Abstract
Rapid pollen tube growth places unique demands on energy production and biosynthetic capacity. The aim of this work is to understand how primary metabolism meets the demands of such rapid growth. Aerobically grown pollen produce ethanol in large quantities. The ethanolic fermentation pathway consists of two committed enzymes: pyruvate decarboxylase (PDC) and alcohol dehydrogenase (ADH). Because adh mutations do not affect male gametophyte function, the obvious question is why pollen synthesize an abundant enzyme if they could do just as well without. Using transposon tagging in Petunia hybrida, we isolated a null mutant in pollen-specific Pdc2. Growth of the mutant pollen tubes through the style is reduced, and the mutant allele shows reduced transmission through the male, when in competition with wild-type pollen. We propose that not ADH but rather PDC is the critical enzyme in a novel, pollen-specific pathway. This pathway serves to bypass pyruvate dehydrogenase enzymes and thereby maintain biosynthetic capacity and energy production under the unique conditions prevailing during pollen–pistil interaction.
INTRODUCTION
Fertilization in flowering plants relies on the elongation of the pollen tubes through the style toward the ovary. Often, the total number of pollen grains on a stigma exceeds the number of grains necessary to fertilize all the ovules, leading to competition between growing pollen tubes in the style (Howden et al., 1998). Consequently, rapid elongation of the pollen tube is essential for male reproductive success, and the pollen tube is indeed the fastest growing plant cell known. Pollen tube growth rate can reach 1 cm/h in maize (Zea mays), and pollen respires 10 times faster than vegetative tissue (Dickinson, 1965; Tadege and Kuhlemeier, 1997; Taylor and Hepler, 1997). Pollen development in the anther and the growth of the pollen tube are highly energy-demanding processes, and pollen contains ∼20 times more mitochondria per cell than normal vegetative tissues in maize (Lee and Warmke, 1979). The key mitochondrial enzyme pyruvate dehydrogenase (PDH), which catalyzes the conversion of pyruvate to acetyl-CoA, was shown to be present at high levels in pollen (Thelen et al., 1999).
Among other mechanisms, oxygen has been proposed as a possible cue for pollen tube guidance. Blasiak et al. (2001) showed a clear tropic response in pollen tubes of several species to an oxygen gradient in a colloidal medium. Indeed, the existence of an oxygen gradient in the unpollinated style was shown in Hipeastrum hybridum. Oxygen pressure is high in the stigma and style but suddenly decreases at the base of the style, approaching zero values in the ovary. Moreover, pollen tube growth itself creates hypoxic regions within the style (Linskens and Schrauwen, 1966). Thus, energy metabolism may influence pollen tube growth both directly and, possibly, indirectly through the generation of guidance signals.
Fermentation serves to regenerate NAD+ from NADH, a function that is of vital importance under low oxygen conditions, when oxidative phosphorylation is impaired. Plants conduct ethanolic fermentation, an ancient metabolic pathway that involves two dedicated enzymes. Pyruvate decarboxylase (PDC) is responsible for the irreversible conversion of pyruvate into acetaldehyde and CO2. Alcohol dehydrogenase (ADH) subsequently converts acetaldehyde into ethanol, with the concomitant regeneration of NAD+ (Figure 1). These two enzymes are present at low levels in normoxic tissues and are strongly induced under anoxia (Laszlo and Lawrence, 1983). Their levels may be higher in tissues, such as seeds and root tips, that experience reduced oxygen availability during normal development.
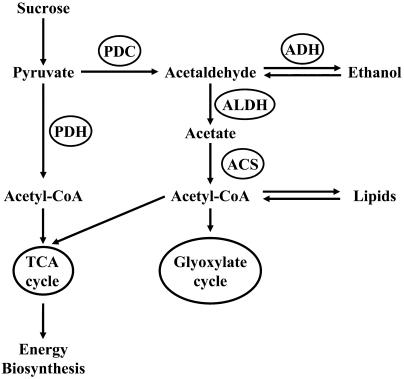
Figure 1.
Proposed Model for the Use of Pyruvate in Pollen.
Pyruvate can be directly converted to acetyl-CoA by PDH and enter the tricarboxylic acid (TCA) cycle, or it can be converted to acetaldehyde by PDC. Acetaldehyde can be reduced to ethanol by ADH, or it can be converted into acetate by the action of ALDH. Acetate can be used by ACS to produce additional acetyl-CoA. This acetyl-CoA can enter the TCA cycle or the glyoxylate cycle or be used in lipid biosynthesis.
PDC and ADH are major proteins in pollen, suggesting that the male gametophyte performs ethanolic fermentation. Indeed, we previously showed that tobacco (Nicotiana tabacum) pollen grains produce copious amounts of ethanol when germinating in a medium that mimics the stylar environment (Bucher et al., 1995). More than half of the carbohydrate flows through this pathway, emphasizing its metabolic importance. The regulation of this pathway in pollen is unique. In contrast with vegetative tissues such as roots, in pollen ethanol is produced concomitantly with an extremely high respiratory rate, and the flux to ethanol is not regulated by oxygen but rather by sugar availability (Bucher et al., 1995; Tadege and Kuhlemeier, 1997; Tadege et al., 1998).
If ethanolic fermentation is a pathway of major quantitative importance in pollen, one would expect the inactivation of ADH to interfere with pollen function. The available data, however, suggested that this is not the case and that plants mutated in ADH are not impaired in pollen development or pollen tube growth. Adh+/adh− heterozygotes segregate in normal Mendelian ratios (Freeling and Bennett, 1985). Thus, there is a major discrepancy between genetic and physiological data. There are several ways this apparent paradox might be resolved. One is that the pathway is important only during in vitro pollen tube growth and that indeed the enzymes are without in vivo function. A second is that the adh mutants are not nulls. In fact, using sensitive gas chromatography, we were able to detect ethanol in pollen of maize, tomato (Lycopersicon esculentum), and N. plumbaginifolia adh mutants (T. Mandel and C. Kuhlemeier, unpublished data). A third possibility is that the pathway may serve a function distinct from the regeneration of NAD+. We have previously presented evidence for the existence of a complete, pollen-specific metabolic pathway, starting from pyruvate (Figure 1). In this PDH bypass, pyruvate is first converted to acetaldehyde, then oxidized to acetate, and finally converted to acetyl-CoA by the enzymes PDC, aldehyde dehydrogenase (ALDH), and acetyl-CoA synthase (ACS) (Figure 1). High levels of ALDH and ACS mRNA (op den Camp and Kuhlemeier, 1997) are present in pollen, and experiments done by Mellema et al. (2002) demonstrated that the PDH bypass supports energy production and lipid biosynthesis in tobacco pollen.
To clarify the role of the fermentation pathway in pollen and to resolve the above-mentioned paradox, we decided to screen for mutants in the fermentation pathway and/or in the PDH bypass. We chose the Solanaceous species Petunia hybrida, a close relative of N. tabacum. P. hybrida pollen has been studied extensively and readily germinates in vitro. Choosing P. hybrida as our model system also enabled us to benefit from the dTph1 transposon system available in this species. The petunia line W138 contains >200 copies of the 284-bp dTph1 element (Gerats et al., 1990), and their frequent transposition causes a high incidence of mutation among W138 progeny (van Houwelingen et al., 1998). Insertion of a dTph1 transposon in the gene of interest can be identified by a PCR-based strategy widely applied in P. hybrida (Koes et al., 1995).
In the work described here, we isolated a null mutation in the pollen-specific P. hybrida Pdc2 gene. Analysis of the mutant phenotype indicates that the PDH bypass plays an important role during pollen tube elongation.
RESULTS
Isolation of a pdc2 Mutant
To identify mutants in the fermentation pathway and/or in the PDH bypass, we performed a forward genetic screen of the high transposition line P. hybrida W138. Our strategy was based on the assumption that in vitro pollen germination and tube growth could use either the PDH-dependent pathway or the PDH bypass (Figure 1). If this is so, pollen tubes will continue to grow in the absence of functional PDH, as long as ethanolic fermentation and/or the PDH bypass remain functional. We made use of the specific PDH inhibitor (R)-aminoethylphosphinate (AEP) (Laber and Amrhein, 1987). Whereas germinating seeds are highly sensitive to this inhibitor, pollen readily germinates when exposed to AEP concentrations as high as 90 μM (op den Camp and Kuhlemeier, 1997). Pollen samples of homozygous mutant plants are expected to show reduced tube growth in a medium containing AEP compared with a control culture medium containing no AEP. In heterozygous plants, half of the pollen grains are expected to produce shorter or no tubes on AEP.
In a screen of 2400 P. hybrida W138 plants, we identified five candidates that showed reduced pollen germination or tube growth in a medium containing 60 μM AEP. These five plants did not display any apparent physiological or morphological aberrations under normal greenhouse conditions. After a second screening of these plants, as well as of their offspring, we could confirm a clear and persistent mutant pollen tube growth phenotype for a single candidate plant, named CX13 (Figures 2C and 2D).
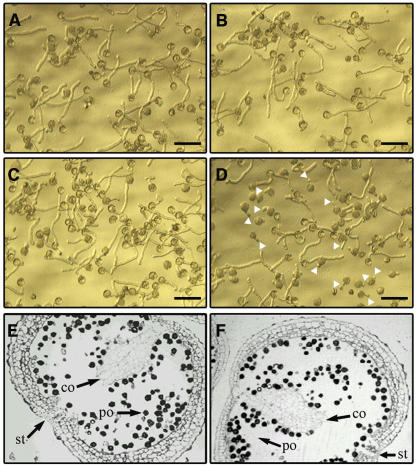
Figure 2.
Light Microscopy Photographs of Pollen Tubes Grown in Vitro with or without AEP and Cross Section of Anthers.
(A) to (D) Wild-type pollen grown in pollen germination medium (A) and in pollen germination medium with 60 μM AEP (B) and CX13 pollen (pdc2/+) grown in pollen germination medium (C) and in pollen germination medium with 60 μM AEP (D). Arrowheads in (D) indicate the nongerminating pollen grains. Heterozygous CX13 pollen germinated at a rate of 85.3 ± 2.1% (se) without AEP and 45.8 ± 1.7% (se) with AEP. The actual germination percentages depended on the batch of pollen, but the effect of AEP was consistent. Bars = 100 μm.
(E) and (F) Cross section of stage 8 anthers according to Koltunow et al. (1990) from pdc2/pdc2 plants (E) and wild-type plants (F). co, connective; st, stomium; po, pollen.
Morphological analysis of stage 8 anthers (Koltunow et al., 1990) was made to check if pollen development had progressed normally in the pdc2 mutant. No obvious morphological differences could be detected between mutant and wild-type anthers. The connective tissues of the anthers were equally degraded, and the stomium was necrotic in both. The anthers also contained similar amounts of pollen that had the same shape and size (Figures 2E and 2F).
Characterization of the Petunia Pdc Gene Family
In parallel experiments, we characterized the P. hybrida Pdc gene family. PDC is the first enzyme and the control step of ethanolic fermentation. The Pdc genes were shown to exist as a small gene family in tobacco (Bucher et al., 1995), and our aim was to isolate a transposon insertion in the Pdc gene by a reverse genetic approach.
The cDNAs of the two petunia Pdc genes were isolated by RT-PCR. A partial Pdc1 cDNA was amplified from mRNA of anoxically treated petunia plants, and the full-length Pdc2 cDNA was amplified from mature pollen mRNA. The Pdc2 cDNA sequence encodes a 589–amino acid protein highly homologous to the tobacco PDC2 protein. A partial sequence of the Pdc2 gene was obtained by PCR on P. hybrida R27 genomic DNA and showed that the Pdc2 coding sequence is composed of six exons and at least five introns (Figure 3A).
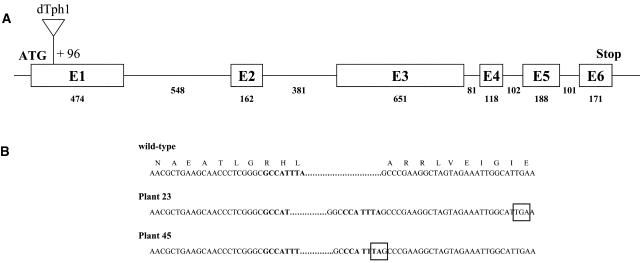
Figure 3.
Structure of the Pdc2 Gene.
(A) Schematic representation of the structure of the Pdc2 gene and position of the dTph1 insertion. Boxes represent the exons, and the lines represent introns. Start and stop codons are indicated. The numbers indicate the length of each exon or intron in nucleotides. The triangle represents the dTph1 insertion.
(B) Pdc2 cDNA and protein sequence in the wild-type plant and at the dTph1 transposon insertion site in plants 23 and 45. Nucleotides in bold represent the ones just before the transposon insertion site. Stop codons are boxed.
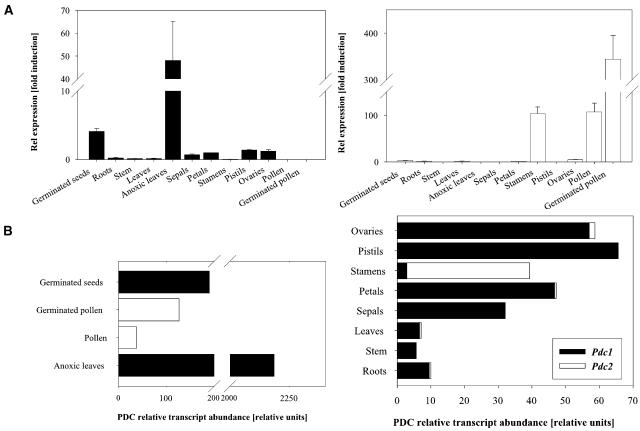
To determine the expression pattern of these two genes, quantitative real-time RT-PCR experiments were performed. RNA from various organs was isolated, and the abundance of the two Pdc genes was quantified based on three independent reverse transcription reactions (Figure 4A). The Pdc1 transcript was detected at low levels in most organs, whereas the expression level was more substantial in germinated seeds and strongly induced in anoxically treated leaves (322-fold induction compared with untreated leaves). No Pdc1 mRNA could be detected in pollen and in germinated pollen. Expression of Pdc2 was detected at high levels in stamens, pollen, and germinated pollen (100- to 340-fold higher than in leaves) (Figure 4A). Low levels of Pdc2 expression were observed in germinated seeds, roots, leaves, petals, and in ovaries. No Pdc2 transcripts could be amplified from stem, leaves treated with anoxia, sepals, and pistils. Comparison of absolute Pdc transcript abundance in each organ showed that Pdc1 was the main transcript in germinated seeds (∼99.5% of total Pdc transcripts), roots, leaves, petals, and ovaries and the only transcript in stem, sepals, pistils, and anoxic leaves (Figure 4B). In stamens, Pdc2 was the major transcript (92.6% of all Pdc transcripts); in pollen and germinated pollen, Pdc2 accounted for 100% of all Pdc transcripts. Pdc2 mRNA was low in roots, leaves, petals, and ovaries, accounting for ∼3% of the total PDC transcript in this organ. These data suggest that Pdc1 operates in anoxic conditions, whereas Pdc2 could have a function in the male gametophyte. The expression pattern of these genes closely resembles that of the Pdc genes in tobacco (Bucher et al., 1995).
Figure 4.
Relative Expression Level and Transcript Abundance of Pdc1 and Pdc2 in Different Organs.
(A) Quantification of mRNA levels was achieved by quantitative real-time PCR (see Methods). Pdc expression levels were normalized with respect to the internal control Actin and are plotted relative to the expression from petals. Data bars represent the mean ± se levels of transcripts from three experiments with independent reverse transcriptions.
(B) Pdc transcript abundance in different organs. Total Pdc mRNA in roots is 10. Contribution from individual genes is represented according to (A).
Screening for Plants with a dTph1 Insertion in the Pdc2 Gene
A PCR-based screening method was used to select for insertion of the dTph1 in Pdc2 using the same petunia collection that was used for our forward genetic screen. Only one heterozygous insertion mutant was detected in a population of 2400 W138 plants. This plant turned out to be the same as the one identified in our forward screen: CX13. The position of the dTph1 element in the gene was determined by cloning and sequencing of the fragments flanking the transposon insertion site. The transposon had inserted in the first exon of the gene, 96 bp downstream of the initiator ATG (Figure 3A). This insertion disrupts the protein coding sequence because the dTph1 transposon contains stop codons in all three reading frames in both orientations. Thus, we assume that CX13 is a pdc2 null.
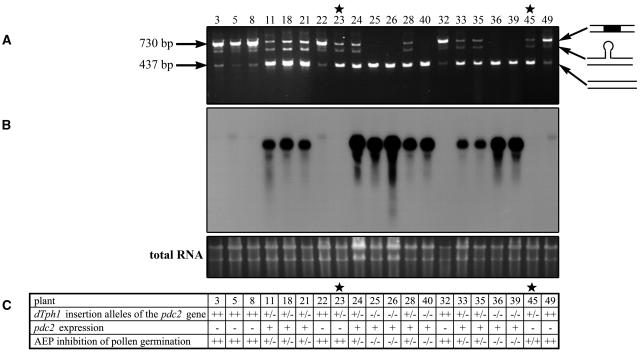
We tested the progeny of CX13 for the presence or absence of the transposon in the F1 progeny. PCR analysis of 20 plants revealed that six were homozygous for the transposon insertion (plants 3, 5, 8, 22, 32, and 49), nine were heterozygous (plants 11, 18, 21, 23, 24, 28, 33, 35, and 45), and five were wild-type (plants 25, 26, 40, 36, and 39) (Figure 5A). Thus, the original CX13 plant was heterozygous for the transposon insertion.
Figure 5.
Cosegregation Analysis for the Phenotype and the Genotype of the Progeny from the Self-Fertilization of CX13.
(A) PCR analysis on genomic DNA of the progeny. Three bands are detected on the gel: the lower one (437 bp) corresponds to the amplification of the wild-type allele of the Pdc2 gene and is ∼300 bp smaller than the higher band corresponding to the amplification of the allele containing the dTph1 insertion (730 bp).
(B) Levels of Pdc2 expression in the pollen of the different progeny plants. The blot was hybridized with a 260-bp specific fragment of the 32P-labeled Pdc2 cDNA. Equal loading was determined by SYBR gold nucleic acid staining of the gel.
(C) Table summarizing the results from the PCR and RNA gel blot analysis and the AEP screen on the progeny. T, the Pdc2 allele carrying the dTph1 insertion; 0, the wild-type Pdc2 allele; + and −, the presence or absence of RNA, respectively; ++, pollen does not germinate on AEP; +/−, approximately half of the pollen germinates on AEP; −/−, the pollen germinates on AEP. Stars indicate the two plants showing abnormal patterns.
Among the amplified fragments corresponding to the wild-type (437 bp) and the mutant allele (730 bp), we detected an additional band of intermediate size. Purification and reamplification of this band resulted in the same three-band pattern, showing that the original intermediate size band was a hybrid between the wild type and mutant allele. A low-intensity band at 437 bp in the homozygous pdc2 plants probably indicates somatic excision of the transposon.
RNA gel blot analysis was performed to determine whether the Pdc2 transcripts were still present in the progeny of the CX13 plant. A 260-bp fragment specific of the Pdc2 cDNA was used to probe total RNA from pollen. A signal was detected in all of the wild-type plants (Figure 5B), but no signal was detected in the plants homozygous for the dTph1 insertion in the Pdc2 gene. The signal detected in the heterozygous plants was weaker, in most of the cases, than in the wild-type plants, indicating that the transcript level of the Pdc2 gene in pollen is probably copy number dependent. The absence of Pdc2 expression in plants 23 and 45, previously identified as heterozygotes, is likely to be caused by imprecise excision of the transposon. The presence of a small remnant of the transposon (footprint) would explain the PCR amplification of a wild-type-size fragment.
To determine the phenotypes of these plants, pollen was germinated on a 60 μM AEP medium (Figure 5C). In this medium, homozygous pdc2 pollen tubes were not able to grow, half of the heterozygous pdc2/+ pollen tubes were still able to grow, and wild-type pollen grains showed a normal tube growth. As expected, except for plants 23 and 45, phenotype and genotype were fully cosegregant. Sequence analysis of the Pdc2 alleles in these two plants confirmed the presence of footprints disrupting the Pdc2 coding sequence. In plant 23, excision of the dTph1 element from the Pdc2 gene caused an addition of 7 bp to the open reading frame in the first exon, creating a stop codon 130 bp downstream of the ATG and abolishing the Pdc2 translation (Figure 3B). This footprint also generated a PCR restriction fragment length polymorphism for NcoI, which is absent in the wild-type Pdc2 allele. In plant 45, the footprint added 8 bp to the open reading frame, creating a stop codon 103 bp downstream of the ATG in the first exon of the Pdc2 gene (Figure 3B). After backcrossing the footprint allele of plant 23 into P. hybrida W115 (also known as cv Mitchell) for five generations, phenotype and genotype still cosegregated, demonstrating that the phenotype is also apparent in this genetic background. Taken together, these results demonstrate that the mutation in the Pdc2 gene is responsible for the reduced in vitro pollen tube growth phenotype in the presence of the PDH inhibitor AEP.
Genetic Consequences of the pdc2 Mutation
We took advantage of the presence of the stable footprint allele in plant 23, which creates a restriction site for NcoI in the pdc2 allele, to test whether an in vivo function of the Pdc2 gene could be revealed by poor transmission through the male parent. After three backcrosses into the stable petunia W115 background, a substantial decrease in dTPh1 transposon copy number was achieved. The progeny of selfed heterozygous (pdc2/+) plants was analyzed by NcoI restriction of the PCR product obtained with two Pdc2-specific primers. A highly significant deviation (P < 0.0001) from the expected 1:2:1 segregation ratio was observed (Table 1). This segregation distortion implies reduced male transmission of the mutant allele.
Table 1.
Effect of pdc2 on Pollen Tube Growth in Vivo
| No. of Offspring |
|||||
|---|---|---|---|---|---|
| Female | Male | +/+ | pdc2/+ | pdc2/pdc2 | Probability |
| pdc2/+ | pdc2/+ | 139 | 202 | 27 | P < 0.0001 |
| pdc2/pdc2 | pdc2/+ | 96 | 12 | P < 0.0001 | |
| pdc2/+ | pdc2/pdc2 | 44 | 64 | P = 0.0543 | |
| +/+ | pdc2/+ | 87 | 20 | P < 0.0001 | |
| pdc2/+ | +/+ | 43 | 63 | P = 0.0521 | |
| +/+ | pdc2/pdc2 | 106 | |||
Table showing offspring ratio from crosses of plants carrying Pdc2 and pdc2 alleles. P, probability of obtaining by chance the observed variation from an expected 1:2:1 ratio in the selfing of a pdc2/+ plant or a 1:1 ratio in the subsequent crosses; boldface indicates significant deviation.
Reciprocal crosses were made between heterozygous plants and both homozygotes for either the wild-type or mutant allele. All crosses produced seed, and we observed a highly significant deviation (P < 0.0001) from the expected male transmission rate (11 and 18% inheritance out of the 50% expected). This was true regardless of the female genotype (Table 1). By contrast, no significant deviation was observed in the reciprocal crosses when pdc2/pdc2 or wild-type pollen was used to pollinate a pdc2/+ pistil. These results indicate that transmission is exclusively affected through the male.
To definitively confirm that the differential transmission of the two alleles was caused by the Pdc2 gene, the pdc2 defect was complemented by introduction of a transgene. We introduced the Zymomonas PDC coding sequence under the control of a 35S promoter in a pdc2/pdc2 plant. RT-PCR on pollen RNA of the transgenic plant revealed that the Zymomonas PDC was expressed in pollen. No significant deviation from the expected segregation ratio was observed in the crosses of wild-type sibling pollen to T0 transgenic pistils (56% with Zymomonas PDC and 44% without Zymomonas PDC), revealing that the T0 plant had one copy of the transgene inserted in its genome (Table 2). However, when pollen of the T0 plant was crossed to a wild-type sibling pistil, we observed a significant deviation from the expected segregation ratio (81% with Zymomonas PDC observed, 50% expected). Thus, the Zymomonas PDC transgene is able to complement the pdc2 defect and confers a competitive advantage to the growing pollen tube.
Table 2.
Effect of the Zymomonas PDC on pdc2 Pollen Tube Growth in Vivo
| No. of Offspring |
||||
|---|---|---|---|---|
| Female | Male | pdc2/+ ZymPDC/− | pdc2/+ −/− | Probability |
| pdc2/pdc2 ZymPDC/− | +/+ | 83 | 61 | P = 0.0668 |
| +/+ | pdc2/pdc2 ZymPDC/− | 116 | 28 | P < 0.0001 |
Offspring ratio from crosses of transgenic plants homozygous for the pdc2 allele and their wild-type siblings. P, probability of obtaining by chance the observed variation from an expected 1:1 ratio in the two crosses; boldface indicates significant deviation.
Effect of pdc2 on Respiration and Fermentation
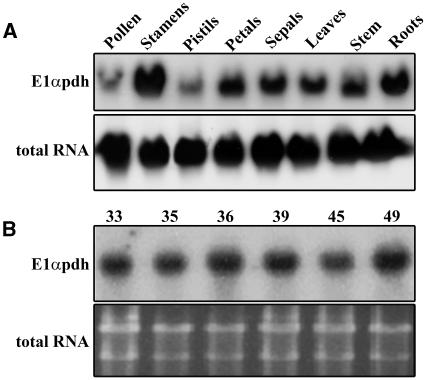
Interference with the fermentation pathway may feedback on the respiratory pathway. In particular, the expression of the exquisitely controlled PDH complex might be affected. We analyzed the expression profile of the gene encoding the E1α subunit of the PDH complex in the wild-type P. hybrida line R27. E1αpdh RNA was present mainly in stamens but also in roots, petals, sepals, leaves, and stem and at a low level in pistils and pollen (Figure 6A). Pollen expression of E1αpdh in a representative sample of the plants used for the progeny analyses was similar to the wild type (Figure 6B). Thus, there is no evidence for feedback, at least at the level of mRNA abundance, on the expression of a critical component of the main respiratory pathway when the Pdc2 gene is not functional.
Figure 6.
Expression of the E1αpdh Gene in Wild-Type Plants and in the Progeny of the CX13 Selfed Plant.
(A) RNA gel blot analysis using total RNA isolated from various organs of wild-type plants. The same blot was hybridized with 32P-labeled 18S rDNA to show relative amounts of RNA samples.
(B) RNA gel blot analysis of total RNA isolated from pollen of the CX13 progeny. Equal loading was determined by SYBR gold nucleic acid staining of the gel. Both blots were hybridized with 32P-labeled E1αpdh-specific cDNA. Genotype of the plants used is as follows: 33 and 35, pdc2/+; 36 and 39, +/+; 45 and 49, pdc2/pdc2.
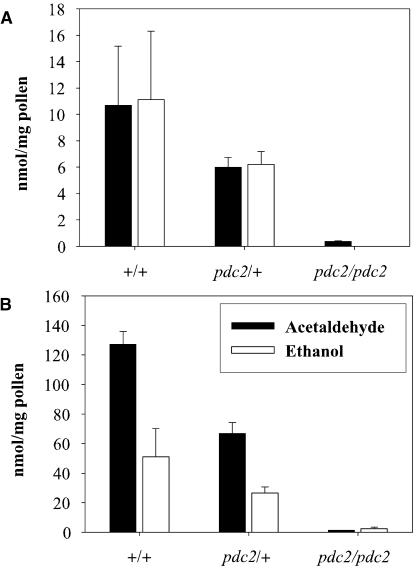
Levels of acetaldehyde and ethanol were measured in germinating pollen samples from plants that were wild-type, heterozygous, or homozygous for the pdc2::dTph1 mutation (Figure 7). Heterozygous pdc2/+ pollen produced ∼50% less acetaldehyde and ethanol than the wild-type pollen after 6 and 24 h, whereas homozygous pdc2 pollen was essentially inactive. Thus, fermentation is highly active in petunia pollen, as it is in tobacco. In combination with our expression data from the P. hybrida Pdc gene family, these results confirm that Pdc2 is the only functional Pdc gene in pollen.
Figure 7.
Acetaldehyde and Ethanol Accumulation in the Gas Phase.
Pollen from wild-type, heterozygous pdc2/+, and homozygous pdc2/pdc2 plants were tested. One-milliliter gas samples were removed from the headspace with a tight gas syringe after 6 h (A) and 24 h (B) and injected in the gas chromatography column. Data bars represent the mean of pollen samples from three individual plants ± se from one representative experiment.
Rescue of pdc2 Pollen Tube Growth
There are two possible explanations for the sensitivity of pdc2 pollen to the PDH inhibitor AEP. The first is that the presence of the inhibitor causes an absolute requirement for ethanolic fermentation. The second is that AEP sensitivity has its basis in an interruption of the PDH bypass. If the first hypothesis is correct, it should be impossible to rescue germination of pollen on the AEP-supplemented medium by adding the nonfermentable ethanol in the pollen germination medium. In a preliminary assay, pdc2 pollen was germinated on a feeder layer of wild-type pollen either in the same medium or in two different vessels separated by a gas phase. Some pdc2 pollen grains present on the AEP-supplemented medium were able to germinate under these conditions, which suggests that one or several volatile compounds could complement the mutant defect.
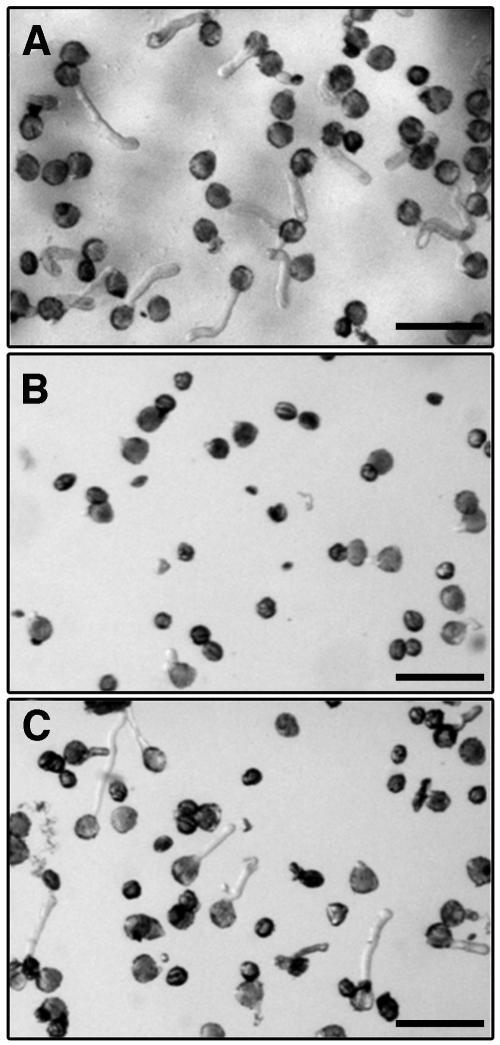
A second assay in which ethanol was applied in the gas phase was undertaken (Figures 8A to 8C). This treatment led to the rescue of pdc2 pollen germination in the presence of AEP, suggesting that ethanol can complement the lack of PDC2 activity in pdc2 pollen. These results clearly show the PDH bypass and not fermentation to be the important pathway in pollen tube growth.
Figure 8.
Light Microscopy Photographs Showing the Effect of AEP and Ethanol on the Growth of pdc2 Pollen Tubes.
(A) Growth of pdc2 pollen with no AEP.
(B) Growth of pdc2 pollen on 20 μM AEP.
(C) Ethanol-rescued growth of pdc2 pollen on 20 μM AEP.
Bars = 100 μm.
Effect of Anoxia and Respiratory Chain Inhibitors on pdc2 Pollen Germination
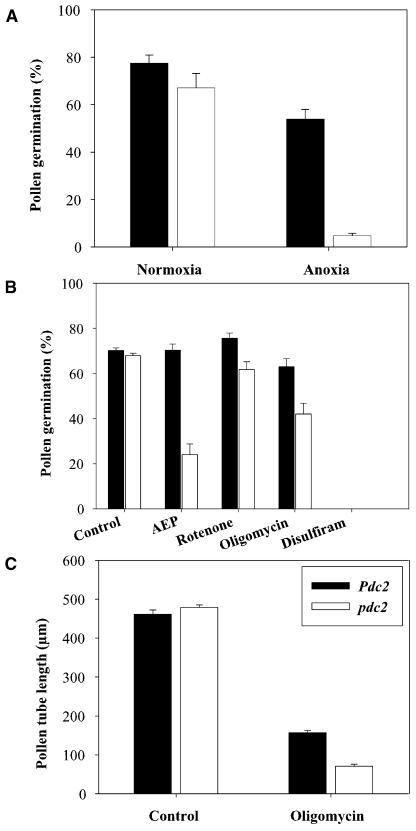
In vegetative tissues, ethanolic fermentation serves to maintain energy production under low oxygen conditions. Therefore, we decided to conduct experiments to determine if tube growth of pdc2 pollen was impaired under anoxia and other stress conditions that interfere with energy production through oxidative phosphorylation. When pollen was germinated under optimal in vitro conditions, no significant differences between the germination percentages of wild-type and pdc2 pollen was observed (Figure 9A). In a parallel experiment using the same batches of pollen, pollen grains were allowed to hydrate for 15 min in germination medium under normoxic conditions and were then transferred to an anoxic workbench for 4 h and 30 min (Figure 9A). The results obtained show that for the wild type, 54% of the pollen grains were able to germinate under anoxic conditions, but pollen tubes were shorter than under normoxic conditions. For pdc2, only ∼5% of the pollen grains were still able to germinate under anoxic conditions. This represents a significant (P < 0.001) decrease of the pdc2 pollen germination rate of ∼92% compared with wild-type pollen germination under anoxic conditions. Thus, wild-type pollen are able to germinate under anoxic conditions, whereas mutant pollen are not.
Figure 9.
Pollen Germination Percentage of Pdc2 and pdc2 Pollen.
(A) Effect of normoxic and anoxic conditions on Pdc2 and pdc2 pollen germination. Germination was scored after 4 h and 30 min (n > 1300 pollen grains for each genotype).
(B) Effects of AEP, rotenone, oligomycin, and disulfiram on Pdc2 and pdc2 pollen germination. Germination was scored after 15 h (n > 1300 pollen grains for each genotype).
(C) Effect of oligomycin on the tube length of Pdc2 and pdc2 pollen. Tube length was scored after 15 h of germination (n > 200 pollen tubes for each genotype). Data bars represent the mean of pollen samples from at least three individual plants ± se from one representative experiment. The same batches of wild-type and pdc2 mutant pollen were used in each set of experiments.
Previous experiments done in our laboratory showed that in germinating pollen, more than half of the dissimilated carbon enters fermentation (Bucher et al., 1995). To investigate the relative physiological importance of ethanolic fermentation and/or of the PDH bypass, compared with respiration during pollen tube growth, in vitro pollen tube germination assays were conducted with wild-type and pdc2 pollen in the presence of four different inhibitors.
In the first experiment, we monitored the pollen germination rate of wild-type and pdc2 pollen in the presence of 60 μM AEP (Figure 9B). Under these conditions, we did not observe a significant alteration in the germination rate or length of wild-type pollen tubes compared with wild-type pollen germinated in a medium without AEP. Consistent with the data obtained in our forward screen, we observed that AEP caused a dramatic effect on pdc2 pollen, resulting in a significant (P < 0.001) decrease of 65% of the germination rate compared with pdc2 pollen germinated in the control medium. The 24% pdc2 pollen grains that were still able to germinate in medium supplemented with AEP produced very short tubes that just exceeded the length of the diameter of the pollen grains.
In the second experiment, we scored the pollen germination rate of wild-type and pdc2 pollen in the presence of 50 μM rotenone in the germination medium (Figure 9B). Rotenone blocks the NAD(P)H dehydrogenase complex I of the respiratory chain. We did not observe a significant effect of this substance on the germination of wild-type and pdc2 pollen. This is consistent with the presence of rotenone-insensitive NAD(P)H dehydrogenases in the mitochondrial membrane of plant mitochondria, which can compensate for the inhibition of complex I.
In the third experiment, we used oligomycin, an inhibitor of phosphorylation that blocks the mitochondrial ATP-synthase (Dickinson, 1966) (Figures 9B and 9C). Germination of wild-type pollen in the presence of 0.36 μg/mL oligomycin resulted in a germination rate of 63%, which was not significantly different from the germination rate obtained in the control experiment. Wild-type pollen exposed to oligomycin had a significant (P < 0.001) reduction of 66% in pollen tube length (on average 156 μm) compared with pollen germinated in a control medium (461 μm). No significant difference was observed between the length of the tubes of wild-type (461 μm) and pdc2 pollen (479 μm) in the control germination medium. However, when pdc2 pollen was germinated in the presence of 0.36 μg/mL oligomycin, we observed a significant (P < 0.001) reduction in the germination rate compared with the control experiment. Only 42% of the pdc2 pollen remained able to germinate, and the length of the tubes was, on average, 70 μm (reduction of 85% compared with pdc2 pollen tubes grown on the control medium). These differences are consistent with the AEP results and are likely due to the lack of ATP production via the fermentation pathway in the pdc2 pollen. Wild-type pollen should still be able to produce ATP via fermentation when exposed to oligomycin. Therefore, the ATP produced from fermentation is apparently sufficient for ∼156 μm of pollen tube growth in the wild type.
In the fourth experiment, we used disulfiram, an inhibitor of ALDH. It has already been shown that adding 30 μM disulfiram to pollen germination media prevents wild-type pollen tubes from growing. By comparison, seedlings grown on 30 μM disulfiram formed fewer roots and were retarded in growth. These results suggested that major differences in metabolism exist between seedlings and pollen and that this might be due to the accumulation of toxic levels of acetaldehyde in pollen (op den Camp and Kuhlemeier, 1997). Because pdc2 pollen does not accumulate acetaldehyde (Figure 7), we decided to test whether pdc2 pollen would germinate on 30 μM of disulfiram. Neither pdc2 nor wild-type pollen was able to germinate on disulfiram (Figure 9B). This result reaffirms the major differences between seedling and pollen metabolism and further suggests that these differences are not solely due to the accumulation of acetaldehyde in pollen. Even though disulfiram is clinically used as an inhibitor of ALDH to treat alcoholism, it is known to be a potent inhibitor of many enzymes, including cytochrome P450s, glutathione S-transferase, 5-lipoxygenase, dopamine-α-hydroxylase, RNA-dependent DNA polymerase of Rous sarcoma virus, 1,4,5-triphosphate 5-phosphatase, caspases, and DNA topoisomerases (Yakisich et al., 2001).
In summary, we conclude that the performance of pdc2 pollen is equivalent to that of the wild type under optimal in vitro germination conditions but suffers under conditions that impair oxidative phosphorylation.
Phenotypic Analysis of the pdc2 Pollen
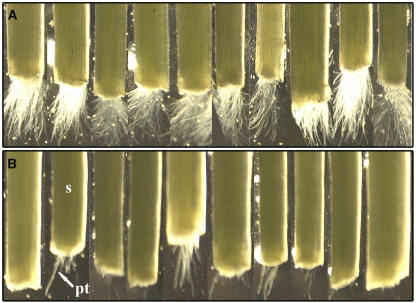
Our in vitro assays showed us that the pdc2 mutation only affected pollen performance when the normal respiratory pathway was compromised (Figure 9). Such conditions might exist in the style during in vivo pollen tube growth (Linskens and Schrauwen, 1966; Blasiak et al., 2001). To examine the pollen tube growth rate under in vivo conditions, both types of pollen grains were germinated on the stigma of wild-type W115 plants, and the exit of the pollen tubes from the cut style was monitored (Figures 10A and 10B). In this semi–in vivo pollination assay, most of the wild-type pollen tubes (32 out of 42 assays) were visible at the cut end of the style after ∼15 h. By contrast, pdc2 pollen tubes rarely reached the cut end of the style after 15 h, and when they did (14 out of 42 assays), only a few tubes emerged from the style after 15 h. It took the pdc2 pollen tubes ∼22 h to protrude from the cut end of the style. Analogous experiments with pdc2/pdc2 styles in the W115 background gave comparable results. This suggests that wild-type pollen tubes grow faster in the style than pdc2 pollen tubes and that this is independent of style genotype.
Figure 10.
Effect of pdc2 on Pollen Tube Growth in Semi–in Vivo Conditions.
Pollen grains from wild-type (A) and pdc2/pdc2 (B) lines were germinated under semi–in vivo conditions on W115 wild-type pistils as described in Methods. Pollen tube growth was examined every 30 min after 12 h of incubation. pt, pollen tube; s, style.
DISCUSSION
The ADH Paradox
The development of the pollen and its adhesion to the stigma, the growth of the pollen tube through the style and its guidance toward the ovule, and fertilization place unique demands on energy production, biosynthetic capacity, and regulatory circuits. This is exemplified by the many mutants that specifically affect male gametophytic development and growth. Pollen tubes grow at extremely high rates and consequently have a very high demand for energy. Because in many circumstances pollen is in excess, individual pollen tubes will compete for access to the ovules (Howden et al., 1998). Thus, rapid growth and efficient energy metabolism are of prime importance for male reproductive success.
Because both PDC and ADH are very abundant in pollen, it seems obvious that their function is to provide the pollen with ATP. This makes additional sense in light of the fact that oxygen tension in the style may be low (Linskens and Schrauwen, 1966). However, to our knowledge, adh mutations have never been reported to affect male gametophyte function. Here, we present a solution to this paradox. We propose that not ADH but rather PDC is the critical enzyme in a novel pathway of crucial importance in pollen.
The pdc2 Mutant Is a Male Gametophytic Progamic Phase Mutant
The petunia Pdc gene family consists of two members. Pdc2 is highly and exclusively expressed in anthers and pollen. In vitro–grown petunia pollen produced large quantities of acetaldehyde and ethanol, as is the case in tobacco. This ethanol production was entirely dependent on the presence of Pdc2. Pdc1 was widely expressed in vegetative organs and induced during oxygen limitation, but it was not found in the male gametophyte nor could it compensate for loss of Pdc2 in pollen.
Mutant pdc2 pollen grew through the style more slowly than the corresponding wild-type pollen, and the mutant allele was transmitted through the male at a reduced frequency, when it was in competition with wild-type pollen. It has been calculated that mutant alleles that measurably reduce allele frequency in fewer than 10 generations have essentially the same fate as a lethal mutation because they will inevitably be lost from the population (Gilliland et al., 1998). The fact that in the progeny of pdc2 heterozygous plants the wild-type genotype has a clear advantage over the mutant suggests that PDC2 plays an essential role in pollen tube growth and is under strong selective pressure.
The transmission defect became apparent only in crosses involving a pdc2 heterozygous plant as pollen donor. When relieved of competition from wild-type pollen, homozygous pdc2 pollen was competent to produce a full seed set. Furthermore, pdc2 did not affect pollen development, although PDC2 is already detected in stage 9 microspores in tobacco (Tadege and Kuhlemeier, 1997). It also did not affect pollen tube growth in vitro because no significant difference could be detected between the length of wild-type and pdc2 pollen tubes. The reduced male transmission and normal pollen morphology define the pdc2 mutation as a male gametophytic progamic phase mutation that acts upon pollen tube growth. Thus, pdc2 appears to act at a later stage than most of the molecularly identified mutations that affect the male gametophyte (e.g., GEMINI POLLEN/MOR1 [Twell et al., 2002] and AtPTEN1 [Gupta et al., 2002], which both produce aberrant pollen grains, or the double mutant AtPY1 AtPY2, which causes complete inhibition of pollen germination [Steinebrunner et al., 2003]).
Mutations with a defect more similar to pdc2 have been described. The maize rop2 mutation is associated with a low frequency of male-specific transmission and encodes a Rop GTPase (Arthur et al., 2003). The Arabidopsis thaliana Ttd41 (T-DNA transmission defect) mutation leads to a partial male-specific transmission defect and has no apparent abnormalities in vitro (Procissi et al., 2001); the gene(s) responsible for this lesion have not yet been identified molecularly. Pollen tubes carrying the seth9 mutation grow more slowly than their wild-type homologs but in noncompetitive pollinations are able to reach and fertilize the embryo sac. The seth9 mutation is linked to an insertion interrupting a gene with homology to an ADH-like protein of unknown specificity (Lalanne et al., 2004). A similar mutation, hap3, has been mapped close to the gene encoding the SUC1 sucrose transporter (Johnson et al., 2004).
The in Vivo Function of PDC2 Is to Bypass PDH
When cultivated in vitro under conditions of reduced respiration, be it by applying anoxia or various respiratory inhibitors, growth of the mutant was reduced, whereas the wild type was unaffected. The defect in the mutant can be rescued by addition of the nonfermentable carbon source ethanol. Thus, the loss of ATP production through ethanolic fermentation is not the cause for the inhibition of tube growth on AEP. In Saccharomyces cerevisiae, triple pdc knockouts grow normally on ethanol but completely fail to grow on glucose (Flikweert et al., 1996). Because small amounts of C2 compounds can rescue the defect, it was hypothesized that PDC has a biosynthetic function, providing cytosolic acetyl-CoA, in particular for lipid biosynthesis (van Maris et al., 2003).
Under in vitro growth conditions, mutant pollen grew at the same rate as the wild type. By contrast, the growth of pdc2 pollen tubes was markedly reduced compared with the wild type under in vivo growth conditions. Pollen tubes grow faster, longer, and more homogenously in vivo than in vitro (Johnson and Preuss, 2002); thus, optimal in vivo pollen tube growth places additional demands on primary metabolism. If this demand cannot be met in the mutant, additional routes to provide acetyl-CoA may be required under the unique growth conditions prevailing in the growing pollen tube. During in vitro pollen tube growth, the PDH bypass supports respiration as well as lipid biosynthesis (Mellema et al., 2002), and the interruption of the bypass could cause the mutant phenotype by limiting respiratory or biosynthetic capacity or both. PDH is a tightly controlled enzyme (Tovar-Mendez et al., 2003), and this control may interfere with the demands of pollen metabolism. We propose that under conditions of rapid pollen tube growth, the capacity of PDH to metabolize pyruvate becomes limiting, as is the case in yeast. This limitation may both concern the role of plastid PDH in lipid biosynthesis and the role of mitochondrial PDH at the entry into the TCA cycle.
Aerobic ethanol production by S. cerevisiae is thought to depend on the relative capacities of the fermentative and respiratory pathways. When external glucose levels are low, or when hexose import is genetically impaired, S. cerevisiae does not produce ethanol in the presence of oxygen (Pronk et al., 1996; Otterstedt et al., 2004). High glucose levels result in a glycolytic rate exceeding that of the PDH reaction, thereby generating an overflow toward PDC and hence ethanol production. At high pyruvate concentration, the high Km of PDC for pyruvate becomes inconsequential, and instead the high capacity of the enzyme becomes a key factor.
Pollen germinates in a high sugar environment, similar to glucose-grown yeast. This will cause high glycolytic flux and high pyruvate levels, favoring flux through PDC. Under conditions of rapid growth and sufficient oxygen, acetaldehyde will either be used for biosynthetic purposes or respired to CO2. The Km of ALDH for acetaldehyde is two orders of magnitude lower than that of ADH (Pronk et al., 1996; Liu and Schnable, 2002). If this is true in pollen, oxidation of acetaldehyde will be highly favored, and the main function of ADH may be to prevent the accidental accumulation of reactive acetaldehyde. Such a pathway is supported by the ethanol feeding experiments (Figure 8; Mellema et al., 2002).
Transcripts for Pdc1, Aldh, and Adh have been detected in unpollinated pistils, and Adh transcripts were detected in pollinated pistils, suggesting that fermentation and/or the bypass could be functional in this organ (op den Camp and Kuhlemeier, 1997). During pollen tube growth, ethanol produced in the pistil could be taken up by the pollen and used via the PDH bypass (after conversion to acetaldehyde by ADH) as an additional source of energy. The CO2 generated through ethanolic fermentation could be one of the carbon sources for the conversion of phosphoenolpyruvate in malate via phosphoenolpyruvate carboxylase (Jansen et al., 1992). However, we have not been able to detect more than trace levels of ethanol in pollinated pistils (data not shown); therefore, we favor the idea that acetaldehyde normally enters the bypass.
The Role of Oxygen in Pollen Metabolism
In vegetative tissues, ethanol production commences at O2 concentrations as high as 5%. This is much higher than the Km for O2 of cytochrome oxidase, which is in the low micromolar range. Therefore, the switch from respiration to fermentation is likely to be regulated by an oxygen sensor rather than directly by the terminal oxidase (Bucher et al., 1994). Such a sensor may serve to prioritize the different needs of the cell or tissue (Geigenberger, 2003). Pollen tubes have only one priority: to grow as quickly as possible toward the ovule. Very little is known about oxygen sensing in pollen. In vitro–grown tobacco pollen accumulated ethanol even at an oxygen concentration of 40%, and the flux to ethanol was regulated by the sugar concentration in the medium (Bucher et al., 1995; Tadege and Kuhlemeier, 1997). Oxygen levels in the style are likely to be reduced (Linskens and Schrauwen, 1966). It is not known how this limited oxygen availability affects the regulation of respiratory metabolism. During maturation in the anther, pollen accumulates high levels of carbohydrates, sugar concentrations in the stigmatic fluid and in the style are high, and pollen may be exposed to a situation of virtually unlimited fuel that provides the nutritional support they need to grow as quickly as possible (Wu et al., 1995). Under such conditions, it makes sense not to economize on energy. Respiration in pollen is extremely high, and respiratory flux might be solely regulated by the capacity of the cytochrome oxidase.
Conclusion
We propose that during in vivo pollen tube growth a new pathway operates that provides acetyl-CoA, independently of PDH. Thereby, both respiratory carbon flux and biosynthetic capacity are enhanced. The extremely rapid growth of the pollen tube requires unique adaptations of metabolism. Although the regulation of pollen development is under intense study in many laboratories, the subject of primary pollen metabolism and how it differs from standard pathways has received little attention in recent years. We suggest that to understand the regulatory circuits underlying pollen growth, it is necessary to understand the primary pathways that are being regulated.
METHODS
Plant Material
Transposon mutagenesis was performed in Petunia hybrida W138, using the collection of 2400 plants containing ∼60,000 dTph1 insertions as described (Stuurman et al., 2002). The plants were grown in a greenhouse under standard conditions. Anaerobic incubations were performed in an anaerobic workbench as described (Bucher and Kuhlemeier, 1993). P. hybrida R27 is a nontransposing line isogenic to P. hybrida W138.
Cultivation and Observation of Pollen Tubes
Pollen of each W138 plant was collected and distributed over two wells of a microtiter plate containing pollen germination medium, one well with and the other without 60 μM AEP. Germination medium consisted of 1.6 mM H3BO3, 2.9 mM Ca(NO3)2·4H2O, 0.8 mM MgSO4·7H2O, 1 mM KNO3, 20 mM Mes-KOH, pH 6, 12% polyethylene glycol 6000, and 60 mM sucrose.
Tube growth rescue attempts were performed with a feeder layer of wild-type P. hybrida R27 pollen and with ethanol. Mutant pollen was spread on a well containing germination medium with 20 μM AEP in a 96-well plate. Mutant pollen and the feeder layer of R27 pollen grains were either applied in the same medium or separated by a gas phase. In the first case, mutant and feeder layer pollen were incubated in the same medium, and 1 mg of wild-type pollen was first mixed with the medium in an eppendorf tube. Pollen that had sunk to the bottom was transferred to 96-well plates. The plates were centrifuged (Sigma 3K 15; Sigma-Aldrich, Basel, Switzerland) until all grains were pelleted on the bottom of the well. Mutant pollen released from anthers by dipping the anther in the culture medium floated on the medium. In the second case, mutant and feeder pollen were separated by a gas phase, and 1 mg wild-type pollen was cultured in a separate well. Wild-type pollen well and mutant pollen well were sealed and connected by a small air channel through which gases could diffuse.
Ethanol (1.5 M) and mutant pollen were applied to separate wells connected by the gas phase. After 5 h, scoring of pollen tube germination was done under the microscope. Pollen tubes were observed and photographed with an SMZ 1500 microscope coupled to a Sony DKC-5000 digital photo camera (Tokyo, Japan). Pollen was considered to have germinated when the length of the tube exceeded the diameter of the pollen grain.
Inhibitors were added at the beginning of the experiment. Rotenone (dissolved in DMSO) was added at a final concentration of 50 μM. Oligomycin (dissolved in water) was added to a final concentration of 0.36 μg/mL. Disulfiram (dissolved in DMSO) was added to a final concentration of 30 μM.
Scoring of germination rate and pollen tube length was done after 15 h. Images were captured on a Zeiss Axioskop 2 with Axiovision software (Jena, Germany). Pollen tube lengths were measured with ImageJ (http://rsb.info.nih.gov/nih-image/index.html).
For semi–in vivo germination, pollen grains of either wild-type or pdc2 W138 plants were smeared in large excess onto the stigmas of wild-type W115 pistils. The styles were cut (1 cm from the top) at 15 min after pollination, and the cut tip was placed into the liquid pollen germination medium. After 12 h, the pistils were examined every 30 min using the Nikon C-DSD230 microscope (Tokyo, Japan) coupled to a Sony DKC-5000 digital photo camera.
RNA Preparation, Reverse Transcription, PCR Amplification, and Sequence Analysis of PCR Products
Total mRNA was extracted from different P. hybrida R27 tissues using the RNeasy plant mini kit (Qiagen, Hombrechtikon, Switzerland). Poly(A)+ RNA was isolated from R27 petunia pollen using an Oligotex mRNA kit (Qiagen). All cDNA synthesis was performed with AMV reverse transcriptase (Promega, Madison, WI). All PCR fragments were cloned and verified by sequencing.
The Pdc2 cDNA was isolated by RT-PCR with primers specific for the tobacco (Nicotiana tabacum) Pdc2 gene TobPdc2: prP1282 (5′-ATTGCTGAGACAGGGGATTCTT-3′) and prP1603 (5′-TCAATCGTGTAACCACCATTATT-3′). The full-length cDNA was isolated using 5′ and 3′ rapid amplification of cDNA ends based on the manufacturer's protocol (Marathon cDNA amplification kit; Clontech, Palo Alto, CA). Primers were pr227pdc2 (5′-ATGTTCTTTTGGTCGCAACGAAT-3′) and prP1282. Primers P5 Pdc2 (5′-GGGGTAGCCAATGGTGGTA-3′) and P1904 Pdc2 (5′-AAGAAATGGAGGTGCACACAG-3′) were subsequently used to isolate the Pdc2 cDNA.
The partial Pdc1 cDNA was amplified from total RNA isolated from the petunia leaves subjected to anoxia for 4 h with the primers prP1282 and prP1603. The primer pr232pdc1 (5′-TGATAGTCCTCTGCCCGCATCGCAG-3′) was used for the 5′ rapid amplification of cDNA ends.
The E1αpdh cDNA was amplified from pollen mRNA with primers specific for the pea (Pisum sativum) E1αPDH cDNA: prEPS724 (5′-CATGGGATTGTTGGTGCTCA-3′) and prEPS1146 (5′-GTGCTTCCAGGATCAGACAT-3′). The full-length cDNA was isolated using 5′ and 3′ rapid amplification of cDNA, and the primers used were prEPS1146 and prEPS724.
For analyses of the pdc2 footprint alleles, genomic DNA was isolated from leaves of plants 23 and 45. Genomic DNA was PCR amplified with P5 Pdc2 and P442 Pdc2 (5′-TGGCTAAAATCTGGCAAT-3′); PCR products were separated on a 1.5% (w/v) agarose gel, and the smallest band was eluted and sequenced.
To differentiate the wild-type Pdc2 and the pdc2 mutant alleles, PCR analyses were performed with primers P5 Pdc2 and P442 Pdc2 on petunia genomic DNA isolated from leaf tissue. In the case of plants carrying the footprint that originated in plant 23, PCR products were subsequently digested with NcoI to distinguish the pdc2 mutant allele from the wild-type Pdc2 allele. PCR products were separated on a 1.5% (w/v) agarose gel. Primer synthesis and sequencing were done at Microsynth (Balgach, Switzerland).
Transposon Insertion Mutagenesis
The screening for dTph1 insertion mutants was performed as described by Koes et al. (1995). The primers used to screen for dTph1 insertions in Pdc2 were as follows: the dTph1 primer out 1 (5′-GGGAATTCGCTCCGCCCCTG-3′) together with the Pdc2-specific primer pr30pdc2 (5′-GTATTCAAGATTCTAGTTCTGCGT-3′). PCR was performed for 30 cycles of 30 s at 94°C, 1 min at 50°C, and 2 min at 72°C. Amplification products were separated by electrophoresis in 1% agarose gels, blotted to Hybond-N membranes, and hybridized with 32P-labeled Pdc2 cDNA probes generated with primer pr30pdc2 and pr630pdc2r (5′-GGTAATGTATACCTTGGTGAGATA-3′).
RNA Gel Blot Analysis
Total RNA extraction and RNA gel blot analysis were performed as described previously (Caderas et al., 2000) under high stringency conditions (65°C). Probes specific for Pdc2 and E1αpdh were generated by PCR and labeled by random hexamer labeling as described by the manufacturer (Rediprime II; Amersham Biosciences, Otelfingen, Switzerland). Primers used for the Pdc2 amplification were pr227pdc2 and prP1282. E1αpdh-specific primers were prEPS724 and prEPS1146.
Reverse Transcription and Quantitative Real-Time PCR
Total RNA was prepared from the individual organs of petunia W115 plant as described by Caderas et al. (2000).
RNA extraction of germinated pollen grains was done as described (Steiner et al., 2003). Briefly, pollen was germinated in vitro at 1.5 mg pollen per 300 μL germination medium (12% polyethylene glycol 6000, 60 mM sucrose, 1 g/L casein–hydrolysate [w/v] [Difco Laboratories, Chemie Brunschwig, Basel, Switzerland], 15 mM Mes-KOH buffer, pH 5.9, 1 mM CaCl2, 1 mM KCl, 0.8 mM MgSO4, 30 μM CuSO4, and 1.6 mM H3BO4) in small Petri dishes at 25°C for 6 h. For RNA extraction, 10 mL of pollen suspension was concentrated twice by centrifugation at 2000g for 10 min to a final volume of 3 mL and immediately transferred to liquid N2 and either processed further or stored at −80°C.
The quality of the RNA was confirmed by visualization by UV irradiation of ethidium bromide–stained rRNA separated on 1.5% (w/v) agarose gel. Total RNA (1 μg per reaction) was DNase I treated. First-strand cDNA was synthesized with an oligo(dT) primer and reverse transcriptase, avian myeloblastosis virus (Promega).
Transcripts were quantified by quantitative real-time PCR on the LightCycler system (Roche Diagnostics, Mannheim, Germany). The following primers were used for PCR experiments: Pdc1 forward primer, 5′-TCAGAGGATGGTGTCTGCTG-3′, and reverse primer, 5′-GCAAGCACCCTCTTATCTCG-3′; Pdc2 forward primer, 5′-GGCTTAGTTGAAGCAATTCACAA-3′, and reverse primer, 5′-TCCTTCTTTTCTCCAGTTGCTG-3′; Actin forward primer, 5′-TCCATGATTGGAATGGAAGC-3′, and reverse primer, 5′-GACCCACCACTGAGCACAA-3′. Primers were optimized for amplification in a gradient cycler with various annealing temperatures from 54.1 to 63.1°C.
For PCR reactions, a master mix of the following reaction components was prepared (the end concentration is indicated in parentheses): 6 μL of water, 1 μL of forward primer (0.5 μM), 1 μL of reverse primer (0.5 μM), and 10 μL of LightCycler (Quantitect SYBR Green; Qiagen). LightCycler master mix was filled in the LightCycler glass capillaries, and 2 μL of cDNA was added as PCR template.
Thermal cycling conditions were as follows: denaturation 95°C for 15 min followed by amplification and quantification, 60 cycles (94°C for 10 s, annealing temperature of 59°C for 30 s and 72°C for 30 s with a single fluorescence measurement), melting curve program (65 to 95°C with heating rate of 0.1°C s−1 and a continuous fluorescence measurement), and a cooling step at 35°C.
For relative quantification, PCR efficiencies for each gene were determined as follows: Standard curves for each gene were generated using the cDNA with the highest abundance of the gene to cover the range of all template concentrations. Real-time PCR efficiencies (E) were calculated from the given slopes in the LightCycler software of the standard curves according to the equation: E = 10 [−1/slope]. PCR efficiencies for the used target and reference genes are as follows: Pdc1, 1.75; Pdc2, 1.72; Actin, 1.79. Crossing points were determined using the fit point method in the LightCycler software 3.5.3 (Roche Diagnostics). cDNA abundance and induction level of the Pdc genes were calculated in comparison to the reference gene Actin as described previously (Kursteiner et al., 2003).
Acetaldehyde and Ethanol Measurement in the Gas Phase
Pollen samples (≈2 mg) of wild-type, heterozygous (pdc2/+), and homozygous (pdc2/pdc2) mutant plants were placed on a filter in 10-mL vials. Pollen germination medium (150 μL) was added, and the vial was sealed gas tight. After 6 and 24 h of incubation, 1 mL of gas sample was removed from the headspace with a Hamilton syringe and injected into a Sigma 300 gas chromatography column (Perkin-Elmer, Foster City, CA).
Plastic Section of Anthers
Plastic sections of anthers were prepared as described previously (Loreto et al., 2001). Sections were viewed on a Zeiss Axioscop2 equipped with an Axiocam camera.
Plant Transformation
Cloning of the Zymomonas mobilis PDC gene in the pMON505 expression vector under the control of the 35S promoter of Cauliflower mosaic virus has been described previously (Bucher et al., 1994). The construct was transferred via Agrobacterium tumefaciens LBA 4404 to P. hybrida pdc2/pdc2 plants (obtained after three backcrosses of the W138 pdc2 allele containing the footprint in the W115 background) using the standard leaf disc transformation method. Pollen of the primary transformants were tested via RT-PCR for the presence of the Zymomonas PDC RNA with primers ZymF1 (5′-ATCCCTCAATGCAGGTGAAC-3′) and ZymR1 (5′-GCCAGAGCAACCTTGATAGC-3′). Genotyping of the progeny of the crosses using the transgenic plant was done by PCR with the same primers.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers X81855 (TobPDC2), AY928611 (Pdc2), AY928612 (Pdc1), PSU51918 (pea E1αPDH), and AY905543 (E1αPdh).
Supplementary Material
Acknowledgments
We gratefully acknowledge the invaluable advice and critical review of the manuscript by Sheila McCormick (USDA, Albany, CA), Douglas Randall (University of Missouri, Columbia, MO), Lee Sweetlove (University of Oxford, UK), and James Moore (University of Berne). Lothar Willmitzer (Max Planck Institute for Molecular Plant Physiology, Golm, Germany) generously provided the opportunity to do the metabolite analysis. Financial support was from the Canton of Berne, the Swiss National Science Foundation through the National Centres for Competence in Research “Plant Survival”, and the European Union projects Fruta Fresca (FAIR-CT98-4211) and Hybtec (QLK5-CT-1999-30902).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Cris Kuhlemeier (cris.kuhlemeier@ips.unibe.ch).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.033290.
References
- Arthur, K.M., Vejlupkova, Z., Meeley, R.B., and Fowler, J.E. (2003). Maize ROP2 GTPase provides a competitive advantage to the male gametophyte. Genetics 165 2137–2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasiak, J., Mulcahy, D.L., and Musgrave, M.E. (2001). Oxytropism: A new twist in pollen tube orientation. Planta 213 318–322. [DOI] [PubMed] [Google Scholar]
- Bucher, M., Brander, K.A., Sbicego, S., Mandel, T., and Kuhlemeier, C. (1995). Aerobic fermentation in tobacco pollen. Plant Mol. Biol. 28 739–750. [DOI] [PubMed] [Google Scholar]
- Bucher, M., Brandle, R., and Kuhlemeier, C. (1994). Ethanolic fermentation in transgenic tobacco expressing Zymomonas mobilis pyruvate decarboxylase. EMBO J. 13 2755–2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucher, M., and Kuhlemeier, C. (1993). Long-term anoxia tolerance. Multi-level regulation of gene expression in the amphibious plant Acorus calamus L. Plant Physiol. 103 441–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caderas, D., Muster, M., Vogler, H., Mandel, T., Rose, J.K., McQueen-Mason, S., and Kuhlemeier, C. (2000). Limited correlation between expansin gene expression and elongation growth rate. Plant Physiol. 123 1399–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson, D.B. (1965). Germination of lily pollen: Respiration and tube growth. Science 150 1818–1819. [DOI] [PubMed] [Google Scholar]
- Dickinson, D.B. (1966). Inhibition of pollen respiration by oligomycin. Nature 210 1362–1363. [Google Scholar]
- Flikweert, M.T., Van Der Zanden, L., Janssen, W.M., Steensma, H.Y., Van Dijken, J.P., and Pronk, J.T. (1996). Pyruvate decarboxylase: An indispensable enzyme for growth of Saccharomyces cerevisiae on glucose. Yeast 12 247–257. [DOI] [PubMed] [Google Scholar]
- Freeling, M., and Bennett, D.C. (1985). Maize Adh1. Annu. Rev. Genet. 19 297–323. [DOI] [PubMed] [Google Scholar]
- Geigenberger, P. (2003). Response of plant metabolism to too little oxygen. Curr. Opin. Plant Biol. 6 247–256. [DOI] [PubMed] [Google Scholar]
- Gerats, A.G.M., Huits, H., Vrijlandt, E., Marana, C., Souer, E., and Beld, M. (1990). Molecular characterization of a nonautonomous transposable element (Dtph1) of Petunia. Plant Cell 2 1121–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilliland, L.U., McKinney, E.C., Asmussen, M.A., and Meagher, R.B. (1998). Detection of deleterious genotypes in multigenerational studies. I. Disruptions in individual Arabidopsis actin genes. Genetics 149 717–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta, R., Ting, J.T.L., Sokolov, L.N., Johnson, S.A., and Luan, S. (2002). A tumor suppressor homolog, AtPTEN1, is essential for pollen development in Arabidopsis. Plant Cell 14 2495–2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howden, R., Park, S.K., Moore, J.M., Orme, J., Grossniklaus, U., and Twell, D. (1998). Selection of T-DNA-tagged male and female gametophytic mutants by segregation distortion in Arabidopsis. Genetics 149 621–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen, M.A.K., Sessa, G., Malkin, S., and Fluhr, R. (1992). PEPC-mediated carbon fixation in transmitting tract cells reflects style-pollen tube interactions. Plant J. 2 507–515. [Google Scholar]
- Johnson, M.A., and Preuss, D. (2002). Plotting a course: Multiple signals guide pollen tubes to their targets. Dev. Cell 2 273–281. [DOI] [PubMed] [Google Scholar]
- Johnson, M.A., von Besser, K., Zhou, Q., Smith, E., Aux, G., Patton, D., Levin, J.Z., and Preuss, D. (2004). Arabidopsis hapless mutations define essential gametophytic functions. Genetics 168 971–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koes, R., et al. (1995). Targeted gene inactivation in petunia by PCR-based selection of transposon insertion mutants. Proc. Natl. Acad. Sci. USA 92 8149–8153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koltunow, A.M., Truettner, J., Cox, K.H., Wallroth, M., and Goldberg, R.B. (1990). Different temporal and spatial gene expression patterns occur during anther development. Plant Cell 2 1201–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kursteiner, O., Dupuis, I., and Kuhlemeier, C. (2003). The pyruvate decarboxylase1 gene of Arabidopsis is required during anoxia but not other environmental stresses. Plant Physiol. 132 968–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laber, B., and Amrhein, N. (1987). Metabolism of 1-aminoethylphosphinate generates acetylphosphinate, a potent inhibitor of pyruvate dehydrogenase. Biochem. J. 248 351–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalanne, E., Michaelidis, C., Moore, J.M., Gagliano, W., Johnson, A., Patel, R., Howden, R., Vielle-Calzada, J.-P., Grossniklaus, U., and Twell, D. (2004). Analysis of transposon insertion mutants highlights the diversity of mechanisms underlying male progamic development in Arabidopsis. Genetics 167 1975–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laszlo, A., and Lawrence, P. (1983). Parallel induction and synthesis of Pdc and Adh in anoxic maize roots. Mol. Gen. Genet. 192 110–117. [Google Scholar]
- Lee, S.L.J., and Warmke, H.E. (1979). Organelle size and number in fertile and T-cytoplasmic male-sterile corn. Am. J. Bot. 66 141–148. [Google Scholar]
- Linskens, H.F., and Schrauwen, J. (1966). Measurement of oxygen tension changes in style during pollen tube growth. Planta 71 98–106. [DOI] [PubMed] [Google Scholar]
- Liu, F., and Schnable, P.S. (2002). Functional specialization of maize mitochondrial aldehyde dehydrogenases. Plant Physiol. 130 1657–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loreto, F., Mannozzi, M., Maris, C., Nascetti, P., Ferranti, F., and Pasqualini, S. (2001). Ozone quenching properties of isoprene and its antioxidant role in leaves. Plant Physiol. 126 993–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellema, S., Eichenberger, W., Rawyler, A., Suter, M., Tadege, M., and Kuhlemeier, C. (2002). The ethanolic fermentation pathway supports respiration and lipid biosynthesis in tobacco pollen. Plant J. 30 329–336. [DOI] [PubMed] [Google Scholar]
- op den Camp, R.G.L., and Kuhlemeier, C. (1997). Aldehyde dehydrogenase in tobacco pollen. Plant Mol. Biol. 35 355–365. [DOI] [PubMed] [Google Scholar]
- Otterstedt, K., Larsson, C., Bill, R.M., Stahlberg, A., Boles, E., Hohmann, S., and Gustafsson, L. (2004). Switching the mode of metabolism in the yeast Saccharomyces cerevisiae. EMBO Rep. 5 532–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Procissi, A., de Laissardiere, S., Ferault, M., Vezon, D., Pelletier, G., and Bonhomme, S. (2001). Five gametophytic mutations affecting pollen development and pollen tube growth in Arabidopsis thaliana. Genetics 158 1773–1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pronk, J.T., Steensma, H.Y., and vanDijken, J.P. (1996). Pyruvate metabolism in Saccharomyces cerevisiae. Yeast 12 1607–1633. [DOI] [PubMed] [Google Scholar]
- Steinebrunner, I., Wu, J., Sun, Y., Corbett, A., and Roux, S.J. (2003). Disruption of Apyrases inhibits pollen germination in Arabidopsis. Plant Physiol. 131 1638–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner, C., Bauer, J., Amrhein, N., and Bucher, M. (2003). Two novel genes are differentially expressed during early germination of the male gametophyte of Nicotiana tabacum. Biochim. Biophys. Acta 1625 123–133. [DOI] [PubMed] [Google Scholar]
- Stuurman, J., Jaggi, F., and Kuhlemeier, C. (2002). Shoot meristem maintenance is controlled by a GRAS-gene mediated signal from differentiating cells. Genes Dev. 16 2213–2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadege, M., Brandle, R., and Kuhlemeier, C. (1998). Anoxia tolerance in tobacco roots: Effect of overexpression of pyruvate decarboxylase. Plant J. 14 327–335. [Google Scholar]
- Tadege, M., and Kuhlemeier, C. (1997). Aerobic fermentation during tobacco pollen development. Plant Mol. Biol. 35 343–354. [DOI] [PubMed] [Google Scholar]
- Taylor, L.P., and Hepler, P.K. (1997). Pollen germination and tube growth. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48 461–491. [DOI] [PubMed] [Google Scholar]
- Thelen, J.J., Miernyk, J.A., and Randall, D.D. (1999). Molecular cloning and expression analysis of the mitochondrial pyruvate dehydrogenase from maize. Plant Physiol. 119 635–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tovar-Mendez, A., Miernyk, J.A., and Randall, D.D. (2003). Regulation of pyruvate dehydrogenase complex activity in plant cells. Eur. J. Biochem. 270 1043–1049. [DOI] [PubMed] [Google Scholar]
- Twell, D., Park, S.K., Hawkins, T.J., Schubert, D., Schmidt, R., Smertenko, A., and Hussey, P.J. (2002). MOR1/GEM1 has an essential role in the plant-specific cytokinetic phragmoplast. Nat. Cell Biol. 4 711–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Houwelingen, A., Souer, E., Spelt, K., Kloos, D., Mol, J., and Koes, R. (1998). Analysis of flower pigmentation mutants generated by random transposon mutagenesis in Petunia hybrida. Plant J. 13 39–50. [DOI] [PubMed] [Google Scholar]
- van Maris, A.J.A., Luttik, M.A.H., Winkler, A.A., van Dijken, J.P., and Pronk, J.T. (2003). Overproduction of threonine aldolase circumvents the biosynthetic role of pyruvate decarboxylase in glucose-limited chemostat cultures of Saccharomyces cerevisiae. Appl. Environ. Microbiol. 69 2094–2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, H.M., Wang, H., and Cheung, A.Y. (1995). A pollen tube growth stimulatory glycoprotein is deglycosylated by pollen tubes and displays a glycosylation gradient in the flower. Cell 82 395–403. [DOI] [PubMed] [Google Scholar]
- Yakisich, J.S., Siden, A., Eneroth, P., and Cruz, M. (2001). Disulfiram is a potent in vitro inhibitor of DNA topoisomerases. Biochem. Biophys. Res. Commun. 289 586–590. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.