Abstract
A combined genetic and transcriptome analysis was performed to study the molecular basis of the arbuscular mycorrhiza (AM) symbiosis. By testing the AM phenotype of nodulation-impaired mutants and complementation analysis, we defined seven Lotus japonicus common symbiosis genes (SYMRK, CASTOR, POLLUX, SYM3, SYM6, SYM15, and SYM24) that are required for both fungal and bacterial entry into root epidermal or cortical cells. To describe the phenotype of these mutants at the molecular level, we screened for differentiating transcriptional responses of mutant and wild-type roots by large-scale gene expression profiling using cDNA-amplified fragment length polymorphism. Two percent of root transcripts was found to increase in abundance during AM development, from which a set of AM-regulated marker genes was established. A Ser-protease (SbtS) and a Cys-protease (CysS) were also activated during root nodule development. AM-induced transcriptional activation was abolished in roots carrying mutations in common symbiosis genes, suggesting a central position of these genes in a pathway leading to the transcriptional activation of downstream genes. By contrast, AM fungus-induced gene repression appeared to be unaffected in mutant backgrounds, which indicates the presence of additional independent signaling pathways.
INTRODUCTION
Legumes can form root endosymbioses with both phosphorus-acquiring arbuscular mycorrhizal (AM) fungi and nitrogen-fixing rhizobial bacteria. Although ∼80% of all living land plants form AM (Read et al., 2000; Brundrett, 2002), the nitrogen-fixing root nodule symbiosis (RNS) with rhizobia is almost exclusively restricted to legumes. Signal exchange during the early stages of the bacterial interaction includes flavonoids secreted by the plant roots, which stimulate the biosynthesis of bacterial lipochito-oligosaccharides, the so-called Nod factors (NF). In turn, NF are perceived as symbiotic signals by the plant and induce root hair deformation, plant gene expression, and, depending on the host species, activation of cortical cells, leading to the formation of preinfection threads and/or nodule meristems (Oldroyd and Downie, 2004). Despite the importance of AM for plant nutrition, the molecular events that accompany this symbiosis are not well understood, partly because of the obligate biotrophic nature and the complex genetics of the fungal microsymbiont. For example, symbiotic signals from the fungus that induce plant gene expression at a distance have been detected but not characterized (Kosuta et al., 2003). However, the morphological description of the fungal infection process in the legume Lotus japonicus has revealed a series of successive steps that are under the genetic control of the plant (Bonfante et al., 2000; Novero et al., 2002; Demchenko et al., 2004; Parniske, 2004). Upon fungal contact, the anticlinal walls of two adjacent epidermal cells separate and allow the entry of the fungus into the resulting cleft. Genetic analysis suggested that this epidermal opening response is an active process that involves the L. japonicus SYM15 gene (Demchenko et al., 2004). The fungus subsequently enters either one of the neighboring epidermal cells or the underlying hypodermal cells and continues to grow intracellularly through one or two more cell layers before it exits a plant cell to explore the extracellular spaces of the root cortex. Arbuscules are highly branched intracellular fungal structures that are formed upon fungal entry into cortical cells. Expression patterns of plant transporter genes suggest that at least part of the nutrient exchange between the symbiotic partners occurs within arbusculated plant cells (Rausch et al., 2001; Harrison et al., 2002; Paszkowski et al., 2002).
In legumes, the genetic programs for fungal and bacterial symbiosis partially overlap; legume genes have been identified that are required for the establishment of both AM and RNS (Hirsch et al., 2001; Marsh and Schultze, 2001; Stougaard, 2001; Parniske, 2004) and are referred to as the common SYM genes (Kistner and Parniske, 2002). Common symbiosis mutants were identified in Medicago sativa, Pisum sativum, L. japonicus, Medicago truncatula, Phaseolus vulgaris, Vicia faba, and Melilotus alba (Duc et al., 1989; Bradbury et al., 1991; Sagan et al., 1995; Shirtliffe and Vessey, 1996; Schauser et al., 1998; Szczyglowski et al., 1998; Wegel et al., 1998; Catoira et al., 2000; Senoo et al., 2000; Kawaguchi et al., 2002; Lum et al., 2002).
Cloning of a series of key genes required for symbiosis led to the identification of the Nod factor receptor kinase genes NFR1 and NFR5 (Madsen et al., 2003; Radutoiu et al., 2003). Moreover, four common SYM genes have been cloned, and their predicted protein products carry the hallmarks of signal transduction components. L. japonicus SYMRK (previously SYM2 [Schauser et al., 1998] and SYM21 [Szczyglowski et al., 1998]) encodes a receptor kinase (Stracke et al., 2002) the activity of which is regulated by phosphorylation (Yoshida and Parniske, 2005).
Orthologous receptor kinases have been identified from M. sativa (NORK), M. truncatula (DMI2), and P. sativum (SYM19) (Endre et al., 2002; Stracke et al., 2002). Phenotypic analysis of mutant plants has established a role of this receptor kinase in a pathway leading from the perception of the NF signal to the activation of symbiosis-related gene expression (Schneider et al., 1999; Catoira et al., 2000; Stracke et al., 2002). The L. japonicus genes CASTOR (previously SYM4 [Schauser et al., 1998], SYM22 [Szczyglowski et al., 1998], and SYM71 [Kawaguchi et al., 2002]) and POLLUX (previously SYM23 [Szczyglowski et al., 1998] and SYM86 [Imaizumi-Anraku et al., 2004]) encode closely related proteins that are predicted ion channels (Imaizumi-Anraku et al., 2004). The Medicago DMI1 gene (Ane et al., 2004) is probably a POLLUX ortholog (Imaizumi-Anraku et al., 2004), whereas a CASTOR ortholog from Medicago has not been described. Medicago DMI3 encodes a calcium and calmodulin-dependent protein kinase (Levy et al., 2004; Mitra et al., 2004a). These predictions, together with a detailed phenotypic analysis of the corresponding mutants (Catoira et al., 2000; Imaizumi-Anraku et al., 2004; Mitra et al., 2004b), strongly suggest a role for these genes in signaling downstream of NF perception.
Analysis of calcium spiking, one of the earliest responses of root hair cells to NF application (Ehrhardt et al., 1996), placed the pea and Medicago orthologs of SYMRK (PsSYM19, NORK, and DMI2 [Endre et al., 2002; Stracke et al., 2002]) as well as CASTOR and POLLUX upstream of this response (Wais et al., 2000; Walker et al., 2000; Harris et al., 2003; Imaizumi-Anraku et al., 2004), indicating that these genes act at a similar hierarchical level very early in the symbiotic signaling process. By contrast, M. truncatula DMI3 has been placed downstream of the calcium-spiking response (Wais et al., 2000) and has structural features such as EF hands and a calmodulin binding domain that conceptually allow it to interpret the calcium oscillations and convert them to a phosphorylation reaction as a readout (Levy et al., 2004; Mitra et al., 2004a).
Plant genes that are transcriptionally regulated in response to symbiotic stimuli can serve as useful markers to monitor the activity of signaling pathways. There are three classes of these genes, nodulins, mycorrhizins, and symbiosins, that are activated in response to a rhizobial signal, an AM fungal signal, or by either stimulus, respectively. From the large number of nodulins (van Kammen, 1984) and early nodulin (ENOD) genes (Kouchi et al., 2004), a subset has been found to be also activated in response to inoculation with AM fungi (Frühling et al., 1997; van Rhijn et al., 1997; Albrecht et al., 1998; Journet et al., 2001; Manthey et al., 2004). In M. truncatula, DMI1, DMI2, and DMI3 have been placed upstream of symbiosis-induced gene activation, as accumulation of >30 transcripts, including MtENOD11, MtENOD12, and MtRIP1, by either rhizobial inoculation or NF application is impaired in dmi1, dmi2, and dmi3 mutant backgrounds (Catoira et al., 2000; Mitra et al., 2004b).
To further dissect the plant's common program for fungal and bacterial symbiosis, we searched for additional common SYM mutants of L. japonicus. We present the outcome of complementation studies between mutants originating from three independent mutagenesis experiments, which resulted in the identification of seven common SYM genes. To characterize their symbiotic phenotypes in detail, we studied the morphology of the root responses to fungal and bacterial symbionts. Furthermore, a large-scale gene expression profiling experiment in L. japonicus was performed to obtain insight into the changes in the root transcriptome during AM formation and to compare the transcriptome of wild-type roots with that of mutants affected in common SYM genes. We established a central role of the common SYM genes in the transcriptional reprogramming of the root during AM and RNS. Moreover, AM-induced gene repression patterns provided evidence for the existence of signal perception and transduction that occur independently of common SYM genes.
RESULTS
Seven L. japonicus Genes Are Required for RNS and AM
Chemical (ethyl methanesulfonate [EMS]), T-DNA, or transposon-tagging mutagenesis in the legume L. japonicus yielded a large number of nodulation-impaired mutants originating from different laboratories (Schauser et al., 1998; Szczyglowski et al., 1998; Perry et al., 2003). We tested these nodulation mutants for their ability to form symbiosis with the AM fungus, Glomus intraradices, and identified a group of mutants that were also defective in AM formation. Because of their common impairment in both root symbioses, we refer to these lines as common SYM mutants. Complementation analyses of mutants described by Schauser et al. (1998), Szczyglowski et al. (1998), and Novero et al. (2002) were performed, and seven complementation groups were established. The results of complementation and phenotypic analyses are summarized in Table 1. The symbiotic phenotypes in all complementation groups segregated as recessive monogenic traits (data not shown). The mycorrhiza and nodulation phenotypes completely cosegregated in F2 progeny of crosses to the mapping parents L. japonicus ecotypes MG-20 and Funakura or L. filicaulis, providing evidence that mutations in single genes are responsible for the observed deficiencies in both symbioses.
Table 1.
Allelism and Phenotypes of Common Symbiosis Mutants Originating from Independent Mutagenesis Experiments
|
Mesorhizobium loti |
Glomus intraradices |
|||||
|---|---|---|---|---|---|---|
| Common SYM Gene (Previous Designation) |
Mutant Allele Tested (Line Description) |
Infection Threads |
Nodule Meristem |
Balloon-Like Swellings in Outer Cell Layers |
Arbuscule Formation |
|
| 2 wai | 6 to 8 wai | |||||
| Gifu wild type | + | + | − | + | ++ | |
| SYMRK (SYM2, SYM21) | symRK-3 (cac41.5)a | −b | − | + | − | + |
| symRK-7 (EMS61)a | −b | − | + | − | + | |
| CASTOR (SYM4, SYM22, SYM71) | castor-1 (282-227)c | −d | − | + | + | + |
| castor-2 (EMS1749)c | −d | − | + | − | − | |
| castor-4 (EMS46) | −d | − | + | − | + | |
| POLLUX (SYM23, SYM86) | pollux-1 (EMS70) | −d | − | + | − | + |
| pollux-2 (EMS167) | −d | − | + | − | + | |
| SYM3 | sym3-1 (5371-22) | − | −e | + | − | + |
| sym3-3 (EMS247) | − | −e | + | − | + | |
| SYM6 | sym6-1 (10512.9) | − | + | n.d. | n.d. | n.d. |
| sym6-3 (EMS126) | − | + | + | − | +f | |
| SYM15 | sym15-2 (cac57.3)g | − | − | − | − | − |
| SYM24 | sym24-1 (EMS76) | − | −e | + | − | + |
Mutant lines with the prefix EMS were identified after EMS mutagenesis by Szczyglowski et al. (1998), with the exception of EMS1749, which was generated and identified by J.K. Webb (Novero et al., 2002). The remaining lines resulted from an attempt to generate tagged mutants through T-DNA transformation (Schauser et al., 1998). n.d., not determined; wai, weeks after inoculation.
AM phenotypes observed here for symRK mutants confirmed cytological observations by Wegel et al. (1998) and Demchenko et al. (2004).
Absence of infection threads has been described previously for symRK mutants (Stracke et al., 2002).
AM phenotypes oberved here for castor mutants EMS1749 and 282-227 confirmed previous cytological observations using G. margarita as a fungal symbiont (Bonfante et al., 2000; Novero et al., 2002).
Absence of infection threads has been described previously for castor and pollux mutants (Bonfante et al., 2000; Imaizumi-Anraku et al., 2004).
sym3 and sym24-1 mutants formed small ineffective nodules at a low frequency of less than one nodule per plant.
Only one arbuscule was observed in >30 root systems inspected.
The AM phenotype observed here for the sym15-2 mutant cac57.3 confirmed cytological observations by Demchenko et al. (2004).
Fungal Infection Is Aborted in the Outer Cell Layers
In earlier reports, the consequences of mutations in the L. japonicus common SYM genes SYMRK, CASTOR, and SYM15 for the AM symbiosis were described at the cytological level (Bonfante et al., 2000; Novero et al., 2002; Demchenko et al., 2004). The fact that mutations in POLLUX, SYM3, SYM24, or SYM30 cause an AM defect has been stated previously, but with little or no phenotypic detail (Szczyglowski et al., 1998; Wegel et al., 1998; Imaizumi-Anraku et al., 2004). Here, we analyzed fungal infection of the outer root cell layers of representative common sym mutants (Table 1, Figure 1). Fungal hyphae were observed to penetrate between two epidermal cells in the wild type (Figure 1A) and mutants affected in the SYMRK, CASTOR, POLLUX, SYM3, SYM6, or SYM24 genes (Figures 1C to 1H; data not shown), suggesting that the epidermal opening response was unaffected. Furthermore, fungal hyphae were found to continue to explore extracellular spaces between outer cell layers, or sometimes even to penetrate into cells. These exploratory hyphae often show balloon-like swellings or other deformations, which are typically associated with infection arrest and were not observed on wild type roots (Table 1). Interestingly, such exploratory hyphae leading to hyphal deformation below the root surface are not observed on roots of the sym15-2 mutant (Figure 1B). At the morphological level, the fungus is blocked on sym15-2 roots at an earlier stage than on any of the other mutants. We conclude that sym15-2 is impaired in the epidermal opening response as described by Demchenko et al. (2004), whereas the other tested common sym mutants are not.
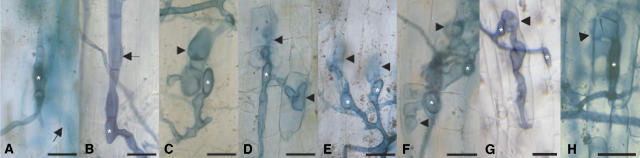
Figure 1.
AM Infection on the Wild Type and Symbiotic Mutants.
Photographs of cleared L. japonicus roots stained with ink were taken after 6 weeks of cocultivation with G. intraradices in chive nurse pot systems. Bars = 25 μm.
(A) B-129 Gifu wild type. G. intraradices has formed an appressorium (asterisk) and entered the root between the longitudinal walls of two epidermal cells. Fungal hyphae (arrow) are directed toward the inner cortex, out of the focal plane.
(B) sym15-2. The fungus has formed an appressorium (asterisk) on the groove between two epidermal cells. The hyphae (arrow) failed to enter the root between epidermal cell walls but grew extraradically along the border of two epidermal cell files.
(C) sym3-1. The fungus has formed an appressorium (asterisk) and entered the root between the longitudinal walls of two epidermal cells. Hyphal growth into the inner cortex did not occur, and the infection process was aborted between the epidermal and exodermal cell layers (arrowhead). This is a typical example of the balloon-like swelling of fungal hyphae (arrowhead) frequently observed in this phenotypic class of mutants (Table 1).
(D) pollux-1. The infection process was as described for (C). One hyphal branch of the hypha has penetrated an epidermal cell, where the infection aborted (bottom arrowhead).
(E) sym24. The infection process was as described for (C).
(F) sym6-3. Two appressoria (asterisks) in close vicinity to each other between epidermal cells. Fungal hyphae originating from these appressoria branched and entered epidermal cells, where the infection process aborted (arrowheads).
(G) castor-2. The infection process was as described for (C). A balloon-like hyphal swelling is indicated by the arrowhead.
(H) symRK-2. The infection process was as described for (C). A balloon-like hyphal swelling is indicated by the arrowhead.
Mutations in Common SYM Genes Affect Arbuscule Formation with Different Severity
A limited ability of certain common symbiosis mutants of L. japonicus to form arbuscules has been described previously (Wegel et al., 1998; Novero et al., 2002; Demchenko et al., 2004). We analyzed the AM phenotypes of the common sym mutants at two different time points to evaluate their potential for, and to determine the timing of, arbuscule formation (Table 1). After 2 weeks of cocultivation with the AM fungus, abundant arbuscules were present in roots of the wild type but not in the symbiotic mutants. The only exception was a mutant carrying the weak castor-1 allele. At later time points, between 6 and 8 weeks of cocultivation with the AM fungus, rare successful infection events of the mutant root cortex by the fungus had occurred and arbuscules could be observed with variable frequency in the root system of mutants affected in the POLLUX, SYM3, or SYM24 genes (Table 1). However, such delayed colonization of the root cortex frequently did not yield arbuscules in all inspected symRK mutants, in sym3-1, in pollux-1, and in the sym24-1 mutant. This differs significantly from the wild-type colonization pattern, in which arbuscules form without major delay after fungal hyphae reach the root cortex. Arbuscules were not observed in roots of >100 castor-2 and sym15-2 mutant individuals, and only a single arbuscule was observed upon inspection of >30 root systems of sym6-3 (Table 1).
Common SYM Genes Are Required for Infection Thread Initiation and Early Stages of Nodule Development
Nodules did not form on roots of castor, pollux, symRK, and sym15-2 mutants and were severely reduced in development on sym6 mutants (Schauser et al., 1998; Szczyglowski et al., 1998). Occasionally, nodules developed on sym24 mutant roots at a frequency of approximately one nodule per seven root systems. Also on sym3, inefficient nodulation was observed at a low frequency (Table 1). We studied the ability of the mutants to form infection threads using a derivative of M. loti strain R7A that constitutively expresses the β-galactosidase reporter gene. Whereas infection threads could be clearly detected in wild-type root hairs, this was not the case in root hairs of most of the mutants listed in Table 1. Exceptions were root hairs of mutants in the SYM3 gene, on which infection threads were observed but at a low frequency.
cDNA-Amplified Fragment Length Polymorphism Transcriptome Analysis Demonstrates That Plant Gene Activation during AM Depends on CASTOR and SYM15
To dissect the phenotypes of common symbiosis mutants at the molecular level, we performed transcriptional expression profiling of wild-type versus mutant roots. The aim was to establish a set of symbiosis-regulated marker genes that can be used to interrogate different mutant backgrounds to refine our picture of the signaling pathways leading to AM symbiosis. We chose the cDNA-amplified fragment length polymorphism (cDNA-AFLP) approach because it allows direct side-by-side comparison of transcript patterns across multiple samples and has the potential to visualize transcripts expressed at low levels (Bachem et al., 1996). Furthermore, it does not require prior sequence information or investment in array technology. Transcript profiles of L. japonicus uninoculated wild-type roots were compared with those of roots inoculated with either the AM fungus G. intraradices or M. loti to identify mycorrhizins and symbiosins.
Approximately 7000 cDNA amplification products were inspected, of which 344 were differentially represented. The majority of differential fragments were exclusively present or strongly enhanced in wild-type mycorrhized roots (Figures 2A and 2F; see Supplemental Table 1 online). A second group of fragments represented genes with a nodulin-like expression pattern (Figure 2E; see Supplemental Table 1 online). Only six fragments were enhanced in both symbiotic interactions (potential symbiosins) (Figure 2F; see Supplemental Table 1 online).
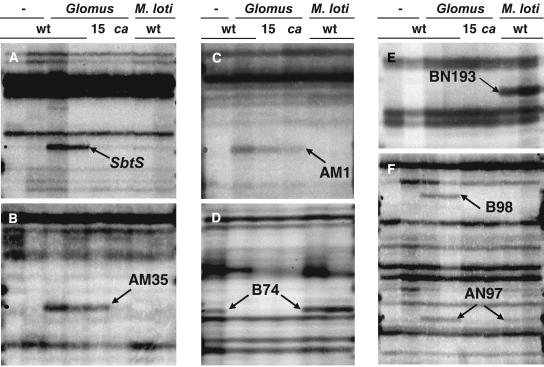
Figure 2.
Transcript Profiling of Symbiotic Roots by cDNA-AFLP.
Autoradiographs of AFLP gels of cDNA samples prepared from 3-week-old L. japonicus roots grown in the absence of symbiont (−), grown for 2 weeks in the presence of G. intraradices (Glomus), or from nodulated roots (M. loti). Genetic backgrounds include wild-type L. japonicus Gifu (wt), mutant sym15-2 (15), and mutant castor-2 (ca). Duplicate samples for G. intraradices–inoculated and noninoculated wild-type roots in (A), (B), (E), and (F) are from two independent experiments to monitor the reproducibility of transcript profiles. Samples from nodulated roots in each panel were taken, from left to right, 14 or 21 d after inoculation of 6-d-old seedlings with M. loti.
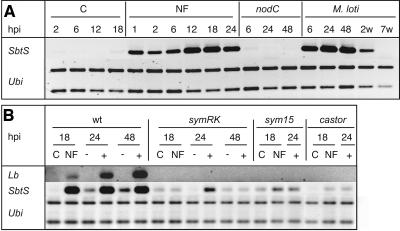
(A) SbtS cDNA fragment enhanced in mycorrhized wild-type roots but not in symbiotic mutants or nodulated roots. The expression of this gene was found subsequently to be transiently induced upon M. loti inoculation (see Figure 4A).
(B) cDNA fragment AM35 was detected in wild-type and sym15-2 mutant roots after G. intraradices inoculation.
(C) cDNA fragment AM1 was detected in wild-type and mutant roots after G. intraradices inoculation.
(D) B74 represents a glutathione S-transferase gene repressed upon G. intraradices inoculation in both wild-type and symbiotic mutant roots.
(E) BN193 was expressed only in nodulated roots (potential nodulin).
(F) B98 was expressed exclusively in mycorrhized wild-type roots (potential mycorrhizin). AN97 was expressed in wild-type roots upon inoculation with either G. intraradices and M. loti (potential symbiosin).
Consistent with the presence of hyphae in fungus-inoculated root samples, several sequenced fragments were likely to be derived from the fungus G. intraradices, as judged from sequence similarity to either fungal DNA or fungal protein sequences. For 10 fragments, the nonplant origin was confirmed experimentally (see Supplemental Table 2 online). At least 50% of the AM-specific cDNA-AFLP fragments were considered to be of plant origin because they exhibited strong sequence similarity to plant ESTs, primarily from L. japonicus, M. truncatula, or Glycine max. Because ∼4% of all displayed cDNA-AFLP fragments were specific to roots infected with AM fungus, the cDNA-AFLP analysis suggests that at least 2% of the plant root transcriptome changes during AM development.
The cDNA-AFLP analysis included castor-2 and sym15-2 mutant roots inoculated with G. intraradices (Figure 2) to visualize the effects of common sym mutations on root transcript profiles. These two reference lines were chosen because arbuscule development was never observed in their roots (Table 1) (Bonfante et al., 2000; Novero et al., 2002; Demchenko et al., 2004). Thus, misleading false-positive expression patterns resulting from rare successful breakthrough events could be avoided. The vast majority of plant cDNA fragments that were induced by AM in wild-type samples were not induced in either of the two mutant lines. Although 40 fragments were enhanced both in the wild type and in one or both mutants (Figures 2B and 2C; see Supplemental Table 1 online), many of them turned out to be of fungal origin. For some plant-derived fragments (e.g., S259; see Supplemental Table 1 online), further investigations confirmed the differential expression pattern but suggested that expression was also influenced by factors other than symbiosis. Therefore, the activation of the majority of plant genes during AM is CASTOR- and SYM15-dependent.
Because the sym15-2 and castor-2 mutants differ in their AM phenotype, one might expect this difference to be reflected by differential gene activation. Indeed, a small number of plant-derived cDNA fragments were enhanced during AM but specifically absent from either castor-2 or sym15-2 root samples (see Supplemental Table 1 online). Two of these fragments represented two L. japonicus GDSL motif–containing lipase genes (Symbiosis-Induced Lipase-Like genes [SILLI1a and SILLI1b; see Supplemental Table 1 online]). Both genes were activated in the wild type and castor-2 but not in sym15-2 roots upon infection with the AM fungus, and this pattern was confirmed by RT-PCR. Sequence analysis of large-insert genomic clones revealed the presence of a lipase gene family in L. japonicus in which at least eight members are arrayed in tandem. The expression of highly similar gene family members was found to be dependent on plant age, and transcripts were also detected in leaves (data not shown). Interestingly, enhanced expression was also observed in nodulated roots (data not shown). Because of this complexity, we did not subject the lipase gene family to a refined expression analysis.
Novel Marker Genes for Symbiotic Root Development
We used semiquantitative RT-PCR with gene-specific primers as an independent method to verify the expression pattern of a subset of differentially displayed cDNA-AFLP fragments. Figure 3 shows a time course of transcript accumulation during AM development for a subset of genes exhibiting a robust and reproducible AM-responsive expression pattern. Most of these genes were analyzed further by determining their corresponding full-length cDNA as well as their genomic sequences (see Supplemental Table 3 online). The latter was done within the context of the L. japonicus genome sequencing project.
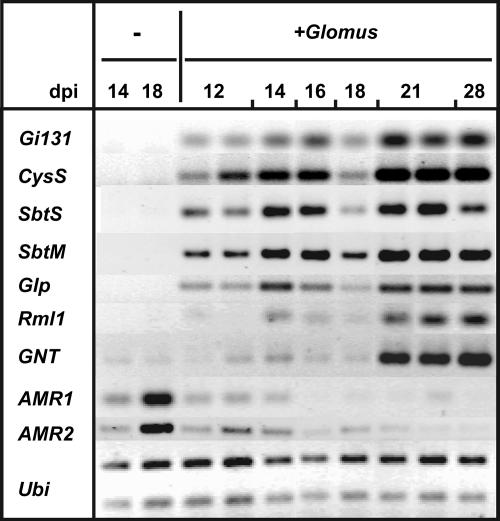
Figure 3.
Time Course of AM-Regulated Gene Expression.
Transcript levels in roots uninoculated (−) or inoculated with G. intraradices (+Glomus) harvested at different days postinoculation (dpi) were analyzed by RT-PCR. Induction of the L. japonicus genes CysS, SbtS, SbtM, Glp, Rml1, and GNT, and repression of AMR1 and AMR2. A cDNA fragment of a fungal aspartate protease gene (Gi131) was amplified as a measure of the degree of colonization by the AM fungus. A fragment of a constitutively expressed L. japonicus polyubiquitin gene (Ubi) was amplified as a control for the amount of RNA template in the reactions. The first arbuscules were already observed at 6 dpi, and the degree of arbuscular colonization increased until 6 weeks after inoculation. In the experiment shown, the 18-dpi sample exhibited an atypically low degree of fungal colonization, as detected by trypan blue staining (data not shown) and the low abundance of fungal transcript Gi131. Both induction and repression of AM-regulated plant genes were lower in this sample, indicating that the level of gene regulation was a function of the degree of fungal colonization and not of plant age. Two independent samples for the 12- and 21-dpi harvest were included to document biological variation.
The abundance of a fungal cDNA fragment from a predicted aspartyl protease gene (Gi131) was found to correlate with the extent of root colonization by the fungus (data not shown). Therefore, Gi131 was used as a marker for the degree of fungal colonization (Figures 3, 4A, and 4C; see Supplemental Table 2 online).
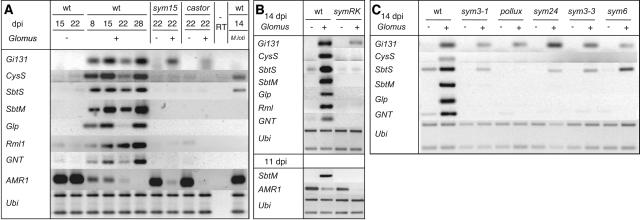
Figure 4.
Expression of AM-Regulated Genes in Symbiotic Mutants of L. japonicus.
RT-PCR analysis of transcript levels in roots of the wild type (wt), sym15-2 (sym15), and castor-2 (castor) (A); roots of the wild type and symRK-3 (symRK) (B); and roots of the wild type, sym3-1 (sym3-1) pollux-1 (pollux), sym24 (sym24), sym3-3 (sym3-3), and sym6-3 (sym6) (C). Noninoculated roots (−) and roots inoculated with G. intraradices (+) or M. loti (M. loti) were harvested at different days after inoculation (dpi) for RNA extraction. −RT indicates control PCR without prior reverse transcription, using as template RNA of mycorrhized wild-type roots at 3 weeks after inoculation. A L. japonicus polyubiquitin gene (Ubi) was amplified as a control for equal amounts of template between samples.
Three Protease Genes and a Germin-Like Protein Gene Are Activated Early during AM Symbiosis
Among the AM-induced genes, SbtM and SbtS show strong similarity to Ser-protease–encoding genes of the subtilase superfamily (Siezen and Leunissen, 1997). L. japonicus mycorrhiza-subtilase (SbtM) transcripts (Figure 3) were not detected in uninoculated roots. Their steady state levels increased with the degree of mycorrhizal colonization, as reflected by the accumulation of the fungal Gi131 RT-PCR product (Figure 3) and observed after trypan blue staining of part of the root samples (data not shown). SbtM gene induction was observed exclusively in wild-type roots inoculated with G. intraradices but not in other plant tissues or under different growth conditions tested. In four independent experiments, SbtM transcript levels in uninoculated roots were at or below the detection limit of RT-PCR. SbtM expression was not detected in roots upon NF treatment or M. loti inoculation (Figure 4A; data not shown), nor was it apparent during phosphate starvation or in powdery mildew–infected leaves (data not shown). No SbtM transcript could be detected in mutant roots affected in either of the common SYM genes with or without G. intraradices at 7, 14, or 21 d after inoculation (Figure 4; data not shown). Therefore, activation of SbtM is specific for AM and requires the function of SYMRK, CASTOR, POLLUX, SYM3, SYM6, SYM15, and SYM24.
The second subtilase gene, SbtS, was also strongly upregulated after inoculation with the AM fungus (Figure 3), and this upregulation was significantly attenuated in mutants (Figures 4A to 4C). Induction was also observed in M. loti–inoculated roots (Figure 4A). Therefore, the gene was named SbtS for L. japonicus symbiosis subtilase. Neither SbtM nor SbtS transcripts were detected in L. japonicus leaves or stems (data not shown).
Two independent cDNA-AFLP fragments with sequence similarity to Cys-proteases were found to be induced during AM. RT-PCR experiments using primers matching one of the fragments revealed gene induction as early as 8 d after inoculation with the AM fungus and also in nodulated roots (CysS; Figures 3 and 4). Amplification of cDNA fragments by 5′ and 3′ rapid amplification of cDNA ends (RACE) as well as genomic sequencing of BAC clones revealed the presence of a tandem array of several very closely related family members in L. japonicus (data not shown). The expression pattern of individual members of this family has yet to be determined. In addition, mRNA corresponding to a cDNA fragment with sequence similarity to germin-like protein (Glp) genes also accumulated early during AM (Figures 3 and 4A).
Induction of SbtS Expression by NF or Rhizobia Is Strongly Attenuated in symRK, castor, and sym15 Mutants
A time course of SbtS activation after inoculation with M. loti strain R7A or treatment with NF was analyzed (Figure 5A). Induced expression was detected at the earliest time points investigated, 1 h after NF treatment and 6 h after bacterial inoculation. Expression remained strong during the first days after inoculation but was lower in roots with 2-week-old nodules and could not be detected at 7 weeks after inoculation. This transient expression pattern is probably responsible for the lack of a detectable SbtS cDNA-AFLP fragment 2 and 3 weeks after inoculation with M. loti (Figure 2A). The full response to bacteria is dependent on NF, as M. loti strain R7AC2, a bacterial nodC mutant unable to produce NF, did not induce the gene expression to a similar extent as wild-type bacteria (Figures 5A and 5B). Full activation of SbtS during the interaction with rhizobia depends on the function of CASTOR, SYM15, and SYMRK, because only a weak and transient activation occurred in roots of the corresponding symbiotic mutants (Figure 5B). Such weak and transient induction was reproducibly observed in three different mutants representing symRK-2, symRK-3, and symRK-7 alleles (Figure 5B; data not shown). A L. japonicus leghemoglobin gene induced early upon NF or M. loti inoculation (Stracke et al., 2002) was included as a reference marker (Figure 5B). The induction of this gene by NF or M. loti was compromised in plants homozygous for the castor-2, sym15-2, or symRK-3 allele (Figure 5B).
Figure 5.
Early Expression of SbtS and Lb in Response to NF or M. loti.
Transcript abundance analyzed by RT-PCR in L. japonicus roots treated with M. loti NF, mock-treated (C), or inoculated with M. loti strain wild-type R7A (M. loti, +) or R7AC2 carrying a nodC mutation (nodC, −). Roots were harvested for RNA extraction hours (hpi) or 2 or 7 weeks (2w, 7w) after inoculation. A L. japonicus polyubiquitin gene (Ubi) was amplified to control for equal amounts of template between samples.
(A) Time course of gene expression in wild-type plants.
(B) Expression pattern in the symbiotic mutants symRK-3 (symRK), sym15-2 (sym15), and castor-2 (castor).
Rml1, a Remorin Gene, and GNT, a N-Acetylglucosaminyltransferase Gene, Are Activated Later during AM Development
L. japonicus RML1 is homologous with remorin from potato (Solanum tuberosum), a protein that is attached to the plasma membrane, binds oligogalacturonids, endogenous elicitors of plant wound responses (reviewed in León et al., 2001), and becomes phosphorylated in a ligand-dependent manner (Reymond et al., 1996). The GNT gene encodes a protein with 80% similarity to N-acetylglucosaminyltransferases, which are involved in glycoprotein modification in the Golgi (β-1,4-mannosyl-glycoprotein β-1,4-N-acetylglucosaminyltransferase). The induction of Rml1 and GNT upon fungal inoculation (Figure 3) is CASTOR-, SYM15-, and SYMRK-dependent (Figures 4A and 4B). The expression of Rml1 and GNT was delayed relative to the other AM-induced genes (Figure 3). Transcript levels of Rml1 were not increased above the basal level at 7 or 8 d after inoculation (Figure 4A) but only at 12 to 14 d after inoculation and became more pronounced at later time points (Figures 3 and 4A). By contrast, both subtilase genes were clearly upregulated at the earliest time points investigated (7 and 8 d after inoculation; Figure 4A; data not shown). Given these differences in the timing of expression, the subtilase genes could be categorized as early and Rml1 and GNT as late AM-induced genes. It will be interesting to correlate the expression of these late and early genes with the development of specific symbiotic structures.
Early and Sustained Activation of the Cbp1 Promoter during AM
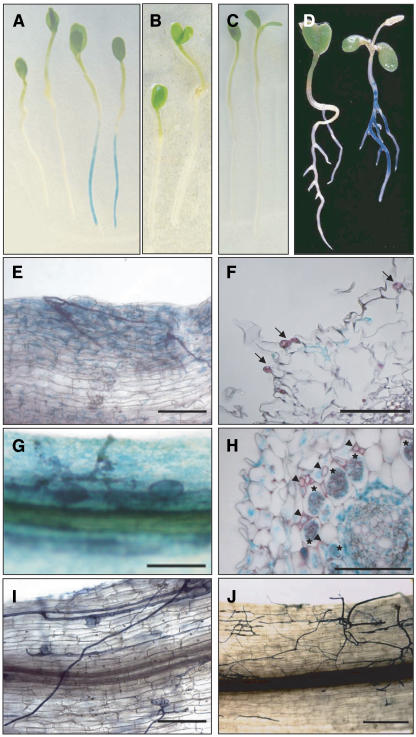
β-Glucuronidase (GUS) activity in roots of a transgenic line harboring a Cbp1 promoter:GUS fusion (T90; Webb et al., 2000) was previously found to be induced rapidly upon treatment with M. loti (Webb et al., 2000) (Figure 6A). Because one of our goals was to identify useful molecular markers, we tested the responsiveness of this promoter during AM development. The T90 line carrying ProCbp1:GUS was inoculated with G. intraradices, and GUS activity was detected after 4 d, the earliest time point analyzed (data not shown). GUS activity was also observed in cells not in direct contact with fungal hyphae (Figures 6E and 6F), and expression persisted during later stages of AM development (Figures 6E to 6H). Strong GUS activity was detected not only in arbuscule-containing cells but also in cells of the pericycle and vascular tissue that are never infected by fungal hyphae (Figure 6H). Our results demonstrate that the Cbp1 promoter is activated early during AM infection and that direct fungal contact with a host cell is not required for ProCbp1:GUS activation within that cell. The T90 line was crossed to castor-2 and sym15-2, and individual F2 mutants homozygous for the ProCbp1:GUS insertion were identified using an insertion-specific PCR marker (see Methods). F3 progeny of the castor and sym15 mutants carrying the ProCbp1:GUS insertion did not show any GUS staining upon either fungal or rhizobial inoculation (Figures 6B, 6C, 6I, and 6J). These results indicate that CASTOR and SYM15 are required for ProCbp1:GUS activation upon stimulation with either the fungal or the bacterial microsymbiont.
Figure 6.
Early Symbiotic Activation of ProCbp1:GUS in AM and RNS Is Abolished in castor-2 and sym15-2 Mutant Backgrounds.
Expression and histochemical localization of ProCbp1:GUS in roots of transgenic line T90 ([A] and [D] to [H]) and F3 progeny of castor-2 × T90 ([B] and [I]) and sym15-2 × T90 ([C] and [J]) after inoculation with M. loti ([A] to [C]) or G. intraradices ([D] to [J]). In (E), (G), (I), and (J), root samples were double stained with black ink for fungal structures and with 5-bromo-4-chloro-3-indolyl-β-glucuronic acid for GUS activity. Bars = 200 μm ([E] and [H] to [J]) and 100 μm ([F] and [G]).
(A) to (C) GUS activity was detected 24 h after inoculation with M. loti in 5-d-old T90 roots ([A], right) but not in T90 noninoculated control roots ([A], left) or in inoculated roots of castor-2 × T90 (B) and sym15-2 × T90 (C).
(D) GUS activity in T90 roots after 2 weeks of cocultivation with G. intraradices (right) but not in noninoculated control roots (left).
(E) and (F) GUS activity in T90 roots after 1 week of cocultivation with G. intraradices. Expression was analyzed in whole roots (E) and semithin cross sections (F). GUS activity was found in epidermal and exodermal cells associated with extraradical hyphae (arrows).
(G) and (H) GUS activity in T90 roots after 4 weeks of cocultivation with G. intraradices. Expression was analyzed in whole roots (G) and semithin cross sections (H). GUS stain localized to various cell types including arbuscule (asterisks) and non-arbuscule-containing cells. The arrowheads indicate intercellular hyphae.
(I) and (J) Extraradical hyphae and appressoria formation on roots of castor-2 × T90 (I) and sym15-2 × T90 (J). No GUS activity was detected.
Downregulation of AMR1 Gene Expression by Fungal Inoculation Occurs in Common sym Mutants
The largest group of differentially displayed cDNA-AFLP fragments represents plant genes that are upregulated in mycorrhized wild-type L. japonicus roots but not in the symbiotic mutants castor-2 and sym15-2 (Figure 2A). However, an additional expression pattern indicative of transcriptional regulation that occurred independently of the CASTOR or SYM15 gene was detected.
Interestingly, for all nine of the AM-repressed cDNA-AFLP fragments, the pattern suggested repressed expression in both wild-type and mutant roots after inoculation with G. intraradices (Figure 2D). Two of the fragments, corresponding to genes AMR1 and AMR2 (for AM repressed), were chosen for analysis in RT-PCR experiments, and the mycorrhiza-dependent repression was confirmed (Figure 3). Database searches did not reveal a function for AMR1, whereas AMR2 is probably a pseudogene. Although the full-length cDNA of AMR2 exhibited sequence similarity to Ser-protease inhibitor genes of the Serpin family, the cDNA suffers from a frameshift within three nucleotides after the start codon, which leads to its translation into a protein completely unrelated to serpins. The open reading frame comprises only 402 bp, less than the first half of the 1.3-kb cDNA. Curiously, the same frameshift was found in ecotypes Gifu and MG-20 and the distantly related species L. filicaulis, indicating that this frameshift predated speciation (data not shown).
AMR1 repression after infection with the AM fungus occurred to the same extent in roots of the wild type and both castor-2 and sym15 mutants (Figure 4A). Downregulation was also observed in roots of a symRK mutant (Figure 4C), although it was less pronounced in an independent experiment (data not shown). A similar expression pattern had been observed for a gene of M. truncatula, Mt4, which is downregulated by AM fungi in wild-type roots of both M. truncatula and M. sativa and in roots of the M. sativa nork mutant MN-1008 (Burleigh and Harrison, 1997). Interestingly, Mt4 is also repressed under moderate and high phosphate conditions (Burleigh and Harrison, 1997, 1998). In L. japonicus roots, a moderate level of phosphate fertilization of 0.2 mM compared with the 0.01 mM used in controls had no detectable effect on AMR1 expression (data not shown). AMR1 transcript abundance exhibited a diurnal rhythm, with transcript levels being highest in the morning (data not shown).
DISCUSSION
Seven L. japonicus Genes Are Required for Intracellular Infection by Fungal and Bacterial Symbionts
Here, we show that in L. japonicus at least seven genetic loci control early steps of both fungal and bacterial symbiosis. The microscopic analysis of their mutant phenotypes indicated that these genes are involved in the successful intracellular infection by microsymbionts. During the interaction with M. loti, infection threads were not formed. Although mutants in SYMRK, CASTOR, POLLUX, SYM3, SYM15, and SYM24 do not form nodules or form nodules at very low frequency (Table 1), small nodule-like structures were observed on mutants defective in the SYM6 gene, indicating that this gene is not required for the induction of cell divisions leading to nodule primordium formation (Schauser et al., 1998). It is unlikely that this finding is attributable to allele leakiness, because the sym6-3 mutation results in a very strong AM phenotype (Table 1). This mutant, therefore, uncouples nodule initiation from infection, which further supports the view that the genetic overlap between the two symbioses is at the level of intracellular infection.
On all common sym mutant roots listed in Table 1, the infection by AM fungi is aborted either before cell entry or within the first penetrated cells. Hyphae are either blocked at the surface (sym15-2) or continue to explore extracellular spaces between outer cell layers or sometimes even penetrate into cells. These exploratory hyphae often show balloon-like swellings or other deformations that are a diagnostic feature of aborted infections. This fact clearly indicates the requirement of the common symbiosis genes for fungal intracellular infection of the outer cell layers.
Old roots of symRK mutants, harboring likely null alleles such as transposon insertions and truncations, developed infrequent but structurally intact arbuscules, indicating that this gene is not absolutely required for the formation of these symbiotic structures (Stracke et al., 2002; Demchenko et al., 2004). By contrast, the CASTOR and SYM15 genes are indispensable for arbuscule development (Novero et al., 2002; Demchenko et al., 2004). A role for the SYM6 gene during arbuscule development is likely because arbuscules were detected at a very low frequency in roots of the sym6-3 mutant (Table 1). The pollux, sym3, and sym24 mutants form arbuscules with a similar delay as symRK mutants (Table 1). However, intraradical infection patches were observed in these mutants that were not associated with arbuscules but only with extracellular hyphae. Therefore, it is possible that these genes make incremental contributions to arbuscule development. Significant phenotypic variation between alleles has been observed in different castor and sym15 mutants (Novero et al., 2002; Demchenko et al., 2004). Therefore, careful side-by-side comparisons of null alleles of POLLUX, SYM3, and SYM24 will be required to determine their precise role during later stages of AM development.
Large-Scale cDNA-AFLP Profiling Reveals Major Transcriptional Reprogramming during AM Symbiosis
To obtain insights into the role of the common SYM genes in fungal AM symbiosis, we performed transcript profiling by cDNA-AFLP, which has the potential to display a large proportion of an mRNA population and therefore is suitable for genome-scale analysis (Bachem et al., 1996). Our cDNA-AFLP profiling experiment revealed that the plant root undergoes substantial transcriptional reprogramming during AM development; ∼2% of all plant transcripts were upregulated, and a small number of transcripts were downregulated.
We compared the sequences obtained from our cDNA-AFLP screen with those described by other laboratories to be AM-regulated. Four L. japonicus AM-induced fragments showed similarity to M. truncatula sequences from a cDNA library enriched for AM-specific root transcripts (MtGIMs; Wulf et al., 2003). These exhibited sequence similarity to a Cys-protease, a germin-like protein, a glutathione S-transferase, and an N-acetylglucosaminyltransferase. A cDNA with sequence similarity to genes for Cys-rich antifungal proteins (S212; see Supplemental Table 1 online) was detected in nodulated L. japonicus roots, whereas an MtGIM clone with high sequence similarity to this fragment was found in mycorrhized roots of M. truncatula (Wulf et al., 2003; Manthey et al., 2004). Transcript profiling in Medicago using cDNA arrays resulted in several AM-regulated genes (Liu et al., 2003; Manthey et al., 2004). Three genes identified by Liu et al. (2003) showed sequence similarity to Lotus cDNA-AFLP fragments: a Cys-protease, a glutathione S-transferase, and a subtilase. MtENOD8.1, a GDSL motif–containing lipase for which a high expression during nodulation was known, was found to increase expression also in AM roots of M. truncatula (Manthey et al., 2004). Like the L. japonicus GDSL lipase genes identified in this study (see Supplemental Table 1 online), MtENOD8.1 is part of a gene cluster. Brechenmacher et al. (2004) and Manthey et al. (2004) identified a putative peptidyl-prolyl cis-trans isomerase gene as upregulated in mycorrhized roots. A L. japonicus cDNA-AFLP fragment with high sequence similarity (B135, S302; see Supplemental Table 1 online) was decreased 2 weeks after inoculation with AM fungi (data not shown).
The conserved activation in both Lotus and Medicago supports the idea that these genes might be of functional relevance during AM. However, because most of these genes are members of larger gene families, it remains to be established whether orthologous (i.e., functionally identical) gene pairs have been identified.
Common SYM Genes Are Required for the Activation of Most AM-Regulated Genes
Not a single plant gene could be identified that was reproducibly activated in a truly symbiosis-specific manner independent of the common SYM genes. Because of the scale of the cDNA-AFLP experiment, only a single time point was analyzed. Thus, it is possible that some transcripts were not detected at all or were missed because of a transient expression pattern, as exemplified by SbtS induction during nodulation (Figures 2A and 4A). However, the cumulative evidence provided by the substantial number of fragments inspected strongly suggests that the sustained transcriptional activation of the majority of genes in response to the AM fungus G. intraradices depends on the function of CASTOR and SYM15 (Figures 2 and 4). This qualitative result from the cDNA-AFLP analysis was confirmed by a detailed RT-PCR analysis of a series of newly identified marker genes (Figure 4) and was extended to include the remaining common SYM genes. We conclude that the common SYM genes tested are key components of the transcriptional reprogramming of the roots during AM symbiosis.
This global effect of common SYM mutations on AM-activated transcription is intriguing given the complex expression patterns of AM-induced genes. The analysis of promoter–reporter fusions and in situ hybridization has revealed that different AM-induced genes are activated in different cell types and at different stages of AM development (Harrison, 1996; Chabaud et al., 2002; Harrison et al., 2002; Doll et al., 2003; Liu et al., 2003). There are several possible mechanisms through which gene expression could be affected in the mutants. The common SYM proteins could be involved directly in a signal transduction process leading to transcriptional gene activation. This is particularly likely for genes such as Cbp1 because the Cbp1 promoter:GUS fusion was activated early during infection and also induced in epidermal cells at a distance from fungal hyphae. However, some AM-responsive genes, such as phosphate and hexose transporter genes that are involved in arbuscule physiology, are activated only after fungal penetration into the root tissue (Harrison, 1996; Harrison et al., 2002). As mutations in the common SYM genes lead to an early arrest of fungal infection attempts at the root surface, the lack of activation of late genes could be an indirect consequence of a lack of arbuscule development. Therefore, it is possible that a direct defect in signaling in combination with the defect of AM development gives rise to the observed global lack of gene activation in the mutant lines analyzed.
Transcripts of PsENOD5 and PsENOD12A do not accumulate in the interaction of roots of pea Pssym8 mutants with Gigaspora margarita (Albrecht et al., 1998). Likewise, in M. truncatula, the AM fungus–induced transcriptional activation of several genes was not observed in a dmi3 mutant background (Weidmann et al., 2004). Although these studies involved only individual mutants, the results can be taken as a first indication that the global effect of common sym mutants on transcriptional activation observed here in the L. japonicus/G. intraradices system may also take place at a similar scale in other legumes.
A role of common SYM genes in signal transduction is further supported by their mutant phenotypes in response to rhizobial signals. Its induction within 1 h or less upon NF treatment places SbtS among the earliest ENOD genes known to date, together with MtRIP1 and MtENOD12 (Pichon et al., 1992; Ramu et al., 2002). Also, the Cbp1 promoter responds rapidly to M. loti (Webb et al., 2000) (data not shown). Because of the impairment of both SbtS and Cbp1 responses in symbiotic mutants, we conclude that the corresponding common SYM genes have a role in the earliest stages of NF-mediated signaling.
Signal Transduction Independent of Common SYM Genes
Signaling through the common SYM genes is usually portrayed as a single pathway, mostly because the currently available tools are not sufficient for a more refined placement. Gene activation by rhizobia or AM fungi depends on common SYM genes, but a single linear pathway does not encompass the specific activation of nodulins and mycorrhizins in only one of the symbioses. Clearly, additional parallel pathways or a signaling network are likely to mediate the development of nodulation and AM. At present, calcium spiking is the only molecular marker that distinguishes between common sym mutants. Therefore, one aim of this study was to use transcriptional gene regulation as a readout to decipher the activity of different signaling pathways and to identify regulons that require only a subset or none of the common SYM genes for their regulation. Indeed, we found that all AM-repressed cDNA-AFLP fragments (including AMR1) in our transcript profiling experiment were also repressed in castor and sym15 mutant roots, providing clear evidence for the existence of signal transduction pathways, leading from the perception of the fungus by the plant to transcriptional gene regulation, that are independent of the common SYM genes.
METHODS
Biological Material and Growth Conditions
All plant mutant lines used were derived from Lotus japonicus ecotype B-129 Gifu, which is referred to as “wild type” throughout this study. The ProCbp1:GUS fusion in line T90 was generated through a promoter-trapping experiment (Webb et al., 2000).
For inoculation of L. japonicus with Mesorhizobium loti R7A (Sullivan et al., 2002) or strain R7AC2 (nodC::Tn5 mutant; J.T. Sullivan and C.W. Ronson, personal communication) and NF treatment, growth conditions and inoculation procedures were as described (Stracke et al., 2002).
Inoculation of seedlings with Glomus intraradices (strain deposited in the Bank of European Glomales as BEG195) was essentially as described (Wegel et al., 1998), except that plants were grown in expanded clay particles (Biosorb; PDI Agrochemical, Essex, UK), watered daily, and treated once per week with half-strength Hoagland solution (Hoagland and Arnold, 1950) with phosphate content reduced to 0.1 mM. Novel mutants were grown side by side with the already described mutants to allow direct comparison and to exclude the possible influence of different experimental conditions on their respective phenotypes.
Root Sectioning and Staining
Ink staining and sectioning of roots were done essentially as described (Demchenko et al., 2004). After staining for GUS activity, roots were cut into 5-mm-long pieces and fixed in 4% formaldehyde and 0.25% glutaraldehyde overnight at 4°C. Roots were washed for 1 h in double-distilled water, dehydrated in a graded ethanol series, equilibrated in Histo-Clear (National Diagnostics, Atlanta, GA), and embedded in Paraplast Plus (Sigma-Aldrich, St. Louis, MO). Serial sections (10 μm) were cut with a rotary microtome, attached to SuperFrost Plus slides (Roth, Karlsruhe, Germany), and deparaffinized in Histo-Clear. The tissue sections were rehydrated in a graded ethanol series, equilibrated in double-distilled water, and incubated in Schiff's reagent for 2 h for Feulgen staining (Demchenko et al., 2004). Then, sections were washed 3 × 15 min with SO3/water (Demchenko et al., 2004) and 2 × 5 min with double-distilled water. Sections were dehydrated in a graded ethanol series, equilibrated in Histo-Clear, and mounted with Eukitt (Kindler, Freiburg, Germany).
cDNA-AFLP
For cDNA-AFLP, RNA was prepared from L. japonicus roots as described (Goormachtig et al., 1995). Of the extracted total RNA, 50 μg was used for poly(A)+ extraction on Dyna beads (Dynal, Oslo, Norway) according to the manufacturer's instructions and was then subjected to cDNA synthesis. The cDNA-AFLP analysis was performed as described (Durrant et al., 2000) using 69 AFLP primer combinations. The 33P-labeled cDNA fragments were separated on 6% polyacrylamide gels and visualized by autoradiography. DNA sequences of 205 reamplified cDNA-AFLP fragments were determined either by direct sequencing or after cloning.
RNA Extraction and RT-PCR
For RNA extraction (except for the cDNA-AFLP; see above), L. japonicus root material was ground in liquid nitrogen, thoroughly mixed with 500 μL of prewarmed extraction buffer (Chang et al., 1993), and extracted twice with 1 volume of phenol:chloroform:isoamyl alcohol (125:24:1, pH 4.7; Sigma-Aldrich). The aqueous supernatant was precipitated with LiCl at a final concentration of 3 M for 4 to 12 h at 4°C. After centrifugation, the pellet was washed with 75% ethanol, dried, and dissolved in water. Contaminating genomic DNA was removed by 30 min of incubation with 1 unit of DNaseI (Amersham Biosciences, Uppsala, Sweden) per microgram of RNA at 37°C in the presence of 10 units of SUPERasIn (Ambion, Austin, TX), followed by phenol:chloroform:isoamyl alcohol extraction and precipitation with sodium acetate:ethanol. Pellets were washed and dissolved in RNA-Secure resuspension solution (Ambion) at 60°C, and 1 μg of total RNA was used for first-strand cDNA synthesis.
Reverse transcription and PCR were performed as described (Stracke et al., 2002). For a control of equal amounts of cDNA template within the different samples, cDNA fragments of a L. japonicus polyubiquitin gene (AW719307) were amplified (Stracke et al., 2002). A reaction using RNA template without RT was routinely included as a control for the presence of genomic DNA contamination.
5′ RACE PCRs were performed using the FirstChoice RLM RACE kit (Ambion), according to the manufacturer's instructions. 3′ RACE was performed using either this kit or primer 10,011 (see Supplemental Table 4 online) as a reverse primer on oligo(dT)-primed cDNA. PCR conditions and primer sequences are listed in Supplemental Tables 3 and 4 online.
To confirm the origin of fungal cDNA fragments, RNA was extracted from chive (Allium schoenoprasum) roots infected with the same fungal isolate and subjected to cDNA synthesis as described above. In cases in which sequence-identical fragments could be amplified from cDNA samples from infected Lotus and chive roots, the fragment was considered to be of fungal origin (see Supplemental Table 2 online).
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers AP004549, AP004985, AP006863, AP006864, AP006865, AP006866, AP006147, AP006148, AJ609244 to AJ609269, and DN652254 to DN652405.
Supplementary Material
Acknowledgments
We thank Martin Groth for help with Figure 1. C.K. received a Deutsche Forschungsgemeinschaft postdoctoral fellowship. This work was supported by grants from the Biotechnology and Biological Sciences Research Council. Research at the Sainsbury Laboratory is funded by the Gatsby Charitable Foundation.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Martin Parniske (parniske@lmu.de).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.032714.
References
- Albrecht, C., Geurts, R., Lapeyrie, F., and Bisseling, T. (1998). Endomycorrhizae and rhizobial Nod factors both require SYM8 to induce the expression of the early nodulin genes PsENOD5 and PsENOD12A. Plant J. 15 605–614. [DOI] [PubMed] [Google Scholar]
- Ane, J.M., et al. (2004). Medicago truncatula DMI1 required for bacterial and fungal symbioses in legumes. Science 303 1364–1367. [DOI] [PubMed] [Google Scholar]
- Bachem, C.W.B., van der Hoeven, R.S., de Bruijn, S.M., Vreugdenhil, D., Zabeau, M., and Visser, R.G.F. (1996). Visualization of differential gene expression using a novel method of RNA fingerprinting based on AFLP: Analysis of gene expression during potato tuber development. Plant J. 9 745–754. [DOI] [PubMed] [Google Scholar]
- Bonfante, P., Genre, A., Faccio, A., Martini, I., Schauser, L., Stougaard, J., Webb, J., and Parniske, M. (2000). The Lotus japonicus LjSym4 gene is required for the successful symbiotic infection of root epidermal cells. Mol. Plant Microbe Interact. 13 1109–1120. [DOI] [PubMed] [Google Scholar]
- Bradbury, S.M., Peterson, R.L., and Bowley, S.R. (1991). Interaction between three alfalfa nodulation genotypes and two Glomus species. New Phytol. 119 115–120. [DOI] [PubMed] [Google Scholar]
- Brechenmacher, L., Weidmann, S., van Tuinen, D., Chatagnier, O., Gianinazzi, S., Franken, P., and Gianinazzi-Pearson, V. (2004). Expression profiling of up-regulated plant and fungal genes in early and late stages of Medicago truncatula-Glomus mosseae interactions. Mycorrhiza 14 253–262. [DOI] [PubMed] [Google Scholar]
- Brundrett, M. (2002). Coevolution of roots and mycorrhizas of land plants. New Phytol. 154 275–304. [DOI] [PubMed] [Google Scholar]
- Burleigh, S.H., and Harrison, M.J. (1997). A novel gene whose expression in Medicago truncatula roots is suppressed in response to colonization by vesicular-arbuscular mycorrhizal (VAM) fungi and to phosphate nutrition. Plant Mol. Biol. 34 199–208. [DOI] [PubMed] [Google Scholar]
- Burleigh, S.M., and Harrison, M.J. (1998). Characterization of the Mt4 gene from Medicago truncatula. Gene 216 47–53. [DOI] [PubMed] [Google Scholar]
- Catoira, R., Galera, C., De Billy, F., Penmetsa, R.V., Journet, E.P., Maillet, F., Rosenberg, C., Cook, D., Gough, C., and Dénarié, J. (2000). Four genes of Medicago truncatula controlling components of a Nod factor transduction pathway. Plant Cell 12 1647–1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabaud, M., Venard, C., Defaux-Petras, A., Becard, G., and Barker, D.G. (2002). Targeted inoculation of Medicago truncatula in vitro root cultures reveals MtENOD11 expression during early stages of infection by arbuscular mycorrhizal fungi. New Phytol. 156 265–273. [DOI] [PubMed] [Google Scholar]
- Chang, S., Puryear, J., and Cairney, J. (1993). A simple and efficient method for isolating RNA from pine trees. Plant Mol. Biol. Rep. 11 113–116. [Google Scholar]
- Demchenko, K., Winzer, T., Stougaard, J., Parniske, M., and Pawlowski, K. (2004). Distinct roles of Lotus japonicus SYMRK and SYM15 in root colonization and arbuscule formation. New Phytol. 163 381–392. [DOI] [PubMed] [Google Scholar]
- Doll, J., Hause, B., Demchenko, K., Pawlowski, K., and Krajinski, F. (2003). A member of the germin-like protein family is a highly conserved mycorrhiza-specific induced gene. Plant Cell Physiol. 44 1208–1214. [DOI] [PubMed] [Google Scholar]
- Duc, G., Trouvelot, A., Gianinazzi-Pearson, V., and Gianinazzi, S. (1989). First report of non-mycorrhizal plant mutants (Myc−) obtained in pea (Pisum sativum L.) and fababean (Vicia faba L.). Plant Sci. 60 215–222. [Google Scholar]
- Durrant, W.E., Rowland, O., Piedras, P., Hammond-Kosack, K.E., and Jones, J.D. (2000). cDNA-AFLP reveals a striking overlap in race-specific resistance and wound response gene expression profiles. Plant Cell 12 963–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrhardt, D., Wais, R., and Long, S. (1996). Calcium spiking in plant root hairs responding to Rhizobium nodulation signals. Cell 85 673–681. [DOI] [PubMed] [Google Scholar]
- Endre, G., Kereszt, A., Kevei, Z., Mihacea, S., Kalo, P., and Kiss, G.B. (2002). A receptor kinase gene regulating symbiotic nodule development. Nature 417 962–966. [DOI] [PubMed] [Google Scholar]
- Frühling, M., Roussel, H., Gianinazzi-Pearson, V., Pühler, A., and Perlick, A.M. (1997). The Vicia faba leghemoglobin gene VfLb29 is induced in root nodules and in roots colonized by the arbuscular mycorrhizal fungus Glomus fasciculatum. Mol. Plant Microbe Interact. 10 124–131. [DOI] [PubMed] [Google Scholar]
- Goormachtig, S., Valerio-Lepiniec, M., Szczyglowski, K., Van Montagu, M., Holsters, M., and de Bruijn, F. (1995). Use of differential display to identify novel Sesbania rostrata genes enhanced by Azorhizobium caulinodans infection. Mol. Plant Microbe Interact. 8 816–824. [DOI] [PubMed] [Google Scholar]
- Harris, J.M., Wais, R., and Long, S.R. (2003). Rhizobium-induced calcium spiking in Lotus japonicus. Mol. Plant Microbe Interact. 16 335–341. [DOI] [PubMed] [Google Scholar]
- Harrison, M. (1996). A sugar transporter from Medicago truncatula: Altered expression pattern in roots during vesicular-arbuscular (VA) mycorrhizal associations. Plant J. 9 491–503. [DOI] [PubMed] [Google Scholar]
- Harrison, M.J., Dewbre, G.R., and Liu, J. (2002). A phosphate transporter from Medicago truncatula involved in the acquisition of phosphate released by arbuscular mycorrhizal fungi. Plant Cell 14 2413–2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch, A.M., Lum, M.R., and Downie, J.A. (2001). What makes the rhizobia-legume symbiosis so special? Plant Physiol. 127 1484–1492. [PMC free article] [PubMed] [Google Scholar]
- Hoagland, D., and Arnold, D. (1950). The water culture method for growing plants without soil. Calif. Agric. Exp. Stn. Circ. 347 1–39. [Google Scholar]
- Imaizumi-Anraku, H., et al. (2004). Plastid proteins crucial for symbiotic fungal and bacterial entry into plant roots. Nature 433 527–531. [DOI] [PubMed] [Google Scholar]
- Journet, E.P., El-Gachtouli, N., Vernoud, V., de Billy, F., Pichon, M., Dedieu, A., Arnould, C., Morandi, D., Barker, D.G., and Gianinazzi-Pearson, V. (2001). Medicago truncatula ENOD11: A novel RPRP-encoding early nodulin gene expressed during mycorrhization in arbuscule-containing cells. Mol. Plant Microbe Interact. 14 737–748. [DOI] [PubMed] [Google Scholar]
- Kawaguchi, M., Imaizumi-Anraku, H., Koiwa, H., Niwa, S., Ikuta, A., Syono, K., and Akao, S. (2002). Root, root hair, and symbiotic mutants of the model legume Lotus japonicus. Mol. Plant Microbe Interact. 15 17–26. [DOI] [PubMed] [Google Scholar]
- Kistner, C., and Parniske, M. (2002). Evolution of signal transduction in intracellular symbiosis. Trends Plant Sci. 7 511–518. [DOI] [PubMed] [Google Scholar]
- Kosuta, S., Chabaud, M., Lougnon, G., Gough, C., Dénarié, J., Barker, D.G., and Becard, G. (2003). A diffusible factor from arbuscular mycorrhizal fungi induces symbiosis-specific MtENOD11 expression in roots of Medicago truncatula. Plant Physiol. 131 952–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouchi, H., et al. (2004). Large-scale analysis of gene expression profiles during early stages of root nodule formation in a model legume, Lotus japonicus. DNA Res. 11 263–274. [DOI] [PubMed] [Google Scholar]
- León, J., Rojo, E., and Sanchez-Serrano, J.J. (2001). Wound signalling in plants. J. Exp. Bot. 52 1–9. [DOI] [PubMed] [Google Scholar]
- Levy, J., et al. (2004). A putative Ca2+ and calmodulin-dependent protein kinase required for bacterial and fungal symbioses. Science 303 1361–1364. [DOI] [PubMed] [Google Scholar]
- Liu, J., Blaylock, L.A., Endre, G., Cho, J., Town, C.D., VandenBosch, K.A., and Harrison, M.J. (2003). Transcript profiling coupled with spatial expression analyses reveals genes involved in distinct developmental stages of an arbuscular mycorrhizal symbiosis. Plant Cell 15 2106–2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lum, M.R., Li, Y., LaRue, T.A., David-Schwartz, R., Kapulnik, Y., and Hirsch, A.M. (2002). Investigation of four classes of non-nodulating white sweetclover (Melilotus alba annua Desr.) mutants and their responses to arbuscular-mycorrhizal fungi. Integr. Comp. Biol. 42 295–303. [DOI] [PubMed] [Google Scholar]
- Madsen, E.B., Madsen, L.H., Radutoiu, S., Olbryt, M., Rakwalska, M., Szczyglowski, K., Sato, S., Kaneko, T., Tabata, S., Sandal, N., and Stougaard, J. (2003). A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature 425 637–640. [DOI] [PubMed] [Google Scholar]
- Manthey, K., Krajinski, F., Hohnjec, N., Firnhaber, C., Pühler, A., Perlick, A.M., and Küster, H. (2004). Transcriptome profiling in root nodules and arbuscular mycorrhiza identifies a collection of novel genes induced during Medicago truncatula root endosymbioses. Mol. Plant Microbe Interact. 17 1063–1077. [DOI] [PubMed] [Google Scholar]
- Marsh, J.F., and Schultze, M. (2001). Analysis of arbuscular mycorrhizas using symbiosis-defective plant mutants. New Phytol. 150 525–532. [Google Scholar]
- Mitra, R.M., Gleason, C.A., Edwards, A., Hadfield, J., Downie, J.A., Oldroyd, G.E.D., and Long, S.R. (2004. a). A Ca2+/calmodulin-dependent protein kinase required for symbiotic nodule development: Gene identification by transcript-based cloning. Proc. Natl. Acad. Sci. USA 101 4701–4705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra, R.M., Shaw, S.L., and Long, S.R. (2004. b). Six nonnodulating plant mutants defective for Nod factor-induced transcriptional changes associated with the legume-rhizobia symbiosis. Proc. Natl. Acad. Sci. USA 101 10217–10222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novero, M., Faccio, A., Genre, A., Stougaard, J., Webb, K.J., Mulder, L., Parniske, M., and Bonfante, P. (2002). Dual requirement of the LjSym4 gene for mycorrhizal development in epidermal and cortical cells of Lotus japonicus roots. New Phytol. 154 741–749. [DOI] [PubMed] [Google Scholar]
- Oldroyd, G.E.D., and Downie, J.A. (2004). Calcium, kinases and nodulation signalling in legumes. Nat. Rev. Mol. Cell Biol. 5 566–576. [DOI] [PubMed] [Google Scholar]
- Parniske, M. (2004). Molecular genetics of the arbuscular mycorrhizal symbiosis. Curr. Opin. Plant Biol. 7 414–421. [DOI] [PubMed] [Google Scholar]
- Paszkowski, U., Kroken, S., Roux, C., and Briggs, S.P. (2002). Rice phosphate transporters include an evolutionarily divergent gene specifically activated in arbuscular mycorrhizal symbiosis. Proc. Natl. Acad. Sci. USA 99 13324–13329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry, J.A., Wang, T.L., Welham, T.J., Gardner, S., Pike, J.M., Yoshida, S., and Parniske, M. (2003). A TILLING reverse genetics tool and a web-accessible collection of mutants of the legume Lotus japonicus. Plant Physiol. 131 866–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichon, M., Journet, E.-P., Dedieu, A., De Billy, F., Truchet, G., and Barker, D.G. (1992). Rhizobium meliloti elicits transient expression of the early nodulin gene ENOD12 in the differentiating root epidermis of transgenic alfalfa. Plant Cell 4 1199–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radutoiu, S., Madsen, L.H., Madsen, E.B., Felle, H.H., Umehara, Y., Gronlund, M., Sato, S., Nakamura, Y., Tabata, S., Sandal, N., and Stougaard, J. (2003). Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 425 585–592. [DOI] [PubMed] [Google Scholar]
- Ramu, S.K., Peng, H.M., and Cook, D.R. (2002). Nod factor induction of reactive oxygen species production is correlated with expression of the early nodulin gene rip1 in Medicago truncatula. Mol. Plant Microbe Interact. 15 522–528. [DOI] [PubMed] [Google Scholar]
- Rausch, C., Daram, P., Brunner, S., Jansa, J., Laloi, M., Leggewie, G., Amrhein, N., and Bucher, M. (2001). A phosphate transporter expressed in arbuscule-containing cells in potato. Nature 414 462–470. [DOI] [PubMed] [Google Scholar]
- Read, D.J., Ducket, J.G., Francis, R., Ligron, R., and Russell, A. (2000). Symbiotic fungal associations in ‘lower’ land plants. Philos. Trans. R. Soc. Lond. B Biol. Sci. 355 815–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond, P., Kunz, B., Paul-Pletzer, K., Grimm, R., Eckerskorn, C., and Farmer, E.E. (1996). Cloning of a cDNA encoding a plasma membrane-associated, uronide binding phosphoprotein with physical properties similar to viral movement proteins. Plant Cell 8 2265–2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagan, M., Morandi, D., Tarenghi, E., and Duc, G. (1995). Selection of nodulation and mycorrhizal mutants in the model plant Medicago trunculata (Gaertn.) after γ-ray mutagenesis. Plant Sci. 111 63–71. [Google Scholar]
- Schauser, L., Handberg, K., Sandal, N., Stiller, J., Thykjær, T., Pajuelo, E., Nielsen, A., and Stougaard, J. (1998). Symbiotic mutants deficient in nodule establishment identified after T-DNA transformation of Lotus japonicus. Mol. Gen. Genet. 259 414–423. [DOI] [PubMed] [Google Scholar]
- Schneider, A., Walker, S.A., Poyser, S., Sagan, M., Ellis, T.H.N., and Downie, J.A. (1999). Genetic mapping and functional analysis of a nodulation-defective mutant (sym19) of pea (Pisum sativum L.). Mol. Gen. Genet. 262 1–11. [DOI] [PubMed] [Google Scholar]
- Senoo, K., Solaiman, M.Z., Kawaguchi, M., Imaizumi-Anraku, H., Akao, S., Tanaka, A., and Obata, H. (2000). Isolation of two different phenotypes of mycorrhizal mutants in the model legume plant Lotus japonicus after EMS-treatment. Plant Cell Physiol. 41 726–732. [DOI] [PubMed] [Google Scholar]
- Shirtliffe, S.J., and Vessey, J.K. (1996). A nodulation (Nod+/Fix−) mutant of Phaseolus vulgaris L. has nodule-like structures lacking peripheral vascular bundles (Pvb−) and is resistant to mycorrhizal infection (Myc−). Plant Sci. 118 209–220. [Google Scholar]
- Siezen, R.J., and Leunissen, J.A.M. (1997). Subtilases: The superfamily of subtilisin-like serine proteases. Protein Sci. 6 501–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stougaard, J. (2001). Genetics and genomics of root symbiosis. Curr. Opin. Plant Biol. 4 328–335. [DOI] [PubMed] [Google Scholar]
- Stracke, S., Kistner, C., Yoshida, S., Mulder, L., Sato, S., Kaneko, T., Tabata, S., Sandal, N., Stougaard, J., Szczyglowski, K., and Parniske, M. (2002). A plant receptor-like kinase required for both fungal and bacterial symbiosis. Nature 417 959–962. [DOI] [PubMed] [Google Scholar]
- Sullivan, J.T., et al. (2002). Comparative sequence analysis of the symbiosis island of Mesorhizobium loti strain R7A. J. Bacteriol. 184 3086–3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczyglowski, K., Shaw, R.S., Wopereis, J., Copeland, S., Hamburger, D., Kasiborski, B., Dazzo, F.B., and de Bruijn, F.J. (1998). Nodule organogenesis and symbiotic mutants of the model legume Lotus japonicus. Mol. Plant Microbe Interact. 11 684–697. [Google Scholar]
- van Kammen, A. (1984). Suggested nomenclature for plant genes involved in nodulation and symbiosis. Plant Mol. Biol. Rep. 2 43–45. [Google Scholar]
- van Rhijn, P., et al. (1997). Expression of early nodulin genes in alfalfa mycorrhizae indicates that signal transduction pathways used in forming arbuscular mycorrhizae and Rhizobium-induced nodules may be conserved. Proc. Natl. Acad. Sci. USA 94 5467–5472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wais, R.J., Galera, C., Oldroyd, G., Catoira, R., Penmetsa, R.V., Cook, D., Gough, C., Dénarié, J., and Long, S.R. (2000). Genetic analysis of calcium spiking responses in nodulation mutants of Medicago truncatula. Proc. Natl. Acad. Sci. USA 97 13407–13412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker, S., Viprey, V., and Downie, J. (2000). Dissection of nodulation signaling using pea mutants defective for calcium spiking induced by nod factors and chitin oligomers. Proc. Natl. Acad. Sci. USA 97 13413–13418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb, K.J., Skøt, L., Nicholson, M.N., Jørgensen, B., and Mizen, S. (2000). Mesorhizobium loti increases root-specific expression of a calcium-binding protein homologue identified by promoter tagging in Lotus japonicus. Mol. Plant Microbe Interact. 13 606–616. [DOI] [PubMed] [Google Scholar]
- Wegel, E., Schauser, L., Sandal, N., Stougaard, J., and Parniske, M. (1998). Mycorrhiza mutants of Lotus japonicus define genetically independent steps during symbiotic infection. Mol. Plant Microbe Interact. 11 933–936. [Google Scholar]
- Weidmann, S., Sanchez, L., Descombin, J., Chatagnier, O., Gianinazzi, S., and Gianinazzi-Pearson, V. (2004). Fungal elicitation of signal transduction-related plant genes precedes mycorrhiza establishment and requires the dmi3 gene in Medicago truncatula. Mol. Plant Microbe Interact. 17 1385–1393. [DOI] [PubMed] [Google Scholar]
- Wulf, A., Manthey, K., Doll, J., Perlick, A.M., Linke, B., Bekel, T., Meyer, F., Franken, P., Kuster, H., and Krajinski, F. (2003). Transcriptional changes in response to arbuscular mycorrhiza development in the model plant Medicago truncatula. Mol. Plant Microbe Interact. 16 306–314. [DOI] [PubMed] [Google Scholar]
- Yoshida, S., and Parniske, M. (2005). Regulation of plant symbiosis receptor kinase (SYMRK) through serine and threonine phosphorylation. J. Biol. Chem. 280 9203–9209. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.