Abstract
Innate immunity is the first line of defense against invading pathogens. Toll-like receptors (TLRs) act as sentinels of the innate immune system, sensing a variety of ligands from lipopolysaccharide to flagellin to dsRNA through their ligand-binding domain that is composed of leucine-rich repeats (LRRs). Ligand binding initiates a signaling cascade that leads to the up-regulation of inflammation mediators. In this study, we have expressed and crystallized the ectodomain (ECD) of human TLR3, which recognizes dsRNA, a molecular signature of viruses, and have determined the molecular structure to 2.4-Å resolution. The overall horseshoe-shaped structure of the TLR3-ECD is formed by 23 repeating LRRs that are capped at each end by specialized non-LRR domains. The extensive β-sheet on the molecule's concave surface forms a platform for several modifications, including insertions in the LRRs and 11 N-linked glycans. The TLR3-ECD structure indicates how LRR loops can establish distinct pathogen recognition receptors.
Keywords: dsRNA, innate immunity, pathogen recognition receptor
The interaction of pathogens with Toll-like receptors (TLRs) initiates the host immune response to infection (1, 2). Homologues of TLRs serve immune functions in insects and plants, as well as vertebrates, indicating that the TLR family predates the divergence of plants and animals some two billion years ago (3, 4). In mammals, activation of TLRs by pathogens sets in motion an innate immune inflammatory process that prevents pathogen dissemination and, through TLRs on dendritic cells, directs the development of acquired immunity (5, 6). In contrast to the antigen receptors of the acquired immune system, which are generated by somatic recombination and mutational events, the TLRs are encoded by a limited number of genes in the germline, 10 known in humans (2). These 10 receptors recognize a remarkably wide variety of pathogen-derived molecular signatures, including glycolipids such as lipopolysaccharide (7), proteins such as flagellin (8), and nucleic acids such as dsRNA (9). The TLRs are type I integral membrane glycoproteins, consisting of pathogen-binding ectodomains (ECD) and cytoplasmic signaling domains, known as Toll IL-1 receptor (TIR) domains, joined by a single transmembrane helix (10). The molecular structures of several TIR domains are known (11, 12), but the detailed atomic resolution structures of TLR ECDs have yet to be determined. The primary structures of the 10 human TLR-ECDs contain 19-25 leucine-rich repeats (LRRs) capped on each end by characteristic N-terminal (LRR-NT) and C-terminal (LRR-CT) motifs. From known LRR-containing structures, the TLR-ECDs are predicted to be horseshoe-shaped with extensive β-sheets on their concave surfaces (10, 13). However, it is not known how these LRR-based platforms are adapted structurally to the recognition of pathogens.
Human TLR3 responds to dsRNA (9), a molecular signature of RNA viruses (14). It is located intracellularly in dendritic cells (15), where it serves as a sensor for viral infection of phagocytosed cells (16). Upon activation, TLR3 signals, in a MyD88-independent manner, through Toll/IL-1R domain-containing adaptor-inducing IFN-β (TRIF) (17), to up-regulate type I IFN. To gain structural insight into TLR function, we have produced the ECD of human TLR3 in high yield. Here we describe the 2.4-Å crystal structure of the TLR3-ECD.
Materials and Methods
Expression, Purification, and Crystallization. Human TLR3 (residues 22-702, NCBI accession no. NP_003256) fused to an N-terminal GP67 secretion signal sequence and a C-terminal TEV cleavage site followed by FLAG and 6×His tags was inserted into a baculovirus expression system by the Protein Expression Laboratory (Science Applications International Corp., Frederick, MD) and expressed in High Five cells (Invitrogen) by using a multiplicity of infection of 3. Supernatant was harvested at 48 h postinfection, concentrated, and diafiltered into binding buffer [PBS/0.35 M NaCl/5% glycerol/1 mM 2-mercaptoethanol (BME)]. Protein was purified by sequential HisTrap (Amersham Biosciences) and anti-FLAG M2 mAB (Sigma-Aldrich) affinity columns, followed by gel filtration (Superdex 200) in crystallization buffer, 0.02 M Tris, pH 7.5/0.5 M NaCl/5% glycerol/1 mM BME. Human TLR3-ECD was purified to homogeneity; glycan content was determined by deglycosylation with PNGase F (New England Biolabs). The molecular weight was determined by MALDI-TOF mass spectrometry (Protein Chemistry Laboratory, Science Applications International Corp.). Crystallization conditions were identified from Hampton Research, Riverside, CA, and NexTal Technologies, Montreal factorial screens at a protein concentration of 2 mg·ml-1. Plate-like crystals, 0.1 × 0.1 × 0.05 mm, grew from 0.1 M NaAcetate, pH 4.2-4.8/0.2 M lithium or ammonium sulfate/22.5-26% polyethylene glycol 4000 at 17°C. Native crystals were cryoprotected by sequential soaks in 15 and 30% d-glucose in mother liquor and flash-frozen in liquid nitrogen. Derivative crystals were crosslinked in 25% glutaraldehyde for 30 min (18), transferred to mother liquor at pH 5, and then soaked in a variety of heavy atom solutions prepared in the final cryoprotectant solution as described (19). After the heavy atom soak, crystals were flash-frozen in liquid nitrogen.
Size-Exclusion Chromatography. TLR3-ECD in 0.02 M NaAcetate, pH 5/0.1 M NaCl was mixed in a 1:1.5 ratio with dsRNA [5′-AACCUAAUAAUUAUCAAAAUG-3′ and its complement (Dharmacon Research, Lafayette, CO), and 50 μl was applied to a 3-ml Superdex 200 column (GE Healthcare). Elution, using an Amersham Pharmacia SMART System, was monitored at 260 and 280 nm. For poly(I):(C)-binding studies, TLR3-ECD (84 μg) was mixed with 70 μg of poly(I):(C) and assayed as above.
Structure Determination and Refinement. Data were collected at Southeast Regional Collaborative Access Team 22-ID beamline at the Advanced Photon Source, Argonne National Laboratory, Argonne, IL. Supporting institutions may be found at www.sercat.org/members.html. hkl2000 (20) was used to process and scale data. Native crystals were orthorhombic C2221 with one molecule per asymmetric unit, whereas the derivative crystals were monoclinic P21 with two molecules per asymmetric unit. The twofold screw axis was conserved between the two crystal forms (see Table 2, which is published as supporting information on the PNAS web site, for data collection parameters). The rhenium data were isomorphous with the uranyl and samarium derivative data but, of themselves, gave no anomalous or isomorphous Patterson peaks. Therefore, the Re data were used as the native monoclinic data. sharp (21) was used to identify and refine the uranyl and samarium sites. Four uranyl sites and two samarium sites were input into solve/resolve (22). The partial phases and initial model were recycled through resolve for identification of the noncrystallographic symmetry and further model building. One complete molecule, built into the phased monoclinic map using o (23), was used as the search model for molecular replacement in phaser-ccp4 Suite (24) for the high-resolution native data. The resulting model underwent iterative rounds of rebuilding using o and refinement using both refmac-ccp4 (24) and cns 1.1 (25). Sugars were added as density (1σ, 2 Fo-Fc) became obvious at the predicted glycosylation sites. Solvent was added by the cns water-pick module (25) at 1-3 σ. The model was assessed by both procheck (26) and molprobity (27). Refinement statistics are listed in Table 3, which is published as supporting information on the PNAS web site. dsRNA:TLR3-ECD complexes were modeled by using o (23) and molmol (28). Figures were prepared in spock (29), pymol (DeLano Scientific, San Carlos, CA), grasp (30), and povray (Persistence of Vision Raytracer, Williamstown, Australia).
Results and Discussion
Structure of the TLR3-ECD. Human TLR3-ECD, produced in insect cells, contained ≈20 kDa of N-linked glycan. The TLR3-ECD was monomeric by gel filtration, and N-terminal sequence analysis indicated that the mature protein began at Ala-22 (Table 1). The molecular structure was determined to a resolution of 2.4 Å with a working R factor of 20.7% (Rfree = 23.4%).
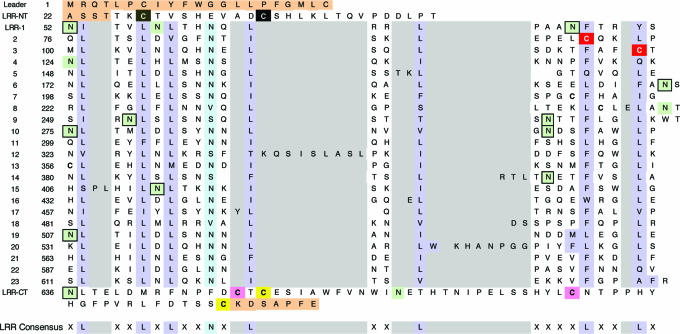
Table 1. Location of structural motifs within the TLR3-ECD amino acid sequence.
The TLR3-ECD sequence (NCBI accession no. NP_003256) contains an N-terminal capping motif (LRR-NT), 23 LRRs, and a C-terminal cap (LRR-CT). Orange highlighting indicates residues not included in the current structure. Cysteines forming disulfide bonds are color coded in pairs. All potential glycosylation sites are highlighted in light green with black boxes indicating N-linked glycosylation observed in the structure. The hydrophobic residues in the LRR consensus are colored in purple with the position for conserved asparagines shown in cyan. Insertions are indicated in the gray boxes.
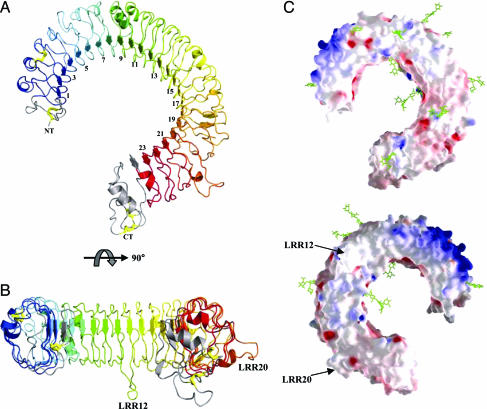
The overall structure of the TLR3-ECD is that of a large curved solenoid with an inner diameter of ≈42 Å, an outer diameter of 90 Å, and a thickness of 35 Å (Fig. 1 A and B). The concave inner surface consists of a large parallel β-sheet with each β-strand roughly perpendicular to the solenoid axis and linked to the next strand by an irregular loop. This extended β-sheet has an accessible molecular surface of 4,026 Å2. As with all LRR structures, the consensus hydrophobic residues (Table 1) point toward the interior of the molecule, forming a hydrophobic core (13). The observed curvature of the TLR3-ECD is similar to that of other smaller LRR proteins. A structure homology search of the Protein Data Bank [using dali (31)] shows greatest similarity with glycoprotein IB α [1M0Z (32)], internalin A [1O6S (33)], and YopM [1JL5 (34)]. However, the TLR3-ECD differs from other LRR structures by a striking lack of twist across the solenoid, as seen in a side view (Fig. 1B). The flatness of TLR3-ECD can be attributed to the presence of irregular LRRs (LRRs 8, 12, 14, 18, and 20; Fig. 5, which is published as supporting information on the PNAS web site), which disrupt the twist introduced by the succession of identical LRRs in other LRR structures. The lack of twist yields a flat surface that may facilitate receptor oligomerization and therefore signaling.
Fig. 1.
Overview of TLR3-ECD structure. (A) TLR3-ECD is shown in a ribbon diagram with the N- and C-terminal capping motifs colored in gray, the LRRs colored from blue to red beginning at LRR1, and the four disulfide bonds depicted by yellow ball-and-stick. (B) Side view colored as in A shows the two insertions at LRRs 12 and 20, respectively. (C) Electrostatic potential surface shows positive (blue) and negative (red) charges at neutral pH. The N-linked glycans are shown as green ball-and-stick. One surface is covered in glycan (Upper), whereas the opposite face (Lower) is devoid of glycan with a pocket of basic residues bounded by the two LRRs containing insertions 12 and 20, respectively.
At its N terminus, the TLR3 LRR-N-terminal (LRR-NT) motif consists of a hairpin loop anchored by a disulfide bond between Cys-28 and Cys-38. The LRR-NT is followed by 23 LRRs. Most strikingly, two of the LRRs, LRR12 and LRR20, contain large insertions following the β-strand (Figs. 1 and 5). The LRR12 insertion forms a long protrusion parallel with the β-sheet, whereas the insertion of LRR20 extends outward from the convex side of the TLR3-ECD (Fig. 1 A and B). Because these insertions are unique to TLR3 and conserved in all known mammalian TLR3 orthologs (human, mouse, rat, and cow), it is likely that they play important roles in TLR3 function, perhaps in ligand binding. Also of note, the loop portion of LRR8 contains a very regular α-helix (Fig. 5), which could potentially alleviate the twist seen in other LRR structures. The TLR3-ECD contains 11 Cys residues (Table 1). Of these, eight form disulfide bonds, and three are free. Free Cys-356 is modified by a 2-mercaptoethanol molecule from the crystallization buffer, whereas the other free Cys residues in LRRs 7 and 8 remain reduced. Following the LRRs is a C-terminal capping domain (LRR-CT), which forms a compact structure stabilized by two disulfide bridges (Table 1), typical of LRR-CT motifs seen in both glycoprotein 1B α and Nogo receptor ligand binding domain [1OZN (35)]. Seven residues (unobserved in this structure) connect the C-terminal Cys residue of the LRR-CT to the transmembrane domain, limiting the flexibility of the ECD relative to the membrane. This short tether between the compact LRR-CT and the vesicular membrane may play a role in positioning the TLR3-ECD relative to the membrane.
The molecular surface of the TLR3-ECD is abundantly and unevenly populated with N-linked carbohydrates (Fig. 1C). Of the 15 predicted N-glycosylation sites, 11 are observed to be glycosylated (Table 1). Because TLR3-ECD protein was expressed in insect cells, the glycans lack the high mannose content characteristic of N-linked carbohydrate in mammalian cells (36). All 11 glycans contain the first β (1→4) linked GlcNAc, and 10 are further glycosylated with a second β(1→4) linked GlcNAc ring. Two sites also have an α(1→6) linked l-Fucopyranose (Fuc) branch attached to their first GlcNAc ring. Additional α-d-mannopyranose (Man) rings are visible on N398, N413, and N507. MALDI-TOF mass spectrometry of TLR3-ECD indicated a molecular weight of 95,525 Da, of which 80,084 Da constitutes the protein core and ≈5,450 Da the visible carbohydrates. The 15,441 Da difference, unobserved in the electron density, can be accounted for by the homogenous glycosylation of all 15 sites with a full-length paucimannose type N-glycan: α-Man-(1→3)-[α-Man-(1→6)]-β-Man-(1→4)-β-GlcNAc-(1→4)-[α-Fuc-(1→6)]-β-GlcNAc, the most highly processed glycan found on most insect cell-expressed proteins.
Two of the observed glycans are located on the β-sheet of the solenoid, at Asn-252 (LRR 9) and Asn-413 (LRR 15), a surface that in other LRR proteins is involved in ligand binding and recognition (10, 37). The presence of glycan on the concave surface may, therefore, affect and direct by occlusion ligand binding by TLR3. Interestingly, the surface of the solenoid proximal to the C-terminal ends of the β-strands is completely devoid of glycosylation (Fig. 1C Lower). As a result, this flat surface offers the largest surface area available to participate in intermolecular interactions.
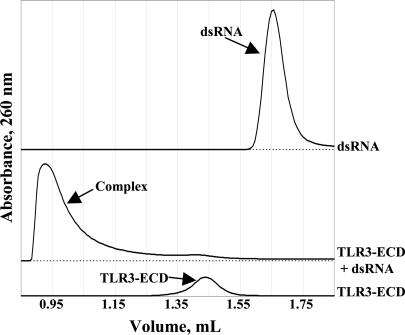
Recombinant TLR3-ECD Binds dsRNA. TLR3 initiates a signaling cascade in response to dsRNA; poly (I):(C), viral RNA, mRNA, and short interfering RNA have all been reported as potential ligands (9, 38, 39). Our recombinant human TLR3-ECD shows effective binding to a short dsRNA oligonucleotide (Fig. 2). The TLR3-ECD:dsRNA oligonucleotide complex forms a large aggregate that migrates in the column void volume. This result suggests that ligand binding promotes multimerization of the receptor, offering a potential mechanism by which the ligand-binding signal is transmitted to the cytosolic signaling domain. Similar results were observed with poly(I):(C), a surrogate for viral dsRNA (data not shown).
Fig. 2.
dsRNA binding to TLR3-ECD. TLR3-ECD and dsRNA form a large, aggregated complex, as observed by gel filtration.
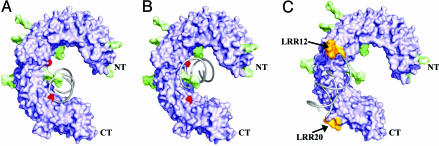
Possible Ligand-Binding Sites. At present, we do not have direct evidence for the location of the dsRNA-binding site on the TLR3-ECD. However, examination of the structure suggests potential binding sites. Two sulfate ions from the crystallization medium are observed bound to the concave surface of TLR3-ECD, via hydrogen bonds to His, Asn, Tyr, and Arg side chains (Fig. 6 A and B, which is published as supporting information on the PNAS web site). Sulfate ions can mimic the phosphate of the nucleotide backbone and provide clues to the potential ligand-binding site. Therefore, a 19-mer RNA duplex (40) was docked onto the receptor by aligning a phosphate from the duplex backbone onto each of the sulfate ions in turn (Fig. 3 A and B). The concave surface proximal to the C terminus coordinates the first sulfate ion (Fig. 3B). The RNA molecule in this model presents its major groove to the β-sheet surface, whereas the minor groove, rich in negative charge, aligns well with the positively charged edges of the β-sheet.
Fig. 3.
Proposed modes of ligand recognition for TLR3-ECD. Surface rendering of the TLR3-ECD shows the protein core in purple, glycan in green, sulfate ions in red, and LRR12 and -20 insertions in orange. A 19-mer of dsRNA shown in gray ribbons was docked into two potential binding sites (A and B) based on the location of sulfate ions that may mimic nucleic phosphate backbone interactions. (C) The unglycosylated surface of the TLR3-ECD offers a third potential docking site with a shallow groove bounded by the insertions in LRRs 12 and 20.
The second sulfate site falls between the two glycans on the concave surface (Fig. 3B). The presence of these glycans sterically hinders dsRNA binding. However, it should be noted that the ECD in these crystals was expressed in insect cells, and the glycosylation could be different in human cells. In vivo, if under some conditions one or both sites are not glycosylated, dsRNA could occupy this site, whereby glycosylation provides a mechanism to control dsRNA binding by TLR3. In these two models, the duplex RNA, when bound on the concave surface, extends beyond the edge of a single TLR3-ECD and could bind to a second ECD.
A third potential ligand-binding site was identified on the TLR3-ECD surface near the C terminus that lacks glycosylation (Fig. 1C Lower). A shallow groove between the two insertions on LRRs 12 and 20 (indicated by the yellow surface) can accommodate dsRNA. This region of the surface also contains a number of positively charged residues that could potentially interact with the phosphate groups of the RNA. This model of RNA binding to the exterior of the ECD presents a mode of oligomerization where ECDs could bind to the two “faces” of the RNA duplex.
In several type I transmembrane receptors, such as human growth hormone receptor (41, 42), the ligand-binding signal is transmitted across the membrane by receptor oligomerization. We note that the crystal lattice packing of the molecule reveals two TLR3 molecules in contact through their nonglycosylated faces proximal to the C terminus (Fig. 4). The two molecules share a buried surface area of ≈1,150 Å2 with four hydrogen bonds between them. Although we have not demonstrated that this dimer is physiologically relevant, this close interaction supports that this face of the receptor is clearly available for interaction with either protein or ligand and may provide a site for receptor dimerization. The third binding site described above lies in this region of the surface.
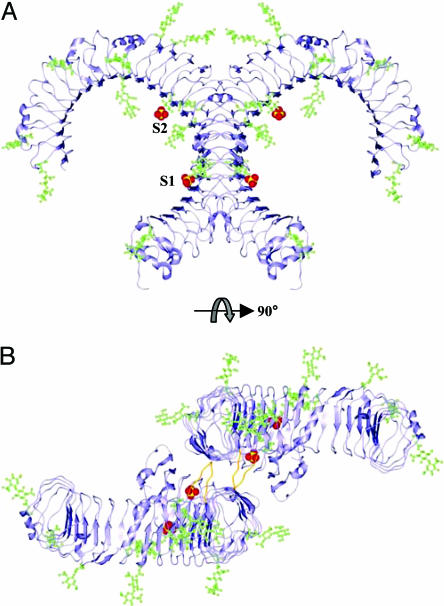
Fig. 4.
Crystal packing of TLR3-ECD. Ribbon diagram of the TLR3-ECD symmetry-related dimer found in the crystal lattice. N-glycosylation is shown in green ball-and-stick, and the two sulfate ions are shown in yellow and red space-filling (S1 and S2). The side view (A) shows the predominance of glycan decorating the faces of the molecule. Looking down the crystallographic twofold between the molecules (B), a surface near the C terminus that is devoid of glycan allows for close contact between molecules. The interface contains four hydrogen bonds, one of which (Asp-575) is contributed by the insertion in LRR20 (shown in orange).
Conclusions
The structure of TLR3-ECD provides an atomic view of how the LRR scaffold can accommodate the broad diversity of ligand structures required of the TLR family. The horseshoe-shaped protein core provides a large surface area for ligand interaction and recognition. The extensive β-sheet on the concave surface of TLR3-ECD forms a stable platform upon which loops, insertions, and short helices are anchored. These irregular features are highly conserved among mammalian TLR3 orthologs and apparently evolved over time to facilitate recognition of pathogens that pose a specific threat to the host. The diversity of the loops connecting the β-strands can also introduce relaxation points in the protein core that translates into a reduction in the overall twist of the molecule, which in turn could also affect ligand binding. In addition, as seen in TLR3-ECD, differential glycosylation among the TLRs could control access to specific interaction surfaces. A striking feature of the TLR structure is the large spatial separation of many of these features on the protein core, raising the possibility that each TLR paralog might recognize several different ligands at different sites, thus helping to explain the ability of TLRs to recognize a variety of structurally unrelated ligands (1).
Note. We were informed by Ian Wilson, Scripps Research Institute, La Jolla, CA, that their group had also determined the structure of TLR3-ECD (43).
Supplementary Material
Acknowledgments
We thank the Southeast Regional Collaborative Access Team staff and Thang Chiu for help with the data collection. We thank Alison Burgess-Hickman for careful reading of the manuscript, Marcin Nowotny for help with modeling the RNA ligand, and Don Ronning for assistance in preparation of the figures. We gratefully acknowledge the valuable assistance of Drs. D. Esposito, R. Hopkins, and W. Gillette of the Protein Expression Laboratory, Science Applications International Corp., Frederick, MD, in developing protein expression systems. Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract W-31-109-Eng-38. This research was supported in part by an intramural National Institute of Allergy and Infectious Diseases Biodefense Award.
Author contributions: J.K.B., D.M.S., and D.R.D. designed research; J.K.B., I.B., and P.R.H. performed research; J.A. and J.S. contributed new reagents/analytical tools; J.K.B., I.B., P.R.H., D.M.S., and D.R.D. analyzed data; and J.K.B., I.B., P.R.H., D.M.S., and D.R.D wrote the paper.
Abbreviations: TLRs, Toll-like receptors; ECD, ectodomain; LRR, leucine-rich repeats; LRR-CT, LRR-C-terminal.
Data deposition: The atomic coordinates for TLR3-ECD have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2A0Z).
References
- 1.Takeda, K., Kaisho, T. & Akira, S. (2003) Annu. Rev. Immunol. 21, 335-376. [DOI] [PubMed] [Google Scholar]
- 2.Takeda, K. & Akira, S. (2005) Int. Immunol. 17, 1-14. [DOI] [PubMed] [Google Scholar]
- 3.Lemaitre, B., Nicolas, E., Michaut, L., Reichhart, J. M. & Hoffmann, J. A. (1996) Cell 86, 973-983. [DOI] [PubMed] [Google Scholar]
- 4.Beutler, B. & Rehli, M. (2002) Curr. Top. Microbiol. Immunol. 270, 1-21. [DOI] [PubMed] [Google Scholar]
- 5.Akira, S. & Takeda, K. (2004) Nat. Rev. Immunol. 4, 499-511. [DOI] [PubMed] [Google Scholar]
- 6.Iwasaki, A. & Medzhitov, R. (2004) Nat. Immunol. 5, 987-995. [DOI] [PubMed] [Google Scholar]
- 7.Poltorak, A., He, X., Smirnova, I., Liu, M. Y., Huffel, C. V., Du, X., Birdwell, D., Alejos, E., Silva, M., Galanos, C., et al. (1998) Science 282, 2085-2088. [DOI] [PubMed] [Google Scholar]
- 8.Hayashi, F., Smith, K. D., Ozinsky, A., Hawn, T. R., Yi, E. C., Goodlett, D. R., Eng, J. K., Akira, S., Underhill, D. M. & Aderem, A. (2001) Nature 410, 1099-1103. [DOI] [PubMed] [Google Scholar]
- 9.Alexopoulou, L., Holt, A. C., Medzhitov, R. & Flavell, R. A. (2001) Nature 413, 732-738. [DOI] [PubMed] [Google Scholar]
- 10.Bell, J. K., Mullen, G. E. D., Leifer, C. A., Mazzoni, A., Davies, D. R. & Segal, D. M. (2003) Trends Immunol. 24, 528-533. [DOI] [PubMed] [Google Scholar]
- 11.Khan, J. A., Brint, E. K., O'Neill, L. A. & Tong, L. (2004) J. Biol. Chem. 279, 31664-31670. [DOI] [PubMed] [Google Scholar]
- 12.Xu, Y., Tao, X., Shen, B., Horng, T., Medzhitov, R., Manley, J. L. & Tong, L. (2000) Nature 408, 111-115. [DOI] [PubMed] [Google Scholar]
- 13.Kajava, A. V. & Kobe, B. (2002) Protein Sci. 11, 1082-1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bowie, A. G. & Haga, I. R. (2005) Mol. Immunol. 42, 859-867. [DOI] [PubMed] [Google Scholar]
- 15.Matsumoto, M., Funami, K., Tanabe, M., Oshiumi, H., Shingai, M., Seto, Y., Yamamoto, A. & Seya, T. (2003) J. Immunol. 171, 3154-3162. [DOI] [PubMed] [Google Scholar]
- 16.Schulz, O., Diebold, S. S., Chen, M., Naslund, T. I., Nolte, M. A., Alexopoulou, L., Azuma, Y. T., Flavell, R. A., Liljestrom, P. & Reis e Sousa (2005) Nature 433, 887-892. [DOI] [PubMed] [Google Scholar]
- 17.Yamamoto, M., Sato, S., Mori, K., Hoshino, K., Takeuchi, O., Takeda, K. & Akira, S. (2002) J. Immunol. 169, 6668-6672. [DOI] [PubMed] [Google Scholar]
- 18.Lusty, C. (1999) J. Appl. Crystallogr. 32, 106-112. [Google Scholar]
- 19.Sun, P. D., Radaev, S. & Kattah, M. (2000) Acta Crystallogr. D 58, 1092-1098. [DOI] [PubMed] [Google Scholar]
- 20.Otwinowski, Z. & Minor, W. (1997) Methods Enzymol. 276, 306-326. [DOI] [PubMed] [Google Scholar]
- 21.de La Fortelle, E. & Bricogne, G. (1997) Methods Enzymol. 276, 472-494. [DOI] [PubMed] [Google Scholar]
- 22.Terwilliger, T. C. (2003) Methods Enzymol. 374, 22-37. [DOI] [PubMed] [Google Scholar]
- 23.Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard. (1991) Acta. Crystallogr. A 47 (Pt. 2), 110-119. [DOI] [PubMed] [Google Scholar]
- 24.Collaborative Computational Project, No. 4 (1994) Acta. Crystallogr. D 50, 760-763.15299374 [Google Scholar]
- 25.Brunger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta Crystallogr. D 54 (Pt. 5), 905-921. [DOI] [PubMed] [Google Scholar]
- 26.Laskowski, R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. M. (1993) J. Appl. Crystallogr. 26, 283-291. [Google Scholar]
- 27.Simon, C., Lovell, I. W., Davis, W., Arendall, B., de Bakker, P. I., Word, J. M., Prisant, M. G., Richardson, J. S. & Richardson, D. C. (2003) Proteins Struct. Funct. Genet. 50, 437-450. [DOI] [PubMed] [Google Scholar]
- 28.Koradi, R., Billeter, M. & Wuthrich, K. (1996) J. Mol. Graphics 14, 51-32. [DOI] [PubMed] [Google Scholar]
- 29.Christopher, J. A. (1998) spock (Center for Macromolecular Design, Texas A&M University, College Station, TX).
- 30.Nicholls, A., Sharp, K. A. & Honig, B. (1991) Proteins 11, 281-296. [DOI] [PubMed] [Google Scholar]
- 31.Holm, L. & Sander, C. (1996) Science 273, 595-603. [DOI] [PubMed] [Google Scholar]
- 32.Huizinga, E. G., Tsuji, S., Romijn, R. A., Schiphorst, M. E., de Groot, P. G., Sixma, J. J. & Gros, P. (2002) Science 297, 1176-1179. [DOI] [PubMed] [Google Scholar]
- 33.Schubert, W. D., Urbanke, C., Ziehm, T., Beier, V., Machner, M. P., Domann, E., Wehland, J., Chakraborty, T. & Heinz, D. W. (2002) Cell 111, 825-836. [DOI] [PubMed] [Google Scholar]
- 34.Evdokimov, A. G., Anderson, D. E., Routzahn, K. M. & Waugh, D. S. (2001) J. Mol. Biol. 312, 807-821. [DOI] [PubMed] [Google Scholar]
- 35.He, X. L., Bazan, J. F., McDermott, G., Park, J. B., Wang, K., Tessier-Lavigne, M., He, Z. & Garcia, K. C. (2003) Neuron 38, 177-185. [DOI] [PubMed] [Google Scholar]
- 36.Tomiya, N., Narang, S., Lee, Y. C. & Betenbaugh, M. J. (2004) Glycoconj. J. 21, 343-360. [DOI] [PubMed] [Google Scholar]
- 37.Kobe, B. & Kajava, A. V. (2001) Curr. Opin. Struct. Biol. 11, 725-732. [DOI] [PubMed] [Google Scholar]
- 38.Kariko, K., Bhuyan, P., Capodici, J. & Weissman, D. (2004) J. Immunol. 172, 6545-6549. [DOI] [PubMed] [Google Scholar]
- 39.Kariko, K., Ni, H., Capodici, J., Lamphier, M. & Weissman, D. (2004) J. Biol. Chem. 279, 12542-12550. [DOI] [PubMed] [Google Scholar]
- 40.Klosterman, P. S., Shah, S. A. & Steitz, T. A. (1999) Biochemistry 38, 14784-14792. [DOI] [PubMed] [Google Scholar]
- 41.de Vos, A. M., Ultsch, M. & Kossiakoff, A. A. (1992) Science 255, 306-312. [DOI] [PubMed] [Google Scholar]
- 42.Grotzinger, J. (2002) Biochim. Biophys. Acta 1592, 215-223. [DOI] [PubMed] [Google Scholar]
- 43.Choe, J., Kelker, M. S. & Wilson, I. A. (June 22, 2005) Science, 10.1126/science 1115253. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.