Abstract
Pluripotent stem cells are localized in specialized microenvironments, called stem cell niches, where signals from surrounding cells maintain their undifferentiated status. In the Arabidopsis thaliana shoot meristem, the homeobox gene WUSCHEL (WUS) is expressed in the organizing center underneath the stem cells and integrates regulatory information from several pathways to define the boundaries of the stem cell niche. To investigate how these boundaries are precisely maintained within the proliferating cellular context of the shoot meristem, we analyzed the transcriptional control of the WUS gene. Our results show that the WUS promoter contains distinct regulatory regions that control tissue specificity and levels of transcription in a combinatorial manner. However, a 57-bp regulatory region is all that is required to control the boundaries of WUS transcription in the shoot meristem stem cell niche, and this activity can be further assigned to two adjacent short sequence motifs within this region. Our results indicate that the diverse regulatory pathways that control the stem cells in the shoot meristem converge at these two short sequence elements of the WUS promoter, suggesting that the integration of regulatory signals takes place at the level of a central transactivating complex.
INTRODUCTION
Plants produce most of their organs postembryonically from stem cells at the shoot and root apices. Similar to animal stem cells, plant stem cells are located in niches where neighboring cells provide signals to maintain them in an undifferentiated state (Spradling et al., 2001; Weigel and Jürgens, 2002; Laux, 2003). Cells that leave the stem cell niche initiate differentiation and give rise to lateral organs such as leaves and flowers. In general, the decision between stem cell fate and differentiation can be regulated in two ways: either in a lineage mechanism in which each stem cell divides asymmetrically to give rise to one stem cell and one cell prone to undergo differentiation, or in a population-based mechanism by which the outcome of an individual division cannot be predicted; rather, the stem cell population as a whole is kept constant by external cues (Spradling et al., 2001). The shoot meristem stem cell niche operates in a population mode and thus requires precise spatial regulation of stem cell–inducing signals to maintain the correct position and number of stem cells (Bäurle and Laux, 2003). Because all cells in the shoot meristem, including the signaling niche cells, continuously divide, a longstanding question of plant development is how the position and the boundaries of the stem cell niche are stably maintained, or, as Newman (1965) put it several decades ago, how the pattern (of the shoot apex) can be maintained while the matter (the constituent cells) constantly changes.
The regulation of transcriptional domains of regulatory genes plays a pivotal role for many developmental processes; thus, the analysis of transcriptional control is crucial to gain insight into the mechanisms that govern spatial and temporal patterning in development (Watanabe and Okada, 2003). In Arabidopsis thaliana shoot and floral meristems, transcriptional regulation of the WUSCHEL (WUS) homeobox gene controls the stem cell pool. WUS is expressed in a small group of cells underneath the stem cells termed the organizing center and is required to keep the stem cells in an undifferentiated state, indicating that the organizing center cells act as signaling cells of the shoot meristem stem cell niche (Laux et al., 1996; Mayer et al., 1998). Ectopic expression of WUS inhibits differentiation and can result in the formation of ectopic stem cells or even somatic embryos, indicating the necessity to locally restrict WUS activity (Schoof et al., 2000; Brand et al., 2002; Zuo et al., 2002; Gallois et al., 2004). Recent findings indicate that the regulation of WUS transcription is a central checkpoint in stem cell control, integrating information from several regulatory pathways. First, the size of the stem cell population is controlled through the size of the WUS expression domain. This is achieved by a dynamic feedback loop, with WUS indirectly activating the expression of the signaling peptide CLAVATA3 (CLV3) in the stem cells and CLV3 repressing WUS transcription through the CLV1 receptor kinase signaling pathway (Brand et al., 2000; Schoof et al., 2000; Rojo et al., 2002; Lenhard and Laux, 2003). Second, temporal control of stem cell activity in the determinate floral meristem is achieved by the repression of WUS transcription through AGAMOUS (AG) activity (Lenhard et al., 2001; Lohmann et al., 2001). Furthermore, based on changes of its expression domain in shoot meristem mutants, several other regulatory pathways have been implicated in the control of WUS gene expression (Laufs et al., 1998; Kaya et al., 2001; Stuurman et al., 2002; Bertrand et al., 2003; Ueda et al., 2004; Zhao et al., 2004; Carles et al., 2005; Wu et al., 2005). In addition to the inhibitory CLV3 signal, a positive signal originating from the stem cells has been postulated that would activate WUS transcription and anchor the organizing center to the shoot tip (Schoof et al., 2000). However, no direct regulator of WUS transcription has been identified to date.
Therefore, understanding how the boundaries of the WUS transcriptional domain are regulated is central to gaining insight into how the position and size of the stem cell niche are maintained at the tip of the shoot meristem. Here, we have identified two short sequence motifs within the WUS promoter that act as central integrating elements in stem cell control.
RESULTS
Regulatory Elements That Control WUS Transcription in the Stem Cell Niche
To delimit the control region of the WUS gene, we analyzed the expression pattern of β-glucuronidase (GUS) driven by WUS promoter fragments. The putative transcription start site of the WUS gene was determined by two independent rapid amplification of cDNA ends (RACE) experiments as being 126 nucleotides upstream of the ATG start codon (Figure 1A). As a starting point for promoter analysis, we chose an 8.7-kb WUS genomic fragment, HindBst (−5685/2970; referring to the putative transcriptional start site), in which the WUS coding region was replaced with the coding region of the GUS gene (Figure 1B). This reporter comprised 5.7-kb upstream and 1.3-kb downstream sequences and mimicked the described mRNA expression pattern of WUS in meristems (Mayer et al., 1998), showing strong GUS expression in young floral meristems and weaker GUS expression in the organizing center of the vegetative and inflorescence meristems (Figure 2A). The construct also recapitulated ovule-specific mRNA expression (Figure 2A), in which WUS is expressed in the apical nucellus during early developmental stages and is required for ovule patterning (Gross-Hardt et al., 2002; Sieber et al., 2004). In addition, GUS staining in stamens was detected in a pattern similar to the mRNA expression (Wellmer et al., 2004) but was very weak and was not analyzed further. We focused our analysis on inflorescence and floral meristems but obtained corresponding results for the vegetative meristem of the seedling where analyzed (Figures 2 and 3E).
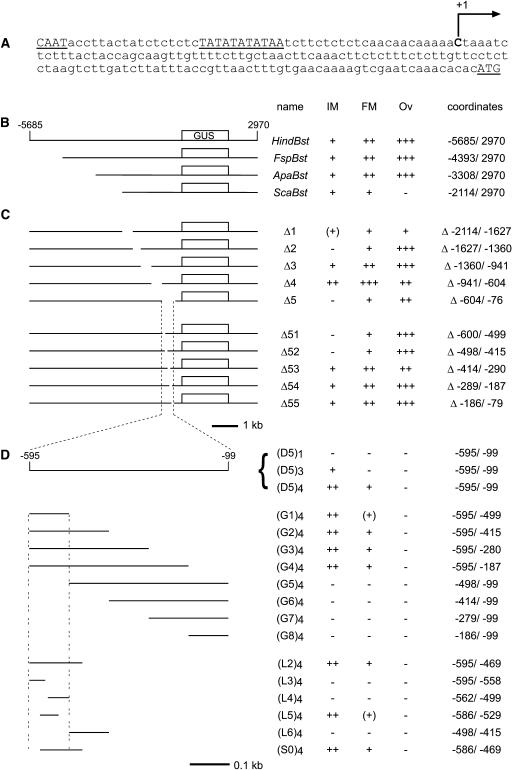
Figure 1.
Diagram of the Reporter Constructs Used in the WUS Promoter Analysis.
(A) The putative WUS transcription start site (+1) was determined by RACE PCR. Putative CAAT and TATA boxes and the start codon are underlined.
(B) to (D) For each construct, a scheme is shown at left. At right, the name, the relative staining intensities in the inflorescence meristem (IM), the floral meristem (FM), and the ovule (Ov) [−, none; (+), very faint; +, weak; ++, moderate; +++, strong], and the exact coordinates of the WUS promoter fragments or deletions (Δ) are given. The WUS coding region was replaced by the GUS coding sequence (box).
(B) Diagram of the truncation constructs analyzed.
(C) Diagram of the internal deletion constructs analyzed.
(D) Diagram of the constructs used during the functional definition of cis-regulatory sequences. To generate the reporter constructs, the monomers were multimerized as indicated and fused to -60 CaMV:GUS. D (deletion), G (gain of function), L (little deletion), and S (linker scanning) denote series of constructs.
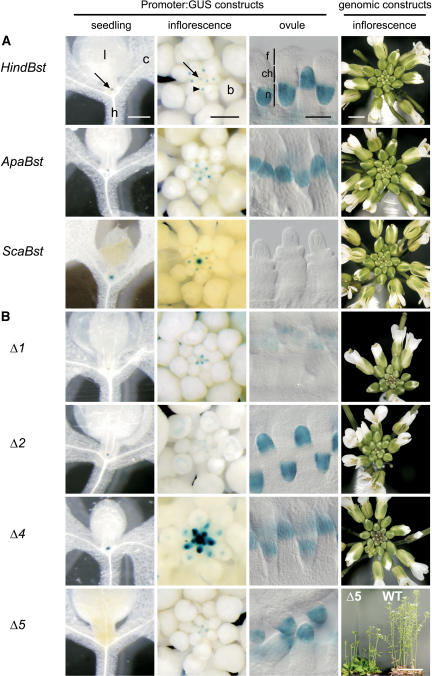
Figure 2.
Expression Patterns of WUS:GUS Constructs and Complementation of the Inflorescence Phenotype with Corresponding Genomic Fragments.
Each row shows the expression pattern of the indicated WUS promoter fragment in (from left to right) seedlings, inflorescences, and ovules and the complementation of a homozygous wus-1 mutant plant with the corresponding genomic fragment (right column). Seedlings and inflorescences were stained with GUS with 2 mM Fe-cyanide for 1 d except ScaBst (2 mM, 2 d) and Δ4 (5 mM, 1 d). Ovules were stained with GUS with 5 mM Fe-cyanide for 1 d.
(A) Truncation constructs.
(B) Deletion constructs.
b, floral bud; c, cotyledon; ch, chalaza; f, funiculus; h, hypocotyl; l, leaf; n, nucellus. Arrows indicate the shoot meristem, and the arrowhead indicates the floral meristem. Bars = 0.5 mm (seedling, inflorescence), 30 μm (ovule), 2 mm (genomic constructs, except Δ5), and 5 cm (genomic construct Δ5 and the wild type).
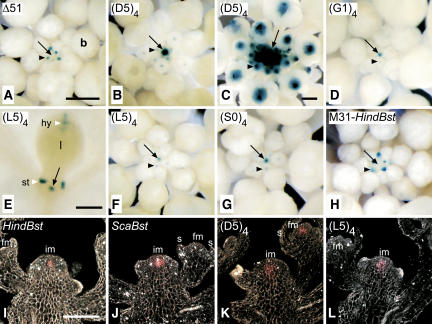
Figure 3.
Expression Patterns of WUS:GUS Reporter Constructs in Inflorescences.
Inflorescences ([A] to [D] and [F] to [L]) or seedling (E) were stained with either 2 mM Fe-cyanide ([A], [B], and [D] to [H]) or 5 mM Fe-cyanide ([C] and [I] to [L]) in the staining buffer for 1 d ([A] and [H] to [J]) or 3 d ([B] to [G], [K], and [L]).
(A) to (H) Whole-mount views with bright-field optics. GUS activity is visualized by blue color.
(A) Δ51.
(B) (D5)4.
(C) (D5)4 clv1-4.
(D) (G1)4.
(E) (L5)4. Staining in hydathodes (hy) and stipules (st) is attributable to background activity of the included minimal promoter.
(F) (L5)4.
(G) (S0)4.
(H) M31-HindBst (−566/−564 mutated; see Figure 4).
(I) to (L) Eight-micrometer sections viewed with dark-field optics. GUS activity is visualized by pink color.
(I) HindBst.
(J) ScaBst.
(K) (D5)4.
(L) (L5)4.
b, floral bud; fm, floral meristem; im, inflorescence meristem; s, sepal. Arrows indicate the shoot meristem, and arrowheads indicate the floral meristem. Bars = 0.5 mm ([A] and [E]), 2 mm (C), and 60 μm (I). Magnification of (B), (D), and (F) to (H) is as in (A); magnification of (J) to (L) is as in (I).
Progressive truncations from the 5′ end revealed that the region upstream of position −3308 was dispensable for promoter activity (Figures 1B and 2A, ApaBst). By contrast, the region between −3308 and −2114 was essential for expression in ovules and for high-level expression in floral meristems (Figures 1B and 2A, ScaBst). In plants carrying the ScaBst (−2114/2970) reporter construct, the spatial GUS expression pattern in inflorescence and floral meristems was unchanged, indicating that the regulatory sequences controlling the boundaries of WUS expression in the stem cell niche are present (Figures 2A, 3I, and 3J). Corresponding WUS genomic versions of these promoter truncations were able to complement the inflorescence and floral meristem defects observed in wus mutants (Figure 2A, Table 1). In accordance with the loss of GUS activity in ovules, seed set in wus mutants complemented with the ScaBst genomic construct was strongly reduced.
Table 1.
Complementation of wus-1 Mutant Flowers with Promoter Truncations and Internal Deletion Constructs
| Whorl |
|||||
|---|---|---|---|---|---|
| Construct | Sepals | Petals | Stamens | Carpels | n |
| HindBst | 4.0 ± 0 | 4.0 ± 0 | 5.3 ± 0.4 | 2.0 ± 0 | 20 |
| FspBst | 4.0 ± 0 | 4.0 ± 0 | 5.6 ± 0.5 | 2.0 ± 0 | 20 |
| ApaBst | 4.0 ± 0 | 4.0 ± 0 | 5.8 ± 0.4 | 2.0 ± 0 | 20 |
| ScaBst | 4.0 ± 0 | 4.0 ± 0 | 5.9 ± 0.3 | 2.0 ± 0 | 20 |
| Δ1 | 4.0 ± 0 | 4.0 ± 0 | 5.7 ± 0.5 | 2.0 ± 0 | 20 |
| Δ2 | 4.0 ± 0 | 4.0 ± 0 | 4.5 ± 1.2 | 1.3 ± 0.8 | 20 |
| Δ3 | 4.0 ± 0 | 4.0 ± 0 | 5.9 ± 0.4 | 2.0 ± 0 | 20 |
| Δ4 | 4.0 ± 0 | 4.0 ± 0 | 5.9 ± 0.2 | 2.0 ± 0 | 18 |
| Δ5 | 4.5 ± 1.0 | 4.4 ± 0.8 | 5.1 ± 1.3 | 0.1 ± 0.3 | 20 |
| wus-1a | 3.7 ± 0.7 | 3.6 ± 0.9 | 0.9 ± 0.5 | 0.0 ± 0 | 53 |
| Ler | 4.0 ± 0 | 4.0 ± 0 | 6.0 ± 0.3 | 2.0 ± 0 | 20 |
The average floral organ number and the sd in n flowers of the line that showed the best complementation are indicated.
Data for wus-1 are taken from Laux et al. (1996).
To further characterize the WUS regulatory regions, we analyzed the effects of internal deletions of 267 to 528 bp in length within the 2.1-kb upstream region (Figure 1C). We performed the experiments in the context of the complete HindBst reporter to minimize positional effects attributable to different integration sites of the transgenes. Deletion Δ1 (Δ−2114/−1627) resulted in qualitatively unaltered but generally weaker GUS expression (Figure 2B), indicating that the deleted sequence harbors general transcriptional enhancer element(s). Deletion Δ3 (Δ−1360/−941) did not alter the GUS expression pattern (Figure 1C), and deletion Δ4 (Δ−941/−604) gave stronger GUS expression in the inflorescence and floral meristems (Figure 2B). However, replacement of the latter region by an unrelated DNA fragment of the same length produced normal GUS expression levels (data not shown), suggesting that the increased expression strength in Δ4 was caused by spacing effects rather than by the excision of a negative regulatory element. Again, the corresponding WUS genomic deletion constructs (Δ1, Δ3, and Δ4) complemented the inflorescence and floral meristem defects of the wus mutant, and seed set was restored to wild-type levels (Figure 2B, Table 1; data not shown).
Deletions Δ2 (Δ−1627/−1360) and Δ5 (Δ−604/−76) completely abolished GUS expression in the inflorescence meristem and reduced expression in floral meristems (Figure 2B), indicating the presence of essential regulatory elements in these regions. However, the genomic fragment carrying the Δ2 deletion complemented the inflorescence and flower phenotype conferred by the wus mutant (Figure 2B, Table 1). This finding suggests that the Δ2 deletion reduced transcription below the detection limit of the GUS assay but still allowed for sufficient transcription of WUS to rescue stem cell maintenance in the shoot meristem and to a reduced extent in the floral meristem. Because ovule-specific expression was not affected in Δ2 (Figures 1C and 2B), the region between −1627 and −1360 presumably contains a meristem-specific enhancer (Figure 4).
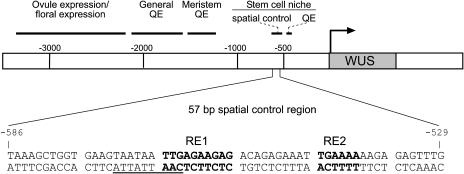
Figure 4.
Regulatory Architecture of the WUS Promoter.
The approximate positions of regulatory domains are indicated. At bottom, nucleotides within the 57-bp spatial control region essential for promoter activity in the stem cell niche of the inflorescence meristem (RE1 and RE2) are indicated in boldface letters; the predicted HD-ZIP binding site is underlined. QE, quantitative element required for enhanced expression levels.
By contrast, the genomic fragment carrying the Δ5 deletion did not rescue the phenotype conferred by the wus mutant (Figure 2B, Table 1), suggesting the presence of essential control elements in the region between −604 and −76. Therefore, we further analyzed this sequence by introducing 100-bp deletions within this region in the context of the HindBst GUS reporter (Figure 1C). Deletions Δ51 (Δ−600/−499) and Δ52 (Δ−499/−415) both abolished GUS expression in the inflorescence meristem and reduced it in floral meristems (Figures 1C and 3A), very similar to what was observed in deletion Δ5. By contrast, deletions Δ53 to Δ55 (covering −414 to −79) did not affect GUS expression patterns (Figure 1C).
Collectively, these results suggest that the regulation of WUS transcription is mediated through several distinct cis-regulatory regions. However, the sequences between −600 and −415 are the only ones that are absolutely necessary for WUS expression in the inflorescence meristem stem cell niche; therefore, we focused our further analysis on this region.
A 57-bp Element Controls WUS Expression Boundaries in Shoot and Floral Meristem Stem Cell Niches
We next asked whether the identified sequences required for the correct expression of WUS in the shoot stem cell niche are also sufficient. For this purpose, we fused tandem repeats of the region deleted in Δ5 (−604/−76), designated D5, to the minimal promoter of the Cauliflower mosaic virus (CaMV) 35S gene (−60 CaMV) followed by the GUS coding sequence, and tested whether these synthetic promoters were able to drive GUS expression in transgenic Arabidopsis plants. We did not detect any GUS expression in plants carrying the reporter with only a single copy of D5 [(D5)1:GUS; Figure 1D]. By contrast, three tandem copies of D5 provided moderate levels of GUS expression in the organizing center of the inflorescence meristem [(D5)3:GUS; Figure 1D], and four copies conferred strong GUS staining in the inflorescence meristem and weaker staining in young floral meristems [(D5)4:GUS; Figures 1D, 3B, and 3K]. Importantly, the spatial expression pattern provided by these multimers precisely recapitulated the one observed with the ScaBst reporter (cf. Figures 3J and 3K) and in WUS RNA in situ hybridizations (Mayer et al., 1998; Schoof et al., 2000), indicating that the D5 promoter fragment contains all of the necessary sequences for correct transcriptional control in the stem cell niche. Similar to the endogenous WUS gene, the (D5)4:GUS reporter displayed strongly enhanced expression in a clv1-4 mutant background (Figure 3C) (Schoof et al., 2000). In addition, expression of the (D5)4:GUS reporter in wild-type floral meristems was terminated approximately when carpel primordia emerged (data not shown), similar to the time when endogenous WUS expression is terminated (Mayer et al., 1998). Together, these findings suggest that the repression of WUS transcription mediated by CLV3 (Brand et al., 2000; Schoof et al., 2000) and AG (Lenhard et al., 2001; Lohmann et al., 2001) acts through the D5 control region. However, we did not find an AG consensus binding site (Shiraishi et al., 1993) anywhere in the WUS promoter, suggesting that AG acts indirectly to repress WUS expression. It is plausible that multimers of these short fragments are required for detectable levels of transcription because of the lack of transcriptional enhancers present in the natural promoter context, such as those identified by the internal deletions in the region −2114 to −1360.
Notably, the portion of GUS-positive plants among all primary transformants was lower with these short synthetic WUS promoter constructs than with full-length or nearly full-length WUS promoter constructs (see Supplemental Table 1 online). This finding might reflect a stronger influence of the transgene's integration site in the genome on the expression levels of short compared with longer constructs.
We subsequently analyzed a series of reporter genes that carried tetrameric tandem repeats of D5 subfragments (Figure 1D) and identified a 57-bp region named L5 (−586/−529), which provided the correct spatial WUS expression pattern in the stem cell niche of the inflorescence meristem (Figure 3L), the vegetative meristem (Figure 3E), and in floral meristems, albeit at a lower level (Figure 3F). Comparison of the expression levels provided by subfragments G1, L2, and L6 (Figures 1D and 3D) revealed that the presence of a 29-bp interval (−498/−469) adjacent to the 57-bp region is required for increased expression but is not sufficient to confer expression on its own (Figures 1D and 4).
Thus, a 57-bp promoter fragment (−586/−529) provides all of the spatial and temporal information necessary for WUS transcription in the stem cell niche of shoot and floral meristems.
Two Distinct Sequence Motifs within the 57-bp Regulatory Region Are Essential for WUS Transcription in the Stem Cell Niche
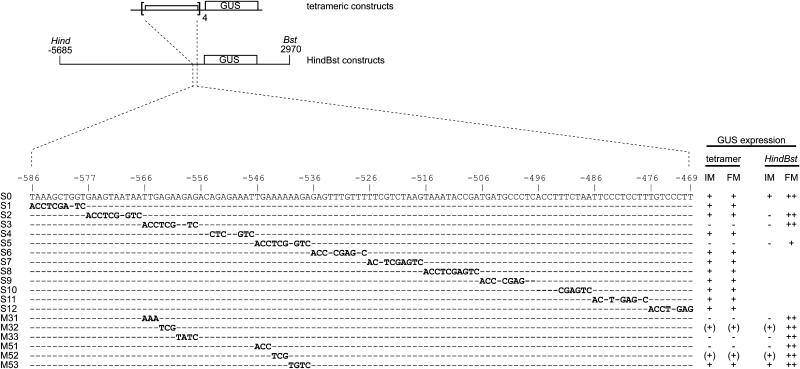
To further define the regulatory sequences present in the 57-bp fragment, we performed linker-scanning mutagenesis using the −586/−469 fragment of the WUS promoter, named S0 (for scanning fragment 0), which contained both the 57-bp fragment and the neighboring putative enhancer element (Figure 1D). The S0 fragment was permutated by substituting 10-bp elements with always the same unrelated 10-bp sequence. The resulting mutated promoter fragments were used to create 12 tetrameric GUS reporter constructs: (S1)4:GUS to (S12)4:GUS (Figure 5). Plants carrying the unmutated (S0)4:GUS reporter or any 1 of 10 of the mutated reporters [(S1)4:GUS, (S2)4:GUS, (S4)4:GUS, (S6)4:GUS to (S12)4:GUS] displayed GUS expression in the inflorescence and floral meristems similar to that of the (D5)4:GUS reporter gene (Figures 3G and 5). By contrast, two mutated constructs, (S3)4:GUS and (S5)4:GUS, in which sequences −566 to −557 and −546 to −537, respectively, had been exchanged, had completely lost promoter activity (Figure 5). By introducing the respective mutations into the HindBst reporter gene (S3-HindBst and S5-HindBst; Figure 5), we confirmed that the two decamers mutated in these constructs are also necessary for GUS expression in the inflorescence meristem in the context of the full-length WUS promoter.
Figure 5.
Linker Scanning Analysis.
Schemes of the constructs analyzed. The 118-bp WUS promoter fragment S0 (−586/−469) was permutated with the decamer sequence ACCTCGAGTC, generating the mutated fragments S1 to S12. The −566/−557 and −546/−537 regions were also scanned with trinucleotide/tetranucleotide exchanges (M31 to M33 and M51 to M53). For the reporter constructs, each mutated fragment was tetramerized and fused to −60 CaMV:GUS. Unaltered nucleotides are indicated with dashes. Relative staining intensities in inflorescence meristems (IM) and floral meristems (FM) are indicated at right for tetrameric (tetramer) and full-length WUS promoter constructs (HindBst). All full-length promoter constructs additionally showed strong staining in ovules unaffected by the indicated mutations.
Next, we replaced trinucleotide and tetranucleotide motifs within the two decamers in the context of the tetrameric (S0)4:GUS gain-of-function construct to further restrict the essential cis-regulatory sequences (Figure 5). Nucleotides −566/−564 (M31; Figure 3H), −560/−557 (M33), and −546/−544 (M51) were absolutely essential for reporter gene activity in the shoot and floral meristems. Mutating the nucleotides −563/−561 (M32) and −543/−541 (M52) reduced reporter expression in the shoot and floral meristems but did not abolish it, indicating that these nucleotides are important but not essential. By contrast, replacing the nucleotides −540/−537 (M53) did not affect reporter activity. When we introduced the same mutations into the 8.7-kb HindBst reporter construct, we obtained analogous results, confirming that the identified nucleotides are also essential for expression in the shoot meristem in the context of the full-length WUS promoter (Figure 5). However, these constructs retained expression in the floral meristems, as expected from the presence of a redundant floral meristem–specific control element between −3308 and −2114 (see above).
Together, these results indicate that two distinct sequence motifs that we named RE1 (for regulatory element 1; −566/−557) and RE2 (−546/−541) regulate the boundaries of the WUS expression domain in the stem cell niche of shoot and floral meristems (Figure 4).
We noticed that the region mutated in S2 (−576/−567) and S3 (−566/−557) contains a sequence motif (TAATAATTG, −572/−564; Figure 4) similar to the consensus binding site for several HD-ZIP proteins (CAATNATTG) (Johannesson et al., 2001). Because only the S3 but not the S2 mutation affected promoter activity of the tetrameric promoter constructs, we introduced the S2 mutation, which covers the major part of this putative binding site, into the full-length WUS promoter. Indeed, no expression in the inflorescence meristem was detected with this S2-HindBst:GUS reporter gene (Figure 5), suggesting an essential function of the S2 region in the context of the full-length promoter.
DISCUSSION
The ability to stably maintain multipotent stem cells is crucial for the postembryonic production of new cells in plants and animals. In the plant shoot meristem, the stem cells are specified by WUS-dependent signals from underlying organizing center cells, and transcriptional control of the WUS gene within the proliferating shoot apex is a key regulatory switch in stem cell regulation. To gain insight into the mechanisms of how the boundaries and the position of the stem cell niche are stably maintained, we identified sequences within the WUS promoter that control the spatial and temporal transcription pattern.
Regulatory Domains of the WUS Promoter in Stem Cell Control
Our results show that the WUS promoter contains distinct regulatory regions that control tissue specificity and levels of transcription in a combinatorial manner (Figure 4). Among them, a 57-bp region ∼550 bp upstream of the putative transcription start provides all information necessary for the correct spatial and temporal transcriptional pattern in the stem cell niches of shoot and floral meristems. Because all other nucleotides within this region were dispensable, two short sequence motifs, RE1 and RE2, mediate this control. In fact, these elements are highly conserved in WUS promoter sequences throughout the Brassicaceae family, supporting a central role in stem cell niche transcription (E. Tucker and T. Laux, unpublished results).
Interestingly, the first three nucleotides (TTG) of RE1 overlap with an HD-ZIP consensus binding site–like motif. HD-ZIP proteins can form heteromeric combinations involved in a variety of developmental processes (Johannesson et al., 2001), raising the possibility that HD-ZIP proteins might be involved in the spatial control of WUS transcription in the stem cell niche. In accordance with this possibility, a novel HD-ZIP–related protein was isolated in further analysis that specifically binds to this consensus sequence in the WUS promoter (I. Bäurle and T. Laux, unpublished results). However, because this consensus-like sequence is only necessary in the context of the full-length promoter but not in multimerized short promoter fragments, we hypothesize that factors binding to it do not mediate the spatial regulation of WUS promoter activity but rather enhance transcription levels, the requirement of which can be bypassed by increased copy numbers of RE1 and RE2.
Notably, the regulation of the stem cell niche in shoot and floral meristems, which are homologous systems and share several regulatory mechanisms (Steeves and Sussex, 1989; Schoof et al., 2000), involves not only common but also meristem-type–specific regulatory regions. For example, the 57-bp regulatory region containing RE1 and RE2 is sufficient for the spatial expression of WUS in both meristems, but its deletion from the full-length promoter abolishes WUS expression only in the shoot meristem, indicating the presence of redundant cis elements that function exclusively in the stem cell niche of the floral meristem. It is conceivable that such differentially used promoter elements might account for differences in growth dynamics, gene expression levels, and temporal control of shoot and floral meristems and have evolved during diversification of their developmental programs.
The expression levels of synthetic tandem repeat promoter constructs containing RE1 and RE2 appear to be highly dependent on the site of integration within the genome, suggesting that their efficacy requires a favorable chromatin state at the integration site. Because this is not the case for constructs with the full-length promoter, some of the identified regulatory regions could act in chromatin organization, such as scaffold attachment (Breyne et al., 1992), nucleosome position and conformation, or recruitment of histone-modifying enzymes (Wagner, 2003) at the WUS locus. In fact, the boundaries of the WUS expression domain are deregulated in a mutant with compromised histone acetyltransferase activity, although it has yet to be determined whether the WUS promoter is directly affected in this case (Bertrand et al., 2003).
Integration of Regulatory Inputs in Stem Cell Control
The spatial and temporal control of the stem cell niche requires the integration of different cues at the level of WUS gene expression. One surprising result of this study is that all of these regulatory cues converge at two adjacent small regulatory sites, RE1 and RE2, of the WUS promoter. How is this achieved? Two mutually nonexclusive mechanisms can be envisioned. First, different combinations of transcription factors mediating independent regulatory inputs could bind to these motifs alternately or in a combinatorial way. Conversely, different cues could modify the activity of a common central transcription complex. The repression of WUS transcription via the stem cell–borne CLV3 signal could be an example of the latter case. In clv3 loss-of-function mutants, the WUS expression domain is increased from the embryo stage on (Brand et al., 2000; Schoof et al., 2000). However, we did not find a promoter mutation that altered WUS expression as expected for a construct lacking CLV3-dependent repression or any duplicated sequence motif that would suggest redundancy of such a putative negative regulatory element. Thus, a plausible mechanism for CLV3 action on WUS transcription could involve the phosphorylation of RE1- or RE2-specific transcription factors by the intracellular receptor kinase signaling pathway activated by CLV3 (Clark, 2001). The knowledge of WUS-regulating cis elements reported here provides the basis for the search for direct upstream regulators that will eventually allow insight into how transcriptional domains are stably maintained within a changing cellular context of the proliferating shoot apex.
METHODS
Plant Material and Plant Transformation
Arabidopsis thaliana growth conditions and the wus-1 allele used for the complementation experiments have been described (Laux et al., 1996). The clv1-4 allele was also described previously (Clark et al., 1993). All plasmids were introduced into Agrobacterium tumefaciens strain GV3101(pMP90) (Koncz and Schell, 1986) by electroporation and transformed into Landsberg erecta (Ler) wild-type plants by the floral dip method (Clough and Bent, 1998).
GUS Staining
GUS staining was performed as described (Schoof et al., 2000). Material was cleared in 70% ethanol before taking photographs using a Leica MZ12 binocular and a Leica DC300 camera (Leica Microsystems, Wetzlar, Germany). For sections, tissue was dehydrated in an ethanol series up to 50%, postfixed in FAA (50% ethanol:5% formaldehyde:10% acetic acid) for 30 min at room temperature, dehydrated completely, and embedded in Paraplast (Sigma-Aldrich, Taufkirchen, Germany).
Cloning Details
Cloning details are available upon request. For simplicity, fragment names derived from restriction sites are abbreviated as follows: HindIII as Hind, Bst1107I as Bst, ScaI as Sca.
Supplementary Material
Acknowledgments
We thank Michael Lenhard and members of our laboratory for helpful comments on the manuscript. We thank Klaus Mayer for performing the RACE experiment. This work was supported by a grant from the Deutsche Forschungsgemeinschaft (SFB 592) to T.L.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Thomas Laux (laux@biologie.uni-freiburg.de).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.032623.
References
- Bäurle, I., and Laux, T. (2003). Apical meristems: The plant's fountain of youth. Bioessays 25 961–970. [DOI] [PubMed] [Google Scholar]
- Bertrand, C., Bergounioux, C., Domenichini, S., Delarue, M., and Zhou, D.X. (2003). Arabidopsis histone acetyltransferase AtGCN5 regulates the floral meristem activity through the WUSCHEL/AGAMOUS pathway. J. Biol. Chem. 278 28246–28251. [DOI] [PubMed] [Google Scholar]
- Brand, U., Fletcher, J.C., Hobe, M., Meyerowitz, E.M., and Simon, R. (2000). Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 289 617–619. [DOI] [PubMed] [Google Scholar]
- Brand, U., Grunewald, M., Hobe, M., and Simon, R. (2002). Regulation of CLV3 expression by two homeobox genes in Arabidopsis. Plant Physiol. 129 565–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breyne, P., van Montagu, M., Depicker, A., and Gheysen, G. (1992). Characterization of a plant scaffold attachment region in a DNA fragment that normalizes transgene expression in tobacco. Plant Cell 4 463–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carles, C.C., Choffnes-Inada, D., Reville, K., Lertpiriyapong, K., and Fletcher, J.C. (2005). ULTRAPETALA1 encodes a SAND domain putative transcriptional regulator that controls shoot and floral meristem activity in Arabidopsis. Development 132 897–911. [DOI] [PubMed] [Google Scholar]
- Clark, S.E. (2001). Cell signalling at the shoot meristem. Nat. Rev. Mol. Cell Biol. 2 276–284. [DOI] [PubMed] [Google Scholar]
- Clark, S.E., Running, M.P., and Meyerowitz, E.M. (1993). CLAVATA1, a regulator of meristem and flower development in Arabidopsis. Development 119 397–418. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Gallois, J.L., Nora, F.R., Mizukami, Y., and Sablowski, R. (2004). WUSCHEL induces shoot stem cell activity and developmental plasticity in the root meristem. Genes Dev. 18 375–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross-Hardt, R., Lenhard, M., and Laux, T. (2002). WUSCHEL signaling functions in interregional communication during Arabidopsis ovule development. Genes Dev. 16 1129–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannesson, H., Wang, Y., and Engström, P. (2001). DNA-binding and dimerization preferences of Arabidopsis homeodomain-leucine zipper transcription factors in vitro. Plant Mol. Biol. 45 63–73. [DOI] [PubMed] [Google Scholar]
- Kaya, H., Shibahara, K.I., Taoka, K.I., Iwabuchi, M., Stillman, B., and Araki, T. (2001). FASCIATA genes for chromatin assembly factor-1 in Arabidopsis maintain the cellular organization of apical meristems. Cell 104 131–142. [DOI] [PubMed] [Google Scholar]
- Koncz, C., and Schell, J. (1986). The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel Agrobacterium binary vector. Mol. Gen. Genet. 204 383–396. [Google Scholar]
- Laufs, P., Dockx, J., Kronenberger, J., and Traas, J. (1998). MGOUN1 and MGOUN2: Two genes required for primordium initiation at the shoot apical and floral meristems in Arabidopsis thaliana. Development 125 1253–1260. [DOI] [PubMed] [Google Scholar]
- Laux, T. (2003). The stem cell concept in plants: A matter of debate. Cell 113 281–283. [DOI] [PubMed] [Google Scholar]
- Laux, T., Mayer, K.F.X., Berger, J., and Jürgens, G. (1996). The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development 122 87–96. [DOI] [PubMed] [Google Scholar]
- Lenhard, M., Bohnert, A., Jürgens, G., and Laux, T. (2001). Termination of stem cell maintenance in Arabidopsis floral meristems by interactions between WUSCHEL and AGAMOUS. Cell 105 805–814. [DOI] [PubMed] [Google Scholar]
- Lenhard, M., and Laux, T. (2003). Stem cell homeostasis in the Arabidopsis shoot meristem is regulated by intercellular movement of CLAVATA3 and its sequestration by CLAVATA1. Development 130 3163–3173. [DOI] [PubMed] [Google Scholar]
- Lohmann, J., Huong, R., Hobe, M., Busch, M., Parcy, F., Simon, R., and Weigel, D. (2001). A molecular link between stem cell regulation and floral patterning in Arabidopsis. Cell 105 793–803. [DOI] [PubMed] [Google Scholar]
- Mayer, K.F.X., Schoof, H., Haecker, A., Lenhard, M., Jürgens, G., and Laux, T. (1998). Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95 805–815. [DOI] [PubMed] [Google Scholar]
- Newman, I.V. (1965). Patterns in the meristems of vascular plants. III. Pursuing the patterns where no cell is a permanent cell. J. Linn. Soc. Lond. Bot. 59 185–214. [Google Scholar]
- Rojo, E., Sharma, V.K., Kovaleva, V., Raikhel, N.V., and Fletcher, J.C. (2002). CLV3 is localized to the extracellular space, where it activates the Arabidopsis CLAVATA stem cell signaling pathway. Plant Cell 14 969–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoof, H., Lenhard, M., Haecker, A., Mayer, K.F.X., Jürgens, G., and Laux, T. (2000). The stem cell population of Arabidopsis shoot meristems is maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100 635–644. [DOI] [PubMed] [Google Scholar]
- Shiraishi, H., Okada, K., and Shimura, Y. (1993). Nucleotide sequences recognized by the AGAMOUS MADS domain of Arabidopsis thaliana in vitro. Plant J. 4 385–398. [DOI] [PubMed] [Google Scholar]
- Sieber, P., Gheyselinck, J., Gross-Hardt, R., Laux, T., Grossniklaus, U., and Schneitz, K. (2004). Pattern formation during early ovule development in Arabidopsis thaliana. Dev. Biol. 273 321–334. [DOI] [PubMed] [Google Scholar]
- Spradling, A., Drummond-Barbosa, D., and Kai, T. (2001). Stem cells find their niche. Nature 414 98–104. [DOI] [PubMed] [Google Scholar]
- Steeves, T.A., and Sussex, I.M. (1989). Patterns in Plant Development. (Cambridge, UK: Cambridge University Press).
- Stuurman, J., Jaggi, F., and Kuhlemeier, C. (2002). Shoot meristem maintenance is controlled by a GRAS-gene mediated signal from differentiating cells. Genes Dev. 16 2213–2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda, M., Matsui, K., Ishiguro, S., Sano, R., Wada, T., Paponov, I., Palme, K., and Okada, K. (2004). The HALTED ROOT gene encoding the 26S proteasome subunit RPT2a is essential for the maintenance of Arabidopsis meristems. Development 131 2101–2111. [DOI] [PubMed] [Google Scholar]
- Wagner, D. (2003). Chromatin regulation of plant development. Curr. Opin. Plant Biol. 6 20–28. [DOI] [PubMed] [Google Scholar]
- Watanabe, K., and Okada, K. (2003). Two discrete cis elements control the abaxial side-specific expression of the FILAMENTOUS FLOWER gene in Arabidopsis. Plant Cell 15 2592–2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel, D., and Jürgens, G. (2002). Stem cells that make stems. Nature 415 751–754. [DOI] [PubMed] [Google Scholar]
- Wellmer, F., Riechmann, J.L., Alves-Ferreira, M., and Meyerowitz, E.M. (2004). Genome-wide analysis of spatial gene expression in Arabidopsis flowers. Plant Cell 16 1314–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, X., Dabi, T., and Weigel, D. (2005). Requirement of homeobox gene STIMPY/WOX9 for Arabidopsis meristem growth and maintenance. Curr. Biol. 15 436–440. [DOI] [PubMed] [Google Scholar]
- Zhao, Y., Medrano, L., Ohashi, K., Fletcher, J.C., Yu, H., Sakai, H., and Meyerowitz, E.M. (2004). HANABA TARANU is a GATA transcription factor that regulates shoot apical meristem and flower development in Arabidopsis. Plant Cell 16 2586–2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo, J., Niu, Q.W., Frugis, G., and Chua, N.H. (2002). The WUSCHEL gene promotes vegetative-to-embryonic transition in Arabidopsis. Plant J. 30 349–359. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.