Abstract
The phytohormone abscisic acid (ABA) modulates the expression of many genes important to plant growth and development and to stress adaptation. In this study, we found that an APETALA2/EREBP-type transcription factor, AtERF7, plays an important role in ABA responses. AtERF7 interacts with the protein kinase PKS3, which has been shown to be a global regulator of ABA responses. AtERF7 binds to the GCC box and acts as a repressor of gene transcription. AtERF7 interacts with the Arabidopsis thaliana homolog of a human global corepressor of transcription, AtSin3, which in turn may interact with HDA19, a histone deacetylase. The transcriptional repression activity of AtERF7 is enhanced by HDA19 and AtSin3. Arabidopsis plants overexpressing AtERF7 show reduced sensitivity of guard cells to ABA and increased transpirational water loss. By contrast, AtERF7 and AtSin3 RNA interference lines show increased sensitivity to ABA during germination. Together, our results suggest that AtERF7 plays an important role in ABA responses and may be part of a transcriptional repressor complex and be regulated by PKS3.
INTRODUCTION
The phytohormone abscisic acid (ABA) is an important regulator of plant growth and development and of plant responses to environmental stress such as drought, cold, and salt (Finkelstein et al., 2002). Drought and salt stress lead to the accumulation of ABA, which initiates many adaptive responses (Leung and Giraudat, 1998). Under water-deficit conditions, for example, ABA-induced stomatal closure reduces transpirational water loss from plants (Assmann and Wang, 2001; Schroeder et al., 2001).
Although ABA receptors have yet to be found, many components involved in ABA signaling have been identified. The type 2C protein phosphatases ABI1 and ABI2 are central regulators of ABA responses (Koornneef et al., 1984; Merlot et al., 2001). The dominant abi1-1 and abi2-1 mutations render Arabidopsis thaliana plants insensitive to ABA in seed germination, root growth, stomatal closure, and gene regulation (Koornneef et al., 1984; Leung et al., 1997; Allen et al., 1999; Hoth et al., 2002). The Ser/Thr protein kinase OST1, an Arabidopsis ortholog of Vicia faba AAPK, is activated by ABA and plays a positive role in guard cell ABA responses (Mustilli et al., 2002; Assmann, 2003). The only heterotrimeric G protein present in Arabidopsis is also a positive regulator of ABA responses in guard cells (Wang et al., 2001). PKS3, another Ser/Thr protein kinase, interacts physically with ABI2 and is a global negative regulator of ABA responses (Guo et al., 2002). Other negative regulators of ABA responses include the farnesyl transferase ERA1 (Cutler et al., 1996), inositol phosphatase FRY1 (Xiong et al., 2001b), and several proteins involved in RNA metabolism (Lu and Fedoroff, 2000; Hugouvieux et al., 2001; Xiong et al., 2001a).
Several transcriptional activators are involved in ABA responses (Himmelbach et al., 2003). Arabidopsis ABI3 and maize (Zea mays) VP1 encode transcription factors of the B3 domain family (McCarty et al., 1991; Giraudat et al., 1992). ABI4 is a transcription factor in the APETALA2 (AP2) domain family, whereas ABI5 is a basic domain/leucine zipper protein that binds to the ABA-responsive element G box (Finkelstein et al., 1998; Finkelstein and Lynch, 2000). Mutations in ABI3, ABI4, or ABI5 cause ABA insensitivity specifically in seeds (Koornneef et al., 1984; Finkelstein and Lynch, 2000; Finkelstein et al., 2002). Besides transcriptional activators, ABA responses in plants may also involve transcriptional repressors. The activity and/or expression of the transcription factors presumably are regulated by some of the ABA signaling components. In fact, the homeodomain protein ATHB6 was found to interact with ABI1 and play a negative role in ABA signaling (Himmelbach et al., 2002).
In yeast and human cells, histone deacetylation plays an important role in transcriptional repression (Hassin et al., 1997; Heinzel et al., 1997; Knoepfler and Eisenman, 1999; Murphy et al., 1999). Sin3 can repress transcription when tethered to a promoter through a heterologous DNA binding domain (Wang and Stillman, 1993), thereby providing the specificity of signal transduction accompanying transcriptional repression (Knoepfler and Eisenman, 1999). The transcription corepressors N-CoR and SMRT interact with unliganded nuclear receptors and Sin3 proteins, which appear to be required for transcription repression by unliganded receptors and by the Mad/Mxi protein (Pazin and Kadonaga, 1997). Recent studies have revealed a wide range of DNA binding transcription factors (REST/NRSF, MNF-β, p53, and MeCP2) associated with Sin3 (Knoepfler and Eisenman, 1999; Brubaker et al., 2000). Although yeast Sin3 does not possess DNA binding activity, it can repress transcription when coupled to a heterologous DNA binding domain; thus, it may repress transcription by tethering to DNA by a sequence-specific DNA binding protein (Ayer et al., 1995). Recent studies suggested important roles of histone deacetylase in plant development (Lusser et al., 2001; Meyer, 2001). AtHD1 is the Arabidopsis homolog of yeast RPD3 and human HD1. Reduction of AtHD1 (HDA19) mRNA levels by an antisense approach resulted in the accumulation of tetra-acetylated H4 and pleiotropic developmental defects (Tian and Chen, 2001). Similarly, antisense inhibition of AtRPD3A delayed flowering in transgenic Arabidopsis plants, suggesting that histone acetylation may play a role in controlling genes involved in the transition from the vegetative to the reproductive phase of development (Wu et al., 2000). More recently, HDA19 was reported to be involved in jasmonic acid and ethylene signaling during the pathogen response in Arabidopsis (Zhou et al., 2005).
In this study, we identified AtERF7, a member of the ethylene-responsive element binding factor family, as an important transcriptional repressor in ABA responses. AtERF7 interacts with PKS3 and can be phosphorylated by PKS3 in vitro. AtERF7 also interacts with the transcriptional corepressor, AtSin3, which in turn may interact with the histone deacetylase HDA19. AtSin3 and HDA19 enhance the transcription repression activity of AtERF7. Overexpression of AtERF7 in transgenic Arabidopsis plants reduced ABA responses in guard cells and decreased drought tolerance, whereas reductions in AtERF7 expression caused ABA hypersensitivity in guard cells, seed germination, and seedling growth. Our findings reveal a potential link between ABA signaling and chromatin remodeling.
RESULTS
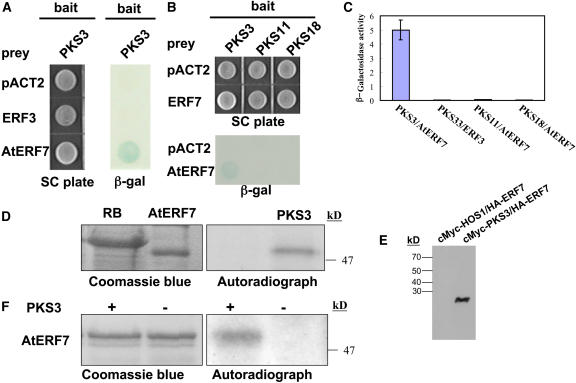
AtERF7 Interacts with PKS3
In a yeast two-hybrid screen using the protein kinase SOS2 as a bait to screen for proteins that may interact with SOS2 and possibly with other protein kinases in the SOS2 family, we found AtERF7 (At3g20310) as a very weak SOS2-interacting protein (U. Halfter and J.-K. Zhu, unpublished results). We tested other protein kinases in the SOS2 family and found that PKS3 shows a strong interaction with AtERF7 (Figures 1A and 1B). Other kinases related to PKS3, such as PKS11 and PKS18 (Gong et al., 2002a, 2002b), do not interact with AtERF7 (Figures 1B and 1C). These results indicate that AtERF7 preferentially interacts with PKS3.
Figure 1.
PKS3 Specifically Interacts with AtERF7.
(A) PKS3 interacts with AtERF7 but not AtERF3 in the yeast two-hybrid assay. Yeast strains containing pAS-PKS3 as bait and pACT-AtERF7 as prey were grown on SC medium lacking Trp and Leu for 48 h (left panel) and were assayed for LacZ expression by a filter lift assay (right panel). pACT-AtERF3 and the empty prey vector were used as negative controls. Blue color indicates interaction. β-gal, β-galactosidase activity.
(B) AtERF7 interacts with PKS3 but not PKS11 or PKS18 in the yeast two-hybrid assay.
(C) Quantitative analysis of β-galactosidase activity of the yeast strains in liquid culture showing the interaction between AtERF7 and PKS3 as well as with the control partners (ERF3, PKS11, and PKS18). Values are the means of data taken from three independent experiments. Error bars indicate standard deviation.
(D) PKS3 interacts with AtERF7 in vitro. Radiolabeled PKS3 was pulled down by GST-AtERF7 but not by GST-RB.
(E) AtERF7 interacts with PKS3 in vivo. AtERF7-HA and PKS3-Myc were transiently coexpressed in wild-type protoplasts, and then PKS3-Myc was precipitated with anti-Myc. The precipitated anti-Myc samples were subjected to anti-HA protein gel blot analysis. AtERF7 did not coimmunoprecipitate with HOS1.
(F) In vitro phosphorylation of AtERF7 by PKS3.
AtERF7 is a putative transcriptional repressor in the AP2/EREBP family (Ohta et al., 2001). AtERF3 is a closely related protein in the transcriptional repressor subfamily. However, as shown in Figures 1A and 1C, AtERF3 does not interact with PKS3. Therefore, PKS3 appears to interact specifically with AtERF7.
To confirm the interaction between AtERF7 and PKS3, we performed a glutathione S-transferase (GST) pulldown assay. GST-AtERF7 or GST-RB (for the maize retinoblastoma protein) (Halfter et al., 2000) bound to glutathione–agarose beads was incubated with PKS3, obtained by in vitro translation and labeled using [35S]Met. After extensive washing, the bound PKS3 was detected by SDS-PAGE. [35S]Met-labeled PKS3 protein was pulled down by GST-AtERF7 but not by GST-RB (Figure 1D).
To test whether AtERF7 and PKS3 can also interact in vivo, Myc-tagged PKS3 (PKS3-MYC) and hemagglutinin (HA)-tagged AtERF7 (AtERF7-HA) were transfected into Arabidopsis protoplasts either separately or together. PKS3-Myc was then precipitated with anti-Myc antibodies. Figure 1E shows that AtERF7-HA was detected in the proteins precipitated by anti-Myc from protoplasts cotransfected with both PKS3-Myc and AtERF7-HA. The results demonstrate that PKS3 and AtERF7 could be coimmunoprecipitated. Control experiments show that AtERF7 did not coimmunoprecipitate with HOS1 (a RING finger protein encoded by the high expression of osmotically responsive gene l locus of Arabidopsis) (Lee et al., 2001) from protoplasts cotransfected with AtERF7-HA and HOS1-Myc (Figure 1E). These results suggest that AtERF7 and PKS3 interact in vivo in plant cells.
To determine whether AtERF7 can be a substrate of PKS3, we expressed the PKS3 and AtERF7 proteins in Escherichia coli as GST fusion proteins. The kinase activity was assayed by incubation of AtERF7 and PKS3 in the presence of [γ-32P]ATP in a kinase buffer. Figure 1F shows that PKS3 phosphorylated AtERF7, suggesting that AtERF7 can be a substrate of PKS3, at least in vitro.
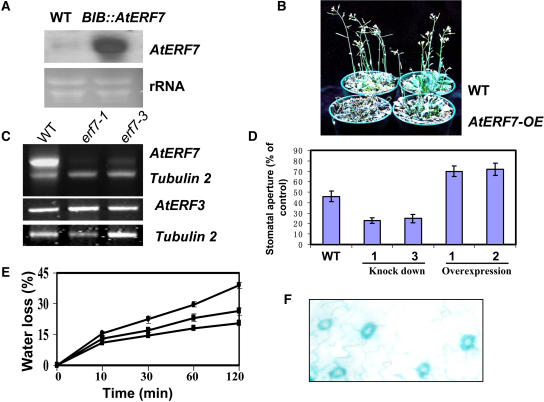
AtERF7 Overexpression or Knockdown Increases or Reduces ABA Sensitivity in Guard Cells, Respectively
To investigate the in vivo function of AtERF7, we overexpressed it by fusing its coding region to the constitutive super promoter (Narasimhulu et al., 1996) and introducing the construct into Arabidopsis. Twenty-three T3 homozygous transgenic lines were recovered, and a representative line with high AtERF7 expression (Figure 2A) was used for detailed analysis. We tested whether drought tolerance is affected by AtERF7 overexpression. After withholding water for 2 weeks, wild-type leaves remained green and turgid, but AtERF7 overexpression transgenic plants wilted (Figure 2B). To determine whether the overexpression of AtERF7 affects the sensitivity of guard cells to ABA, we measured the changes in stomatal aperture after ABA treatment. We exposed wild-type and AtERF7 overexpression transgenic plants to strong light and high humidity to induce full stomatal opening and then excised the leaves and placed them in stomatal-opening buffer for 2 h. After ABA treatment, wild-type plants showed greatly reduced stomatal aperture (48%), whereas AtERF7 overexpression plants showed stomatal aperture reduction of only 8% (Figure 2D). These data suggest that stomatal closure is less sensitive to ABA in AtERF7 overexpression plants.
Figure 2.
Role of AtERF7 in Controlling Transpirational Water Loss and Guard Cell Sensitivity to ABA.
(A) Expression of AtERF7 in one AtERF7 overexpression transgenic line (line 1).
(B) Increased wilting of transgenic plants with AtERF7 overexpression (OE) under drought stress. Both wild-type and transgenic plants were grown under normal conditions for 15 d and then subjected to drought stress.
(C) RT-PCR results showing AtERF7 (top gel) and AtERF3 (bottom gel) expression in the wild type and two independent lines of aterf7 RNAi lines. Tubulin primers were used in PCR as an internal control.
(D) Stomatal behavior in the wild type and AtERF7 RNAi lines (aterf7-1 and aterf7-3) and overexpression plants in response to ABA. Stomata were opened by exposing plants for 12 h to light and high humidity, and leaves were incubated for 1.5 h in stomatal-opening solution containing 50 mM KCl, 10 μM CaCl2, and 10 mM Mes, pH 6.15. Stomatal apertures were measured 1.5 h after adding 2 μM ABA. Data represent means ± sd (n = 140 to 150 stomata).
(E) Water loss from excised leaves of the wild type and AtERF7 RNAi lines and transgenic plants with AtERF7 overexpression. Data represent means ± sd of three independent experiments (12 to 15 measurements per point).
(F) Expression of AtERF7 promoter–GUS in guard cells.
To assess the functional consequence of the loss of function of AtERF7, we generated aterf7 RNA interference (RNAi) lines using gene-specific sequences (Chuang and Meyerowitz, 2000). The expression of AtERF7 was examined by RT-PCR in two randomly chosen independent aterf7 (aterf7-1 and aterf7-3) RNAi lines. The result shows that AtERF7 expression was knocked down in the RNAi lines (Figure 2C). Control experiments showed that AtERF3, the most closely related gene to AtERF7, was not affected in the AtERF7 RNAi lines. Twenty-two aterf7 RNAi lines were germinated on MS nutrient agar plates (Sigma-Aldrich, St. Louis, MO) supplemented with 0.5 μM ABA. All but two of the aterf7 lines were more sensitive to ABA. The ABA-hypersensitive phenotypes of aterf7 cosegregated with the hygromycin resistance marker (data not shown).
We further analyzed two representative T3 homozygous lines. Stomatal apertures in response to ABA were also investigated in the AtERF7 RNAi lines. As shown in Figure 2D, 1 μM ABA caused more stomatal closure in the AtERF7 RNAi lines compared with wild-type plants. Thus, stomatal closing in the AtERF7 RNAi lines is more sensitive to ABA than in the wild type. These results suggest that AtERF7 plays an important role in controlling ABA sensitivity in guard cells.
To determine whether the decreased stomatal sensitivity to ABA in the overexpession lines affects plant transpirational water loss, we compared the rate of water loss in excised leaves from wild-type and AtERF7 overexpression transgenic plants. The AtERF7 overexpression plants showed faster water loss than wild-type plants (Figure 2E). This result is consistent with the rapid wilting in overexpression plants upon drought treatment. To determine whether AtERF7 might be expressed in guard cells, we fused the AtERF7 promoter region to the β-glucuronidase (GUS) reporter and analyzed GUS activity in transgenic plants. As shown in Figure 2F, the AtERF7 promoter was highly active in leaf guard cells, suggesting that AtERF7 is expressed in guard cells.
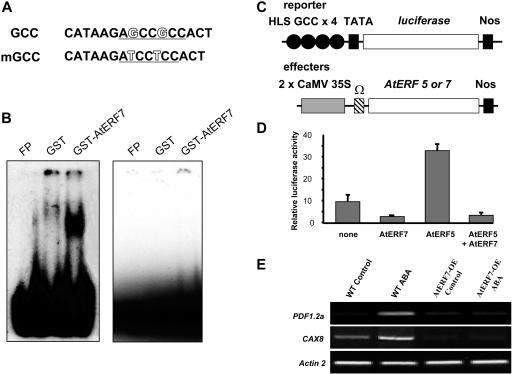
AtERF7 Binds to the GCC Box and Acts as a Repressor of GCC Box–Mediated Transcription
The amino acid sequence of AtERF7 suggests that it is a transcription factor (Ohta et al., 2001). Consistent with AtERF7 being a putative transcription factor, an AtERF7–green fluorescent protein (GFP) fusion protein was localized to the nucleus (data not shown). To analyze the DNA binding activity of AtERF7, we expressed the protein as a GST fusion in E. coli. Electrophoretic mobility shift assay results shown in Figure 3B reveal that the recombinant AtERF7 fusion protein binds to a 16-bp oligonucleotide probe containing the GCC box (Figure 3A). By contrast, AtERF7 does not bind to the oligonucleotide probe containing the mutant GCC box (Figure 3B). The mutant GCC box (Figure 3A) contains two point mutations that were shown to eliminate or reduce GCC box activity in vivo (Ohme-Takagi and Shinshi, 1995). In addition, the control protein GST is unable to bind the wild-type GCC box (Figure 3B).
Figure 3.
AtERF7 Binds to the GCC Box and Acts as a Transcriptional Repressor.
(A) GCC box sequences used in the experiments. The GCC box sequence is underlined. GCC, wild-type GCC box sequence; mGCC, GCC box with two mutations. The two nucleotides mutated in mGCC are outlined.
(B) AtERF7 binding to the GCC box. Electrophoretic mobility shift assay involved the use of a GST-AtERF7 fusion protein and a 16-bp wild-type GCC box oligonucleotide (left gel) or a mutated version (right gel). Free probe (FP), GST alone, or 0.1 μg of GST-AtERF7 was used in the reaction as indicated.
(C) Schemes of the reporter and effector plasmids. The Gal4 binding site and GCC box sequence were fused to a minimal TATA box and the LUC gene. The AtERF7 and AtERF5 effector plasmids were under the control of the CaMV 35S promoter. Ω, translational enhancer of tobacco mosaic virus; Nos, the terminator signal of the gene for nopaline synthase.
(D) Repression of reporter gene activity by AtERF7 and suppression of AtERF5-mediated transactivation by AtERF7. To normalize values obtained after each transfection, a gene for luciferase from Renilla was used as an internal control. Luciferase activity is expressed in arbitrary units relative to the activity of Renilla luciferase as described by Ohta et al. (2001). Values shown are means of data taken from three independent experiments; error bars indicate sd.
(E) AtERF7 overexpression reduces the expression of genes containing the GCC box in their promoter regions under ABA treatment. Wild-type and AtERF7 overexpression plants were treated with 100 μM ABA for 6 h, and total RNAs were isolated from the treated and untreated wild-type and transgenic plants. Actin2 primers were used in PCR as an internal control.
AtERF7 is highly similar to AtERF3, which has been shown to be a repressor of GCC box–mediated transcription (Fujimoto et al., 2000; Ohta et al., 2001). To test whether AtERF7 is also a transcriptional repressor, we performed transient expression experiments in Arabidopsis leaves using a reporter gene that has four tandem copies of the GCC box sequence around the HOOKLESS promoter, 4 × HLS GCC-LUC (Fujimoto et al., 2000). Expression of AtERF7 resulted in a substantial reduction of the expression of 4 × HLS GCC-LUC (Figure 3D). AtERF5 was known to act as an activator of GCC box–mediated transcription (Fujimoto et al., 2000). AtERF5 induced an activation of 4 × HLS GCC-LUC by fivefold, but coexpression of AtERF7 prevented this activation (Figure 3D). The observed repression was specific, as a reporter lacking the GCC box was not affected by AtERF7 (data not shown).
To begin to assess the role of AtERF7 in ABA-mediated gene expression in plants, we used semiquantitative RT-PCR analysis to compare two ABA-induced genes in the wild type and the AtERF7 overexpression line. Previous studies showed that the PDF1.2a (At5g44420) gene, encoding a plant defensin, has a typical GCC box (GCCGCC) in the promoter region (Brown et al., 2003). Our database survey found that the CAX8 (At5g17850) gene also has a typical GCC box in the promoter region. These genes were induced by ABA (Figure 3E), consistent with previous findings (H. Okamoto and M. Knight, unpublished data). We found that the ABA upregulation of these genes was blocked in the AtERF7 overexpression line compared with wild-type plants, suggesting that AtERF7 may have a role in ABA-mediated gene expression in plants.
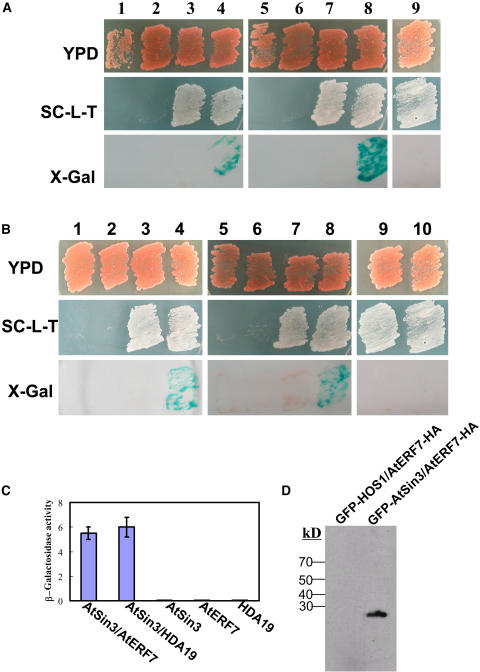
Interaction between AtERF7, AtSin3, and HDA19
There is growing evidence that transcriptional repression events, critical to all biological processes, may result from regulated alterations of chromatin remodeling (Wolffe, 1996; Goodrich and Tweedie, 2002). Both yeast and mammalian Sin3 can interact with DNA binding proteins (Wang and Stillman, 1990; Ayer et al., 1995), and Sin3-mediated repression of transcription may involve the recruitment of histone deacetylase (Alland et al., 1997; Hassin et al., 1997; Heinzel et al., 1997; David et al., 1998; Knoepfler and Eisenman, 1999). We hypothesized that the repression mechanism of AtERF7 in ABA responses might involve corepressors such as Sin3 and histone deacetylase to effect transcriptional repression via the modification of chromatin structure. To test this hypothesis, we surveyed the Arabidopsis genome and found a Sin3-like protein (At1g24190) that is highly similar to animal and yeast Sin3 and that possesses three putative paired amphipathic helix (PAH) domains. We tested potential interactions among the Arabidopsis AtSin3, AtERF7, and HDA19 (At4g38130) (Tian and Chen, 2001) using a yeast two-hybrid system. An AtSin3-GAL4 DNA binding domain fusion construct was used to produce the bait protein, with pACT-AtERF7 and pACT-HDA19 as preys. Cotransformation of yeast Y190 with these constructs (pAS-AtSin3 with pACT-AtERF7, pAS-AtSin3 with pACT-HDA19) yielded His/Trp/Leu auxotrophs that were also positive for β-galactosidase expression (Figure 4A). Separate transformation of Y190 with either pAS-AtSin3 or pACT-AtERF7 alone did not allow yeast growth on selective plates, and transformants did not express β-galactosidase. In addition, with yeast colonies from the cotransformation of pAS-AtSin3 with an empty prey vector or with the control prey protein (an RNA binding protein), β-galactosidase activity was not detected. These results show that AtSin3 interacts with both AtERF7 and HDA19 in the yeast two-hybrid system (Figures 4A and 4C).
Figure 4.
AtSin3 Interacts with AtERF7 and HDA19 in the Yeast Two-Hybrid System, and Coimmunoprecipitation between AtSin3 and AtERF7.
(A) AtSin3 interacts with AtERF7 or HDA19 but not with SOS1. Yeast strains containing pAS-AtSin3 bait and/or pACT-AtERF7 and pACT-HDA19 prey were assayed for LacZ expression. Yeast grown on yeast peptone dextrose (YPD) is shown in the top panel; β-galactosidase filter assay results are shown in the bottom panel. The middle panel shows yeast growth on SC medium lacking Trp and Leu (SC-L-T). 1, pAS-AtSin3; 2, pACT-AtERF7; 3, pAS-AtSin3 + pACT; 4, pAS-AtSin3 + pACT-AtERF7; 5, pAS-AtSin3; 6, pACT-HDA19; 7, pAS-AtSin3 + pACT; 8, pAS-AtSin3 + pACT-HDA19; 9, pAS-AtSin3 + pACT-RBP.
(B) The PAH domain of AtSin3 (amino acid residues 1 to 336) is required for interactions with AtERF7 and HDA19. 1, pAS-PAH; 2, pACT-AtERF7; 3, pAS-PAH + pACT; 4, pAS-PAH + pACT-AtERF7; 5, pAS-PAH; 6, pACT-HDA19; 7, pAS-PAH + pACT; 8, pAS-PAH + pACT-HDA19; 9, pAS-PAH + pACT-SOS1; 10, pAS-PAH + pACT-RBP. SOS1 and RBP (an RNA binding protein) were used as negative controls.
(C) Quantitative analysis of β-galactosidase activity of the yeast strains in liquid culture showing the interaction between AtSin3 and AtERF7 or HDA19. Values are the means of data taken from three independent experiments. Error bars indicate standard deviation.
(D) AtERF7 interacts with AtSin3 in vivo. AtERF7-HA and GFP-AtSin3 were transiently coexpressed in wild-type protoplasts, and then GFP-AtSin3 was precipitated with anti-GFP. The precipitated anti-GFP samples were subjected to anti-HA protein gel blot analysis. AtERF7 did not coimmunoprecipitate with HOS1.
The PAH domains of Sin3 are important for its function as a transcriptional repressor and for protein–protein interactions (Wang and Stillman, 1993; Ayer et al., 1995). To determine whether the PAH regions of Arabidopsis AtSin3 are important for the interaction with AtERF7 or HDA19, we constructed pAS-PAH (amino acid residues 1 to 336) as a bait. After transforming Y190 with pAS-PAH and pACT-AtERF7 or pACT-HDA19, we found that the PAH domain interacted with AtERF7 and HDA19 (Figure 4B). No interaction was detected between AtERF7 or HDA19 with AtSin3 fragments that lacked the PAH domains (data not shown).
To test whether AtERF7 and AtSin3 can also interact in vivo, coimmunoprecipitation experiments were performed. GFP-tagged AtSin3 (GFP-AtSin3) and HA-tagged AtERF7 (AtERF7-HA) were transfected into Arabidopsis protoplasts together. GFP-AtSin3 was then precipitated with anti-GFP antibodies. The results show that AtERF7-HA was detected in the proteins precipitated by anti-GFP from protoplasts cotransfected with both GFP-AtSin3 and AtERF7-HA, demonstrating that AtSin3 and AtERF7 could be coimmunoprecipitated. Control experiments show that AtERF7 did not coimmunoprecipitate with HOS1 from protoplasts cotransfected with AtERF7-HA and GFP-HOS1 (Figure 4D). These results suggest that AtERF7 and AtSin3 interact in vivo in plant cells.
AtSin3 and HDA19 Enhance AtERF7-Mediated Transcriptional Repression
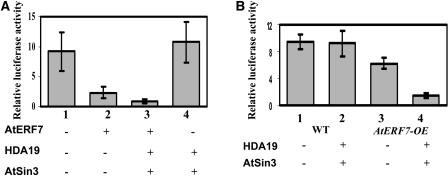
To investigate the role of AtSin3 and HDA19 in AtERF7-mediated repression of transcription, we tested the effect of AtSin3 and HDA19 on the ability of AtERF7 to regulate transcription in plant cells with the use of transient assays. Coexpression of AtERF7 with the 4 × HLS GCC-LUC reporter resulted in a 74% reduction in basal LUC transcription activity (Figure 5A, column 1 versus column 2; significant difference according to the Steel-Dwass test, P < 0.05). Coexpression of AtERF7, AtSin3, HDA19, and the 4 × HLS GCC-LUC reporter resulted in a further reduction in transcription activity (Figure 5A, column 2 versus column 3; significant difference according to the Steel-Dwass test, P < 0.05). Importantly, the transcription was not substantially reduced by coexpression of AtSin3 and HDA19 and the 4 × HLS GCC-LUC reporter without AtERF7 (Figure 5A, column 1 versus column 4), which suggests that the effect of AtSin3 and HDA19 on transcription repression is dependent on AtERF7. Consistent with these results, transfection of plasmids encoding AtSin3 and HDA19 in AtERF7 overexpression transgenic leaves resulted in increased transcription repression (Figure 5B, column 4 versus column 3; significant difference according to the Steel-Dwass test, P < 0.05). This effect is most likely attributable to the high level of AtERF7 protein present in the AtERF7 overexpression transgenic line. Thus, the results suggest that AtERF7 may function in transcriptional repression together with AtSin3 and HDA19.
Figure 5.
AtERF7-Mediated Transcription Repression Is Enhanced by HDA19 and AtSin3.
(A) Repression of reporter gene activity by AtERF7, AtSin3, and HDA19.
(B) Comparison of AtSin3- and HDA19-mediated repression activity in wild-type and AtERF7 overexpression (OE) leaves.
Values shown are means of data from three independent experiments; error bars indicate sd.
Reduced Expression of AtERF7 or AtSin3 Causes ABA Hypersensitivity
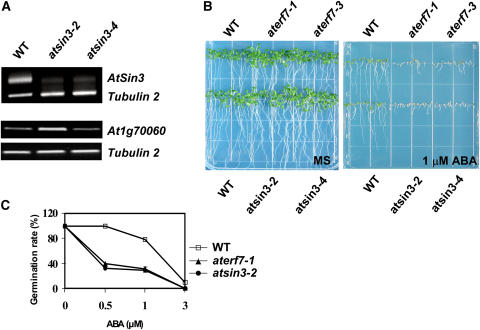
AtSin3 was knocked down by RNAi. RT-PCR analysis shows that AtSin3 expression was knocked down in the RNAi lines (Figure 6A). We tested the seed germination of representative T3 homozygous RNAi lines of AtERF7 and AtSin3 (aterf7-1, aterf7-3, atsin3-2, and atsin3-4) in response to ABA. When seeds were planted in MS agar medium, the aterf7 and atsin3 RNAi seeds germinated and the seedlings grew as well as the wild-type seedlings (Figure 6B). By contrast, when seeds were planted in MS medium supplemented with 1 μM ABA, the RNAi seeds germinated later than the wild-type seeds. Even though the RNAi seeds eventually germinated, their growth and development were delayed by ABA (data not shown). The ABA dose–response curves are presented in Figure 7C for lines aterf7-1 and atsin3-2. In our quantitation of seed germination, only seedlings with green cotyledons were scored as germinated. These results show that reduced expression of AtERF7 or AtSin3 leads to ABA hypersensitivity in germination.
Figure 6.
The aterf7 and atsin3 RNAi Lines Show ABA Hypersensitivity in Seed Germination.
(A) RT-PCR results showing AtSin3 (top gel) and an AtSin3 homolog (At1g70060; bottom gel) expression in the wild type and two independent atsin3 RNAi lines. Tubulin primers were used in PCR as an internal control.
(B) Germination sensitivity to ABA. Seeds from the wild type, two aterf7 RNAi lines, and two atsin3 RNAi lines were germinated and grown on MS agar medium (left panel) or MS agar supplemented with 1 μM ABA (right panel) for 10 d.
(C) Comparisons of germination rates of wild-type and aterf7 and atsin3 RNAi seeds after exposure to different concentrations of ABA for 5 d. Data represent means ± sd of three independent experiments (>110 seeds per point).
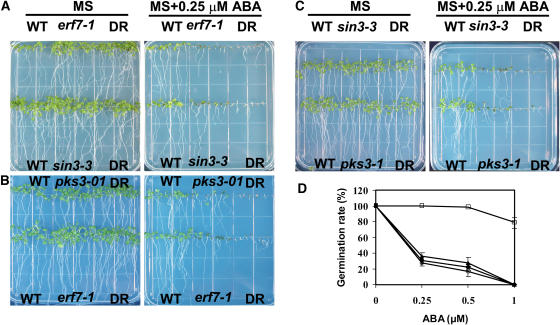
Figure 7.
ABA Sensitivity in aterf7 atsin3, aterf7 pks3, and atsin3 pks3 Double RNAi Lines.
(A) Fifteen-day-old seedlings of the wild type, two single RNAi lines (aterf7 and atsin3), and a double RNAi line grown in MS medium with or without 0.25 μM ABA. DR, the double RNAi line aterf7-1 atsin3-2.
(B) Fifteen-day-old seedlings of the wild type, two single RNAi lines (aterf7 and pks3), and a double RNAi line grown in MS medium with or without 0.25 μM ABA. DR, the double RNAi line aterf7-1 pks3-1.
(C) Fifteen-day-old seedlings of the wild type, two single RNAi lines (atsin3 and pks3), and a double RNAi line grown in MS medium with or without 0.25 μM ABA. DR, the double RNAi line atsin3-2 pks3-1.
(D) Germination of wild-type (open squares), aterf7 atsin3 (closed circles), aterf7 pks3 (open triangles), and pks3 atsin3 (open circles) seeds in the presence of different concentrations of ABA at d 4 after imbibition. Results are averages of three replicates ± sd.
We constructed the double RNAi lines aterf7-1 atsin3-2, aterf7-1 pks3-1, and atsin3-2 pks3-1 by crossing the respective single RNAi lines. On MS medium without ABA, the double or single RNAi lines germinated and grew similarly to the wild type. However, when they were germinated and grown on MS medium supplemented with 0.25 μM ABA, it is clear that the single RNAi lines were more sensitive to ABA, and the double RNAi lines were even more sensitive than the single RNAi lines (Figure 7). The greater ABA sensitivity of the double RNAi lines was indicated by the lack of cotyledon expansion on 0.25 μM ABA (Figure 7). Unlike the wild-type or single RNAi lines (Figure 7B), the double RNAi lines could not germinate on MS agar medium containing 1 μM ABA (Figure 7D).
DISCUSSION
In this study, we identified a transcriptional repressor that is important for ABA responses and provided evidence that suggests a connection between ABA signaling and gene repression by chromatin remodeling. Our genetic analysis showed that AtERF7 and AtSin3 play important roles in mediating ABA responses in Arabidopsis. Transgenic lines overexpressing AtERF7 showed reduced sensitivity to ABA in stomatal closure and increased sensitivity to drought stress (Figure 2). Consistent with this finding, Arabidopsis RNAi lines with AtERF7 expression knocked down displayed increased sensitivity to ABA in stomatal closing as well as seed germination. The effects observed with AtERF7 overexpression or RNAi plants are not necessarily specific for AtERF7, because we did not test the effect of overexpression or RNAi of AtERF3 or AtERF4. AtSin3 RNAi lines also were more sensitive to ABA in seed germination (Figure 6B). Our results indicate that AtERF7 overexpression may downregulate the expression of some ABA-induced genes. It appears that the role of AtERF7 may be to attenuate ABA responses in Arabidopsis. It is unknown at present whether AtERF7 also functions in other signaling pathways, such as ethylene and defense responses, that are known to involve the ERF family of transcription factors and their binding to the GCC box of target gene promoters (Fujimoto et al., 2000).
AtERF7 is very similar to AtERF3 and AtERF4, both of which have been shown to be transcription repressors (Fujimoto et al., 2000). Sequence analysis revealed that AtERF7 possesses the repression domain FDLNFXP (Ohta et al., 2001), which could bind to the GCC box in gene promoters. Our survey of the Arabidopsis genome database found ∼400 genes containing the GCC box sequence in their promoter regions (P. Wang, C.-P. Song, and J.-K. Zhu, unpublished data). AtERF7 may regulate at least a subset of these genes, and some or all of the AtERF7 target genes may function in the ABA pathway. We showed that AtERF7 can indeed repress the basal transcription level of a reporter gene and the transactivation activities of transcriptional activators (Figure 3D). This finding suggests that AtERF7 uses an active repression mechanism. There are several possible mechanisms of active repression (Cowell, 1994; Pazin and Kadonaga, 1997). One is that the transcription factor might recruit corepressors such as histone deacetylase, which modifies chromatin structure and prevents transcription activators from binding to the cis-elements (Pazin and Kadonaga, 1997). Yeast two-hybrid assays and coimmunoprecipitation suggested that AtERF7 interacts with AtSin3, which in turn may interact with HDA19 (Figure 4). Thus, in Arabidopsis, a repressor complex consisting possibly of AtSin3 and HDA19 may be targeted to relevant gene promoters via their association with AtERF7. At present, it is unclear whether AtSin3 is specific for the AtERF7 pathway, because we do not have data on the specificity of interaction between AtSin3 and the various AtERF proteins.
Several protein kinases, including PKS3, have been implicated in ABA signaling (Sheen, 1996; Li et al., 2000, Guo et al., 2002, Jiang et al., 2003). Reduced expression of Arabidopsis PKS3 by RNAi causes ABA hypersensitivity in seed germination, seedling growth, and stomatal closure. PKS3 interacts with calcium sensors (SCaBP5 and others) and with ABI2 (Guo et al., 2002). We found that AtERF7 interacted with PKS3 (Figure 1) and can be phosphorylated by PKS3 in vitro (Figure 1F). Furthermore, the aterf7 and pks3 RNAi lines were similar in their ABA hypersensitivity. The aterf7 pks3 double RNAi line showed a more dramatic ABA phenotype compared with the single RNAi lines and wild-type plants (Figure 7). These results suggest that AtERF7 could be a target protein of PKS3 in the ABA signal transduction pathway. Phosphorylation by PKS3 may increase the DNA binding and/or repression activity of AtERF7.
Our results suggest a model in which AtERF7, bound to the GCC box of ABA-induced genes and regulated by PKS3-mediated phosphorylation, may recruit an AtSin3/HDA19 corepressor complex to repress further gene transcription as part of the mechanism of attenuation of ABA signaling. Although we still do not know whether AtERF7 is phosphorylated in vivo by PKS3 and how phosphorylation might affect the activity of AtERF7, our observation of the interaction between AtERF7 and PKS3 suggests that the DNA binding and/or transcriptional repressor activity of AtERF7 may be regulated by PKS3 via phosphorylation. Our model provides a conceptual framework in which to integrate the molecular mechanisms of transcription repression caused by stresses and stress hormones (e.g., ABA) and may be relevant to the molecular mechanism of plant memory of stress or ABA treatment (Goh et al., 2003). Our results, together with those obtained in yeast and mammalian cells (Hassin et al., 1997; Nagy et al., 1997), suggest that Sin3–histone deacetylase is an evolutionarily conserved corepressor complex that represses the transcription of specific genes using sequence-specific DNA binding proteins to target a chromatin-modifying activity to gene-specific promoters. Future studies are needed to determine the spectrum of genes regulated by chromatin remodeling via sequence-specific transcriptional repressors such as AtERF7 and how the repressor complex may be regulated by ABA signaling.
METHODS
RNAi
Gene-specific cDNA fragments of AtERF7 and AtSin3 were amplified by PCR with the following primer pairs: forward primer 5′-CGGGATCCATTTAAATCCTGGAAATTACCTCCGCCG-3′ and reverse primer 5′-GGACTAGTGGCGCGCCGGTGGATCTGTGAAGGAGCC-3′ for AtERF7, forward primer 5′-CGGGATCCATTTAAAGAGCGTGCACACGATTCAACTGC-3′ and reverse primer 5′-GGACTAGTGGCGCGCCAAATCATCCTCCGGATCTTCACC-3′ for AtSin3. The forward primers contain BamHI and SwaI restriction sites, and the reverse primers contain SpeI and AscI restriction sites (underlined). The resulting PCR product was digested with AscI and SwaI and ligated to the AscI-SwaI–cleaved vector pFGC1008 (http://Ag.Arizona.Edu/chromatin/chromatin.html). The cloned fragments were sequenced to ensure that the correct cDNA was amplified and cloned. This plasmid served as a template to generate a second PCR fragment to complete the inverted repeat construct. The primers used to isolate the first PCR fragment were also used for the second fragment. This second PCR fragment was cleaved with BamHI and XbaI and inserted into the BamHI-XbaI sites of the template plasmid, producing an inverted repeat interrupted by the 335-bp GUS sequence. The double-strand RNA construct was introduced into Agrobacterium tumefaciens strain GV3101 and transformed into wild-type Arabidopsis thaliana (ecotype Columbia) plants by floral infiltration.
Total RNAs isolated from 2-week-old seedlings of the wild type and RNAi lines erf7-1, erf7-3, atsin3-2, and atsin3-4 were used as templates for reverse transcription by use of the SuperScript preamplification system for first-strand cDNA synthesis (Gibco BRL, Grand Island, NY), run for 50 min at 42°C. After cDNA synthesis, 1-μL aliquots were taken for PCR detection of transcripts with the forward primers for RNAi lines 5′-ATGAGGAAAGGGAGAGGCTC-3′ for AtERF7 and 5′-GCATCATGTGCATCAGCCATA-3′ for AtSin3. The reverse primer sequences are not present in the gene-specific AtERF7 and AtSin3 fragments in the RNAi constructs; therefore, they amplify only the respective gene transcripts. The reaction mixture (25 μL) contained 1× PCR buffer, 2 mM MgCl2, 0.2 mM deoxynucleotide triphosphates, 1.2 units of Taq polymerase, and 0.2 μM each of the primer pairs described above. PCR consisted of 30 cycles of 94°C for 1 min, 55°C for 1 min, and 72°C for 1 min. Cycling was preceded by an initial denaturation step (94°C for 2 min) followed by a final extension step (72°C for 10 min). The amplification products were then analyzed on 1% agarose gels.
Histochemical Analysis of GUS Activity
A DNA fragment containing 1627 bp upstream of the AtERF7 start codon was amplified by PCR with oligonucleotides incorporating the BamHI and SalI sites. The PCR fragment was digested with BamHI and SalI and then cloned into the SalI-BamHI site of the promoterless GUS expression vector pCMB1381. The construct ERF7-GUS was transferred from Escherichia coli DH5α into A. tumefaciens GV3101. Arabidopsis plants were transformed by floral infiltration with A. tumefaciens containing pAtERF7-GUS. Histochemical localization of GUS activities in the transgenic plants was analyzed after incubating the transgenic plants in X-gluc buffer (50 mM sodium phosphate buffer, pH 7.0, 10 mM EDTA, 0.1% Triton X-100, 0.5 mM potassium ferrocyanide, and 2 mg/mL 5-bromo-4-chloro-3-indolyl glucuronide [X-gluc]) at 37°C for 12 h.
Overexpression of AtERF7
For AtERF7 overexpression, the open reading frame of AtERF7 was amplified by PCR using the following primers: 5′-TCTAGATGAGGAAAGGGAGAGGCTCTTC-3′ (the XbaI site is underlined) and 5′-GGTACCTCAGAGACGTAGATCGGTACA-3′ (the KpnI site is underlined). The PCR products were first cloned into pBluescript SK− (Stratagene, La Jolla, CA) and confirmed by sequencing. Then, the AtERF7 open reading frame was released by digesting with XbaI and KpnI and subcloned into pBIB vector under the control of the superpromoter, which consists of three copies of the octopine synthase upstream activation sequence in front of the manopine synthase promoter (Narasimhulu et al., 1996). The construct was transformed into Arabidopsis plants (ecotype Columbia), and the resulting T3 transgenic lines were tested for ABA and drought sensitivity.
Semiquantitative RT-PCR
Five micrograms of total RNA from each sample was used to synthesize the first-strand cDNA with the SuperScript preamplification system using the First Strand cDNA Synthesis kit (Gibco BRL). The yield of cDNA was measured according to the PCR signal generated from the internal standard, the housekeeping gene β-actin, amplified from 18 to 24 cycles starting with 0.1 μL of the cDNA solution. The volume of each cDNA pool was adjusted to give the same exponential phase PCR signal strength for β-actin after 20 cycles. The resulting cDNAs were subjected to PCR with primers designed to amplify PDF1.2a and CAX8 DNAs. The expression of actin was used as an internal control. The RT-PCR product was analyzed by electrophoresis on a 1.5% agarose gel.
Yeast Two-Hybrid Assay
For yeast two-hybrid assays, the cDNA coding regions of AtSin3, PKS3, PKS11, and Pka18 were amplified by PCR with primers containing restriction sites for BamHI and PstI, and the amplified fragment was inserted into plasmid pAS2 (Clontech, Palo Alto, CA) to form pAS-AtSin3, pAS-PKS3, pAS-PKS11, and pAS-PKS11 as the baits. AtERF7, ERF3, and HDA19 were cloned in frame in the pACT2 vector between the BamHI-EcoRI (ERF3 and ERF4) and NcoI-XhoI (HDA19) sites to create plasmids pACT-AtERF7, pACT-ERF3, and pACT-HDA19, respectively. The yeast two-hybrid interaction assay was performed as described (Halfter et al., 2000; Guo et al., 2002). Transformants of yeast strain Y190 harboring both bait and prey were cultured in 2 mL of yeast peptone dextrose overnight. A 5-mL suspension containing 10,000 cells was dropped onto synthetic complete (SC) medium lacking Trp and Leu and incubated at 30°C for 48 h. Plasmid containing the C-terminal portion of SOS1 (Shi et al., 2000), pAS-SOS1C, and a RNA binding protein (C.-P. Song and J.-K. Zhu, unpublished data), pAS-RBP, were used as negative controls.
In Vitro and in Vivo Protein Interaction Assay
To produce bacterially expressed GST-ERF7 and GST-RB (Halfter et al., 2000), the coding regions of AtERF7 and RB (Halfter et al., 2000) cDNAs were cloned in-frame into the BamHI-EcoRI sites of pGEX-2TK. The constructs were introduced into E. coli BL21 DE3 cells. Recombinant proteins were affinity purified from bacterial lysates with glutathione–Sepharose (Amersham Pharmacia Biotech, Buckinghamshire, UK). Radiolabeled PKS3 proteins were produced from pET14b-PKS3 by use of an in vitro transcription and translation assay kit (TNT Coupled Reticulocyte Lysate System; Promega, Madison, WI) with [35S]Met as the sole source of Met, according to the manufacturer's instructions. Protein pulldown assays were performed as described previously (Guo et al., 2002).
In vivo coimmunoprecipitation experiments were performed as described (Guo et al., 2002).
Epidermal Strip Bioassay and Water Loss Measurement
Stomatal bioassay experiments were performed as described (Hugouvieux et al., 2001; Zhang et al., 2001), with slight modifications. To study the promotion of stomatal closure by ABA, stomata were opened by exposing plants for 12 h to light and high humidity and incubating the leaves for 1.5 h in stomata-opening solution containing 50 mM KCl, 10 μM CaCl, and 10 mM Mes, pH 6.15, in a growth chamber at 22 to 25°C under a photon flux density of 0.20 to 0.30 mmol·m−2·s−1. Stomatal apertures were measured 1.5 h after adding 2 μM ABA.
For water loss measurement, rosette leaves of the wild type, the RNAi mutant, and overexpression lines were detached from their roots, placed in weighing dishes, and incubated on the laboratory bench. Loss in fresh weight was monitored at the indicated times.
Transient Expression Assay
A plasmid for the expression of AtERF7 was constructed as follows. The Cauliflower mosaic virus (CaMV) 35S promoter and the GUS reporter gene in pBI121 was replaced by 2× CaMV 35S promoter to generate 2× 35S-Nos. The cDNAs for AtERF7, AtSin3, and HDA19 were amplified by PCR, digested with SalI, and inserted into SmaI and SalI sites of 2× 35S-Nos. 35S-AtERF5 and 4 × HLS GCC-LUC were constructed previously (Fujimoto et al., 2000; Ohta et al., 2001).
Transient expression was analyzed in Arabidopsis leaves by particle bombardment as described (Fujimoto et al., 2000). In brief, 510 μg of 1-μm gold biolistic particles (Bio-Rad, Hercules, CA) was coated with 1.6 μg of reporter plasmid and 1.2 μg of effector plasmid or blank plasmid. As a reference, the Renilla LUC gene (Promega) under the control of the CaMV 35S promoter was used at a reference gene:reporter gene ratio of 1:5. Bombardment was done using a PDS-1000/He particle bombardment system (DuPont, Wilmington, DE) at 11,000 p.s.i. After bombardment, the samples were floated on 50 mM phosphate buffer, pH 7.0, incubated in a plant growth chamber at 22°C overnight, frozen in liquid nitrogen, and quantified for LUC activity.
Protein Kinase Assay
PKS3-GST (Guo et al., 2002) and AtERF7-GST proteins were affinity purified with glutathione–Sepharose. The kinase buffer included 20 mM Tris-HCl, pH 8.0, 5 mM MgCl2, 50 μM ATP, and 1 mM DTT. The kinase reaction was in a total volume of 20 μL and was started by adding 100 ng of eluted PKS3 protein and 0.5 μL of [γ-32P]ATP (5 μCi), and the mixture was transferred to 30°C for 30 min. The reaction contained 100 ng of ERF7 protein on the glutathione–Sepharose beads. The reaction was stopped by adding 0.5 μL of 0.5 M EDTA. The beads were washed three times with kinase buffer, and samples were denatured by boiling for 4 min, then run on a 10% SDS-PAGE gel.
Electrophoretic Mobility Shift Assay
Individual synthetic single-stranded DNA molecules corresponding to the 16-bp GCC box fragment 5′-CATAAGAGCCGCCACT-3′ and its mutant 5′-CATAAGATCCTCCACT-3′ were annealed with their complementary oligonucleotides. The resulting double-stranded oligonucleotides were end labeled with [γ-32P]ATP (Amersham Pharmacia Biotech) and T4 polynucleotide kinase according to the manufacturer's instructions (Invitrogen, Carlsbad, CA). DNA binding reactions were performed as described previously (Ohme-Takagi and Shinshi, 1995). Briefly, 0.1 μg each of the GST-AtERF7 and GST or GST-RB proteins was added to a total volume of 10 μL in a binding buffer containing 10 mM Tris, pH 8.0, 1 mM EDTA, 100 mM NaCl, 2 mM DTT, 10% glycerol, and 10 fmol of the wild-type or mutant forms of the 16-bp oligonucleotide. After being incubated for 15 min, the reaction mixture was analyzed by electrophoresis on 5% polyacrylamide gels prepared in 0.5× Tris-Borate-EDTA under nondenaturing conditions.
Acknowledgments
We thank R.A. Stevenson for help with the preparation of the manuscript. This work was supported by National Science Foundation Grant IBN-0212346, National Institutes of Health Grant R01 GM-59138, the National Natural Science Foundation of China (30370765), and the National Key Basic Special Funds (2003CB114305).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Jian-Kang Zhu (jian-kang.zhu@ucr.edu).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.033043.
References
- Alland, L., Muhle, R., Hou, H., Potes, J., Chin, L., Schreiber-Agus, N., and DePinho, R.A. (1997). Role for NcoR and histone deacetylase in Sin3-mediated transcriptional repression. Nature 387 49–55. [DOI] [PubMed] [Google Scholar]
- Allen, G.J., Kuchitsu, K., Chu, S.P., Murata, Y., and Schroeder, J.I. (1999). Arabidopsis abi1-1 and abi2-1 phosphatase mutations reduce abscisic acid-induced cytoplasmic calcium rises in guard cells. Plant Cell 11 1785–1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assmann, S.M. (2003). OPEN STOMATA1 opens the door to ABA signaling in Arabidopsis guard cells. Trends Plant Sci. 8 151–153. [DOI] [PubMed] [Google Scholar]
- Assmann, S.M., and Wang, X.Q. (2001). From milliseconds to millions of years: Guard cells and environmental responses. Curr. Opin. Plant Biol. 4 421–428. [DOI] [PubMed] [Google Scholar]
- Ayer, D.E., Lawrence, Q.A., and Eisenman, N. (1995). Mad-max transcriptional repression is mediated by ternary complex formation with mammalian homologs of yeast repressor Sin3. Cell 80 767–776. [DOI] [PubMed] [Google Scholar]
- Brown, R.L., Kazan, K., McGrath, K.C., Maclean, D.J., and Manners, J.M. (2003). A role for the GCC-box in jasmonate-mediated activation of the PDF1.2 gene of Arabidopsis. Plant Physiol. 132 1020–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brubaker, K., Cowley, S.M., Huang, K., Loo, L., Yochum, G.S., Ayer, D.E., Eisenman, R.N., and Radhakrishnan, I. (2000). Solution structure of the interacting domains of the Mad-Sin3 complex: Implications for recruitment of a chromatin-modifying complex. Cell 103 655–665. [DOI] [PubMed] [Google Scholar]
- Chuang, C.F., and Meyerowitz, E.M. (2000). Specific and heritable genetic interference by double-stranded RNA in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 97 4985–4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowell, I.G. (1994). Repression versus activation in the control of gene transcription. Trends Biochem. Sci. 19 38–42. [DOI] [PubMed] [Google Scholar]
- Cutler, S., Ghassemian, M., Bonetta, D., Cooney, S., and McCourt, P. (1996). A protein farnesyl transferase involved in abscisic acid signal transduction in Arabidopsis. Science 273 1239–1241. [DOI] [PubMed] [Google Scholar]
- David, G., Alland, L., Hong, S.H., Wong, C.W., De Pinho, R.A., and Dejean, A. (1998). Histone deacetylase associated with mSin3A mediates repression by the acute promyelocytic leukemia-associated PLZF protein. Oncogene 14 2549–2556. [DOI] [PubMed] [Google Scholar]
- Finkelstein, R.R., Gampala, S.S., and Rock, C.D. (2002). Abscisic acid signaling in seeds and seedlings. Plant Cell 14 (suppl.), S15–S45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein, R.R., and Lynch, T.J. (2000). The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12 599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein, R.R., Wang, M.L., Lynch, T.J., Rao, S., and Goodman, H.M. (1998). The Arabidopsis abscisic acid response locus ABI4 encodes an APETALA2 domain protein. Plant Cell 10 1043–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto, S.Y.M., Ohta, M., Ushi, A., Shinshi, H., and Ohme-Takagi, M. (2000). Arabidopsis ethylene-responsive element binding factors act as transcriptional activators of GCC box-mediated gene expression. Plant Cell 12 393–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraudat, J., Hauge, B.M., Valon, C., Smalle, J., Parcy, F., and Goodman, H.M. (1992). Isolation of the Arabidopsis ABI3 gene by positional cloning. Plant Cell 4 1251–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh, C.H., Nam, H.G., and Park, Y.S. (2003). Stress memory in plants: A negative regulation of stomatal response and transient induction of rd22 gene to light in abscisic acid-entrained Arabidopsis plants. Plant J. 36 240–255. [DOI] [PubMed] [Google Scholar]
- Gong, D., Gong, Z., Guo, Y., Chen, X., and Zhu, J.-K. (2002. a). Biochemical and functional characterization of PKS11, a novel Arabidopsis protein kinase. J. Biol. Chem. 277 28340–28350. [DOI] [PubMed] [Google Scholar]
- Gong, D., Gong, Z., Guo, Y., and Zhu, J.-K. (2002. b). Expression, activation, and biochemical properties of a novel Arabidopsis protein kinase. Plant Physiol. 129 225–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich, J., and Tweedie, S. (2002). Remembrance of things past: Chromatin remodeling in plant development. Annu. Rev. Cell Dev. Biol. 18 707–746. [DOI] [PubMed] [Google Scholar]
- Guo, Y., Xiong, L., Song, C.-P., Gong, D., Halfter, U., and Zhu, J.-K. (2002). A calcium sensor and its interacting protein kinase are global regulators of abscisic acid signaling in Arabidopsis. Dev. Cell 3 233–244. [DOI] [PubMed] [Google Scholar]
- Halfter, U., Ishitani, M., and Zhu, J.-K. (2000). The Arabidopsis SOS2 protein kinase physically interacts with and is activated by the calcium-binding protein SOS3. Proc. Natl. Acad. Sci. USA 97 3735–3740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassin, C.A., Fleisscher, T.C., Billin, A.N., Schreiber, S.L., and Ayer, D.E. (1997). Histone deacetylase activity is required for full transcription repression by mSin3A. Cell 89 341–347. [DOI] [PubMed] [Google Scholar]
- Heinzel, T., et al. (1997). A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature 387 43–48. [DOI] [PubMed] [Google Scholar]
- Himmelbach, A., Hoffmann, T., Leube, M., Hohener, B., and Grill, E. (2002). Homeodomain protein ATHB6 is a target of the protein phosphatase ABI1 and regulates hormone responses in Arabidopsis. EMBO J. 21 3029–3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himmelbach, A., Yang, Y., and Grill, E. (2003). Relay and control of abscisic acid signaling. Curr. Opin. Plant Biol. 6 470–479. [DOI] [PubMed] [Google Scholar]
- Hoth, S., Morgante, M., Sanchez, J.P., Hanafey, M.K., Tingey, S.V., and Chua, N.-H. (2002). Genome-wide gene expression profiling in Arabidopsis thaliana reveals new targets of abscisic acid and largely impaired gene regulation in the abi1-1 mutant. J. Cell Sci. 115 4891–4900. [DOI] [PubMed] [Google Scholar]
- Hugouvieux, V., Kwak, J.M., and Schroeder, J.I. (2001). An mRNA cap binding protein, ABH1, modulates early abscisic acid signal transduction in Arabidopsis. Cell 106 477–487. [DOI] [PubMed] [Google Scholar]
- Jiang, J., An, G., Wang, P., Wang, P., Han, J., Jia, Y., and Song, C.-P. (2003). MAP kinase specifically mediates the ABA-induced H2O2 generation in guard cells of Vicia faba L. Chin. Sci. Bull. 48 1919–1926. [Google Scholar]
- Knoepfler, P.S., and Eisenman, R.N. (1999). Sin meets NuRD and other tails of repression. Cell 99 447–450. [DOI] [PubMed] [Google Scholar]
- Koornneef, M., Reuling, G., and Karssen, C.M. (1984). The isolation and characterization of abscisic acid-insensitive mutants of Arabidopsis thaliana. Physiol. Plant. 61 377–383. [Google Scholar]
- Lee, H., Xiong, L., Gong, Z., Ishitani, M., Stevenson, B., and Zhu, J.-K. (2001). The Arabidopsis HOS1 gene negatively regulates cold signal transduction and encodes a RING finger protein that displays cold-regulated nucleo–cytoplasmic partitioning. Genes Dev. 15 912–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung, J., and Giraudat, J. (1998). Abscisic acid signal transduction. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49 199–222. [DOI] [PubMed] [Google Scholar]
- Leung, J., Merlot, S., and Giraudat, J. (1997). The Arabidopsis ABSCISIC ACID-INSENSITIVE 2 (ABI2) and ABI1 genes encode homologous protein phosphatases 2C involved in abscisic acid signal transduction. Plant Cell 9 759–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J., Wang, X.Q., Watson, M.B., and Assmann, S.M. (2000). Regulation of abscisic acid-induced stomatal closure and anion channels by guard cell AAPK kinase. Science 287 300–303. [DOI] [PubMed] [Google Scholar]
- Lu, C., and Fedoroff, N. (2000). A mutation in the Arabidopsis HYL1 gene encoding a dsRNA binding protein affects responses to abscisic acid, auxin, and cytokinin. Plant Cell 12 2351–2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusser, A., Kolle, D., and Loidl, P. (2001). Histone acetylation: Lessons from the plant kingdom. Trends Plant Sci. 6 59–65. [DOI] [PubMed] [Google Scholar]
- McCarty, D.R., Hattori, T., Carson, C.B., Vasil, V., Lazar, M., and Vasil, I.K. (1991). The Viviparous-1 developmental gene of maize encodes a novel transcriptional activator. Cell 66 895–905. [DOI] [PubMed] [Google Scholar]
- Merlot, S., Gosti, F., Guerrier, D., Vavasseur, A., and Giraudat, J. (2001). The ABI1 and ABI2 protein phosphatases 2C act in a negative feedback regulatory loop of the abscisic acid signalling pathway. Plant J. 25 295–303. [DOI] [PubMed] [Google Scholar]
- Meyer, P. (2001). Chromatin remodelling. Curr. Opin. Plant Biol. 4 457–462. [DOI] [PubMed] [Google Scholar]
- Murphy, M., Ahn, J., Walker, K.K., Hoffman, W.H., Evans, R.M., Levine, A.J., and George, D.L. (1999). Transcriptional repression by wild-type p53 utilizes histone deacetylases, mediated by interaction with mSin3A. Genes Dev. 13 2490–2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustilli, A.C., Merlot, S., Vavasseur, A., Fenzi, F., and Giraudat, J. (2002). Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. Plant Cell 14 3089–3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy, L., Kao, H.-Y., Chakravati, D., Lin, R.J., Hassig, C.A., Ayer, D.E., Schreiber, S.L., and Evans, R.M. (1997). Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell 89 373–380. [DOI] [PubMed] [Google Scholar]
- Narasimhulu, S.B., Deng, X., Sarri, R., and Gelvin, S.B. (1996). Early transcription of Agrobacterium T-DNA genes in tobacco and maize. Plant Cell 8 873–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohme-Takagi, M., and Shinshi, H. (1995). Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. Plant Cell 7 173–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta, M., Masui, K., Hiratsu, K., Shinshi, H., and Ohme-Takagi, M. (2001). Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell 13 1959–1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazin, M.J., and Kadonaga, J.T. (1997). What's up and down with histone deacetylation and transcription? Cell 89 325–328. [DOI] [PubMed] [Google Scholar]
- Schroeder, J.I., Kwak, J.M., and Allen, G.J. (2001). Guard cell abscisic acid signalling and engineering drought hardiness in plants. Nature 410 327–330. [DOI] [PubMed] [Google Scholar]
- Sheen, J. (1996). Specific Ca2+-dependent protein kinase in stress signal transduction. Science 274 1900–1902. [DOI] [PubMed] [Google Scholar]
- Shi, H., Ishitani, M., Kim, C., and Zhu, J.-K. (2000). The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc. Natl. Acad. Sci. USA 97 6896–6901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, L., and Chen, Z.J. (2001). Blocking histone deacetylation in Arabidopsis induces pleiotropic effects on plant gene regulation and development. Proc. Natl. Acad. Sci. USA 98 200–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H., and Stillman, D.J. (1990). In vitro regulation of a SIN3-dependent DNA-binding activity by stimulatory and inhibitory I factors. Proc. Natl. Acad. Sci. USA 87 9761–9765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H., and Stillman, D.J. (1993). Transcriptional repression in Saccharomyces cerevisiae by a Sin3-LexA fusion protein. Mol. Cell. Biol. 13 1805–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X.Q., Ullah, H., Jones, A.M., and Assmann, S.M. (2001). G protein regulation of ion channels and abscisic acid signaling in Arabidopsis guard cells. Science 292 2070–2072. [DOI] [PubMed] [Google Scholar]
- Wolffe, A.P. (1996). Histone deacetylase: A regulator of transcription. Science 272 371–372. [DOI] [PubMed] [Google Scholar]
- Wu, K., Malik, K., Tian, L., Brown, D., and Miki, B. (2000). Functional analysis of a RPD3 histone deacetylase homologue in Arabidopsis thaliana. Plant Mol. Biol. 44 167–176. [DOI] [PubMed] [Google Scholar]
- Xiong, L., Gong, Z., Rock, C.D., Subramanian, S., Guo, Y., Xu, W., Galbraith, D., and Zhu, J.-K. (2001. a). Modulation of abscisic acid signal transduction and biosynthesis by an Sm-like protein in Arabidopsis. Dev. Cell 1 771–781. [DOI] [PubMed] [Google Scholar]
- Xiong, L., Lee, B.-h., Ishitani, M., Lee, H., Zhang, C., and Zhu, J.-K. (2001. b) FIERY1 encoding an inositol polyphosphate 1-phosphatase is a negative regulator of abscisic acid and stress signaling in Arabidopsis. Genes Dev. 15 1971–1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X., Zhang, L., Dong, F., Gao, J., Galbraith, D.W., and Song, C.-P. (2001). Hydrogen peroxide is involved in abscisic acid-induced stomatal closure in Vicia faba. Plant Physiol. 126 1438–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, C., Zhang, L., Duan, J., Miki, B., and Wu, K. (2005). Histone deacetylase 19 is involved in jasmonic acid and ethylene signaling of pathogen response in Arabidopsis. Plant Cell 17 1196–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]