Abstract
We have identified a rice (Oryza sativa) brassinosteroid (BR)-deficient mutant, BR-deficient dwarf2 (brd2). The brd2 locus contains a single base deletion in the coding region of Dim/dwf1, a homolog of Arabidopsis thaliana DIMINUTO/DWARF1 (DIM/DWF1). Introduction of the wild-type Dim/dwf1 gene into brd2 restored the normal phenotype. Overproduction and repression of Dim/dwf1 resulted in contrasting phenotypes, with repressors mimicking the brd2 phenotype and overproducers having large stature with increased numbers of flowers and seeds. Although brd2 contains low levels of common 6-oxo-type BRs, the severity of the brd2 phenotype is much milder than brd1 mutants and most similar to d2 and d11, which show a semidwarf phenotype at the young seedling stage. Quantitative analysis suggested that in brd2, the 24-methylene BR biosynthesis pathway is activated and the uncommon BR, dolichosterone (DS), is produced. DS enhances the rice lamina joint bending angle, rescues the brd1 dwarf phenotype, and inhibits root elongation, indicating that DS is a bioactive BR in rice. Based on these observations, we discuss an alternative BR biosynthetic pathway that produces DS when Dim/dwf1 is defective.
INTRODUCTION
Brassinosteroids (BRs), a class of steroidal plant hormones, are structurally defined as C27, C28, and C29 steroids with substitutions on the A- and B-rings and side chains (Fujioka and Sakurai, 1997; Yokota, 1997) and are widely distributed in both lower and higher plants. Since brassinolide (BL), the most biologically active BR, was initially isolated and its structure determined in 1979 (Grove et al., 1979), a variety of BRs have been identified in a broad range of species. To date, >50 BRs have been isolated and identified from the plant kingdom (Bajguz and Tretyn, 2003). A detailed study of the biosynthesis of BL, a C28 BR, in Catharanthus roseus and Arabidopsis thaliana revealed that two parallel routes, the early and late C-6 oxidation pathways, are connected at multiple steps and are linked to the early C-22 oxidation pathway (Fujioka and Yokota, 2003). In parallel with biochemical studies on BR, molecular genetic approaches using BR-deficient and BR-insensitive mutants of Arabidopsis, pea (Pisum sativum), and tomato (Lycopersicon esculentum) have greatly contributed to our knowledge of the molecular mechanisms of BR biosynthesis and signaling (Clouse and Sasse, 1998; Bishop, 2001; Bishop and Koncz, 2002; Fujioka and Yokota, 2003).
In contrast with the rapid advances in research on BRs in dicots, little is known on the molecular function of BRs in monocots. So far, three types of BR-deficient mutants, d11, d2, and brd1, and one BR-insensitive mutant, d61, have been isolated. Molecular biological studies have demonstrated that the genes containing the d11, d2, and brd1 mutations encode cytochrome P450 enzymes and are involved in the late steps of BR biosynthesis (Hong et al., 2002, 2003; Tanabe et al., 2005), and the D61 gene encodes a putative protein kinase that is very similar to the Arabidopsis BR receptor BRASSINOSTEROID INSENSITIVE1 (BRI1) (Yamamuro et al., 2000). These studies have revealed that the monocot rice (Oryza sativa) plant contains mechanisms for BR biosynthesis and perception that are similar to those in dicots.
However, no rice mutation has yet been identified in the biosynthetic pathways of sterols, which are precursors of BRs; consequently, it is not yet known whether the sterol biosynthetic pathway in rice or other monocots is similar to that in dicots. Recently, a maize (Zea mays) gene, dwf1, was cloned, and its high sequence similarity to the Arabidopsis homolog DIMINUTO/DWARF1 (DIM/DWF1) suggested that monocots and dicots have similar sterol biosynthetic pathways (Tao et al., 2004). However, the absence of a monocot sterol biosynthesis mutant has thus far hampered the understanding of BR and other sterol-based phytohormones in agriculturally important monocot plants. The sterol mutants of dicots fall into two groups. Members of the first group contain a lesion in the late steps of the sterol biosynthetic pathway. The phenotypes of these mutants, such as ste1/bul1/dwf7 (Choe et al., 1999b; Catterou et al., 2001), le/cro6/dwf5 (Serrano-Cartagena et al., 1999; Choe et al., 2000), and dim/dwf1 (Takahashi et al., 1995; Klahre et al., 1998), cannot be distinguished from BR-deficient mutants and can be rescued by treatment with exogenous BL. By contrast, members of the second group are defective in the early steps of sterol biosynthesis. These mutants, such as cph/smt1 (Diener et al., 2000; Schrick et al., 2002), cvp1/smt2 (Carland et al., 1999, 2002), and ell/hyd2/fackel (Jang et al., 2000; Schrick et al., 2000; Souter et al., 2002), share some of the characteristics of BR-deficient mutants but also have unique defects during embryogenesis that cannot be rescued by BR treatment. If rice sterol biosynthesis is similar to that of dicots and rice sterols have biological functions similar to dicot sterols, it should be possible to isolate mutants defective in sterol biosynthesis from rice mutant stocks by screening for BR-related phenotypes.
Here, we report the isolation and characterization of a rice BR-deficient mutant, BR-deficient dwarf2 (brd2). Molecular genetic studies have revealed that brd2 is a loss-of-function allele of Dim/dwf1, a gene homologous to the Arabidopsis DIM/DWF1. Although the level of common endogenous BRs is severely reduced in brd2, the abnormalities of the mutant are less severe in the early vegetative stage. Feeding experiments and quantitative analysis of sterols and BR intermediates have revealed that the brd2 mutant accumulates an active BR, dolichosterone (DS), which is not detected in the wild type. Our data suggest that the weak phenotype of brd2 is caused by the production of DS in a Dim/dwf1-free BR biosynthetic pathway.
RESULTS
Characterization of brd2
Since the identification of the first rice BR-related mutant, d61, which is defective in the rice Bri1 gene (Yamamuro et al., 2000), we have collected rice dwarf mutants with phenotypes similar to d61 to investigate the mechanisms of BR biosynthesis and signaling in rice. We have also previously isolated three other BR-related mutants, brd1, d2, and d11, all of which are defective in BR biosynthesis owing to mutations in BR C-6 oxidase (Cyp85), Cyp90D2, and Cyp724B1, respectively (Hong et al., 2002, 2003; Tanabe et al. 2005). Recently, we identified a dwarf mutant, brd2, which displays the typical BR-deficient phenotype but also has characteristics not observed in the other mutants. Figure 1A shows the gross morphology of the mutant in an early vegetative stage. At this stage, the mutant plant (right) displays a moderate dwarf phenotype with a height ∼70% of the wild-type plant (left) but no other obvious abnormalities. However, as development progresses, the phenotype becomes more severe with characteristics typical of BR-related mutants: dark-green, erect leaves and shortened leaf sheaths. After flowering, the height of the plant reaches only ∼40% of the wild type (Figure 1B). We compared the internode elongation patterns of brd2, the wild type, and other BR-deficient mutants (Figure 1C). In the wild type, the five uppermost internodes elongate; the internodes are numbered from top to bottom such that the uppermost, just below the panicle, is designated as the first internode (Figure 1C). By contrast, the three BR-deficient mutants show different elongation patterns: d2-1 has a specific inhibition in the second internode, brd1-1 is completely defective in internode elongation, and the first internode of brd2 elongates, but the lower internodes do not (Figure 1C). d61-2, which is defective in the rice Bri1 gene, also exhibits dwarfism with a lack of internode elongation except for the first internode (Yamamuro et al., 2000). The brd2 mutant flowers and produces malformed panicles, but the numbers of spikelets and rachis branches, as well as fertility, are severely reduced (Figure 1D). The lengths of the first and second rachis branches in the panicle are also severely reduced. In contrast with these severe defects, the neck internode of the mutant is longer than that of the wild type (Figure 1D). This phenomenon is a common feature in rice BR-related mutants because long neck internodes are also observed in d2 and d61-1. In addition, the brd2 mutant also exhibits shortened grains (Figure 1E) and defective root elongation and development (Figure 1F). Although one seminal root develops normally during embryogenesis, after germination the seminal root elongates to only two-thirds of the wild-type root and formation of the crown root is inhibited, but lateral root formation is not as severely affected as crown root formation.
Figure 1.
Phenotype of the brd2 Mutant.
(A) Gross morphology of wild-type (left) and brd2 (right) seedlings grown for 40 d in soil. Bar = 10 cm.
(B) Gross morphology of field-grown wild type (left) and brd2 (right) at the flowering stage. Bar = 15 cm.
(C) Comparison of the internode elongation pattern of brd2 with previously characterized rice BR-deficient mutants. The five uppermost internodes (I to V) of the wild type were elongated. Bar = 10 cm.
(D) Panicle structure. The wild-type plant (left) developed ∼120 seeds per panicle, and the brd2 mutant panicle developed ∼30 seeds with poor germination rates. Arrows indicate the positions of the nodes. Bar = 10 cm.
(E) Morphology of unhulled grains (top) and brown rice (bottom). The mutant (right) produced shortened grains. Bar = 1 cm.
(F) Root morphology. Seedlings were grown at 30°C in water for 1 week in the light. Bar = 1 cm.
brd2 Is a Knockout Allele of the Dim/dwf1 Gene
To isolate Brd2 by positional cloning, we first performed rough mapping of brd2 using 30 F2 progeny with the dwarf phenotype. Using 62 molecular markers, the brd2 locus was mapped to rice chromosome 10 between 21.8 and 30.2 centimorgan. Because a gene homologous to Arabidopsis DIM1/DWF1 is present in this area, we suspected that brd2 was identical to Dim/dwf1 (Figure 2A). The predicted Dim/dwf1 protein consists of 561 amino acids with 80% identity to the Arabidopsis DIM/DWF1 sequence, and comparison of the genomic and full-length cDNA sequences showed that Dim/dwf1 contains three predicted exons (Figure 2B). Direct sequencing of the brd2 mutant allele revealed a single base deletion (guanine) in exon 2, which leads to a frameshift and produces a premature stop codon near the mutation site (Figure 2B). The mutation causes the loss of the majority of a conserved FAD binding domain and the C-terminal region in Dim/dwf1 (Figure 2C).
Figure 2.
Molecular Characterization of the brd2 Mutation.
(A) The brd2 mutation was mapped at ∼25 centimorgan on chromosome 10, near the map position of Dim/dwf1.
(B) Schematic diagram of the structure of Dim/dwf1 and the site of the mutation in the brd2 allele. Closed boxes represent exons, and open boxes indicate introns. The first Met (ATG) and the stop codon (TAA) are located in exons 2 and 3, respectively. The single base deletion in the brd2 allele is indicated by an arrow, and the premature stop codon is marked with an asterisk.
(C) The deduced amino acid sequence of Dim/dwf1. The mutation site is marked with a closed triangle, and the FAD binding domain is underlined.
(D) Levels of Dim/dwf1 transcripts in wild-type (left) and brd2 (right) plants. Ten micrograms of total RNA was loaded per lane. Ethidium bromide–stained rRNA bands were monitored as a loading control.
(E) Molecular complementation. Transgenic plants derived from mutant callus transformed with the Dim/dwf1 gene had the wild-type phenotype, in contrast with the dwarf phenotype of the mutant (right), and transformants containing the empty vector had the dwarf phenotype (left).
(F) Organ-specific expression of the Dim/dwf1 gene in the wild type. RT, root; SA, shoot apex; LB, leaf blade; LS, leaf sheath; ST, stem; PA, panicle; FL, flower. Five micrograms of total RNA was loaded per lane, and ethidium bromide–stained rRNA bands were monitored as a loading control.
(G) The effect of BL on Dim/dwf1 expression. Total RNA was isolated from wild-type, brd1-1, and d61-2 seedlings either treated for 10 d with a final concentration of 1 μM BL (+) or untreated (−); 7.5 μg was loaded per lane.
We used gel blot analysis to examine the level of Dim/dwf1 transcripts in the mutant. RNA from 10-d-old light-grown wild-type and mutant seedlings was isolated and probed with the ∼800-bp 3′ fragment of the Dim/dwf1 cDNA. The Dim/dwf1 transcript was detected in wild-type seedlings but not in the mutant (Figure 2D). A wild-type genomic DNA fragment containing the entire Dim/dwf1 was transformed into brd2, which restored the wild-type phenotype in all plants that were resistant to the selection marker, hygromycin (Figure 2E, right). Transformation with the empty vector had no effect on the dwarf phenotype (Figure 2E, left). Based on these results, we predict that brd2 is a knockout allele of Dim/dwf1.
The Dim/dwf1 expression pattern in various organs was studied using RNA gel blot analysis. Dim/dwf1 transcripts were detected in all organs examined, albeit at different levels. The highest expression was in stems; intermediate expression was seen in roots, the shoot apex, and flowers; and expression was low in leaves and panicles (Figure 2F). RNA gel blot analysis was also used to examine the effect of exogenously applied BL on Dim/dwf1 expression (Figure 2G). Similar Dim/dwf1 expression levels were observed in the wild type, the BR-deficient mutant brd1, and d61-2, which is partially defective in the BR signaling pathway (Yamamuro et al., 2000), and expression was not affected by treatment with BL. These results indicate that Dim/dwf1 expression is not regulated by BL, in contrast with D2 and BRD1, which are downregulated by BL in a feedback manner (Hong et al., 2002, 2003).
Overexpression and Suppression of Dim/dwf1 in Transgenic Rice Plants
Because only one brd2 mutant was isolated in the first screening, we attempted to isolate other brd2 alleles from our mutant stocks and the rice Tos17 insertion mutant library, but no other alleles were found in these libraries. Therefore, we produced transgenic plants expressing antisense and sense Dim/dwf1 transcripts under the control of the rice Actin constitutive promoter (McElroy et al., 1990). Antisense and sense Dim/dwf1 transgenic plants showed contrasting phenotypes in plant height (dwarf versus tall) (Figure 3A), lamina joint bending (erect versus bent) (Figure 3B), the panicle (decreased versus increased spikelets) (Figure 3C), and internode elongation (stunted versus elongated) (Figure 3D), respectively. The overall phenotype of antisense plants was similar to brd2, although the abnormalities of the antisense plants were less severe than in brd2. The gross morphologies of the transformant lines confirm that the brd2 phenotype is caused by a defect in the functioning of Dim/dwf1. We performed RNA gel blot analysis using two different probes to confirm changes in the levels of Dim/dwf1 mRNA in the transgenic plants (Figure 3E). When the entire cDNA sequence was used as a probe, the intensity of the hybridized band in both antisense and sense plants was greater than in wild-type plants, indicating that the antisense and sense RNAs are highly expressed in the transgenic lines. When the 3′-untranslated sequence, which was not included in the transgene in the antisense or sense constructs, was used as a probe, either no band or a band of very low intensity was observed in the antisense plant samples, and a strong band of an intensity similar to the wild type was observed in the sense plant samples (Figure 3E). The RNA gel blot analysis confirms that the phenotypes observed in the antisense and sense plants are caused by lower and higher expression of Dim/dwf1, respectively.
Figure 3.
Phenotypes of Dim/dwf1 Antisense and Sense Transgenic Plants.
(A) Gross morphology of an antisense plant (left), a control plant containing the empty vector (middle), and a sense plant (right) 3 months after transplantation. Bar = 20 cm.
(B) Leaf morphology: a wild-type leaf (center) bends away from the vertical axis of the leaf toward the abaxial side, an antisense leaf (left) is more erect, and that of an overproducer (right) is more bent.
(C) Panicle structure: the antisense plant (left) generates poor panicles with lower spikelet numbers than the wild type (center), whereas the overproducer (right) generates superior panicles with increased spikelet numbers.
(D) Internode elongation pattern. The antisense plant (left) is severely inhibited in the elongation of internodes II through V, whereas the overproducer has increased numbers and lengths of elongated internodes in comparison with the wild type (center). Bar = 20 cm.
(E) RNA gel blot analysis of the endogenous and transformed Dim/dwf1 transcripts. Two micrograms of total RNA was loaded per lane. An 800-bp cDNA fragment containing the coding sequence was used as a probe to detect both sense and antisense Dim/dwf1 transcripts (top), and a 200-bp probe containing only the 3′-noncoding sequence of the Dim/dwf1 transcript was used to detect endogenous Dim/dwf1 expression (middle). Ethidium bromide–stained rRNA bands were monitored as a loading control (bottom).
Common BR Levels Are Much Lower in brd2 Than in d2 or d11, but the Dwarfism of brd2 Is Most Similar to That of d2 and d11
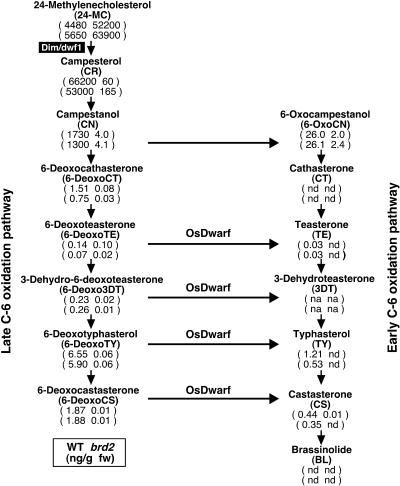
We compared the levels of BR intermediates in brd2 and wild-type seedlings to identify the metabolic step that is blocked in BR biosynthesis. As shown in Figure 4, intermediates from campesterol (CR) to castasterone (CS), in either the early or late C-6 oxidation pathways, were dramatically decreased in the brd2 mutant. No BL was detected in the shoots of either mutant or wild-type plants. The precursor of BL, CS, which is predicted to be the most bioactive BR in rice because BL has not yet been detected in this species (Yamamuro et al., 2000; Hong et al., 2002, 2003), was detected in the wild type, whereas no or a very low level of CS was detected in brd2 in the two independent experiments, confirming that the mutant is deficient in the biosynthesis of the active BR. Because Arabidopsis DIM/DWF1 has been proposed to catalyze the conversion of 24-methylenecholesterol (24-MC) to CR (Klahre et al., 1998), we also measured the levels of 24-MC and CR in wild-type and brd2 plants. In the mutant, the level of CR was decreased >100-fold, and the level of 24-MC was increased >10-fold relative to the wild type, demonstrating that rice Dim/dwf1 catalyzes the conversion of 24-MC to CR, as does Arabidopsis DIM/DWF1.
Figure 4.
Quantitative Analysis of Endogenous BR Intermediates in Wild-Type and brd2 Plants.
Shoots of wild-type and mutant plants were harvested after growth in a greenhouse for 2 months. BR levels (ng/g FW) in wild-type (left) and brd2 (right) plants are shown below each product. BR levels were measured in two independent experiments. na, not analyzed; nd, not detected.
Although the Arabidopsis and rice DIM/DWF1 proteins catalyze the same step in the BR biosynthetic pathway, the severity of the knockout mutants of these genes differs, with the Arabidopsis DIM/DWF1 knockout mutant showing a severely abnormal phenotype (Takahashi et al., 1995) and the rice Dim/dwf1 knockout mutant showing a moderate phenotype at an early vegetative stage (Figure 1A). Genomic DNA gel blot analysis was performed to examine the possibility that the rice genome contains an additional Dim/dwf1 homolog. Even under low-stringency conditions, only bands derived from Dim/dwf1 itself hybridized with the Dim/dwf1 cDNA probe. In addition, no homolog of Dim/dwf1 in the rice genome was identified in a tBLAST search for proteins with similarities to Arabidopsis DIM/DWF1 and rice Dim/dwf1 (data not shown).
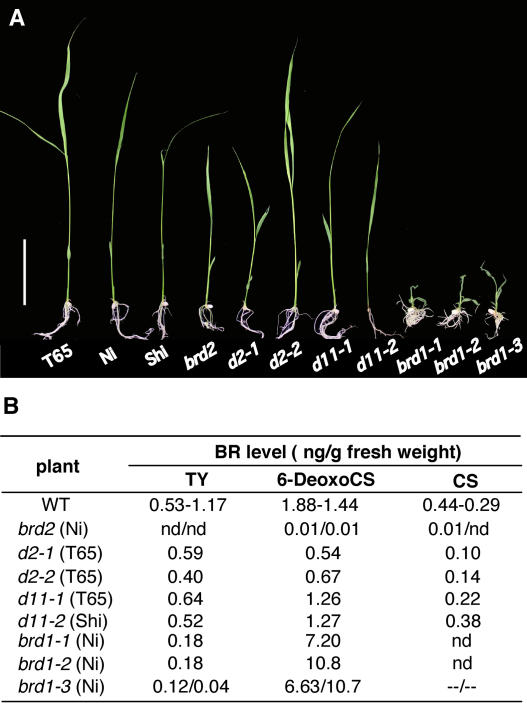
To reconcile the inconsistency between the phenotype and the level of the active BR, CS, in brd2, we also compared the levels of BR intermediates and the phenotypic severity of the rice BR-deficient mutants previously isolated (Figure 5). Mutants showing a mild phenotype, such as d2-1, d2-2, d11-1, and d11-2, contained some CS (0.10 to 0.38 ng/g fresh weight [FW]), 6-deoxocastasterone (6-DeoxoCS) (0.54 to 1.27 ng/g FW), and typhasterol (TY) (0.40 to 0.64 ng/g FW), which corresponded from a quarter to a similar level as in the wild type. By contrast, mutants showing severe phenotype, such as brd1-1, brd1-2, and brd1-3, contained undetectable levels of CS and 0.04 to 0.18 ng/g FW of TY, whereas the level of 6-DeoxoCS was much higher than in the wild type because these mutants are defective in BR C6-oxidase. When we compared brd2 with these BR-deficient mutants, the dwarfism of brd2 was most similar to d2-1 and d11-2, whereas the amounts of three BRs were 0.01 or undetectable (Figure 5).
Figure 5.
Comparison of Dwarfism and Level of BRs between brd2 and Other Rice BR-Deficient Mutants.
(A) Two-week-old seedlings are shown. The mutants were germinated and grown in MS agar. The background of d2-1, d2-2, and d11-1 is T65 (Taichung 65); brd2, brd1-1, brd1-2, and brd1-3 are Ni (Nipponbare) background; and that of d11-2 is Shi (Shiokari). Bar = 5 cm.
(B) Comparison of the level of TY, 6-DeoxoCS, and CS in the BR-deficient mutants. The background plant of each mutant is written in parentheses. The level of BRs in d2, d11, brd1-1, and brd1-2 are adopted from the results previously reported (Hong et al., 2002, 2003; Tanabe et al., 2005). CS level in brd1-3 was not determined because of contamination with unknown compound(s).
An Alternative BR Biosynthetic Pathway Produces an Uncommon Bioactive BR in Rice
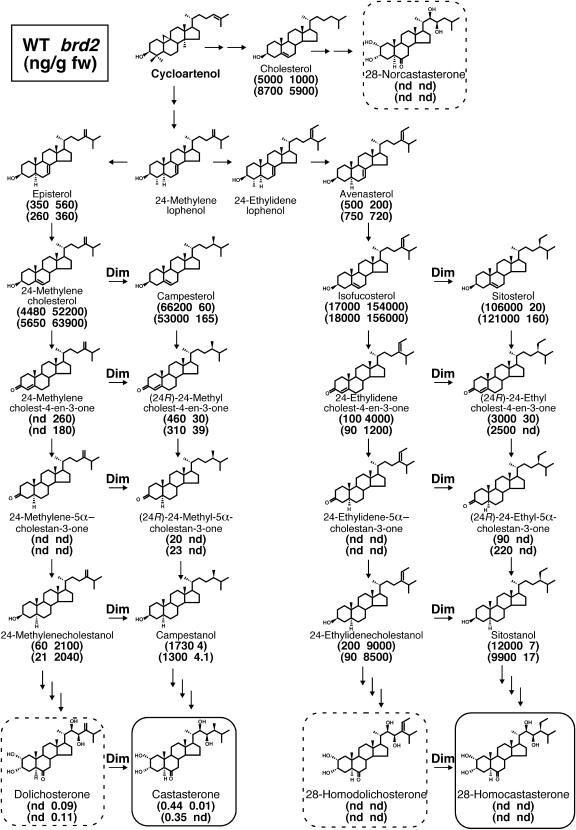
The trace levels of CS in the brd2 mutant, which shows a moderately abnormal phenotype, suggest that an alternative BR biosynthetic pathway may function in the mutant and may produce one or more alternative bioactive BRs that attenuate the severity of the phenotype. Figure 6 shows the proposed normal and alternate BR biosynthesis pathways. The bioactive CS is synthesized via a Dim/dwf1-requiring pathway with the conversion of 24-MC to CR, as mentioned above. Measurements of the levels of all BR-related compounds in these pathways showed that Dim/dwf1 was also necessary for the isomerization and reduction of the Δ24(28) bond of isofucosterol to produce sitosterol. Three alternative pathways are expected to be activated when Δ24(28) isomerization and reduction are defective: the 28-nor–type pathway with 28-norcastasterone as the end product, the 24-methylene–type pathway with DS as the end product, and the 24-ethylidene–type pathway with 28-homodolichosterone as the end product (Figure 6). The first possibility, the 28-nor–type pathway, is less likely because in brd2, the level of a key precursor, cholesterol, was less than the level in the wild type, and the end product of this pathway, 28-norcastasterone, was undetectable. The 24-ethylidene–type pathway, the last possibility, was found to be activated in brd2, and the levels of intermediates in this pathway, such as isofucosterol, 24-ethylidenecholest-4-en-3-one, and 24-ethylidenecholestanol, were greatly increased, but the expected end product, 28-homodolichosterone, could not be detected. Thus, it is difficult to conclude that the 24-ethylidene–type pathway contributes to the production of an alternative bioactive BR in the mutant.
Figure 6.
The Sterol Biosynthetic Pathway and Sterol Contents in the Wild Type and brd2.
Proposed biosynthetic pathways of the wild type (left) and brd2 (right) are shown. The end products of the Dim/dwf1-requiring pathways are enclosed in boxes with solid lines, and those of the Dim/dwf1-free pathway are enclosed in boxes with dashed lines. Sterol levels were measured by two independent experiments. The level of DS was not corrected by an internal standard. The number of arrows does not represent the number of chemical reaction steps. nd, not detected.
The most likely pathway for the production of an alternative bioactive BR is the 24-methylene–type pathway. The levels of intermediates in this pathway were strongly increased in brd2, and the end product of this pathway, DS, which was previously reported to be present in rice (Abe et al., 1984), also accumulated in the mutant but was undetectable in the wild type. Because we did not have an internal standard for DS, we could not estimate its intrinsic level. The endogenous amount of DS should be higher than the raw value, which is presented as the amount of DS in Figure 6. If the recovery rate of DS is the same as that of BL (55%), the expected level of DS in brd2 is estimated to be ∼0.20 ng/g FW, which is approximately half of the CS (∼0.40 ng/g FW) found in the wild type.
DS Has Biological Activity Similar to CS in Rice
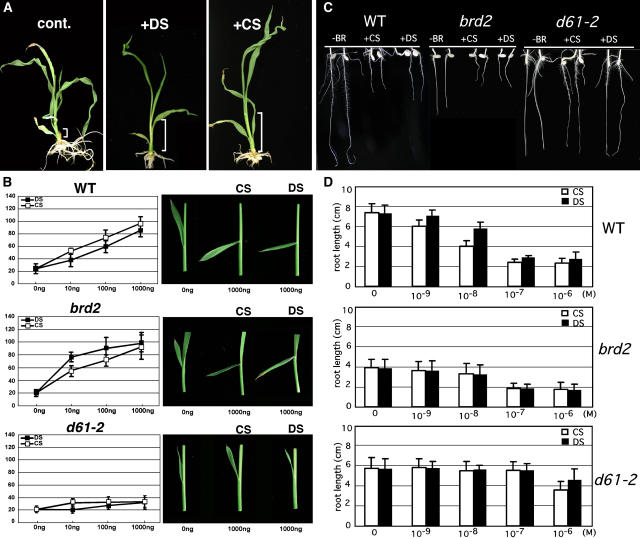
To examine the possibility that the bioactivity of DS compensates for decreased levels of CS in the brd2 mutant, DS feeding experiments were performed using wild-type and BR-related mutants. We first compared the effects of DS and CS on the growth of brd1-1, a knockout mutant of BR C-6 oxidase. CS was found to promote the elongation of leaf sheaths and to rescue the abnormal morphology of newly expanded leaves after a 2-week exposure. Mutant plants treated with DS also grew elongated leaf sheaths, though the effect of DS was somewhat weaker than that of CS. Similarly, the morphology of newly expanded leaves was rescued by treatment with DS (Figure 7A).
Figure 7.
BR Biological Activity of DS.
(A) Phenotypic rescue of the abnormal morphology of brd1-1 by treatment with DS and CS. The malformed phenotype and shortened leaf sheaths of a brd1-1 plant grown for 4 weeks were not rescued by the mock treatment (cont.), whereas 1 μM DS or CS restored the sheath elongation and morphology of newly expanded leaves. White brackets indicate the relative length of the uppermost leaf sheaths.
(B) Lamina joint bending test. DS increased the lamina joint bending of the wild type and brd2 in a dose-responsive manner but not of the BR-perception mutant d61-2. CS treatment had an effect similar to DS treatment, although the efficiency of CS was slightly higher than that of DS in the wild type and lower in brd2. Shown at right are typical lamina joint bending results after treatment with 1 μg CS or DS.
(C) Inhibition of root elongation by treatment with DS or CS. Plants were germinated on agar lacking (−BR) or containing 10−7 M CS or DS. Seedlings were examined 4 d after germination.
(D) Effect of CS and DS on root elongation in wild-type, brd2, and d61-2 seedlings. The plants were germinated as in (A) with the indicated concentrations of CS or DS. The data presented are the means of results from six plants. Error bars = sd.
Lamina joint bending tests were also performed using wild-type, brd2, and d61-2 leaves (Figure 7B). This assay exploits the high sensitivity of the degree of bending between the rice leaf sheath and blade to exogenously applied bioactive BRs. In the wild type, both CS and DS increased lamina joint bending in a dose-dependent manner, with CS being slightly more active than DS. DS was more effective than CS at increasing the bending of the lamina joints in the brd2 mutant. In d61-2, a loss-of-function mutant of the BRI1 receptor kinase, neither CS nor DS increased lamina joint bending, indicating that DS and CS are similarly perceived by the BRI1 receptor.
The effects of CS and DS on root elongation in wild-type, brd2, and d61-2 seedlings were also examined (Figures 7C and 7D). Treatment of wild-type plants with 10−7 M CS or DS severely inhibited root elongation, although the inhibitory effect of DS was slightly weaker than that of CS (Figure 7C). Both compounds also inhibited root elongation in brd2. By contrast, the elongation of d61-2 roots was not inhibited by CS or DS. The inhibition of wild-type root elongation by CS and DS was dose dependent from 10−9 to 10−7 M, and the inhibitory activity of CS was always higher than DS at ≥10−9 M. In brd2, CS and DS also inhibited root elongation in a dose-dependent manner from 10−9 to 10−6 M, but the inhibitory effects were similar at the same concentrations. However, neither CS nor DS inhibited root elongation in the BR-insensitive mutant at concentrations of 10−7 M or lower, whereas both compounds slightly inhibited root elongation at 10−6 M, with CS having a slightly stronger effect than DS. These results support the observation in the lamina joint bending test that DS is a bioactive BR with slightly less activity than CS in the wild type, but in brd2, DS is as bioactive as CS.
DISCUSSION
This study demonstrates that a knockout mutation, brd2, of the rice gene encoding a protein similar to the Arabidopsis DIM/DWF1 causes a semidwarf phenotype in the vegetative stage, with severe defects in internode elongation and panicle and seed development. Quantitative analysis of the endogenous sterol levels showed that brd2 is defective in the conversion of 24-MC to CR, as are the Arabidopsis dim/dwf1 and pea lkb mutants (Nomura et al., 1999; Schultz et al., 2001). The weak phenotype of brd2 during vegetative growth cannot be explained by the residual levels of CS but is probably due to DS produced in the absence of adequate BRs from other pathways. DS has BR activity in rice plants, such as promoting the angle of bending of the lamina joint and inhibiting root elongation.
brd2 Exhibits Features Not Observed in Previously Isolated Rice BR-Related Mutants
Rice BR-related mutants can be distinguished from other rice dwarf mutants by their distinctive internode elongation features (Figure 1C). The group of BR-related mutants that includes the mutants d2, d11, and d61-1 had weak phenotypes and a specific reduction in the second internode. Mutants brd2 and d61-2, with intermediate phenotypes, had a specific reduction from the second to the fourth internodes. Mutants with severe phenotypes, brd1-1 and brd1-2, only elongated at the neck internode. The overall severity of rice BR-related mutants can be represented by these unique internode elongation patterns. For example, the mutants of the d2, d11, and d61-1 group show mild dwarfism of leaves in the vegetative stage and slightly shortened yet fertile grains. By contrast, the mutants of the brd1-1 and brd1-2 group form severely stunted and malformed leaves at the vegetative stage and very small numbers of flowers, which are malformed and infertile. However, the overall phenotype of brd2 does not match either group, although this mutant has a specific reduction in the second to fourth internodes. For example, brd2 has mildly dwarfed leaves but no other abnormal morphologies. The phenotype is similar to that of the d2, d11, and d61-1 group, all of which form erect leaves with slightly stunted sheaths. In contrast with its mild phenotype in the early seedling stage, brd2 is much more severely affected at the developing flower stage than the d61-2 group, approaching that of brd1-1 and brd1-2. The fertility of brd2 is low, as it is difficult to obtain seeds from plants homozygous for brd2, but d61-2 forms fertile flowers and develops viable seeds. Therefore, the phenotype of brd2 is intermediate between mild and severe as growth progresses, a feature that is unique to brd2. It is possible that the brd2 phenotype is caused by an unusual mechanism related to BR biosynthesis.
The brd2 Mutation Is a Knockout of Rice Dim/dwf1
We concluded that the brd2 mutation is caused by a knockout of Dim/dwf1 for several reasons. First, Dim/dwf1 in brd2 contains a single base deletion in exon 2, which leads to a premature stop codon approximately two-thirds of the way through the coding region, causing the production of a nonfunctional protein (Figure 2B). Second, the Dim/dwf1 transcript is not detected in the mutant but is in the wild type (Figure 2D). Third, transformation with wild-type Dim/dwf1 rescues the mutant phenotype of brd2 (Figure 2E). Fourth, the phenotype of plants transformed with the antisense cDNA mimics the abnormal phenotype of brd2 (Figure 3). Finally, in the mutant, the level of 24-MC is greater than in the wild type, whereas the level of CR is dramatically decreased (Figure 4), as in the Arabidopsis dim mutant and the pea lkb mutant.
Klahre et al. (1998) studied the function of Arabidopsis DIM/DWF1 by analyzing the dim mutant, revealing that dim accumulates 24-MC and shows a deficiency in CR. Using deuterium-labeled sterols, the authors also showed that DIM/DWF1 is involved in both the isomerization and reduction of the Δ24(28) bond. A similar accumulation of 24-MC and deficiency in CR was observed in the Arabidopsis dwf1 mutant, which is allelic to dim (Choe et al., 1999a), and also in the pea lkb mutant (Nomura et al., 1999). Taking all of these observations together, although there is no direct biochemical evidence that the DIM/DWF1 protein catalyzes the isomerization and reduction of the Δ24(28) bond, it is possible that the Arabidopsis and pea DIM/DWF1 proteins catalyze the conversion of 24-MC to CR in the synthesis of bioactive BRs in these plants. By analogy, rice Dim/dwf1 should also be involved in the isomerization and reduction of the Δ24(28) bond to convert 24-MC to CR in rice (Figure 4).
DS Functions as a Bioactive BR in Rice
There are three possible hypothetical pathways for sterol metabolism in brd2, in which the isomerization and reduction of the Δ24(28) bond are impaired: the 28-nor–type, the 24-methylene–type, and the 24-ethylidene–type pathways (Figure 6). Nomura et al. (1999) previously discussed the possibility that plants defective in DIM/DWF1 may not show a clear dwarf phenotype because they can synthesize C27-type BRs, probably from cholesterol, which would compensate for decreases in CR-derived BRs, such as BL and CS. This postulated pathway, derived from cholesterol, is the same as the first hypothesized pathway but is not functional in the brd2 mutant. Actually, the level of cholesterol is decreased, not increased, in the mutant. In contrast with the 28-nor–type pathway, the 24-methylene– and 24-ethylidene–type pathways are activated in the mutant, and the levels of 24-methylene–type and 24-ethylidene–type sterols are greatly increased (Figure 6). Similarly, the Arabidopsis dim/dwf1 and pea lkb mutants accumulate relatively large amounts of 24-MC and isofucosterol (Klahre et al., 1998; Nomura et al., 1999), indicating that sterol metabolism of the 24-methylene and 24-ethylidene types is activated in various plant species that are defective in isomerization and reduction of the Δ24(28) bond. Klahre et al. (1998) proposed the activation of a 24-methylene–type pathway, which converts 24-MC to 24-methylenecholestanol, in the Arabidopsis dim mutant by analogy with the pathway from CR to campestanol. Evidence in brd2 suggests that the rice BR-metabolizing enzymes also catalyze the steps from 24-MC to DS. By analogy, it is possible that the enzymes in other plants also catalyze the steps from 24-MC to DS to accumulate DS in dim/dwf1 mutants of other plant species. Further analysis of the sterol content in dim/dwf1 mutants should help to elucidate this hypothesis.
The high levels of sterols, such as 24-methylenecholestanol and 24-ethylidenecholestanol, in the brd2 mutant suggest that BRs containing 24-methylene and 24-ethylidene groups also accumulate in the mutant. This was found to be true in the case of the 24-methylene–type BR, and the level of DS was increased in the mutant, whereas 28-homodolichosterone, the end product of the 24-ethylidene BR pathway, was not detected. The lack of production of 28-homodolichosterone in the mutant may be caused by a low or complete lack of affinity of BR-metabolizing enzymes for 24-ethylidene–type BRs because no 28-homocastasterone was detected in wild-type plants, even though high levels of its precursor, sitostanol, were detected (Figure 6).
The biological activity of DS in rice was confirmed by feeding experiments using three different BR-responding phenomena in rice: the rescue of leaf sheath elongation in a BR-deficient mutant, an increase in the lamina bending angle, and the inhibition of root elongation (Figure 7). In each of these experiments, the wild type was more sensitive to CS than to DS, indicating that CS has a higher specific BR activity than DS. DS and CS had approximately equal activity in the root inhibition assays in both the wild type and the brd2 mutant, but brd2 was more sensitive to DS than to CS in lamina bending assays. These results suggest a difference in tissue-specific BR sensing.
It is noteworthy that in d61-2, which contains a lesion in rice Bri1 kinase, BR-responsive events are not enhanced by treatment with either DS or CS. This observation demonstrates that Bri1 kinase is essential for perception of the signals triggered by DS and CS; consequently, the signaling pathway triggered by DS should be the same as that triggered by CS after perception by the Bri1 kinase. At present, the reason for the higher sensitivity of the brd2 mutant to DS than to CS in the lamina bending assay is unclear. Because lamina joint bending is a unique and hypersensitive phenomenon in BR-triggering events, higher sensitivity to DS does not directly suggest that the brd2 mutant always has higher sensitivity to DS in other tissues or organs. For example, the root inhibition assay showed that there is no difference in the sensitivity between DS and CS in brd2 (Figure 7C). This also suggests the possibility that lamina joint bending of rice may be regulated by a unique mechanism triggered by BR. In fact, its bending is regulated not only by BR but also by auxin in a synergistic manner (Takeno and Pharis, 1982; Cao and Chen, 1995).
METHODS
Plant Materials and Growth Conditions
The brd2 mutant was obtained from rice (Oryza sativa) plants regenerated from a suspension cell culture derived from a japonica strain, Nipponbare. The plants were grown to maturity in the research field, and transgenic plants were grown in a greenhouse under 16 h of illumination at 28°C. For germination, seeds were surface sterilized as described previously (Hong et al., 2003) and plated on MS medium containing 0.8% agar. The plates were then incubated at 30°C under continuous light.
Isolation, Sequencing, and Mapping of the brd2 Gene
The Dim/dwf1 gene was identified in a BLAST search of all available rice genomic databases. The corresponding cDNA sequence was entered as CA757254 and AU030326, which contain the 5′- and 3′-flanking regions, respectively. To identify the site of the mutation in the brd2 gene, we amplified Dim/dwf1 by PCR and sequenced the amplified DNA fragment with appropriate primers and without cloning. The full-length cDNA clone was produced by RT-PCR using 1 μg of total RNA and the Omniscript RT kit (Qiagen, Hilden, Germany). The isolated cDNA was sequenced to confirm that no nucleotide substitutions occurred during the PCR. A clone with the correct sequence was used to prepare probes and the sense and antisense constructs. To map the brd2 mutant, F2 populations derived from a cross between a plant heterozygous for brd2 and an indica strain, kasalath, were used. Thirty plants homozygous for brd2 were identified using molecular markers.
RNA Isolation and RNA Gel Blot Analysis
Total RNA was isolated using the RNeasy plant mini kit (Qiagen). After incubation at 65°C for 5 min, the total RNA was subjected to electrophoresis on a 1% denaturing gel and then transferred to a Hybond N+ membrane for RNA gel blot analysis. The probe used was a 3′-terminal cDNA fragment unless specified otherwise in the figure legends, and the subsequent hybridization and washing were performed using the conditions described by Ueguchi-Tanaka et al. (2000).
Complementation and Antisense Analyses
To isolate a genomic Dim/dwf1 clone, a BAC library was screened using PCR. The XBH43-2B02 clone was identified as containing the entire Dim/dwf1 gene. An 8.8-kb EcoRI fragment encompassing the full-length Dim/dwf1 sequence and the promoter domain were fused in the SmaI site of the hygromycin-resistant binary vector pBI-Hm12, which was kindly provided by Hiroyuki Hirano (Tokyo University, Tokyo, Japan). The construct was introduced into brd2 cells by Agrobacterium tumefaciens–mediated transformation as described by Hiei et al. (1994). Control plants were transformed with the empty vector.
To produce antisense and overexpression constructs, the full-length cDNA was fused downstream of the rice Actin promoter in the pAct-Nos/Hm2 vector. The orientation of the cDNA clone was determined by restriction enzyme digestion.
Quantification of Sterols and BRs
Shoots of wild-type and mutant plants were harvested after growth in a greenhouse for 2 months and lyophilized immediately at −80°C. Extraction and purification for BRs and sterols were performed according to the method described by Fujioka et al. (2002) and He et al. (2003). For BR analysis, plants (20 g FW equivalent) were extracted. [2H6]Brassinolide, [2H6]castasterone, [2H6]typhasterol, [2H6]teasterone, [2H6]cathasterone, [2H6]6-deoxocastasterone, [2H6]6-deoxotyphasterol, [2H6]3-dehydro-6-deoxoteasterone, and [2H6]6-deoxoteasterone (each 1 ng/g FW) were added to the extract as internal standards. For the analysis of 6-deoxocathasterone and sterols, plants (0.5 g FW equivalent) were used. [2H6]episterol (100 ng), [2H7]24-methylenecholesterol (2.5 μg), [2H7]24-methylenecholest-4-en-3-one (500 ng), [2H7]24-methylene-5α-cholestan-3-one (500 ng), [2H7]24-methylenecholestanol (500 ng), [2H6]campesterol (10 μg), [2H6](24R)-24-methylcholest-4-en-3-one (500 ng), [2H6](24R)-24-methyl-5α-cholestan-3-one (50 ng), [2H6]campestanol (250 ng), [2H6]6-oxocampestanol (25 ng), [2H3]cholesterol (2.5 μg), and [2H6]6-deoxocathasterone (2.5 ng) were added to the extract as internal standards. Purified fractions were derivatized and analyzed by gas chromatography–mass spectrometry (GC-MS). BRs and sterols were identified by full-scan GC-MS. The endogenous levels of episterol, 24-MC, 24-methylenecholest-4-en-3-one, 24-methylene-5α-cholestan-3-one, 24-methylenecholestanol, CR, (24R)-24-methylcholest-4-en-3-one, (24R)-24-methyl-5α-cholestan-3-one, campestanol, cholesterol, and 6-oxocampestanol were calculated from the peak area ratios of molecular ions of the internal standard and the endogenous sterol. The endogenous level of 6-deoxocathasterone was calculated from the peak area mass-to-charge (m/z) ratios of 193 for the internal standard and m/z 187 for the endogenous one. The endogenous levels of C29 sterols were roughly estimated from the peak area ratios of molecular ions of internal standards of the corresponding C28 sterols and the endogenous ones. For instance, the level of (24R)-24-ethylcholest-4-en-3-one was calculated from the peak area ratios of molecular ions of m/z 404 for [2H6](24R)-24-methylcholest-4-en-3-one (C28 sterol, internal standard) and m/z 412 for (24R)-24-ethylcholest-4-en-3-one.
For purification and semiquantitative analysis of DS, we purified DS according to the method described by He et al. (2003). Using this method, the chromatographic behavior of DS was the same as BL. Purified HPLC fraction (BL fraction: retention time, 8 to 10 min) was subjected to GC-MS analysis after methaneboronation. Based on the full mass spectrum and retention time in GC, DS was definitely identified from the BL fraction of brd2 mutant. Characteristic ions and retention time of DS methaneboronate were as follows: m/z 510 (M+, 11), 327 (20), 153 (64), 124 (76), 82 (100); GC retention time of DS was 10.95 min (CS, 11.00 min, and BL, 11.70 min, under the same GC conditions).
The raw level was calculated from peak area at m/z 510 using a calibration curve of authentic DS. In the first experiment, only the raw level was shown (0.09 ng/g FW). In the second experiment, the raw level was 0.11 ng/g FW. Because DS was detected in the BL fraction, its endogenous level was corrected using the recovery rate of [2H6]brassinolide (55%). The endogenous level was estimated to be 0.20 ng/g FW.
Lamina Joint Inclination Assay
DS and CS were dissolved in ethanol at final concentrations of 10 ng, 100 ng, and 1000 ng/μL. The seeds were germinated, and the seedlings used in this assay were selected as described by Fujioka et al. (1998). Six plants were used for each treatment.
Sequence data from this article may be found in the EMBL/GenBank data libraries under the following accession numbers: rice Dim/dwf1 (AAM01136), Arabidopsis DIM/DWF1 (Q39085), and pea LKB (AAK15493).
Acknowledgments
We thank Masayo Sekimoto and Makoto Kobayashi (both of RIKEN) for their technical assistance. Youichi Morinaka, Ikuko Aichi, and Hiroko Ohmiya (all of Nagoya University) helped with some preliminary preparation. This work was supported in part by a Grant-in-Aid from the Program for the Promotion of Basic Research Activities for Innovative Biosciences (M.M., M.U.-T., and H.K.) and a Grant-in-Aid from the Center of Excellence (M.M. and M.A.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Makoto Matsuoka (makoto@nuagr1.agr.nagoya-u.ac.jp).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.030973.
References
- Abe, H., Nakamura, K., Morishita, T., Uchiyama, M., Takatsuto, S., and Ikekawa, N. (1984). Endogenous brassinosteroids of the rice plant: Castasterone and dolichosterone. Agric. Biol. Chem. 48 1103–1104. [Google Scholar]
- Bajguz, A., and Tretyn, A. (2003). The chemical characteristic and distribution of brassinosteroids in plants. Phytochemistry 62 1027–1046. [DOI] [PubMed] [Google Scholar]
- Bishop, G.J. (2001). Plants steroid hormone, brassinosteroids: Current highlights of molecular aspects on their synthesis/metabolism, transport, perception and response. Plant Cell Physiol. 42 114–120. [DOI] [PubMed] [Google Scholar]
- Bishop, G.J., and Koncz, C. (2002). Brassinosteroids and plant steroid hormone signaling. Plant Cell 14 (suppl.), S97–S110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, H., and Chen, S. (1995). Brassinosteroid-induced rice lamina joint inclination and its relation to indole-3-acetic acid and ethylene. Plant Growth Regul. 16 189–196. [Google Scholar]
- Carland, F.M., Berg, B.L., FitzGerald, J.N., Jinamornphongs, S., Nelson, T., and Keith, B. (1999). Genetic regulation of vascular tissue patterning in Arabidopsis. Plant Cell 14 2045–2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carland, F.M., Fujioka, S., Takatsuto, S., Yoshida, S., and Nelson, T. (2002). The identification of CVP1 reveals a role for sterols in vascular patterning. Plant Cell 14 2045–2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catterou, M., Dubois, F., Schaller, H., Aubanelle, L., Vilcot, B., Sangwan-Norreel, B.S., and Sangwan, R.S. (2001). Brassinosteroids, microtubules and cell elongation in Arabidopsis thaliana. I. Molecular, cellular and physiological characterization of the Arabidopsis bul1 mutant, defective in the Δ7-sterol-C5-desaturation step leading to brassinosteroid biosynthesis. Planta 212 659–672. [DOI] [PubMed] [Google Scholar]
- Choe, S., Dilkes, B.P., Gregory, B.D., Ross, A.S., Yuan, H., Noguchi, T., Fujioka, S., Takatsuto, S., Tanaka, A., Yoshida, S., Tax, F.E., and Feldmann, K.A. (1999. a). The Arabidopsis dwarf1 mutant is defective in the conversion of 24-methylenecholesterol to campesterol in brassinosteroid biosynthesis. Plant Physiol. 119 897–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe, S., Noguchi, T., Fujioka, S., Takatsuto, S., Tissier, C.P., Gregory, B.D., Ross, A.S., Tanaka, A., Yoshida, S., Tax, F.E., and Feldmann, K.A. (1999. b). The Arabidopsis dwf7/ste1 mutant is defective in the Δ7 sterol C-5 desaturation step leading to brassinosteroid biosynthesis. Plant Cell 11 207–221. [PMC free article] [PubMed] [Google Scholar]
- Choe, S., Tanaka, A., Noguchi, T., Fujioka, S., Takatsuto, S., Ross, A.S., Tax, F.E., Yoshida, S., and Feldmann, K.A. (2000). Lesions in the sterol Δ7 reductase gene of Arabidopsis cause dwarfism due to a block in brassinosteroid biosynthesis. Plant J. 21 431–443. [DOI] [PubMed] [Google Scholar]
- Clouse, S.D., and Sasse, J.M. (1998). Brassinosteroids: Essential regulators of plant growth and development. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49 427–452. [DOI] [PubMed] [Google Scholar]
- Diener, A.C., Li, H.X., Zhou, W.X., Whoriskey, W.J., Nes, W.D., and Fink, G.R. (2000). STEROL METHYLTRANSFERASE 1 controls the level of cholesterol in plants. Plant Cell 12 853–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka, S., Noguchi, T., Takatsuto, S., and Yoshida, S. (1998). Activity of brassinosteroids in the dwarf rice lamina inclination bioassay. Phytochemistry 49 1841–1848. [Google Scholar]
- Fujioka, S., and Sakurai, A. (1997). Brassinosteroids. Nat. Prod. Rep. 14 1–10. [DOI] [PubMed] [Google Scholar]
- Fujioka, S., Takatsuto, S., and Yoshida, S. (2002). An early C-22 oxidation branch in the brassinosteroid biosynthetic pathway. Plant Physiol. 130 930–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka, S., and Yokota, T. (2003). Biosynthesis and metabolism of brassinosteroids. Annu. Rev. Plant Biol. 54 137–164. [DOI] [PubMed] [Google Scholar]
- Grove, M.D., Spencer, G.F., Rohwedder, W.K., Mandava, N.B., Worley, J.F., Warthen, J.D., Steffens, G.L., Flippen-Anderson, J.L., and Cook, J.C. (1979). Brassinolide, a plant growth-promoting steroid isolated from Brassica napus pollen. Nature 281 216–217.16066206 [Google Scholar]
- He, J.X., Fujioka, S., Li, T.C., Kang, S.G., Seto, H., Takatsuto, S., Yoshida, S., and Jang, J.C. (2003). Sterols regulate development and gene expression in Arabidopsis. Plant Physiol. 131 1258–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiei, Y., Ohta, S., Komari, T., and Kumashiro, T. (1994). Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6 271–282. [DOI] [PubMed] [Google Scholar]
- Hong, Z., et al. (2002). Loss-of-function of a rice brassinosteroid biosynthetic enzyme, C-6 oxidase, prevents the organized arrangement and polar elongation of cells in the leaves and stem. Plant J. 32 495–508. [DOI] [PubMed] [Google Scholar]
- Hong, Z., Ueguchi-Tanaka, M., Umemura, K., Uozu, S., Fujioka, S., Takatsuto, S., Yoshida, S., Ashikari, M., Kitano, H., and Matsuoka, M. (2003). A rice brassinosteroid-deficient mutant, ebisu dwarf (d2), is caused by a loss of function of a new member of cytochrome P450. Plant Cell 15 2900–2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang, J.C., Fujioka, S., Tasaka, M., Seto, H., Takatsuto, S., Ishii, A., Aida, M., Yoshida, S., and Sheen, J. (2000). A critical role of sterols in embryonic patterning and meristem programming revealed by the fackel mutants of Arabidopsis thaliana. Genes Dev. 14 1485–1497. [PMC free article] [PubMed] [Google Scholar]
- Klahre, U., Noguchi, T., Fujioka, S., Takatsuto, S., Yokota, T., Nomura, T., Yoshida, S., and Chua, N.H. (1998). The Arabidopsis DIMINUTO/DWARF1 gene encodes a protein involved in steroid synthesis. Plant Cell 10 1677–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElroy, D., Zhang, W., Cao, J., and Wu, R. (1990). Isolation of an efficient actin promoter for use in rice transformation. Plant Cell 2 163–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura, T., Kitasaka, Y., Takatsuto, S., Reid, J.B., Fukami, M., and Yokota, T. (1999). Brassinosteroid/sterol synthesis and plant growth as affected by Ika and Ikb mutations of pea. Plant Physiol. 119 1517–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrick, K., Mayer, U., Horrichs, A., Kuhnt, C., Bellini, D., Dangl, J., Schmidt, J., and Jurgens, G. (2000). FACKEL is a sterol C-14 reductase required for organized cell division and expansion in Arabidopsis embryogenesis. Genes Dev. 14 1471–1484. [PMC free article] [PubMed] [Google Scholar]
- Schrick, K., Mayer, U., Martin, G., Bellini, C., Kuhnt, C., Schmidt, J., and Jurgens, G. (2002). Interactions between sterol biosynthesis genes in embryonic development of Arabidopsis. Plant J. 31 61–73. [DOI] [PubMed] [Google Scholar]
- Schultz, L., Kerckhoffs, L.H.J., Klahre, U., Yokota, T., and Reid, J.B. (2001). Molecular characterization of the brassinosteroid-deficient lkb mutant in pea. Plant Mol. Biol. 47 491–498. [DOI] [PubMed] [Google Scholar]
- Serrano-Cartagena, J., Robles, P., Ponce, M.R., and Micol, J.L. (1999). Genetic analysis of leaf form mutants from the Arabidopsis Information Service collection. Mol. Gen. Genet. 261 725–739. [DOI] [PubMed] [Google Scholar]
- Souter, M., Topping, J., Pullen, M., Friml, J., Palme, K., Hackett, R., Grierson, D., and Lindsey, K. (2002). hydra mutants of Arabidopsis are defective in sterol profiles and auxin and ethylene signaling. Plant Cell 14 1017–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, T., Gasch, A., Nishizawa, N., and Chua, N.H. (1995). The DIMINUTO gene of Arabidopsis is involved in regulating cell elongation. Genes Dev. 9 97–107. [DOI] [PubMed] [Google Scholar]
- Takeno, K., and Pharis, R.P. (1982). Brassinosteroid-induced bending of the leaf lamina of dwarf rice seedlings: An auxin mediated phenomenon. Plant Cell Physiol. 23 1275–1281. [Google Scholar]
- Tanabe, S., Ashikari, M., Fujioka, S., Takatsuto, S., Yoshida, S., Yano, M., Yoshimura, A., Kitano, H., Matsuoka, M., Fujisawa, Y., Kato, H., and Iwasaki, Y. (2005). A novel cytochrome P450 is implicated in brassinosteroid biosynthesis via the characterization of a rice dwarf mutant, dwarf11, with reduced seed length. Plant Cell 17 776–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao, Y., Zheng, J., Xu, Z., Zhang, X., Zhang, K., and Wang, G. (2004). Functional analysis of ZmDWF1, a maize homolog of the Arabidopsis brassinosteroid biosynthetic DWF1/DIM gene. Plant Sci. 167 743–751. [Google Scholar]
- Ueguchi-Tanaka, M., Fujisawa, Y., Kobayashi, M., Ashikari, M., Iwasaki, Y., Kitano, H., and Matsuoka, M. (2000). Rice dwarf mutant d1, which is defective in the α-subunit of the heterotrimeric G protein, affects gibberellin signal transduction. Proc. Natl. Acad. Sci. USA 97 11638–11643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamuro, C., Ihara, Y., Wu, X., Noguchi, T., Fujioka, S., Takatsuto, S., Ashikari, M., Kitano, H., and Matsuoka, M. (2000). Loss of function of a rice brassinosteroid insensitive1 homolog prevents internode elongation and bending of the lamina joint. Plant Cell 12 1591–1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota, T. (1997). The structure, biosynthesis and function of brassinosteroids. Trends Plant Sci. 2 137–143. [Google Scholar]