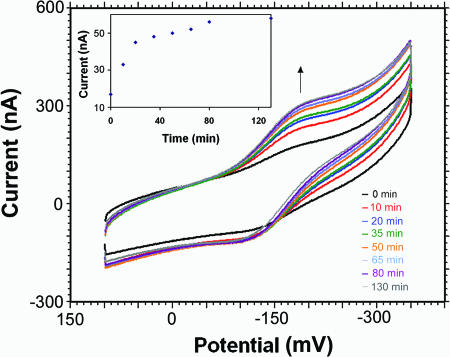
T<>T Repair Probed Electrochemically. Photoactivation of photolyase bound to DNA containing T<>T lesions leads to the repair of T<>Ts, and, as shown in Fig. 5, this repair process can be monitored electrically by irradiating photolyase bound to DNA-modified surfaces containing T<>T. Irradiation either with a black light (λmax = 366 nm) or with a blue LED (λmax = 450 nm) leads to a rapid growth of peak current at 40 mV over several minutes, followed by a plateau. Without irradiation, no significant changes in peak current are evident over this time period. Nor are changes evident upon irradiation of photolyase on surfaces passivated with mercaptohexanol and DNA lacking T<>Ts or simply passivated with mercaptohexanol but no DNA. Photolyase, light, and T<>T are all requisite to observe these electrochemical changes. After the light is turned off, the signal seen for photolyase bound to T<>T decays slowly over a period of several hours; the presence of competing T<>T-modified DNA (10 μM) in solution hastens this signal decay.
Fig. 5.
Effect of irradiation with black light over a period of 130 min on the electrochemistry of photolyase (60 μM) on a T<>T-modified gold electrode. (Inset) Plot of current versus irradiation time. As a function of irradiation, the photolyase signal increases, reaching a plateau at ≈40 min (consistent with T<>T repair) to yield a well stacked DNA duplex.
The light-dependent increase in signal that we observe until a plateau is reached is attributed to T<>T repair. The magnitude of this increase in protein signal as a result of irradiation is also meaningful. Before irradiation, the protein signal is weak, corresponding to a surface coverage of ≈1 pmol/cm2, which we attribute to the poor coupling of the flavin to the DNA-modified electrode owing to the intervening destacked T<>T. After 130 min of irradiation, as a function of repair, the observed signal increases to a calculated surface coverage of 8 pmol/cm2, much closer to the expected 12 pmol/cm2, for the surface density of DNA helices. Thus, the efficiency of DNA-mediated CT to the flavin is restored as the T<>T lesion is repaired. The increase in the flavin signal appears to parallel the photoinitiated repair process.
We also may compare T<>T repair on the surface and in solution under the same experimental conditions (protein and DNA concentrations, light intensity, etc.). Repair in the DNA films appears to be somewhat (≈50%) more efficient compared with repair in solution; this difference could be attributed to local concentration effects at the electrode surface.
Local concentration effects are also evident at the endpoint of the reaction. It is important to note that the flavin signal plateaus to a high value associated with repair, despite the fact that photolyase has a lower affinity for undamaged DNA and therefore might be expected to dissociate from the DNA film. Nevertheless, the signal increases rapidly upon irradiation, and the flavin signal is only lost very slowly once repair is completed or if competing T<>T-containing DNA is added to the solution. Protein dissociation from the DNA-modified surface therefore appears to be slower than in bulk solution (28).
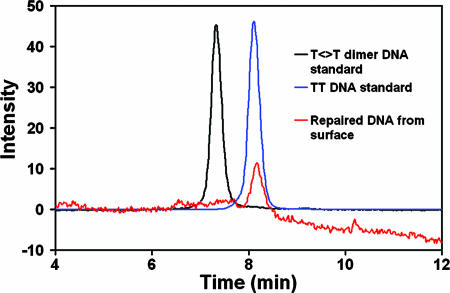
Chemical analysis confirms that the electrochemical changes we observe are associated with T<>T repair. HPLC traces of DNA taken from the electrode surface after irradiation may be compared with those of DNA of the same sequence with and without T<>T damage (Fig. 6). The surface DNA has the same retention time as undamaged DNA, indicating that the T<>T damage has been repaired on the surface. Furthermore, HPLC traces of light (irradiation of T<>T without photolyase) and dark (T<>T with photolyase but without irradiation) controls indicate that both photolyase and light are required for T<>T repair. Notably, T<>T repair does not occur by electrochemical reduction, consistent with earlier results (29); the potentials used to reduce the flavin are insufficient to trigger T<>T repair without photolyase.
Fig. 6.
HPLC chromatograms of DNA from the electrode surface (red) after irradiation, compared with T<>T (black) and undamaged (blue) DNA standards.
Flavin Reduction as a Prerequisite for Repair. Mechanistic studies of T<>T repair by photolyase have underscored the need for the flavin to be fully reduced to effect catalysis (23, 30). When photolyase is isolated and purified, the flavin is oxidized to the catalytically inactive neutral radical form (31), and thus, commonly, an external reductant such as DTT is used for in vitro photorepair studies. Our experiments, however, preclude the use of DTT because free thiols can degrade the surface modified with thiolated DNA. In some experiments, we have used glycyl-l-tyrosine as an external reductant, but in the experiments described here, no external reductant was used. We clearly observe repair of T<>T on the DNA-modified electrodes in the absence of DTT, and, interestingly, we find comparable efficiency in repair in the presence or absence of 10 mM glycyl-l-tyrosine.
If flavin photoreduction is also a prerequisite for repair in these electrochemical experiments, an internal reductant is needed. It has been shown (32) that the neutral flavin radical can also be photoreduced to FADH- in a protein-mediated CT reaction using Trp-306 as the terminal electron donor. To explore whether Trp-306 might serve as the internal reductant, we therefore examined the electrochemistry of the photolyase mutant, W306F. The dramatic increase in the flavin redox signal typically seen upon repair by wild-type photolyase is not observed when the W306F is irradiated on the electrode modified with T<>T (data not shown). Hence, it appears that photoreduction is a necessary step preceding T<>T repair also in the electrochemical experiments.
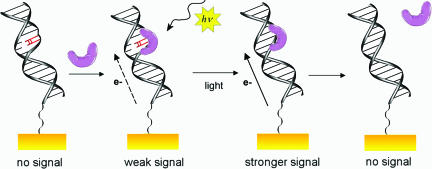
Model for Electrochemical Monitoring of Repair. The photoreaction of photolyase with T<>T lesions in DNA can be monitored electrically because of the sensitivity of DNA electrochemistry to perturbations in base stacking. Fig. 7 illustrates our interpretation of the electrochemical observations. In the absence of photolyase, no signal is observed on the electrodes modified with T<>T-containing DNA. The potentials we probe are insufficient to reduce or oxidize the DNA bases. When the protein is added to the DNA-modified surface, binding to the T<>T site occurs, and a weak signal is observed as a result of the electrochemical reduction of the DNA-bound flavin cofactor. The low signal intensity is the result of poor coupling of the flavin on the DNA-modified electrode owing to the intervening T<>T; the destacked, presumably flipped-out T<>T interferes with communication between the electrode and the protein. Upon irradiation, photoreduction of the flavin cofactor triggers the T<>T repair process; the integrity of the π-stack is restored, and the flavin signal grows in intensity. At this point, the protein is bound nonspecifically and eventually dissociates, leading to a slower loss of the redox signal. Thus, the DNA-modified electrodes provide a platform for monitoring the entire repair process, from binding to catalysis to dissociation.
Fig. 7.
Schematic illustration of the electrochemical results. From left to right: In the absence of photolyase, no signal is observed. When the protein (purple) is added, it binds to the T<>T site, and a weak signal is observed. The low signal intensity is due to the disruption of the π-stack, which interrupts communication between the electrode and the protein. Then, upon photoreactivation, the integrity of the π-stack is restored, and the signal grows in intensity. Finally, because the protein has a lower affinity for undamaged DNA, it dissociates, leading to the slow loss of the redox signal.
Implications. Cyclobutane pyrimidine dimer repair by photolyase can be monitored indirectly based on UV absorbance changes (33), cleavable molecular beacons (34), and aminopurine fluorescence quenching (35), as well as gel electrophoresis (28, 36-38) and HPLC analysis (34); transformation assays also provide an in vivo assay (22). In our experiments, DNA repair is easily observed electrochemically as an increase in the redox signal of the flavin cofactor associated with the repair of the T<>T. The growing electrochemical signal reflects the increasing integrity of the DNA base stack. No exogenous probes are needed. The sensitivity and ease of this assay furthermore offers the possibility of monitoring this reaction in real time.
Thus, DNA-modified electrodes provide powerful tools for studying the reactions of proteins with DNA. It is the exquisite sensitivity of DNA CT chemistry to perturbations in base pair stacking that underpins this methodology. DNA-based electrochemistry now provides a strategy for examining the DNA conformational changes associated with a variety of protein reactions: those involving DNA base flipping and kinking, as well as repair reactions of DNA lesions, such as the photorepair of T<>Ts by photolyase.
Acknowledgments
We thank Eric Han and Carrie Stentz for help with protein expression and purification. This work was supported by National Institutes of Health Grants GM61077 (to J.K.B.) and GM31082 (to A.S.) and a Natural Science and Engineering Research Council of Canada Postdoctoral Fellowship (to M.C.D.).
Author contributions: M.C.D. and J.K.B. designed research; M.C.D. performed research; A.S. contributed new reagents/analytic tools; M.C.D. analyzed data; and M.C.D. and J.K.B. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: T<>T, thymine dimer; CT, charge transport.
References
- 1.Drummond, T. G., Hill, M. G. & Barton, J. K. (2003) Nat. Biotechnol. 21, 1192-1199. [DOI] [PubMed] [Google Scholar]
- 2.Kelley, S. O., Jackson, N. M., Hill, M. G. & Barton, J. K. (1999) Angew. Chem. Int. Ed. 38, 941-945. [DOI] [PubMed] [Google Scholar]
- 3.Boon, E. M., Ceres, D. M., Drummond, T. G., Hill, M. G. & Barton, J. K. (2000) Nat. Biotechnol. 18, 1096-1100. [DOI] [PubMed] [Google Scholar]
- 4.Ceres, D. M., Barton, J. K. (2003) J. Am. Chem. Soc. 125, 14964-14965. [DOI] [PubMed] [Google Scholar]
- 5.Kelley, S. O., Barton, J. K., Jackson, N. M. & Hill, M. G. (1997) Bioconjugate Chem. 8, 31-37. [DOI] [PubMed] [Google Scholar]
- 6.Drummond, T. G., Hill, M. G. & Barton, J. K. (2004) J. Am. Chem. Soc. 126, 15010-15011. [DOI] [PubMed] [Google Scholar]
- 7.Boon, E. M., Jackson, N. M., Wightman, M. D., Kelley, S. O., Hill, M. G. & Barton, J. K. (2003) J. Phys. Chem. B 107, 11805-11812. [Google Scholar]
- 8.Kelley, S. O., Boon, E. M., Barton, J. K., Jackson, N. M. & Hill, M. G. (1999) Nucleic Acids Res. 27, 4830-4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boal, A. K. & Barton, J. K. (2005) Bioconjugate Chem. 16, 312-321. [DOI] [PubMed] [Google Scholar]
- 10.Boon, E. M., Salas, J. E. & Barton, J. K. (2002) Nat. Biotechnol. 20, 282-286. [DOI] [PubMed] [Google Scholar]
- 11.Rajski, S. R., Kumar, S., Roberts, R. J. & Barton, J. K. (1999) J. Am. Chem. Soc. 121, 5615-5616. [Google Scholar]
- 12.Boon, E. M., Livingston, A. L., Chmiel, N. H., David, S. S. & Barton, J. K. (2003) Proc. Natl. Acad. Sci. USA 100, 12543-12547, and correction (2004) 101, 4718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boal, A. K., Yavin, E., Lukianova, O. A., O'Shea, V. L., David, S. S. & Barton, J. K. (2005) Biochemistry 44, 8397-8407. [DOI] [PubMed] [Google Scholar]
- 14.Yavin, E., Boal, A. K., Stemp, E. D. A., Boon, E. M., Livingston, A. L., O'Shea, V. L., David, S. S. & Barton, J. K. (2005) Proc. Natl. Acad. Sci. USA 102, 3546-3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sancar, A. (2003) Chem. Rev. 103, 2203-2237. [DOI] [PubMed] [Google Scholar]
- 16.Heelis, P. F., Okamura, T. & Sancar, A. (1990) Biochemistry 29, 5694-5698. [DOI] [PubMed] [Google Scholar]
- 17.Park, H. W., Kim, S. T., Sancar, A. & Deisenhofer, J. (1995) Science 268, 1866-1872. [DOI] [PubMed] [Google Scholar]
- 18.Mees, A., Klar, T., Gnau, P., Hennecke, U., Eker, A. P. M., Carell, T. & Essen, L.-O. (2004) Science 306, 1789-1793. [DOI] [PubMed] [Google Scholar]
- 19.Ghisla, S. & Massey, V. (1989) Eur. J. Biochem. 181, 1-17. [DOI] [PubMed] [Google Scholar]
- 20.Dandliker, P. J., Holmlin, R. E. & Barton, J. K. (1997) Science 275, 1465-1467. [DOI] [PubMed] [Google Scholar]
- 21.Dandliker, P. J., Nunez, M. E. & Barton, J. K. (1998) Biochemistry 37, 6491-6502. [DOI] [PubMed] [Google Scholar]
- 22.Sancar, A., Smith, F. W. & Sancar, G. B. (1984) J. Biol. Chem. 259, 6028-6032. [PubMed] [Google Scholar]
- 23.Payne, G., Wills, M., Walsh, C. & Sancar, A. (1990) Biochemistry 29, 5706-5711. [DOI] [PubMed] [Google Scholar]
- 24.Kelley, S. O., Barton, J. K., Jackson, N. M., McPherson, L. D., Potter, A. B., Spain, E. M., Allen, M. J. & Hill, M. G. (1998) Langmuir 14, 6781-6784. [Google Scholar]
- 25.Bard, A. J. & Faulkner, N. R. (2001) in Electrochemical Methods: Fundamentals and Applications (Wiley, New York), 2nd Ed., pp. 580-631.
- 26.Husain, I., Griffith, J. & Sancar, A. (1988) Proc. Natl. Acad. Sci. USA 85, 2558-2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park, H., Zhang, K., Ren, Y., Nadji, S., Sinha, N., Taylor, J.-S. & Kang, C. (2002) Proc. Natl. Acad. Sci. USA 99, 15965-15970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Husain, I. & Sancar, A. (1987) Nucleic Acids Res. 15, 1109-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boussicault, F., Krüger, O., Robert, M. & Wille, U. (2004) Org. Biomol. Chem. 2, 2742-2750. [DOI] [PubMed] [Google Scholar]
- 30.Kim, S.-T., Heelis, P. F. & Sancar, A. (1992) Biochemistry 31, 11244-11248. [DOI] [PubMed] [Google Scholar]
- 31.Jorns, M. S., Sancar, G. B. & Sancar, A. (1984) Biochemistry 23, 2673-2679. [DOI] [PubMed] [Google Scholar]
- 32.Li, Y. F., Heelis, P. F. & Sancar, A. (1991) Biochemistry 30, 6322-6329. [DOI] [PubMed] [Google Scholar]
- 33.Jorns, M. S., Sancar, G. B. & Sancar, A. (1985) Biochemistry 24, 1856-1861. [DOI] [PubMed] [Google Scholar]
- 34.Kundu, L., Burgdorf, L. T., Kleiner, O., Batschauer, A. & Carell, T. (2002) Chembiochem 3, 1053-1060. [DOI] [PubMed] [Google Scholar]
- 35.Christine, K. S., MacFarlane, A. W., Yang, K. & Stanley, R. J. (2002) J. Biol. Chem. 277, 38339-38344. [DOI] [PubMed] [Google Scholar]
- 36.Payne, G. & Sancar, A. (1990) Biochemistry 29, 7715-7727. [DOI] [PubMed] [Google Scholar]
- 37.Ramaiah, D., Kan, Y., Koch, T., Orum, H. & Schuster, G. B. (1998) Proc. Natl. Acad. Sci. USA 95, 12902-12905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dutta, K., Hajmadi, V. S. & Verma, N. C. (1993) J. Photochem. Photobiol. B 18, 211-214. [DOI] [PubMed] [Google Scholar]