Abstract
The progression of mitotic division cycles and synchronous development between and within the male and female reproductive organs are essential for plant sexual reproduction. Little is known about the genetic control of the progression of mitotic cycles of the haploid genome during gametogenesis in higher plants. Here, we report the phenotypic and molecular characterization of an Arabidopsis thaliana mutant, slow walker1 (swa1), in which the progression of the mitotic division cycles of the female gametophyte was disrupted. Confocal microscopy revealed that megagametophyte development was asynchronous in swa1, causing embryo sacs to arrest at two-, four-, or eight-nucleate stages within the same pistil. A delayed pollination experiment showed that a small fraction of the swa1 embryo sacs were able to develop into functional female gametophytes. The swa1 mutation also showed a slight reduction in penetrance through the male gametophyte, although the pollen grains were morphologically normal. Molecular analysis indicates that SWA1 encodes a protein with six WD40 repeats that is localized in the nucleolus in interphase cells. The SWA1 gene is expressed in cells undergoing active cell divisions, including functional megaspores and the female gametophytic cells. RNA interference results indicated that knockout of SWA1 inhibited root growth significantly and led to the accumulation of unprocessed 18S pre-rRNA. These data suggest that SWA1 most likely plays a role in rRNA biogenesis that is essential for the progression of the mitotic division cycles during gametogenesis in plants.
INTRODUCTION
The plant life cycle alternates between a diploid sporophytic phase and a haploid gametophytic phase. The gametophytic generation in higher plants is reduced to several cell cycles. The male and female gametophytes, also called the pollen grain and the embryo sac, respectively, consist of a few cells enclosed within the sexual reproductive organs. In most flowering plants, the female gametophyte comprises seven cells: three antipodals, one large diploid central cell, two synergids, and an egg cell (Johri, 1984; Grossniklaus and Schneitz, 1998; Drews and Yadegari, 2002). During female gametogenesis, one of the four meiotic products (megaspores) survives and undergoes three rounds of nuclear division to give rise to an eight-nucleate, coenocytic embryo sac. The other three megaspores undergo cell death. The coenocytic embryo sac then goes through simultaneous cytokinesis (cellularization) to produce the seven-celled female gametophyte (Misra, 1962; Mansfield et al., 1991; Schneitz et al., 1995; Drews et al., 1998).
Little is known about the molecular mechanisms and genetic control of female gametogenesis in plants, although many mutations that are defective in female gametophytic functions were isolated in Arabidopsis thaliana and maize (Zea mays) (Feldmann et al., 1997; Christensen et al., 1998; Drews et al., 1998; Yang and Sundaresan, 2000). Several mutations that control the progression of the mitotic cycles of the haploid gametophytic cells have been reported (Springer et al., 1995; Moore et al., 1997; Drews et al., 1998). The mitotic division cycles in fem female gametophytes are blocked at either a single or multiple stages (Feldmann et al., 1997; Christensen et al., 1998). For example, female gametophyte development is arrested at the one-nucleate stage in Gf (Redei, 1965; Christensen et al., 1997) and at the four-nucleate stage in the prolifera (prl) mutant (Springer et al., 1995). In the hadad (hdd) mutant, female gametophyte development can be arrested at the two-, four-, or eight-nucleate stage (Moore et al., 1997). In addition, several mutants, such as ig1, lethal ovule1 (lo1), and lo2, which are defective in gametophytic cell cycle progression, have also been isolated in maize (Nelson and Clary, 1952; Kermicle, 1971; Huang and Sheridan, 1996). Although many mutants with impaired gametophytic progression of the mitotic division cycles have been reported, only a few genes, including PRL (Springer et al., 1995, 2000) and NOMEGA (Kwee and Sundaresan, 2003), have been cloned. These data suggested that disruption in cell cycle genes often causes an arrest of female gametophyte development during mitotic divisions (Capron et al., 2003; Kwee and Sundaresan, 2003) or uncontrolled nuclear division (Ebel et al., 2004) during female gametogenesis in Arabidopsis. These findings suggest that genes involved in general cell cycle progression play a role during female gametogenesis in higher plants. Gametophytic mutations also provide us with a unique system in which to study the functions of essential genes, whose mutations are difficult to detect in the sporophyte.
Successful fertilization requires both male and female gametophyte development to be coordinated. Developmental delay of the female gametophyte, for example, results in sterility. Here, we report the isolation and characterization of a semisterile mutant, slow walker1 (swa1), which is defective in mitotic progression of the female gametophyte. SWA1 encodes a nucleolar protein with six WD40 repeats that is most likely involved in 18S rRNA biogenesis. The slow progression of the gametophytic division cycles in swa1 suggested that the SWA1 protein is required for the normal progression of mitotic division cycles through the regulation of cell metabolism.
RESULTS
Isolation of the swa1 Mutant
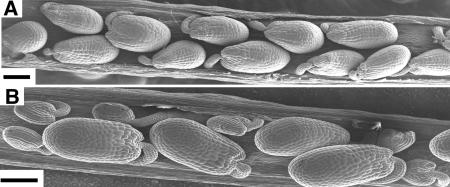
To identify mutations that play a role in gametophyte development, we performed a distorted Mendelian segregation screen (Feldmann et al., 1997; Christensen et al., 1998; Howden et al., 1998; Grini et al., 1999) of the gene trap and enhancer trap lines generated in Arabidopsis ecotype Landsberg erecta (Springer et al., 1995; Sundaresan et al., 1995). One mutant, swa1, showed a nearly 1:2 segregation ratio of kanamycin-resistant to kanamycin-sensitive seedlings (Kanr:Kans = 600:1135) instead of the typical 3:1 ratio, suggesting that it is most likely defective in gametophytic function. To further check the transmission efficiency of the mutation, reciprocal crosses were performed between the mutant heterozygous for the Ds insertion and the wild type. The F1 progeny were all kanamycin-sensitive (Kanr = 0, Kans = 491) when the mutant pistils were pollinated with pollen grains from wild-type plants. However, the F1 progeny showed a Kanr:Kans ratio of 1:1.27 (219:280) when used as male to pollinate wild-type pistils. These data clearly demonstrated that the Ds insertion was transmitted mainly through the male gametophyte and not the female. Therefore, the mutation completely disrupted female gametophyte function and slightly impaired the male function. Furthermore, DNA gel blot analysis showed that a single Ds element was inserted into the mutant genome (data not shown), suggesting that this caused the phenotype or that the Ds insertion and phenotype were tightly linked. Because the mutation can only be transmitted through the male gametophyte and no homozygote was available, we use swa1 to represent the swa1/+ heterozygote genotype throughout this article. The swa1 plants have no visible morphological abnormalities in vegetative organs compared with wild-type plants. The inflorescence and flower exhibited normal external morphology, except that they bore shorter siliques than wild-type plants because of the reduced seed set. The size of the siliques from swa1 plants was approximately two-thirds that of the wild-type at the same age (data not shown). Scanning electron microscopy showed that full seed set was observed in wild-type siliques (Figure 1A). In swa1 siliques, however, approximately half of the ovules (616 of 1196) were aborted 2 or 3 d after pollination (Figure 1B), and the remaining ovules were larger and developed into normal seeds. The difference in ovule size was obvious even at 24 h after pollination (data not shown). Because both wild-type and swa1 plants were grown under the same conditions, we concluded that the aborted ovules were those that carried the swa1 mutation. On the other hand, no obvious morphological difference of pollen grains from the mutant and the wild type was observed (data not shown), which suggested that the mutation most likely affected female gametophyte development or fertilization.
Figure 1.
Phenotype of the swa1/+ Plant.
(A) Scanning electron micrograph showing full seed set of a wild-type silique.
(B) Scanning electron micrograph showing ovule abortion in a swa1/+ silique.
Bars = 100 μm.
Progression of Gametophytic Division Cycles Is Retarded in swa1 Ovules
To further investigate the mutant phenotype, we performed confocal laser scanning microscopy (CLSM) to study ovule development in wild-type and swa1 plants (Christensen et al., 1997). Female gametogenesis in Arabidopsis belongs to the Polygonum type (Misra, 1962; Webb and Gunning, 1994; Schneitz et al., 1995). A single hypodermal archesporial cell of the nucellus directly differentiates into a megaspore mother cell or megasporocyte that undergoes meiosis to produce four haploid megaspores, the tetrad. Three of these, located at the micropylar pole, undergo programmed cell death; only the chalazal-most haploid cell is functional. The functional megaspore proceeds through three rounds of nuclear division to give rise to an eight-nucleate, coenocytic embryo sac. Subsequently, nuclear migration and cellularization take place, resulting in the formation of a seven-celled female gametophyte with three antipodal cells, two synergids, one egg, and a large diploid central cell. Shortly before fertilization, antipodal cells undergo cell death, and finally a four-celled female gametophyte, the female germ unit, is formed when fertilization occurs (Mansfield et al., 1991; Christensen et al., 1997).
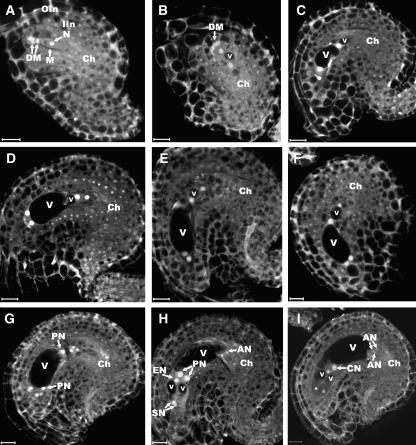
To characterize the mutant ovule phenotype, we first studied female gametogenesis in wild-type plants. Figure 2 shows projected confocal images of ovules at different developmental stages in the wild type. After meiosis, the chalazal-most megaspore enlarges and its nucleus becomes prominent and moves to the center; the micropylar megaspores degenerate, as shown by their strong autofluorescence (Figure 2A). After the functional megaspore reaches a critical size, nuclear division takes place to give rise to a two-nucleate embryo sac (Figure 2B). Concurrently, a big central vacuole is formed and pushes the two nuclei toward the micropylar and chalazal poles, respectively (Figures 2B and 2C). A smaller vacuole is also formed at the chalazal-most end of the embryo sac (Figure 2C). Then, the two nuclei undergo another division simultaneously at each pole. The division plane of the micropylar nucleus is parallel to the chalazal–micropylar axis, whereas the division plane of the chalazal nucleus is perpendicular to the axis (Figure 2D). Subsequently, the chalazal cytoplasm undergoes extensive vacuolation to form several relatively large vacuoles (Figure 2E) that subsequently coalesce to form a larger vacuole to separate the two chalazal nuclei (Figure 2F). The third nuclear division results in the formation of an eight-nucleate, coenocytic embryo sac with four nuclei at each pole (Figure 2G). One of the chalazal nuclei migrates toward the micropyle, and subsequently, cellularization occurs (Figure 2H). Finally, the two polar nuclei fuse to form the central nucleus (Figure 2I).
Figure 2.
Ovule Development Revealed by CLSM in Wild-Type Plants.
(A) A female gametophyte (FG) stage 1 ovule showing the functional (M) and degenerating (DM) megaspores.
(B) An FG2-stage ovule with a two-nucleate embryo sac.
(C) An FG3-stage ovule showing a late two-nucleate embryo sac with an enlarged central vacuole (V) and a small chalazal vacuole (v).
(D) An ovule at early FG4 stage, containing a four-nucleate embryo sac. Note that the division planes of the chalazal and the micropylar nuclei are perpendicular to each other.
(E) An ovule at FG4 stage. Note the appearance of vacuoles (v) at the chalazal pole of the embryo sac.
(F) An ovule with a four-nucleate embryo sac at late FG4 stage, showing the formation of a relatively large vacuole (v) separating the two nuclei at the chalazal pole.
(G) An ovule at early FG5 stage with an eight-nucleate embryo sac in a 4n + 4n configuration. Note that the polar nuclei (PN) are recognizable.
(H) An ovule image showing an embryo sac at FG5 stage. Note that cellularization took place and cell differentiation was completed with the formation of two synergid nuclei (SN), an egg nucleus (EN), three antipodal nuclei (AN; only two of them are seen), and the two prominent polar nuclei (PN), which have not fused yet.
(I) An ovule with a mature seven-celled embryo sac at stage FG6. Note that the polar nuclei fused to form a diploid central nucleus (CN).
All images were projected from multiple 1-μm optical sections. AN, antipodal nucleus; Ch, chalazal end; CN, central cell nucleus; DM, degenerated megaspore; EN, egg cell nucleus; IIn, inner integument; M, megaspore; OIn, outer integument; PN, polar nucleus; SN, synergid nucleus; V, large vacuole; v, small vacuole. The developmental stages are defined according to Christensen et al. (1998). Bars = 10 μm.
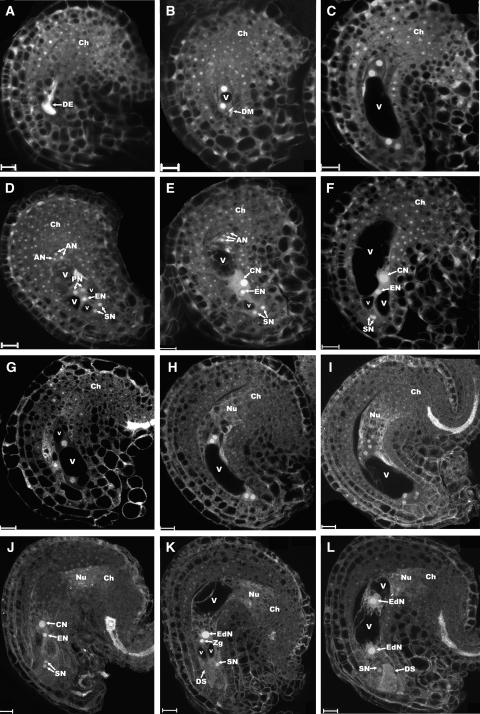
In swa1 pistils, no abnormality was found from archesporial cell development to the formation of the functional megaspore. The archesporial cell enlarges, becomes teardrop-shaped, proceeds to meiosis, and a normal tetrad is formed. Three megaspores at the micropylar end undergo cell death, and the functional megaspore enters stage FG1 (FG stages are determined according to Christensen et al. [1998]). In pistils at flower developmental stage 14 (Smyth et al., 1990) in wild-type plants, the majority of embryo sacs are at the four-celled stage; occasionally, a few are at the seven-celled stage. This indicates that ovule development in Arabidopsis pistils of wild-type plants is synchronous with only a narrow range of variations in development. In the swa1 mutant pistil at the same floral stage, except two for degenerated embryo sacs (Figure 3A), approximately half of the ovules are at the four-celled stage and the other half are at different developmental stages, including the two-nucleate stage, the four-nucleate stage, the eight-nucleate stage, and the seven-celled stage (Figures 3B to 3F). In a swa1 pistil ∼5 h after pollination, we found that 15 ovules out of 34 were fertilized and the endosperm nuclei were visible by confocal microscopy (Figures 3K to 3L), 2 ovules were at the FG7 stage (Figure 3J) and ready for fertilization, and the other 17 ovules were still at the FG3, FG4, or FG5 stage (Figures 3G to 3I). Compared with wild-type ovules, these data indicated that gametophytic cell cycle progression in the mutant ovules was impaired. This suggested that the mutated gene has direct or indirect effects on cell cycle progression.
Figure 3.
Ovule Development in Pistils from a swa1/+ Plant.
(A) to (F) Ovules from the same pistil (sl11) of a swa1/+ mutant plant, showing embryo sacs at different developmental stages from FG1 to FG7.
(G) to (L) Ovules from another pistil (sl12) of the swa1/+ mutant.
(A) An ovule with a degenerated embryo sac (DE) visualized by its strong autofluorescence.
(B) An ovule with an FG3-stage embryo sac.
(C) A four-nucleate embryo sac at early FG4 stage.
(D) An obligate section showing an ovule with a seven-celled embryo sac (FG5 stage). Note that the two polar nuclei (PN) have not fused yet.
(E) An ovule with a seven-celled, FG6-stage embryo sac. The polar nuclei fused to give rise to a central nucleus (CN).
(F) A mature embryo sac of FG7 stage with antipodal cells degenerated.
(G) An ovule with a two-nucleate embryo sac at late FG3 stage.
(H) An early FG4-stage ovule with a four-nucleate embryo sac.
(I) An ovule at early FG5 stage with an eight-nucleate embryo sac.
(J) An ovule with an embryo sac at stage FG7. Vacuoles are not clear because of the projection of multiple optical sections.
(K) A fertilized ovule revealed by the presence of a degenerated synergid (DS).
(L) An ovule showing the first division of the endosperm nucleus (EdN). The zygote is not shown .
All images are projections of multiple 1-μm-thick optical sections. AN, antipodal nucleus; Ch, chalazal end; CN, central cell nucleus; DE, degenerated embryo sac; DM, degenerated megaspore; DS, degenerated synergid; EdN, endosperm nucleus; EN, egg cell nucleus; PN, polar nucleus; SN, synergid nucleus; V, vacuole. Bars = 10 μm.
Synchrony of Female Gametophyte Development Was Impaired in swa1
To investigate the developmental synchrony of female gametophytes in the mutant pistils, a detailed CLSM study was performed according to Christensen et al. (1997). Inflorescences from wild-type and swa1 plants were fixed with glutaraldehyde and cleared with benzyl benzoate and benzyl alcohol before dissection. Then, pistils from the same inflorescence were collected together and opened sequentially. Ovules from each pistil were dissected out and sealed under the cover slip with nail polish before CLSM.
In wild-type pistils, female gametophyte development is synchronous: ovules within the same silique are often at two adjacent developmental stages (Table 1). For example, in pistil SL4, 25 of the 32 ovules checked were at stage FG4 and had four nuclei in the embryo sac, 2 of the 32 ovules were at FG3, and 4 were at FG2. Similarly, in pistil SL5, 24 of the 30 ovules were at stage FG5 and 6 were at FG4, indicating that FG5 was the dominant stage in this pistil (Table 1). In some pistils, such as SL3, most of the ovules were at two adjacent dominant stages. In SL7, a pistil 4 h after pollination, approximately half of the ovules (14 of 32) were fertilized; embryo sacs in 12 ovules were at stage FG7, and the antipodal cells degenerated. Almost all of the megagametophytes (31 of 32) were fertilized by 24 h after pollination (Table 1). Our observation in wild-type pistils is consistent with previous results (Christensen et al., 1997) demonstrating that ovule development within a pistil is synchronous in Arabidopsis.
Table 1.
Synchrony of Female Gametophyte Development in Wild-Type Arabidopsis
| Number of Female Gametophytes at Developmental Stages |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| Pistil Number | FG1 | FG2 | FG3 | FG4 | FG5 | FG6 | FG7 | FG8 | Total FGs |
| SL1 | 31 | 31 | |||||||
| SL2 | 5 | 21 | 26 | ||||||
| SL3 | 1 | 10 | 15 | 26 | |||||
| SL4 | 4 | 2 | 25 | 31 | |||||
| SL5 | 6 | 24 | 30 | ||||||
| SL6 | 1 | 0 | 15 | 16 | |||||
| SL7 | 2 | 1 | 12 | 14 | 29 | ||||
| SL8 | 1 | 0 | 31 | 32 | |||||
Siliques were fixed in 2.5% glutaraldehyde and analyzed with an LSM 510META laser scanning microscope according to Christensen et al. (1997). FG stages were defined according to Christensen et al. (1998).
Similar analysis was performed on swa1 mutant pistils; 21 pistils from three inflorescences from different plants were analyzed by confocal imaging. Consistently, asynchronous development of mutant embryo sacs was observed (Table 2). In pistil sl2, for example, 3 of the 29 ovules were at stage FG3, 10 were at stage FG2, whereas the majority (16) had not progressed past FG1. More strikingly, the female gametophyte cells spanned seven developmental stages, as shown for sl11, in which 12 of the 40 ovules were at either the seven-celled FG6 or four-celled FG7 stage, 16 were at the four-nucleate FG4 stage, and a few stayed at the one- or two-nucleate stage. In pistil sl13, half of the ovules reached the FG7 or FG8 stage, whereas the other half were still at stages FG3 to FG5. It is likely that ovules at FG7 or FG8 in this pistil represented the SWA1 female gametophytes, whereas the ovules at stages FG3 to FG5 were swa1 mutant female gametophytes. These observations suggest that the synchrony of female gametophyte development was impaired in swa1 pistils.
Table 2.
Synchrony of Female Gametophyte Development in swa1/+ Mutants
| Number of Female Gametophytes at Developmental Stages |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| Pistil Number | FG1 | FG2 | FG3 | FG4 | FG5 | FG6 | FG7 | FG8 | Total FGs |
| sl1 | 6 | 6 | 1 | 13 | |||||
| sl2 | 16 | 10 | 3 | 29 | |||||
| sl3 | 1 | 2 | 4 | 13 | 10 | 3 | 33 | ||
| sl4 | 5 | 11 | 16 | 4 | 36 | ||||
| sl5 | 1 | 1 | 16 | 6a | 12 | 36 | |||
| sl6 | 1 | 1 | 17 | 4 | 10 | 33 | |||
| sl7 | 1 | 11 | 8a | 11 | 31 | ||||
| sl8 | 2 | 15 | 9a | 4 | 6 | 36 | |||
| sl9 | 2 | 14 | 6a | 14 | 6 | 42 | |||
| sl10 | 2 | 2 | 15 | 2 | 11 | 3 | 35 | ||
| sl11 | 1 | 1 | 1 | 16 | 9 | 9 | 3 | 40 | |
| sl12 | 1 | 12 | 4 | 2 | 15 | 34 | |||
| sl13 | 8 | 6 | 1 | 16 | 31 | ||||
FG stages were defined according to Christensen et al. (1998).
FG5 here includes both cellularized and uncellularized embryo sacs. The number of uncellularized embryo sacs are one in sl5, two in sl7, five in sl8, and two in sl9.
The Mutant Female Gametophytes Could Be Fertilized by Delayed Pollination
The observations described above suggested that the developmental synchrony of gametophytic nuclear division was impaired in swa1/+ embryo sacs. To further investigate whether the swa1 female gametophytes are able to reach the final stage and become functional, or may simply miss the time for fertilization as a result of slow development, we performed a delayed pollination test in which the time of emasculation and pollination was strictly controlled. Pistils of swa1/+ plants were emasculated at floral stage 12c (Smyth et al., 1990) and pollinated with pollen from wild-type plants after 12 h (stage 13), 24 h (stage 14), 48 h (stage 15), 72 h (stage 17), or 96 h (stage 17). Seeds from three independent plants of each group were collected together. For comparison, pollination at 12 h after emasculation (corresponding to floral stage 13, at which natural pollination occurs) was set as the control (group 1). F1 seeds from each time point were plated on MS medium supplemented with 50 μg/mL kanamycin. The results are summarized in Table 3. No Kanr progeny were obtained in group 1. However, Kanr seedlings were obtained when pollination was postponed. Eleven of 1125 F1 progeny were Kanr in group 2, in which pollination was performed at 24 h after emasculation. Similarly, 8 of 1072 were Kanr when pollination was performed at 48 h after emasculation (group 3). In groups 4 and 5, siliques were obtained through pollination at 72 and 96 h after emasculation; in these cases, 7 of 898 and 13 of 432 in the F1 progeny, respectively, displayed kanamycin resistance. These results suggested that small fractions of swa1 mutant ovules were fertilized and produced seeds when pollination was delayed. This implies that the mutant female gametophytes develop more slowly than wild-type gametophytes and have the potential, although small, to develop into functional female gametophytes.
Table 3.
Kanr:Kans Ratios of F1 Progeny from the Delayed Pollination Test
| Group | Hours after Emasculation | Progeny | Kanr:Kans | P (Group 1 as Reference) |
|---|---|---|---|---|
| 1 | 12 (stage 13) | Kanr, 0 | 0 | – |
| Kans, 1219 | ||||
| 2 | 24 (stage 14) | Kanr, 11 | 0.98% | <0.01 |
| Kans, 1125 | ||||
| 3 | 48 (stage 15) | Kanr, 8 | 0.78% | <0.01 |
| Kans, 1072 | ||||
| 4 | 72 (stage 17) | Kanr, 7 | 0.78% | <0.01 |
| Kans, 898 | ||||
| 5 |
96 (stage 17) |
Kanr, 13 | 3.01% |
<0.01 |
| Kans, 432 |
Pistils of swa1/+ plants were emasculated at floral stage 12c (Smyth et al., 1990) and pollinated with pollen from wild-type plants at 12 h (stage 13), 24 h (stage 14), 48 h (stage 15), 72 h (stage 17), or 96 h (stage 17) after emasculation. The results from each time point were from three independent plants. Group 1 was a control, because Arabidopsis anthesis occurs naturally at stage 13. Groups 2 to 5 all exhibited significant differences from group 1, indicating that a small fraction of swa1 mutant ovules could be fertilized and produce seeds when pollination was delayed.
SWA1 Encodes a Nucleolar Protein with WD40 Repeats
Genomic sequences flanking the Ds element were obtained by thermal asymmetric interlaced PCR (Liu et al., 1995; Grossniklaus et al., 1998). Sequence analysis indicated that the Ds was inserted at 146 to 151 bp downstream the ATG of At2g47990 (Figure 4A). The Ds insertion resulted in a 6-bp duplication of the host sequence at the insertion site (Figure 4A). The insertion was further confirmed by DNA gel blot hybridization with Ds probe and At2g47990-specific probe (data not shown). At2g47990 is a single-copy gene in the Arabidopsis genome. To obtain its cDNA sequence, RT-PCR was performed using primers designed according to genomic and available EST sequences in GenBank. The At2g47990 cDNA is 1593 bp long (accession number NM_130366). Comparison of the cDNA and genomic sequences revealed that the At2g47990 gene contains no intron.
Figure 4.
Molecular Characterization of the SWA1 Gene.
(A) Scheme of the Ds insertion site. The shaded box indicates the predicted open reading frame of the SWA1 gene. The Ds insertion caused a 6-bp duplication in the swa1 mutant. The nucleotide numbers are consistent with those of BAC clone T9J23.
(B) SWA1 cDNA sequence and its predicted peptide sequence. The Ds insertion site is indicated by an arrow, and the WD40 repeat region in the SWA1 peptide is underlined, with the first and last residues marked by stars. The boxed amino acids show the predicted nuclear localization signal.
(C) Phylogenetic tree of SWA1 with its homologs from other organisms.
(D) Alignment of the SWA1 protein with its homologs from other organisms. Identical amino acids are shown with white letters on black boxes, and similar ones are indicated with shading boxes. AtSWA1, Arabidopsis SWA1 protein; Poptr1 644959, poplar homolog; OSJNBa0066Oryza, rice homolog; Q7ZXZ2Xenopus, Xenopus homolog; SAW, Src-associated WD40 protein from mouse; SPBC428.19c, S. pombe homolog; CG3071Drosophila, D. melanogaster homolog; UTP15, S. cerevisiae homolog.
To confirm whether the Ds insertion into At2g47990 is the cause of the semisterile phenotype of swa1, a complementation experiment was performed. A 4.5-kb genomic fragment containing the predicted promoter, open reading frame, and 3′ untranslated region of the At2g47990 gene, between nucleotides 43,430 and 38,931 of the BAC clone T9J23, was subcloned into pCAMBIA1300 and introduced into swa1/+ heterozygous plants via Agrobacterium tumefaciens–mediated transformation (Bechtold and Pelletier, 1998). In total, 187 transgenic lines were obtained via hygromycin and kanamycin double selection. T1 seeds from 25 lines were germinated on MS plates supplemented with 50 μg/mL kanamycin, seedlings were scored for kanamycin resistance, and the ratio of Kanr to Kans was determined. All primary transformants showed a kanamycin segregation ratio of ∼2:1 compared with the ratio of 1:2 in selfed swa1/+ progeny, indicating that the 4.5-kb fragment completely complemented the semisterile phenotype in the swa1/+ background. Furthermore, the next generations of transgenic lines were tested, and 72 swa1/swa1 homozygous plants were obtained from 299 plants, indicating that the Ds element can be transmitted to the next generation through female gametophytes with the aid of the introduced At2g47990 genomic fragment. In addition, full seed set was observed when the complementation copy of At2g47990 was homozygous in T2 plants in the swa1/swa1 background (data not shown). These data demonstrate that At2g47990 is indeed the SWA1 gene.
Further analysis indicated that SWA1 encodes a protein of 530 amino acids with an estimated pI of 9.62 and a molecular mass of 58.9 kD (Figure 4B). Database searches showed that the SWA1 protein contains six WD40 repeats in the region of amino acids 50 to 295 (Figure 4D). Among the repeats, four of them contain the typically conserved core sequence [GAS]-H-X(3)-[VI]-X-[SAC][VLI]-X-[FWLIVY]-X(2-30)-[LIV][AVLI][ST][GA][SG]-X-D-X-[TS]-[IVL][KR][VLI]-[WFY]-[DN] (Neer et al., 1994). The WD40 repeat is composed of ∼40 amino acid residues with a conserved Gly-His (GH) dipeptide at the N terminus and a Trp-Asp (WD) dipeptide at its C terminus, to form a propeller-like structure (Neer et al., 1994). It has been proposed that the propeller-like structures of WD40 can form a stable platform or anchor for other proteins; thus, a complex can be recruited to accommodate sequential and/or simultaneous interactions involving several sets of proteins. The amino acid sequence of SWA1 shared high homology with several proteins from Populus trichocarpa (Populus gene model grail 3.0034001501, protein number 644959), Oryza sativa (locus OSJNBa0066H10.103), Xenopus laevis (locus Q7ZXZ2), Mus musculus (locus AY151261; SAW), Schizosaccharomyces pombe (locus SPBC428.19c), Saccharomyces cerevisiae (locus YMR093w; UTP15), and Drosophila melanogaster (locus CG3071). Among these proteins, the poplar protein is the most similar to SWA1. They are 59% identical and share a similarity of 73% at the amino acid level. The similarity between SWA1 and its rice homolog is ∼58%, whereas the similarity between yeast proteins SPBC428.19c and UTP15 and SWA1 is ∼46 and 43%, respectively. Phylogenetic analysis showed that the poplar homolog is much closer to SWA1 than is the rice homolog, although three of them are in the same clade. Compared with the homologs from yeast, the animal homologs are farther away from the plant proteins (Figures 4C and 4D). In S. cerevisiae, UTP15 is localized in the nucleolus, and it is a component of the U3 snoRNP complex involved in nucleolar processing of pre-18S rRNA (Dragon et al., 2002).
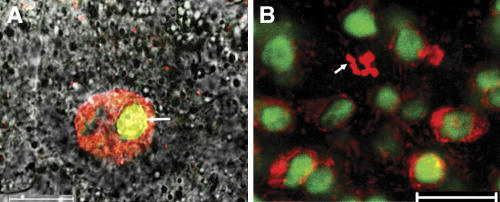
To obtain insight into the subcellular localization of SWA1, a C-terminal translational fusion between SWA1 and GREEN FLUORESCENT PROTEIN (GFP) was made and introduced into wild-type and swa1/+ plants. Transgenic plants expressing the SWA1-GFP fusion gene showed a 2:1 Kanr:Kans segregation and their siliques had full seed set (data not shown), indicating that the fusion gene functionally complemented the ovule phenotype. Confocal microscopy showed that the fusion protein was localized in the nucleolus of root cells at interphase (Figure 5A), and no GFP signal was detected in the cytoplasm or in cells at M phase (Figure 4B). This demonstrated that SWA1 is indeed a nucleolar protein.
Figure 5.
Subcellular Localization of SWA1 in Arabidopsis Transformed with the 35S:SWA1-GFP Gene.
(A) Merged micrograph showing SWA1-GFP localization in the nucleolus (arrow). DNA was visualized with DAPI staining (red). Bar = 10 μm.
(B) Merged photograph showing the nucleolar localization of SWA1-GFP (green) in Arabidopsis root cells at interphase. No SWA1-GFP signal was found in cells at metaphase (arrow). Bar = 10 μm.
SWA1 Is Expressed Ubiquitously throughout the Plant
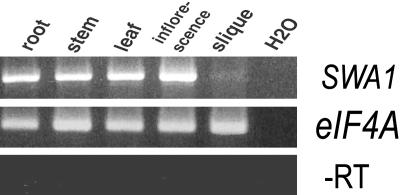
Because SWA1 transcripts were not detectable by RNA gel blot analysis, RT-PCR was performed with gene-specific primers at 5′ and 3′ ends of the cDNA. A single band with expected size was detected in mRNAs from roots, stems, leaves, inflorescences, and siliques (Figure 6), indicating that SWA1 is expressed ubiquitously.
Figure 6.
Expression Pattern of SWA1 Revealed by RT-PCR.
mRNA isolated from different tissues as indicated was used as template and amplified with gene-specific primers by RT-PCR. The eIF4A gene was used as an internal control. Top gel, RT-PCR products showing the presence of SWA1 transcripts in root, stem, leaf, inflorescence, and silique of wild-type plants. Middle gel, RT-PCR products of the eIF4A gene, showing equal amounts of starting mRNAs. Bottom gel, PCR product with mRNA as template directly, showing no DNA contamination in the mRNA sample.
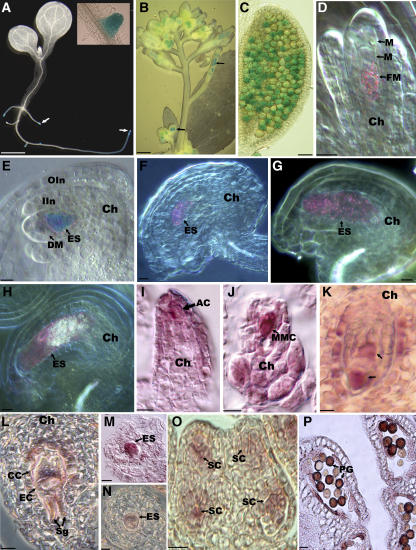
No β-glucuronidase (GUS) activity was detected in the mutant plants, although the GUS reporter gene in the Ds element is inserted in the sense orientation within the gene (Sundaresan et al., 1995). Sequence data indicated that this was attributable to the presence of several in-frame stop codons upstream of the GUS gene. To study the tissue- and cell-specific expression of SWA1, the 1.7-kb promoter region and the 1.2-kb fragment downstream of the SWA1 stop codon were fused on either side of the GUS reporter gene and subcloned into pCAMBIA1300. The resulting construct was transformed into Arabidopsis wild-type plants via Agrobacterium-mediated vacuum transformation (Bechtold and Pelletier, 1998). In T2 transgenic lines, GUS activity was detected in tissues active in cell division. Very strong GUS staining was observed in root tips and lateral root primordia (Figure 7A), and weaker signals were found in young leaf and stem vascular tissues. In reproductive organs, the GUS reporter gene was expressed throughout pollen development from very young floral buds to dehisced anthers, especially in microsporogenous cells, microspores, and mature pollen grains (Figures 7B and 7C), indicating that the SWA1 promoter is active during male sporogenesis and gametogenesis. In female reproductive organs, the reporter gene was expressed in megaspores (Figure 7D) and embryo sacs from one-nucleate (FG1) to seven-celled embryo sacs (FG6) (Figures 7E to 7H). No GUS staining was observed in nonfunctional megaspores.
Figure 7.
Expression Pattern of SWA1 in Arabidopsis Revealed by GUS Reporter and RNA in Situ Hybridization.
(A) to (H) GUS staining of PSWA1:GUS transgenic plants.
(I) to (P) Micrographs from RNA in situ hybridization.
(A) GUS staining in a transgenic seedling showing GUS activity in root tips (arrows) and lateral root primordia (inset). Bar = 0.5 cm.
(B) Micrograph of inflorescence showing GUS activity in anthers and ovules (arrows). Bar = 0.5 cm.
(C) GUS activity in pollen grains of a transgenic plant heterozygous for the transgene.
(D) Nomarski micrograph showing GUS activity (seen as pink dots) in the functional megaspore at the tetrad stage. No staining was observed in nonfunctional megaspores (M).
(E) Micrograph showing GUS staining in an early one-nucleate embryo sac (ES) at FG1 stage. No staining was observed in degenerated megaspores (DM).
(F) Nomarski micrograph showing GUS signal in a two-nucleate embryo sac (ES). No staining was observed in sporophytic tissues.
(G) Nomarski micrograph showing GUS signal in a four-nucleate embryo sac (ES).
(H) Nomarski micrograph showing GUS signal (pink dots) in a mature embryo sac (ES). The bright spots are starch granules typical of the mature embryo sac.
(I) Bright-field micrograph showing SWA1 expression in an archesporial cell (AC).
(J) Micrograph showing SWA1 expression in a megaspore mother cell (MMC).
(K) Micrograph showing SWA1 expression in a two-nucleate embryo sac. Note the strong signal in the polar cytoplasm (arrows).
(L) Nomarski micrograph of an obligate longitudinal section of a mature embryo sac showing SWA1 expression in synergid (Sg), egg cell (EC), and central cell (CC).
(M) Cross section of the micropylar part of a mature embryo sac showing SWA1 expression in the embryo sac (ES).
(N) Nomarski micrograph of a cross section through the middle of a mature embryo sac showing a hybridization signal in the central cell.
(O) Cross section of a young anther showing SWA1 expression in microsporogenous cells (SC).
(P) Micrograph showing SWA1 expression in mature pollen grains (PG).
AC, archesporial cell; Ch, chalazal end; CC, central cell; DM, degenerating megaspore; EC, egg cell; ES, embryo sac; FM, functional megaspore; M, megaspore; MMC, megaspore mother cell; PG, pollen grain; SC, sporogenous cell; Sg, synergid. Bars in (C) to (P) = 10 μm.
To confirm the GUS reporter data, RNA in situ hybridization was also performed using digoxigenin-labeled antisense and sense RNA probes. mRNA of the SWA1 gene was detected in nucellar primordia, archesporial cells (Figure 7I), megaspore mother cells (Figure 7J), and later in functional megaspores (data not shown). Very strong signal was also found in gametophytic cells of the female gametophyte from one-nucleate to seven-celled embryo sacs (Figures 7K to 7N). At the two-nucleate stage, a strong signal was found in the polar cytoplasm of the embryo sac (Figure 7K). In the mature embryo sac, obvious signals were detected in synergid, egg, and central cells (Figures 7L to 7N). After fertilization, the signal was also detected in embryos (data not shown). No signal above background was detected when sense RNA probe was used as a control (data not shown). During anther development, SWA1 transcripts were detected in sporogenous cells (Figure 7O), later in microspore mother cells (data not shown), and mature pollen grains (Figure 7P). At stage 7 of anther development (Sanders et al., 1999), the signal was also obvious in tapetal cells but not detectable in other cell layers (data not shown).
Downregulation of SWA1 by RNA Interference Inhibits Root Growth and Processing of 18S Pre-rRNA
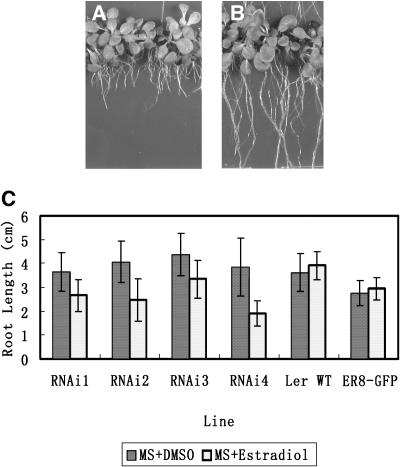
To evaluate the function of the SWA1 gene in plant development, an RNA interference (RNAi) approach was used because swa1/swa1 homozygous plants were not available. The 5′ end region of the SWA1 cDNA was subcloned in the opposite direction separated with an intron into the estrogen-inducible vector pER8 (Zuo et al., 2000). The resulting construct was introduced into Arabidopsis via Agrobacterium-mediated vacuum infiltration, and seven independent transgenic T1 plants were obtained. T2 seeds were germinated and grown on MS plates supplemented with 20 μg/mL hygromycin, and 1 week later, the resistant seedlings were transferred to MS plates with DMSO (control) or 10 μM β-estradiol inducer. No obvious abnormality was observed in leaves and shoot apices of seedlings from these treatments. However, significant differences in root length were observed after 7 d of incubation. It became obvious that seedlings treated with β-estradiol had shorter roots than those on DMSO plates (Figures 8A and 8B). The inhibition of root growth was strong in all transgenic lines checked, especially in lines RNAi2 and RNAi4, in which the average root length of seedlings with β-estradiol treatment was ∼39 and 50% shorter than that of seedlings treated with DMSO (Figure 8C). There was no obvious difference in root length between wild-type and pER8-GFP (Zuo et al., 2000) transgenic plants treated with either DMSO or β-estradiol (Figure 8C). These data indicate that the inhibition of root growth is attributable to the induction of the SWA1 RNAi, which suggests that downregulation of SWA1 significantly inhibits root growth.
Figure 8.
Root Phenotype of SWA1 RNAi Transgenic Plants.
Wild type, pER8-GFP, and RNAi transgenic seedlings were treated with DMSO or β-estradiol as described in Methods.
(A) Micrograph showing seedlings of SWA1-RNAi seedlings (line RNAi4) grown on MS medium supplemented with 10 μM β-estradiol. Note that the roots are shorter than those shown in (B).
(B) Seedlings of line RNAi4 as in (A) grown on MS medium with DMSO as a control.
(C) Statistical comparison of root length for seedlings grown on MS with DMSO control and β-estradiol inducer.
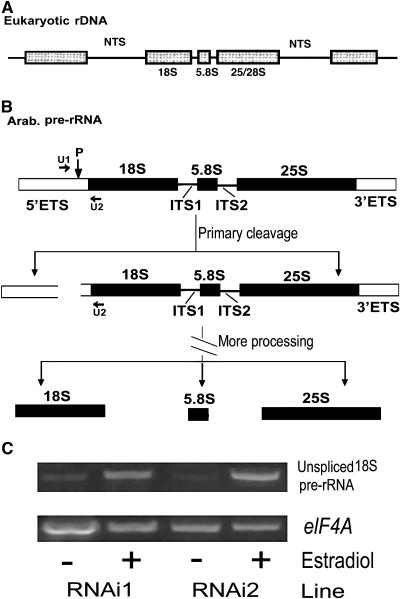
It was reported that UTP15, the homolog of SWA1 from budding yeast, is one of the components of U3 snoRNP and contributes to the processing of 18S pre-rRNA (Dragon et al., 2002). One of the earliest processing events in the pre-rRNA is an endonucleolytic cut in the 5′ external transcript spacer from the 18S rRNA, and this primary pre-RNA cleavage is conserved in all eukaryotes (Venema and Tollervey, 1999). Site P, which is at position +1275 in Arabidopsis 18S pre-RNA, is the primary cleavage site (Figures 9A and 9B), and the U3 snoRNP is essential for this cleavage (Sáez-Vasquez et al., 2004). To investigate whether SWA1 protein is involved in this step of cleavage, we checked the processing of 18S pre-rRNA in the callus derived from roots of RNAi1 and RNAi2 transgenic plants. Total RNA was isolated from callus treated with DMSO (used as a control) or β-estradiol, and DNA contamination was removed with DNase I digestion. RT-PCR was performed with primer U1 (positions +1218 to +1237) and primer U2 (positions +2020 to +2039). The result showed that the amount of unprocessed 18S pre-rRNA increased significantly in the callus induced with β-estradiol compared with the RNAi callus cultured with DMSO (Figure 9C). These data indicate that the knockout of SWA1 expression inhibited the processing of 18S pre-rRNA, suggesting that SWA1, like its yeast homolog, participates in 18S rRNA biogenesis in plants.
Figure 9.
18S Pre-rRNA Processing in RNAi Transgenic Callus.
(A) Structure of eukaryotic rDNA (adapted from Brown and Shaw, 1998). rRNA genes are arranged in tandem, separated by nontranscribed spacers (NTS). 18S, 5.8S, and 25/28S rRNAs are transcribed into pre-rRNA as a unit.
(B) Arabidopsis pre-rRNA transcript contains the 18S, 5.8S, and 25S rRNAs as well as the 5′ and 3′ external transcript spacer (ETS) and two internal transcript spacers between the three rRNAs (ITS1 and ITS2). The positions of primers U1 and U2 used in RT-PCR detection of 18S pre-rRNA in RNAi callus are shown. Site P shows the primary pre-rRNA cleavage site in the 5′ ETS of Arabidopsis. Mature 18S, 5.8S, and 25S rRNAs are generated after additional processing steps that are not described here.
(C) Detection of 18S pre-rRNA in RNAi callus. Top gel, RT-PCR result showing increased amounts of 18S pre-rRNA in RNAi1 and RNAi2 callus induced with β-estradiol compared with DMSO treatment. The data indicate that the knockout of SWA1 expression with RNAi technology leads to the accumulation of unprocessed 18S pre-rRNA. Bottom gel, RT-PCR product of the eIF4A gene, showing equal amounts of starting RNA template.
DISCUSSION
SWA1 Is Essential for the Normal Progression of Mitotic Division Cycles during Female Gametophyte Development in Arabidopsis
The swa1 mutation was first identified in a screen for gametophytic mutants that showed distorted Mendelian segregation. Plants heterozygous for the swa1 mutation had shorter siliques and showed a semisterile phenotype. Outcross experiments further demonstrated that the mutation mainly impaired female gametogenesis and caused a minor defect during male gametogenesis in Arabidopsis. Interestingly, detailed CLSM analysis showed that ovules carrying the Ds insertion were arrested at a variety of developmental stages of two- to eight-nucleate embryo sacs. This indicated that the progression of gametophytic division cycles was affected. A similar phenotype was reported in the hdd mutant in Arabidopsis (Moore et al., 1998). In addition to cell cycle arrest, cellularization of the hdd embryo sacs occurred prematurely after one or two nuclear divisions, and asynchronous nuclear divisions at each pole also occurred in some mutant embryo sacs (Grossniklaus and Schneitz, 1998). In swa1 embryo sacs, neither premature cellularization nor asynchronous nuclear division within the embryo sac occurred, indicating that it is a different mutation from hdd. Clearly, our CLSM analysis showed that the progression of mitotic division cycles during the female gametophyte development was slowed as a result of the swa1 mutation. As demonstrated in the delayed pollination experiment, some of the mutant embryo sacs are able to develop into functional female gametophytes, although at a low frequency. All of these data suggest that the SWA1 gene is essential for the normal progression of the mitotic division cycles during female gametophyte development.
On the other hand, we observed no defects in mitotic cycles during microsporogenesis. Pollen grains were morphologically normal, although the penetration rate of the mutation through the male gametophyte was affected slightly. One possible explanation for this finding is that there are only two divisions in male gametogenesis compared with three required for megagametophyte development. Perhaps residual SWA1 activity from heterozygous pollen mother cells may be sufficient for two divisions. Second, gametophytic cells undergo tremendous growth (at least in size), and the vacuole enlarges during each cycle during female gametogenesis (Figure 2), whereas no or little growth takes place during microsporogenesis. Therefore, a slight change in cell metabolism may not be detrimental to microspore development compared with that in megagametophyte development. Alternatively, there could be genetic redundancy that compensates for the loss of SWA1 activity in microspores.
The SWA1 gene also seems to play a role in the mitotic progression of sporophytic cells, because it is expressed ubiquitously throughout plant development. Strong expression was found in tissues such as root tips and anthers in which active cell division takes place. To investigate its role in these tissues, we used the inducible RNAi technology to downregulate the expression of SWA1. Root growth was inhibited significantly when transgenic seedlings were grown in the presence of the β-estradiol inducer compared with the control sets or treatment. These data indicated that SWA1 plays a role in sporophytic cells as well.
SWA1 Is Involved in the Nucleolar Processing of the 18S Pre-rRNA
Molecular analysis revealed that SWA1 encodes a protein with six WD40 repeats. Proteins containing WD40 repeats play regulatory roles in diverse cellular processes, such as signal transduction, chromatin remodeling, chromosome condensation, cell cycle regulation, transcriptional repression, vesicle trafficking, and RNA processing (Neer et al., 1994; Smith et al., 1999; Vodermaier, 2001; van Nocker and Ludwig, 2003). In SWA1, there are six WD40 repeats instead of the seven in Gβ (Sondek et al., 1996). A three-dimensional remodeling of SWA1 indicates that it forms the typical structure of the WD40 protein and a gross configuration resembling that of the Gβ subunit (data not shown). Both hydrophobic and hydrophilic regions, which are believed to interact with other proteins, are found at the surface of the platform structure. The closest homolog of SWA1 with known function is the yeast UTP15, a component of a large nucleolar U3 complex required for RNA biogenesis (Dragon et al., 2002). Given its similarity in amino acid sequence and nucleolar localization, it is possible that SWA1 plays a role in RNA biogenesis similar to that of UTP15 in yeast. Indeed, our RNAi experiments showed that the processing of the 18S pre-rRNA was indeed inhibited when SWA1 was downregulated. These data indicate that SWA1, like its yeast homolog UTP15, participates in 18S rRNA biogenesis in plants.
In conclusion, our data show that SWA1 is essential for the normal progression of the mitotic cycles during female gametogenesis in Arabidopsis. This effect is most likely caused by the slowdown of general cell metabolism attributable to a defect in 18S rRNA processing.
METHODS
Plant Material and Growth Conditions
Arabidopsis thaliana ecotype Landsberg erecta plants were grown in an air-conditioned room at 22°C under a 16-h-light/8-h-dark cycle. The genetic screen of Ds insertion lines was performed as described previously by Sundaresan et al. (1995). Seeds were sterilized with 20% bleach for 5 min, rinsed five times with sterile water, and germinated on MS plates with or without antibiotics. For the kanamycin plate, 50 mg/L kanamycin (Sigma-Aldrich, St. Louis, MO) was supplemented; for the hygromycin plate, 20 mg/L hygromycin (Roche, Indianapolis, IN) was added. Seeds were stratified in darkness at 4°C for 2 d before being transferred to a greenhouse. Transgenic plants were obtained through Agrobacterium tumefaciens–mediated infiltration (Bechtold and Pelletier, 1998). For β-estradiol treatment, seeds of RNAi transgenic plants were germinated on MS plates supplemented with 20 μg/mL hygromycin and incubated at 22°C with light for 7 d, and the resistant seedlings were transferred to MS plates supplemented with 10 μM β-estradiol (20 mM stock solution in DMSO) or 500 μL of DMSO per liter of MS medium (Sigma-Aldrich). The length of the main root was measured after 7 d.
For callus induction, seeds of lines RNAi1 and RNAi2 were germinated on MS plates supplemented with 20 μg/mL hygromycin and incubated at 22°C with light for 7 d. Roots of hygromycin-resistant plants were cut into 0.5-cm segments and transferred onto callus induction medium containing B5 medium with 0.5 mg/L 2,4-D, 0.05 mg/L kinetin, 30 g/L glucose, and 9 g/L agar. The explants were subcultured every 2 weeks at 22°C with a 16-h light/8-h dark cycle. Callus was collected 2 months later and transferred into flasks with liquid callus medium plus 10 μM β-estradiol (20 mM stock solution in DMSO) or 500 μL of DMSO per liter and incubated at 22°C in the dark for 4 d. Then, the medium was discarded and callus was paper dried before being frozen with liquid N2.
Scanning Electron Microscopy
Pollen grains from dehisced anthers or dissected siliques were stuck onto double-sided tape and immediately frozen in liquid N2 for 3 min before observation with scanning electron microscopy. The frozen samples were observed with a JSM-5310LV scanning electron microscope (JEOL, Tokyo, Japan). Images obtained were edited with Photoshop version 7.0 software (Adobe, San Jose, CA).
CLSM
The confocal observation of ovules was performed according to the method described by Christensen et al. (1997) with slight modifications. Inflorescences were harvested and fixed in 4% glutaraldehyde (in 12.5 mM cacodylate, pH 6.9), and a vacuum was applied for the initial 20 min, after which they were in fixative overnight at room temperature. After fixation, the tissue was dehydrated through a conventional ethanol series with 20 min per step. After the dehydration, the tissue was cleared in 2:1 (v/v) benzyl benzoate:benzyl alcohol for a minimum of 1 h. Pistils or siliques were dissected, mounted with immersion oil, and observed with a Zeiss LSM510 META laser scanning microscope (Zeiss, Jena, Germany) with a 488-nm argon laser and an LP 530 filter. In the study on SWA1 localization, root tips of seedlings were stained with 5 μg/mL 4′,6-diamidino-2′-phenylindole dihydrochloride (DAPI; Roche) solution for 10 min and rinsed briefly with distilled water before observation. The 488-nm argon laser and 507- to 550-nm filter were set for GFP, and the 405-nm diode laser and the band-pass 420- to 480-nm filter were set for DAPI scanning. Images were edited with Zeiss LSM Image Browser software and Photoshop version 7.0 software.
Crosses and Delayed Pollination Test
Wild-type and swa1/+ plants were emasculated at floral stage 12c (Smyth et al., 1990; Christensen et al., 1997) and pollinated at stage 13 (∼12 h after emasculation) in reciprocal crosses. Anthers of heterozygous swa1 plants were removed at stage 12c and pollinated with wild-type pollen after 24 h (stage 14), 48 h (stage 15), 72 h (stage 17), or 96 h (stage 17) in the delayed pollination test. Special care was taken to prevent cross-pollination from unwanted pollen. Seeds of different groups were collected and plated onto MS plates supplemented with 50 μg/mL kanamycin. The seedlings were scored for kanamycin resistance after germination.
Molecular Cloning and Construction
Cloning of SWA1
Genomic DNA was isolated from the swa1 mutant with the plant DNeasy plant mini kit (Qiagen, Valencia, CA) and used as template for the cloning of Ds flanking sequence and SWA1. Thermal asymmetric interlaced PCR was performed as described previously (Liu et al., 1995; Grossniklaus et al., 1998; Yang et al., 1999). Primers 5′-CCAGCCTCCTTCTCTCTCTCTGTT-3′ and 5′-ATCCATCATTTCAAGTTTAGACAT-3′ were designed according to EST sequences available in GenBank (http://www.arabidopsis.org) and used for SWA1 cDNA amplification. Full-length SWA1 cDNA was copied with the One-Step RT-PCR kit (Qiagen) and cloned into pGEM-T vector (Promega, Madison, WI). Fragments were confirmed by sequencing.
RT-PCR Detection of SWA1 Expression
Total RNA was isolated with TRIzol reagent (Invitrogen, Carlsbad, CA) from different tissues of Landsberg erecta wild-type plants. Roots from 10-d-old seedlings were collected. Leaves, stems, inflorescences, and siliques were obtained separately from flowering mature plants. Poly(A)+ was purified from total RNA with the Oligotex mRNA midi kit (Qiagen). The concentration of mRNAs was determined by spectrophotometry. Ten nanograms of mRNA from different tissues was used as template for RT-PCR amplification with the One-Step RT-PCR kit (Qiagen) according to the manufacturer's recommendations. The full-length cDNA of SWA1 was obtained through RT-PCR amplification with the primer combination 5′-CCAGCCTCCTTCTCTCTCTCTGTT-3′ and 5′-ATCCATCATTTCAAGTTTAGACAT-3′ designed according to EST sequences available in GenBank. Similarly, the primer combination 5′-ATGGCAGGACCGCACCGGA-3′ and 5′-GCATGTCAAAAACACGACCGGGAGTTCC-3′ was used for RT-PCR amplification of the EUKARYOTIC TRANSLATION INITIATION FACTOR 4A (eIF4A) gene as an internal control. PCR products were analyzed with 1% agarose gel electrophoresis.
Construction of SWA1 Genomic Clone for Genetic Complementation
A 4.5-kb SWA1 genomic fragment (starting from 1701 bp upstream of the ATG start codon and ending at 1206 bp downstream of the TGA stop codon) was amplified with the ACCu Taq LA DNA polymerase PCR kit (Sigma-Aldrich) and primers 5′-AACTGCAGCCTTGTTTCACGAGTCTGCA-3′ and 5′-GCGAATTCTTGGTATGCATGGACCTGGT-3′. The fragment obtained was subcloned into pCAMBIA1300 (CAMBIA; www.cambia.org.au) at PstI and EcoRI sites, and constructs were verified by sequencing.
Construction of the SWA1-GFP Fusion Gene
To obtain the SWA1-GFP fusion gene, the coding region of SWA1 was amplified with primers 5′-GGACTAGTATGGAGGAAGAGCTTCGTGTTCG-3′ and 5′-CGCGGATCCACTTCTACCCGCAATTCTCA-3′ and inserted into XbaI and BamHI sites of pBI-GFP vector (D.-Q. Shi, unpublished data; the GUS gene of pBI121 was replaced with GFP) to give rise to pBI-35S:SWA1-GFP. Similarly, the coding region was also subcloned into XbaI and BamHI sites of p1300-Pswa1:GFP:Ter (D.-Q. Shi, unpublished data) to yield p1300-Pswa1:SWA1-GFP:Ter.
Construction of the SWA1 Promoter-GUS Reporter
A 1701-bp fragment upstream of the ATG start codon was amplified by PCR with a 5′ primer containing a PstI site (5′-AACTGCAGCCTTGTTTCACGAGTCTGCA-3′) and a 3′ primer with a SalI site (5′-GACGTCGACTATTCTAGAGTACAAGCAGA-3′). Similarly, a 1206-bp fragment downstream of the TGA stop codon was also obtained with primers with SacI and EcoRI, respectively: 5′-CCCGAGCTCTAATCAATATCACAAGTTTTG-3′ and 5′-GCGAATTCTTGGTATGCATGGACCTGGT-3′. PCR fragments of the expected size were sequenced and subcloned into pCAMBIA1300, giving rise to p1300-SWA1PT. A 1.8-kb GUS gene was released with SacI and EcoRI digest from vector pBI121 and inserted into p1300-SWA1PT backbone, giving rise to p1300-PGT.
Construction of the pER8-SWA1-RNAi Vector
The SWA1 coding region was amplified by PCR with primer combination 5′-ATGGAGGAAGAGCTTCGTGTTCG-3′ and 5′-CGGGATCCTCAACTTCTACCCGCAATTC-3′ (containing a BamHI site). The fragment was sequenced and subcloned into the SmaI site of pBluescript SK+ (Stratagene, La Jolla, CA) vector to give rise to pBS-SWA1. The 3′ part of the coding region was replaced with the 5′ part of 516 bp of the coding sequence in the opposite direction by making use of an internal BglII site and a BamHI in the vector, to yield pBS-SWA1-1AWS vector. The second intron (+327 to +740 bp downstream of the ATG) of Adx1 (At4g05450) was obtained by PCR with primers containing the BamHI site and inserted into pBS-SWA1-1AWS, resulting in pBS-SWA1-RNAi. Finally, the complete fragment was cut out with SpeI and ApaI and inserted into pER8 (Zuo et al., 2000) to produce pER8-SWA1-RNAi.
RT-PCR Detection of 18S Pre-rRNA in RNAi Callus
Total RNA was isolated from RNAi callus with TRIzol reagent (Invitrogen) according to the user's manual. RNA was treated with DNase I (TaKaRa Biotechnology, Dalian, China) to remove potential genomic DNA. Reverse transcription was conducted with reverse transcriptase XL (Avian myeloblastosis virus; TaKaRa) according to the manufacturer's instructions. The cDNA was used as template for PCR amplification. Primer U1 (5′-CGTAACGAAGATGTTCTTGGC-3′; +1119 to +1139 bp in 18S pre-rRNA) and primer U2 (5′-ATGCGTCCCTTCCATAAGTC-3′; +2040 to +2021 bp in 18S pre-rRNA) were designed to check the processing of 18S pre-rRNA at site P (Sáez-Vasquez et al., 2004). Amplification of the unprocessed 18S pre-rRNA with these primers yields a 0.8-kb fragment.
In Situ Hybridization
Floral buds and pistils were fixed and processed according to Yang et al. (1999). Eight-micrometer-thick sections were hybridized with digoxigenin-labeled antisense and sense probes. For probe labeling, antisense and sense RNAs were transcribed in vitro with T3 and T7 RNA polymerases, respectively, using the digoxigenin RNA labeling kit (Roche). Half of the labeled product was hydrolyzed to ∼150 bp with alkaline treatment. A mixture of hydrolyzed and nonhydrolyzed RNA probe was used for the hybridization. Hybridized slides were rinsed twice with 2× SSC (1× SSC is 0.15 M NaCl and 0.015 M sodium citrate) at room temperature for 15 min, then with 1× SSC twice at 60°C for 10 min each. Antibody staining and coloring were performed according to the manufacturer's recommendations (Roche). The slides were observed with an Axioskop 2 Plus microscope (Zeiss) with differential interference contrast optics and photographed with a Nikon Coolpix 995 digital camera (Nikon, Tokyo, Japan).
Bioinformatic Methods
BLASTN, BLASTX, and tBLASTN analyses of EST, cDNA, and genomic sequences were conducted with the National Center for Biotechnology Information web service (http://www.ncbi.nih.nlm.gov), using the default E-value cutoff of 1.0 and the default algorithm setting BLOSUM62 as described previously by Altschul et al. (1997). Sequence alignments and protein statistics were determined using DNASTAR software (DNASTAR, Madison, WI).
For the sequence alignment, a homolog from poplar (Populus trichocarpa) was obtained with tBLASTN from PoplarDB (http://poppel.fysbot.umu.se/blast.php); other homologous sequences from different organisms were obtained from the National Center for Biotechnology Information with BLASTP. Multiple sequence alignments were performed with the ClustalW service of NPS@ClustalW multiple alignment web site (http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_clustalw.html). The resulting alignments were used to generate a neighbor-joining tree using Mega 2.1 software (Kumar et al., 2001). For the options, we selected p-distance for the amino acid model, complete deletion for include site, and 500 replacement in bootstrap test of phylogeny.
Acknowledgments
We thank Nam-Hai Chua (The Rockefeller University, New York, NY) for the XVE inducible system and Yang-Sun Chan and Qing-Wen Lin (Temasek Life Sciences Laboratory, Singapore) for technical support in electron microscopy. We also thank Megan Griffith (Institute of Molecular and Cell Biology, Singapore) for critical reading of the manuscript. This work was supported by grants from Temasek Holdings to D.-Q.S. and from the Chinese National Science Foundation (30330310) and the BAI REN JI HUA of the Chinese Academy of Sciences to W.-C.Y.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Wei-Cai Yang (wcyang@genetics.ac.cn).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.033563.
References
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold, N., and Pelletier, G. (1998). In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol. Biol. 82 259–266. [DOI] [PubMed] [Google Scholar]
- Brown, J.W., and Shaw, P.J. (1998). Small nucleolar RNAs and pre-rRNA processing in plants. Plant Cell 10 649–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capron, A., Serralbo, O., Fulop, K., Frugier, F., Parmentier, Y., Dong, A., Lecureuil, A., Guerche, P., Kondorosi, E., Scheres, B., and Genschik, P. (2003). The Arabidopsis anaphase-promoting complex or cyclosome: Molecular and genetic characterization of the APC2 subunit. Plant Cell 15 2370–2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen, C.A., King, E.J., Jordan, J.R., and Drews, G.N. (1997). Megagametogenesis in Arabidopsis wild type and the Gf mutant. Sex. Plant Reprod. 10 49–64. [Google Scholar]
- Christensen, C.A., Subramanian, S., and Drews, G.N. (1998). Identification of gametophytic mutations affecting female gametophyte development in Arabidopsis. Dev. Biol. 202 136–151. [DOI] [PubMed] [Google Scholar]
- Dragon, F., et al. (2002). A larger nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature 417 967–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drews, G.N., Lee, D., and Christensen, C.A. (1998). Genetic analysis of female gametophyte development and function. Plant Cell 10 5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drews, G.N., and Yadegari, R. (2002). Development and function of the angiosperm female gametophyte. Annu. Rev. Genet. 36 99–124. [DOI] [PubMed] [Google Scholar]
- Ebel, C., Mariconti, L., and Gruissem, W. (2004). Plant retinoblastoma homologues control nuclear proliferation in the female gametophyte. Nature 429 776–780. [DOI] [PubMed] [Google Scholar]
- Feldmann, K.A., Coury, D.A., and Christianson, M.L. (1997). Exceptional segregation of a selectable marker (KanR) in Arabidopsis identifies genes important for gametophytic growth and development. Genetics 147 1411–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grini, P.E., Schnittger, A., Schwarz, H., Zimmermann, I., Schwab, B., Jürgens, G., and Hülskamp, M. (1999). Isolation of ethyl methanesulfonate-induced gametophytic mutants in Arabidopsis thaliana by a segregation distortion assay using the multimarker chromosome 1. Genetics 151 849–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossniklaus, U., and Schneitz, K. (1998). The molecular and genetic basis of ovule and megagametophyte development. Semin. Cell Dev. Biol. 9 227–238. [DOI] [PubMed] [Google Scholar]
- Grossniklaus, U., Vielle-Calzada, J.-P., Hoeppner, M.A., and Gagliano, W.B. (1998). Maternal control of embryogenesis by MEDEA, a polycomb group gene in Arabidopsis. Science 280 446–450. [DOI] [PubMed] [Google Scholar]
- Howden, R., Park, S.K., Moore, J.M., Orme, J., Grossniklaus, U., and Twell, D. (1998). Selection of T-DNA-tagged male and female gametophytic mutants by segregation distortion in Arabidopsis. Genetics 149 621–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, B.-Q., and Sheridan, W.F. (1996). Embryo sac development in the maize indeterminate gametophyte1 mutant: Abnormal nuclear behavior and defective microtubule organization. Plant Cell 6 1391–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johri, B.M. (1984). Embryology of Angiosperms. (Berlin: Springer-Verlag).
- Kermicle, J.L. (1971). Pleiotropic effects on seed development of the indeterminate gametophyte gene in maize. Am. J. Bot. 58 1–7. [Google Scholar]
- Kumar, S., Koichiro, T., Ingrid, B.J., and Masatoshi, N. (2001). MEGA2: Molecular Evolutionary Genetics Analysis Software. (Tempe: Arizona State University).
- Kwee, H.-S., and Sundaresan, V. (2003). The NOMEGA gene required for female gametophyte development encodes the putative APC6/CDC16 component of the anaphase promoting complex in Arabidopsis. Plant J. 36 853–866. [DOI] [PubMed] [Google Scholar]
- Liu, Y.-G., Mitsukawa, N., Oosumi, T., and Whittier, R.F. (1995). Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J. 8 457–463. [DOI] [PubMed] [Google Scholar]
- Mansfield, S.G., Briarty, L.G., and Erni, S. (1991). Early embryogenesis in Arabidopsis thaliana. I. The mature embryo sac. Can. J. Bot. 69 447–460. [Google Scholar]
- Misra, R.C. (1962). Contribution to the embryology of Arabidopsis thaliana (Gay and Monn.). Agra. Univ. J. Res. Sci. 11 191–199. [Google Scholar]
- Moore, J.M., Calzada, J.-P.V., Gagliano, W., and Grossniklaus, U. (1997). Genetic characterization of hadad, a mutant disrupting female gametogenesis in Arabidopsis thaliana. Cold Spring Harbor Symp. Quant. Biol. 62 35–47. [PubMed] [Google Scholar]
- Neer, E.J., Schmidt, C.J., Nambudripad, R., and Smith, T.F. (1994). The ancient regulatory-protein family of WD-repeat proteins. Nature 371 297–300. [DOI] [PubMed] [Google Scholar]
- Nelson, O.E., and Clary, G.B. (1952). Genetic control of semi-sterility in maize. J. Hered. 43 205–210. [Google Scholar]
- Redei, G.P. (1965). Non-mendelian megagametogenesis in Arabidopsis. Genetics 51 857–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sáez-Vasquez, J., Caparros-Ruiz, D., Barneche, F., and Echeverria, M. (2004). A plant snoRNP complex containing snoRNAs, fibrillarin, and nucleolin-like proteins is competent for both rRNA gene binding and pre-rRNA processing in vitro. Mol. Cell. Biol. 24 7284–7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders, P.M., Bui, A.Q., Weterings, K., McIntire, K.N., Hsu, Y.-C., Lee, P.Y., Truong, M.T., Beals, T.P., and Goldberg, R.B. (1999). Anther developmental defects in Arabidopsis thaliana male-specific mutants. Sex. Plant Reprod. 11 297–322. [Google Scholar]
- Schneitz, K., Hülskamp, M., and Pruitt, R.E. (1995). Wild-type ovule development in Arabidopsis thaliana: A light microscope study of cleared whole-mount tissue. Plant J. 7 731–749. [Google Scholar]
- Smith, T.F., Gaitatzes, C., Saxena, K., and Neer, E.L. (1999). The WD repeats: A common architecture for diverse functions. Trends Biochem. Sci. 24 181–185. [DOI] [PubMed] [Google Scholar]
- Smyth, D.R., Bowman, J.L., and Meyerowitz, E.M. (1990). Early flower development in Arabidopsis. Plant Cell 2 755–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sondek, J., Bohm, A., Lambright, D.G., Hamm, H.E., and Sigler, P.B. (1996). Crystal structure of a G-protein beta gamma diner at 2.1Å resolution. Nature 379 369–374. [DOI] [PubMed] [Google Scholar]
- Springer, P.S., Holding, D.R., Groover, A., Yordan, C., and Martienssen, R.A. (2000). The essential MCM7 protein PROLIFERA is localized to the nucleus of dividing cells during the G(1) phase and is required maternally for early Arabidopsis development. Development 127 1815–1822. [DOI] [PubMed] [Google Scholar]
- Springer, P.S., McCombie, W.R., Sundaresan, V., and Martienssen, R.A. (1995). Gene trap tagging of PROLIFERA, an essential MCM2-3-5-like gene in Arabidopsis. Science 268 877–880. [DOI] [PubMed] [Google Scholar]
- Sundaresan, V., Springer, P.S., Volpe, T., Haward, S., Jones, J.D.G., Dean, C., Ma, H., and Martienssen, R.A. (1995). Patterns of gene action in plant development revealed by enhancer trap and gene trap transposable elements. Genes Dev. 9 1797–1810. [DOI] [PubMed] [Google Scholar]
- van Nocker, S., and Ludwig, P. (2003). The WD40-repeat protein superfamily in Arabidopsis: Conservation and divergence in structure and function. BMC Genomics 4 50–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venema, J., and Tollervey, D. (1999). Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet. 33 261–331. [DOI] [PubMed] [Google Scholar]
- Vodermaier, H.C. (2001). Cell cycle: Waiters serving the Destruction machinery. Curr. Biol. 11 834–837. [DOI] [PubMed] [Google Scholar]
- Webb, M.C., and Gunning, B.E.S. (1994). Embryo sac development in Arabidopsis thaliana. II. The cytoskeleton during megagametogenesis. Sex. Plant Reprod. 7 153–163. [Google Scholar]
- Yang, W.-C., and Sundaresan, V. (2000). Genetics of gametophyte biogenesis in Arabidopsis. Curr. Opin. Plant Biol. 3 53–57. [DOI] [PubMed] [Google Scholar]
- Yang, W.-C., Ye, D., Xu, J., and Sundaresan, V. (1999). The SPOROCYTELESS gene of Arabidopsis is required for sporogenesis and encodes a novel protein. Genes Dev. 13 2108–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo, J., Niu, Q., and Chua, N.-H. (2000). An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in plants. Plant J. 24 265–273. [DOI] [PubMed] [Google Scholar]