Abstract
This work presents the automated projection spectroscopy (APSY) method for the recording of discrete sets of j projections from N-dimensional (N ≥ 3) NMR experiments at operator-selected projection angles and automatic identification of the correlation cross peaks. The result from APSY is the fully automated generation of the complete or nearly complete peak list for the N-dimensional NMR spectrum from a geometric analysis of the j experimentally recorded, low-dimensional projections. In the present implementation of APSY, two-dimensional projections of the N-dimensional spectrum are recorded by using techniques developed for projection–reconstruction spectroscopy [Kupče,E.& Freeman, R. (2004) J. Am. Chem. Soc. 126, 6429–6440]. All projections are peak-picked with the available automated routine atnos. The previously undescribed algorithm GAPRO (geometric analysis of projections) uses vector algebra to identify subgroups of peaks in different projections that arise from the same resonance in the N-dimensional spectrum, and from these subgroups it calculates the peak positions in the N-dimensional frequency space. Unambiguous identification thus can be achieved for all cross peaks that are not overlapped with other peaks in at least one of the N dimensions. Because of the correlation between the positions of corresponding peaks in multiple projections, uncorrelated noise is efficiently suppressed, so that APSY should be quite widely applicable for correlation spectra of biological macromolecules, which have intrinsically low peak density in the N-dimensional spectral space.
Keywords: GAPRO, multidimensional, NMR, peak picking
In NMR studies of biological macromolecules in solution (1–4), multidimensional NMR data are commonly acquired by sampling the time domain in all dimensions equidistantly at a resolution adjusted to the populated spectral regions (5). With recent advances in sensitivity, due to high field strengths and/or cryogenic detection devices, the time required to explore the time domain in the conventional way typically exceeds by far the time needed for sensitivity considerations, so that the desired resolution in the indirect dimensions determines the duration of the experiment. In this situation of the “sampling limit,” which is common in 3D and higher-dimensional experiments with small and medium-size proteins (6), the desired chemical shift information has been collected by using “unconventional” experimental schemes, such as nonuniform sampling of the time domain (7, 8) or combination of two or more indirect dimensions (9, 10).
The concept of combining indirect dimensions has lead to reduced-dimensionality experiments (9) and G-matrix Fourier transform NMR (11, 12). In G-matrix Fourier transform NMR, several evolution periods of a multidimensional NMR experiment are combined, the data are processed by using a G-matrix, and the resulting set of spectra is analyzed jointly to identify the peaks that arise from the same spin system and to calculate their resonance frequencies (11). In another approach, projection–reconstruction NMR (13–16), the projection–cross-section theorem (17, 18) is combined with reconstruction methods from imaging techniques (19, 20). In particular, a scheme for quadrature detection along tilted planes in the time domain allows the direct recording of orthogonal projections of any multidimensional experiment at arbitrary projection angles (15). In projection–reconstruction NMR, the full multidimensional spectrum then is reconstructed from the projections of the multidimensional spectral data (13–16).
The analysis of complex NMR spectra typically involves intensive human interaction, and automation of NMR spectroscopy with macromolecules is still in development. Thereby the distinction of real peaks from random noise and spectral artifacts as well as peak overlap represent major challenges (21–23). On grounds of principle, automated analysis benefits from higher dimensionality of the spectra (24, 25), because the peaks are then more widely separated, and hence peak overlap is substantially reduced (Fig. 1).
Fig. 1.
Distribution of frequency differences, d, up to 1,000 Hz between pairs of nearest cross peaks in multidimensional NMR spectra of the protein TM1290 for the following triple resonance experiments: 3D HNCO (a), 4D HNCOCA (b), and 5D HACACONH (c). E is the number of times a given value of d is expected. The calculations were based on the published chemical shift assignment of TM1290 (BioMagResBank entry 5560) and assumed a field strength of 750 MHz.
In the present work, we combine technologies to record projections of high-dimensional NMR experiments described by Kupče and Freeman (15) and automated peak-picking using a scheme of Herrmann et al. (22) with a previously undescribed algorithm, GAPRO (geometric analysis of projections). Based on geometrical considerations, GAPRO identifies peaks in the projections that arise from the same resonance in the N-dimensional frequency space and subsequently calculates the resonance frequencies in the N-dimensional spectrum without ever considering the high-dimensional data set itself. This automated analysis of projected spectra, APSY (automated projection spectroscopy), yields a peak list of the original multidimensional experiment without any human interaction. In the following sections, the foundations of APSY are introduced, and characteristic properties of APSY are discussed. Two examples of APSY are a 4D HNCOCA experiment (26, 27) with the 63-residue protein 434-repressor(1–63) (28) and a 5D HACACONH experiment (11) with the 116-residue protein TM1290 (29).
Theoretical Background
Recording of Projection Spectra. The projection–cross-section theorem by Bracewell (17), which was introduced into NMR by Nagayama et al. (18), states that an m-dimensional (m < N) cross section, cm(t), through N-dimensional time domain data is related by an m-dimensional Fourier transformation, 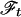 , and its inverse,
, and its inverse, 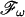 , to an m-dimensional orthogonal projection of the N-dimensional NMR spectrum, Pm(ω), in the frequency domain. Thereby, Pm(ω) and cm(t) are oriented by the same angles with regard to their corresponding coordinate systems (Fig. 2). On this basis, Kupče and Freeman proposed to record projections Pm(ω) by sampling the corresponding time domain data, cm(t), along a straight line (dashed line in Fig. 2). Quadrature detection is obtained from corresponding positive and negative projection angles for the subsequent hypercomplex Fourier transformation (15).
, to an m-dimensional orthogonal projection of the N-dimensional NMR spectrum, Pm(ω), in the frequency domain. Thereby, Pm(ω) and cm(t) are oriented by the same angles with regard to their corresponding coordinate systems (Fig. 2). On this basis, Kupče and Freeman proposed to record projections Pm(ω) by sampling the corresponding time domain data, cm(t), along a straight line (dashed line in Fig. 2). Quadrature detection is obtained from corresponding positive and negative projection angles for the subsequent hypercomplex Fourier transformation (15).
Fig. 2.
Illustration of the projection–cross-section theorem (17–19) for a 2D subspace of an N-dimensional frequency space with two indirect dimensions x and y. A 1D signal 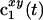 on a straight line in the time domain (tx, ty) (Left) is related to a 1D orthogonal projection
on a straight line in the time domain (tx, ty) (Left) is related to a 1D orthogonal projection 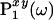 of the spectrum (ωx, ωy) in the frequency domain (Right) by a 1D Fourier transformation,
of the spectrum (ωx, ωy) in the frequency domain (Right) by a 1D Fourier transformation, 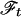 , and the inverse transformation,
, and the inverse transformation, 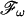 . The projection angle α describing the slope of
. The projection angle α describing the slope of  defines also the slope of
defines also the slope of 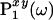 . The cross peak Qi appears at the position
. The cross peak Qi appears at the position 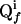 in the projection f. Further indicated are the spectral widths in the two dimensions of the frequency domain, SWx and SWy.
in the projection f. Further indicated are the spectral widths in the two dimensions of the frequency domain, SWx and SWy.
Projections of Cross Peaks. We describe here 2D projections, P2(ω), of an N-dimensional spectrum (N > 2). P2(ω) represents spectral data in a 2D plane, which is spanned by an indirect dimension with unit vector 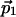 , and the direct dimension, with
, and the direct dimension, with 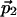 . The indirect dimension is a 1D projection of the N - 1 indirect dimensions. The same projection angles that parametrize the 2D time domain cross section, c2(t), with respect to the N time domain axes, define the position of P2(ω) with respect to the N frequency axes (Table 1). For example, with N = 5, the two unit vectors
. The indirect dimension is a 1D projection of the N - 1 indirect dimensions. The same projection angles that parametrize the 2D time domain cross section, c2(t), with respect to the N time domain axes, define the position of P2(ω) with respect to the N frequency axes (Table 1). For example, with N = 5, the two unit vectors  and
and  are expressed in the coordinate system of the N-dimensional frequency space by
are expressed in the coordinate system of the N-dimensional frequency space by
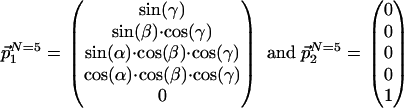 |
[1] |
In the coordinate system of a 2D projection spectrum, P2(ω), a projected cross peak 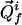 has the position vector
has the position vector 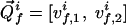 , with
, with 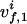 and
and 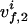 being the chemical shifts along the projected indirect dimension and the direct dimension, respectively. If the origins of both the N-dimensional coordinate system and the 2D coordinate system are in all dimensions in the center of the spectral range, the position vector
being the chemical shifts along the projected indirect dimension and the direct dimension, respectively. If the origins of both the N-dimensional coordinate system and the 2D coordinate system are in all dimensions in the center of the spectral range, the position vector 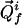 in the N-dimensional frequency space is
in the N-dimensional frequency space is
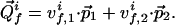 |
[2] |
The N-dimensional cross peak Qi is located in an (N - 2)-dimensional subspace L, which is orthogonal to the projection plane at the point 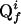 (see Fig. 2). In APSY, a set of j projections is recorded. From each N-dimensional cross peak Qi, a projected peak
(see Fig. 2). In APSY, a set of j projections is recorded. From each N-dimensional cross peak Qi, a projected peak  (f = 1,..., j) appears in each of the j projections. The set of projected peaks that arise from the same N-dimensional peak Qi,
(f = 1,..., j) appears in each of the j projections. The set of projected peaks that arise from the same N-dimensional peak Qi, 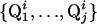 , is defined as a peak subgroup of Qi. Once the subgroup is known and contains a sufficiently large number of elements, the coordinates of Qi can be calculated from the intersections of the subspaces from all subgroup elements. For 2D projections, at least N - 1 elements are needed.
, is defined as a peak subgroup of Qi. Once the subgroup is known and contains a sufficiently large number of elements, the coordinates of Qi can be calculated from the intersections of the subspaces from all subgroup elements. For 2D projections, at least N - 1 elements are needed.
Table 1. Coordinates of the unit vector 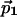 used in this work.
used in this work.
| Dimension | N = 5 | N = 4 | N = 3 |
|---|---|---|---|
| ω1 | sin(γ) | sin(β) | sin(α) |
| ω2 | sin(β)·cos(γ) | sin(α)·cos(β) | cos(α) |
| ω3 | sin(α)·cos(β)·cos(γ) | cos(α)·cos(β) | — |
| ω4 | cos(α)·cos(β)·cos(γ) | — | — |
The APSY Procedure. The procedure is outlined in Fig. 3 and illustrated in Fig. 4. The operator selects the desired N-dimensional NMR experiment, the dimension of the projection spectra, and j sets of projection angles (j ≥ N - 1) and records the projection spectra. These spectra are automatically peak picked by using the program atnos (22), which yields j peak lists. These lists contain the peaks Qgf, where g is an arbitrary numeration of the peaks in each projection f (f = 1,..., j). GAPRO arbitrarily selects N - 1 from among these peak lists and generates the subspaces Lgf that are associated with the peaks Qgf in these projections (Fig. 4a). The intersections of the subspaces Lgf in the N-dimensional space are candidates for the positions of N-dimensional cross peaks (open circles in Fig. 4a), where the number of candidate points typically exceeds the number of peaks in the spectrum. For each of these candidate points, the support, S, is then calculated as the number of subspaces from all j projections that intersect at this point. Thereby at most one subspace from each projection is considered (Fig. 4b; see text below), so that N - 1 ≤ S ≤ j. The peaks 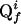 associated with the subspaces that contribute to the support of a given candidate point form a “peak subgroup.” The subgroups are ranked for high S-values, and the top-ranked subgroup is selected (in case of degeneracy, one of the top-ranked subgroups is arbitrarily selected). The subspaces originating from the peaks
associated with the subspaces that contribute to the support of a given candidate point form a “peak subgroup.” The subgroups are ranked for high S-values, and the top-ranked subgroup is selected (in case of degeneracy, one of the top-ranked subgroups is arbitrarily selected). The subspaces originating from the peaks 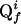 in this subgroup are removed, and new S-values for the residual candidate points are calculated from the remaining subspaces (Fig. 4c). This procedure is repeated until the value of S for all remaining candidate points falls below a user-defined threshold, Smin,1, at which point a list of peak subgroups is generated. The subgroup identification is repeated with k different, randomly chosen starting combinations of N - 1 projections (the user-defined parameter k is a small fraction of the total number of possible combinations of N - 1 projections), and k peak subgroup lists are thus obtained (gray box in Fig. 3). These lists are merged into a single list, which is again subjected to the same type of ranking procedure, so that all subgroups with S < Smin,2 are eliminated. From the resulting “final” list of subgroups, the peak positions in the N-dimensional space are calculated (Fig. 4d). Below, the computational techniques used for individual steps in Fig. 3 are described.
in this subgroup are removed, and new S-values for the residual candidate points are calculated from the remaining subspaces (Fig. 4c). This procedure is repeated until the value of S for all remaining candidate points falls below a user-defined threshold, Smin,1, at which point a list of peak subgroups is generated. The subgroup identification is repeated with k different, randomly chosen starting combinations of N - 1 projections (the user-defined parameter k is a small fraction of the total number of possible combinations of N - 1 projections), and k peak subgroup lists are thus obtained (gray box in Fig. 3). These lists are merged into a single list, which is again subjected to the same type of ranking procedure, so that all subgroups with S < Smin,2 are eliminated. From the resulting “final” list of subgroups, the peak positions in the N-dimensional space are calculated (Fig. 4d). Below, the computational techniques used for individual steps in Fig. 3 are described.
Fig. 3.
Flowchart of APSY. Square boxes indicate processes, and boxes with rounded corners denote intermediate or final results. The steps underlayed in gray are repeated k times and thus generate k lists of peak subgroups.
Fig. 4.
Illustration of the algorithm GAPRO for N = 3, j = 5, k = 1, and Smin,1 = 3 (see Fig. 3). The two indirect dimensions are in the paper plane, and the acquisition dimension ω3 is orthogonal to it. The central part of each image shows a 2D (ω1, ω2)-cross section at the frequency 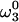 through the 3D spectrum. This 2D cross section is surrounded by five 1D cross sections at
through the 3D spectrum. This 2D cross section is surrounded by five 1D cross sections at 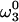 through the five experimental 2D projections of the 3D spectrum with projection angles α = 0°, 90°, 45°, -25°, and -60°. (a) The blue dots mark the result for the cross sections of the automatic peak picking of the 2D projections (Fig. 3). The algorithm then arbitrarily selected N - 1 = 2 of the j projections for the first round of spectral analysis (Fig. 3), with α = 0° and α = -60°. The intersections of the subspaces corresponding to the peaks in these two projections (green lines) identify eight candidate points in the 3D spectrum (open circles). (b) By using the subspaces from all five projections, the support S (number of intersecting subspaces, see text) is calculated for each candidate point. Yellow and red dots indicate S = 2 and S = 5, respectively. (c) One of the three candidate points with the highest support (S = 5) is arbitrarily selected. All peaks in the projections that contribute to the selected candidate point are identified as a peak subgroup (gray dots in the projections labeled with 1). The subspaces from this subgroup are removed (gray dashed lines), and the support S of remaining candidate points is recalculated (there remains one point with S = 5, and another one with S = 4 is shown in orange). (d) After two more rounds of the procedure indicated in c, two additional subgroups are identified and labeled with numbers 2 and 3, respectively. From the three subgroups, the positions of three peaks in the 3D spectrum are calculated (black dots).
through the five experimental 2D projections of the 3D spectrum with projection angles α = 0°, 90°, 45°, -25°, and -60°. (a) The blue dots mark the result for the cross sections of the automatic peak picking of the 2D projections (Fig. 3). The algorithm then arbitrarily selected N - 1 = 2 of the j projections for the first round of spectral analysis (Fig. 3), with α = 0° and α = -60°. The intersections of the subspaces corresponding to the peaks in these two projections (green lines) identify eight candidate points in the 3D spectrum (open circles). (b) By using the subspaces from all five projections, the support S (number of intersecting subspaces, see text) is calculated for each candidate point. Yellow and red dots indicate S = 2 and S = 5, respectively. (c) One of the three candidate points with the highest support (S = 5) is arbitrarily selected. All peaks in the projections that contribute to the selected candidate point are identified as a peak subgroup (gray dots in the projections labeled with 1). The subspaces from this subgroup are removed (gray dashed lines), and the support S of remaining candidate points is recalculated (there remains one point with S = 5, and another one with S = 4 is shown in orange). (d) After two more rounds of the procedure indicated in c, two additional subgroups are identified and labeled with numbers 2 and 3, respectively. From the three subgroups, the positions of three peaks in the 3D spectrum are calculated (black dots).
Intersection of Subspaces. To simplify the mathematical treatment, we describe a (N - l)-dimensional subspace L (1 < l < N) by a point QL in this subspace and a set of orthonormal vectors,  , orthogonal to L. To intersect, for example, four 3D subspaces in 5D frequency space, two of the 3D subspaces can intersect to a 2D subspace, which can then intersect with one of the remaining 3D subspaces to a 1D subspace, which can intersect with the fourth 3D subspace to a point.
, orthogonal to L. To intersect, for example, four 3D subspaces in 5D frequency space, two of the 3D subspaces can intersect to a 2D subspace, which can then intersect with one of the remaining 3D subspaces to a 1D subspace, which can intersect with the fourth 3D subspace to a point.
L and M are two subspaces of dimensionality (N - l) and (N - m), with (1 < l < N) and (1 < m < N). L is described by 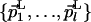 and the point QL; M is described by
and the point QL; M is described by 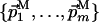 and QM. Both L and M are orthogonal to the direct dimension, and therefore both
and QM. Both L and M are orthogonal to the direct dimension, and therefore both 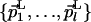 and
and 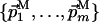 include the unit vector of the direct dimension. If Eqs. 3 and 4 are satisfied, the subspaces L and M intersect in a subspace K of dimensionality (N - k), with k = l + m - 1 as follows:
include the unit vector of the direct dimension. If Eqs. 3 and 4 are satisfied, the subspaces L and M intersect in a subspace K of dimensionality (N - k), with k = l + m - 1 as follows:
 |
[3] |
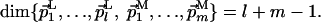 |
[4] |
QL(N) and QM(N) are the Nth coordinates of QL and QM, respectively; Δνmin is a user-defined intersection tolerance in the direct dimension; and dim stands for “dimension of.” Eq. 4 implies that 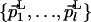 and
and 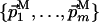 share only the direct dimension. The subspace K is then described by the orthonormal basis
share only the direct dimension. The subspace K is then described by the orthonormal basis 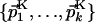 , and by a point QK with its coordinates 1 to (N - 1) given by the l + m scalar products of Eq. 5
, and by a point QK with its coordinates 1 to (N - 1) given by the l + m scalar products of Eq. 5
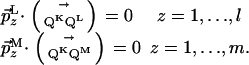 |
[5] |
The Nth coordinate of QK is the arithmetic average of the Nth coordinates of QL and QM.
Distance Between a Point and a Subspace. The distance r between a point Q and a (N - l)-dimensional subspace L, as described by a point QL and an orthonormal set of vectors orthogonal to L, 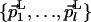 , is given by
, is given by
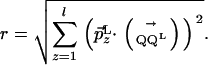 |
[6] |
Peak Positions in the N-Dimensional Space. The fact that the peak positions are generally overdetermined by the experimental data are used to refine the peak coordinates. From each subgroup, N - 1 elements are arbitrarily chosen, and their associated subspaces are intersected to yield the position of the N-dimensional peak (Fig. 4). This procedure is repeated w times, where w is a user-defined parameter. Because of the limited precision of the individual chemical shift measurements, this procedure results in w slightly different peak positions, which then are averaged in each dimension to obtain the final positioning of the N-dimensional peak Qi.
Materials and Methods
Sample Preparation. [U-13C,15N]-labeled 434-repressor(1–63) was produced following procedures described in refs. 28 and 30. For the NMR measurements, a 0.9 mM sample was prepared in 20 mM sodium phosphate buffer (pH 6.5). [U-13C,15N]-labeled TM1290 was produced as described in ref. 31. A NMR sample with 3.2 mM protein concentration in 20 mM phosphate buffer (pH 6.0) was prepared. Both NMR samples contained 95%/5% H2O/D2O and 0.1% NaN3.
NMR Spectroscopy. The NMR experimental schemes used are described in Supporting Materials and Methods, which is published as supporting information on the PNAS web site. A 4D APSY-HNCOCA experiment was recorded with the 434-repressor(1–63) at 30°C on a DRX 750 MHz spectrometer (Bruker, Fällanden, Switzerland) equipped with a z-gradient triple resonance probe. The GAPRO algorithm was applied with the parameters Smin,1 = Smin,2 = 3 (Fig. 3), k = 100 (Fig. 3), w = 400 (Fig. 3), Δνmin = 7.5 Hz (Eq. 3), a signal-to-noise threshold for atnos peak picking of Rmin = 4.0 (22), and a minimal distance for subspace intersection of rmin = 100 Hz. The calculation time for GAPRO on a standard PC running linux with a 2.8-GHz Pentium 4 processor was ≈10 min.
A 5D APSY-HACACONH experiment (11) was recorded with the protein TM1290 at 40°C on a Bruker DRX 500 MHz spectrometer equipped with a z-gradient triple resonance cryogenic probehead. The GAPRO algorithm was applied with the following parameters: Smin,1 = Smin,2 = 4, k = 100, w = 800, Δνmin = 5 Hz, Rmin = 8.0, rmin = 50 Hz. The GAPRO calculation time was ≈30 min.
Peak Picking. Automated peak picking of 2D projection spectra was performed with a peak picking routine derived from atnos (22), which recognizes all local maxima in the spectrum. In the 4D APSY-HNCOCA experiment, peaks from glutamine and asparagine side chains were eliminated by removing all pairs of peaks with the following parameters: proton chemical shifts in the range of 6.5–8.2 ppm, with chemical shift differences in the indirect dimension <10 Hz, and chemical shift differences in the direct dimension between 400 and 700 Hz.
Results
APSY-NMR spectroscopy is illustrated here with a 4D APSY-HNCOCA and a 5D APSY-HACACONH experiment. The 4D APSY-HNCOCA experiment was recorded with the 6.9-kDa protein 434-repressor(1–63). The pulse sequence used and further experimental details are given in Materials and Methods, Supporting Materials and Methods, Figs. 6–9, and Table 2, which are published as supporting information on the PNAS web site. In a total spectrometer time of 4 h, j = 27 2D projections were recorded with the following projection angles (Table 1): (α, β) = (0°, 0°), (0°, 90°), (90°, 0°), (±30°, 0°), (±60°, 0°), (0°, ±30°), (0°, ±60°), (90°, ±30°), (90°, ±60°), (±30°, ±30°), (±60°, ±30°), and (±45°, ±60°). The projection spectra were peak picked with atnos (22) to generate the input for GAPRO. The 4D peak list that resulted after ≈10 min of GAPRO computation time contained 59 peaks, which is to be compared with a total of 60 peaks expected from the chemical structure of the molecule. Although on average 18 ± 9 noise artifacts were picked in each projection, the final 4D peak list generated by the GAPRO algorithm contained 59 cross peaks and not a single artifact. Only the peak that would correlate the residues of the N-terminal dipeptide was missing. It had a signal intensity below the noise level in all projections. The precision of the chemical shifts in the final APSY peak list has been estimated to be 1 Hz in the direct dimension and 8 Hz in each of the three indirect dimensions.
The 5D APSY-HACACONH experiment was recorded with the 12.4-kDa protein TM1290. The pulse sequence used is described in Supporting Materials and Methods, and further experimental details are given in Materials and Methods. In this experiment, 28 2D projections were recorded in 11 h. The projection angles and the spectral widths are listed in Table 2, and the 28 2D projection spectra are shown in Fig. 9. The final 5D peak list produced from the 5D APSY-HACACONH experiment with the protein TM1290 contained all of the peaks that were expected from the chemical structure of the molecule and the previously published NMR assignments (31), and there were no artifacts contained in the final peak list.
Discussion
In this work, we presented the foundations of APSY and introduced the algorithm GAPRO for automated spectral analysis. We then implemented APSY for high-dimensional hetero-nuclear correlation NMR experiments with proteins. In two applications without any human intervention after the initial set-up of the experiments, we obtained complete peak lists with high-precision chemical shifts for 4D and 5D triple resonance spectra. For the future, we anticipate that APSY will be the first step, after protein preparation, in a fully automated process of protein structure determination by NMR. In addition to providing automated peak picking and computation of the corresponding chemical shift lists, as described in this work, APSY is expected to support automated sequential resonance assignment. For these envisaged goals, APSY has the promise of being a valid alternative to related NMR techniques that have recently been introduced for similar purposes. Thus, when compared with projection–reconstruction NMR (13–16), APSY has the advantage of relying exclusively on the analysis of experimental low-dimensional projection spectra, with no need to ever reconstruct the parent high-dimensional spectrum. When compared with G-matrix Fourier transform NMR (11), APSY differs in that there are no restrictions on the selection of the number of projections or the combinations of projection angles. The strongest asset of APSY, however, is that the algorithm GAPRO enables fully automated analysis of the experimental projection spectra. As a primary result, complete peak picking and computation of high-precision chemical shift lists are obtained without any bias that could result from human intervention.
APSY and Protein Size. APSY has so far been applied to the 6.9-kDa protein 434-repressor(1–63) and the 12.4-kDa protein TM1290. To obtain an estimate of possible limitations for APSY applications with larger proteins due to spectral overlap, we analyzed the peak separations in 4D and 5D triple resonance spectra of a sample of 54 proteins with sizes from n = 50 to 300 residues, which were simulated from the BioMagResBank (32) chemical shift deposits. Considered were the average of the distances from each peak to its nearest-by neighbor, dav, and the distance between the two most closely spaced peaks in the entire spectrum, dmin. Fig. 5 represents the data for the two experiments 4D HNCOCA and 5D HACACONH. There is no obvious correlation between n and either dav or dmin, indicating that close approach of peaks is distributed statistically and depends on particular properties of the protein, irrespective of its size. In Fig. 5, the statistical probability to encounter pairs of peaks that could not be resolved by APSY is <1% for protein sizes up to at least 300 residues, which is representative for 4D and 5D triple resonance data sets that contain one peak per residue. Foreseeably, sensitivity of signal detection therefore will be a more stringent limitation for APSY applications than spectral crowding. From our experience to-date, projection spectra with a signal-to-noise ratio of ≈3:1 are required for efficient use of APSY with automated peak picking.
Fig. 5.
Plots of peak separation in hertz vs. protein size (n is the number of residues). dav is the average distance to the closest peak, and dmin is the distance between the closest pair of peaks. (a and b) 4D HNCOCA. (c and d)5D HACACONH. The calculations were based on the BioMagResBank chemical shift deposits of 54 proteins (see Table 3, which is published as supporting information on the PNAS web site) and assumed a 1H frequency of 750 MHz. Gly residues are included only in a and b.
For the few expected closely spaced pairs of peaks, APSY is in a good position to resolve potential difficulties, because it is not required that a given N-dimensional resonance is found in all projection spectra. Peaks with overlap in one or several projections usually will be resolved in many other projections (Fig. 4). Similar to G-matrix Fourier transform NMR (11), APSY is also well prepared to deal with inaccurate peak positions from automated peak picking, which may arise from peak overlap. Because the final N-dimensional APSY peak list is computed as the average of a large number of measurements (Fig. 3), inaccurate peak positions in some of the projections have only a small influence on the overall precision.
APSY and Spectral Artifacts. The positions of the peaks belonging to a peak subgroup (Fig. 3) are correlated in all of the projection spectra, whereas, in contrast, the positions of random noise are uncorrelated. This different behavior of the two types of peaks, which cannot readily be distinguished in the initial automatic peak picking, efficiently discriminates against artifacts. Artifacts are therefore unlikely to pass through the ranking filters (Fig. 3) and thus unlikely to appear in the final peak list. For the two APSY applications presented in this work, the final peak lists contained no artifacts, although the initial peak-picking routine was applied with low signal-to-noise thresholds, Rmin, and included a large number of spurious signals in the initial peak lists. This result supports that the presently used ranking criteria (Fig. 3) were well chosen.
Outlook to Future Implementations of APSY. The present set-up of an APSY experiment (Fig. 3) requires the operator to define seven parameters, in addition to the selection of the N-dimensional experiment, the number and dimension of the projections, and the projection angles. These parameters are the signal-to-noise threshold, Rmin, for atnos peak picking; the thresholds for the two ranking filters, Smin,1 and Smin,2; the number of subgroup list calculations, k; the number of final peak coordinates calculations, w; the minimal distance for intersections, rmin; and the intersection tolerance in the direct dimension, Δνmin. For routine use of APSY, optimized sets of these parameters will emerge, and some of the free variables might be replaced by novel convergence criteria. Further refinements also will apply to selecting optimal numbers of and dimensions for the projections, as well as to identifying optimal combinations of projection angles for given experimental situations. This selection and identification may include consideration of the different relaxation properties of the nuclei involved, leading to the use of projection angles with short evolution times for the most rapidly relaxing spins. Further technical improvements may lead, for example, to higher spectral resolution by linear prediction or to improved spectrometer time usage by concurrent data acquisition and data analysis, with feedback to prevent unneeded data accumulation once the final peak list converges.
Supplementary Material
Acknowledgments
We thank Dr. Torsten Herrmann (Eidgenössische Technische Hochschule Zürich) for providing the software platform radar in which GAPRO was implemented. We also thank Drs. Fred F. Damberger and Touraj Etezady-Esfarjani (both of Eidgenössische Technische Hochschule Zürich) for samples of the two proteins used. This work was supported by the Schweizerischer Nationalfonds and Eidgenössische Technische Hochschule Zürich through the National Center for Competence in Research (NCCR) Structural Biology.
Abbreviations: APSY, automated projection spectroscopy; GAPRO, geometric analysis of projections.
References
- 1.Wüthrich, K. (1986) NMR of Proteins and Nucleic Acids (Wiley, New York).
- 2.Bax, A. & Grzesiek, S. (1993) Acc. Chem. Res. 26, 131-138. [Google Scholar]
- 3.Kay, L. E. & Gardner, K. H. (1997) Curr. Opin. Struct. Biol. 7, 722-731. [DOI] [PubMed] [Google Scholar]
- 4.Wüthrich, K. (2003) Angew. Chem. Int. Ed. 42, 3340-3363. [DOI] [PubMed] [Google Scholar]
- 5.Ernst, R. R., Bodenhausen, G. & Wokaun, A. (1987) Principles of Nuclear Magnetic Resonance in One and Two Dimensions (Oxford Univ. Press, Oxford).
- 6.Szyperski, T., Yeh, D. C., Sukumaran, D. K., Moseley, H. N. B. & Montelione, G. T. (2002) Proc. Natl. Acad. Sci. USA. 99, 8009-8014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Orekhov, V. Y., Ibraghimov, I. & Billeter, M. (2003) J. Biomol. NMR 27, 165-173. [DOI] [PubMed] [Google Scholar]
- 8.Rovnyak, D., Frueh, D. P., Sastry, M., Sun, Z. Y. J., Stern, A. S., Hoch, J. C. & Wagner, G. (2004) J. Magn. Reson. 170, 15-21. [DOI] [PubMed] [Google Scholar]
- 9.Szyperski, T., Wider, G., Bushweller, J. H. & Wüthrich, K. (1993) J. Am. Chem. Soc. 115, 9307-9308. [Google Scholar]
- 10.Freeman, R. & Kupce, E. (2004) Concepts Magn. Reson. 23A, 63-75. [Google Scholar]
- 11.Kim, S. & Szyperski, T. (2003) J. Am. Chem. Soc. 125, 1385-1393. [DOI] [PubMed] [Google Scholar]
- 12.Kozminski, W. & Zhukov, I. (2003) J. Biomol. NMR 26, 157-166. [DOI] [PubMed] [Google Scholar]
- 13.Kupce, E. & Freeman, R. (2003) J. Am. Chem. Soc. 125, 13958-13959. [DOI] [PubMed] [Google Scholar]
- 14.Kupce, E. & Freeman, R. (2003) J. Biomol. NMR 27, 383-387. [DOI] [PubMed] [Google Scholar]
- 15.Kupce, E. & Freeman, R. (2004) J. Am. Chem. Soc. 126, 6429-6440. [DOI] [PubMed] [Google Scholar]
- 16.Kupce, E. & Freeman, R. (2004) Concepts Magn. Reson. 22A, 4-11. [Google Scholar]
- 17.Bracewell, R. N. (1956) Aust. J. Phys. 9, 198-217. [Google Scholar]
- 18.Nagayama, K., Bachmann, P., Wüthrich, K. & Ernst, R. R. (1978) J. Magn. Reson. 31, 133-148. [Google Scholar]
- 19.Mersereau, R. M. & Oppenheim, A. V. (1974) Proc. IEEE 62, 1319-1338. [Google Scholar]
- 20.Lauterbur, P. C. (1973) Nature 242, 190-191. [Google Scholar]
- 21.Koradi, R., Billeter, M., Engeli, M., Güntert, P. & Wüthrich, K. (1998) J. Magn. Reson. 135, 288-297. [DOI] [PubMed] [Google Scholar]
- 22.Herrmann, T., Güntert, P. & Wüthrich, K. (2002) J. Biomol. NMR 24, 171-189. [DOI] [PubMed] [Google Scholar]
- 23.Baran, M. C., Huang, Y. J., Moseley, H. N. B. & Montelione, G. T. (2004) Chem. Rev. 104, 3541-3555. [DOI] [PubMed] [Google Scholar]
- 24.Xia, Y. L., Zhu, G., Veeraraghavan, S. & Gao, X. L. (2004) J. Biomol. NMR 29, 467-476. [DOI] [PubMed] [Google Scholar]
- 25.Moseley, H. N. B., Riaz, N., Aramini, J. M., Szyperski, T. & Montelione, G. T. (2004) J. Magn. Reson. 170, 263-277. [DOI] [PubMed] [Google Scholar]
- 26.Grzesiek, S. & Bax, A. (1992) J. Magn. Reson. 96, 432-440. [Google Scholar]
- 27.Yang, D. W. & Kay, L. E. (1999) J. Am. Chem. Soc. 121, 2571-2575. [Google Scholar]
- 28.Neri, D., Billeter, M. & Wüthrich, K. (1992) J. Mol. Biol. 223, 743-767. [DOI] [PubMed] [Google Scholar]
- 29.Etezady-Esfarjani, T., Herrmann, T., Peti, W., Klock, H. E., Lesley, S. A. & Wüthrich, K. (2004) J. Biomol. NMR 29, 403-406. [DOI] [PubMed] [Google Scholar]
- 30.Neri, D., Szyperski, T., Otting, G., Senn, H. & Wüthrich, K. (1989) Biochemistry 28, 7510-7516. [DOI] [PubMed] [Google Scholar]
- 31.Etezady-Esfarjani, T., Peti, W. & Wüthrich, K. (2003) J. Biomol. NMR 25, 167-168. [DOI] [PubMed] [Google Scholar]
- 32.Seavey, B. R., Farr, E. A., Westler, W. M. & Markley, J. L. (1991) J. Biomol. NMR 1, 217-236. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.







