Abstract
The conversion of castasterone (CS) to brassinolide (BL), a Baeyer-Villiger oxidation, represents the final and rate-limiting step in the biosynthesis of BL in plants. Heterologously expressed Arabidopsis thaliana CYP85A2 in yeast mediated the conversion of CS to BL as well as the C-6 oxidation of brassinosteroids (BRs). This indicated that CYP85A2 is a bifunctional enzyme that possesses BR C-6 oxidase and BL synthase activity. CYP85A2 is thus a cytochrome P450 that mediates Baeyer-Villiger oxidation in plants. Biochemical, physiological, and molecular genetic analyses of Arabidopsis CYP85A2 loss-of-function and overexpression lines demonstrated that CS has to be a bioactive BR that controls the overall growth and development of Arabidopsis plants. Mutant studies also revealed that BL may not always be necessary for normal growth and development but that Arabidopsis plants acquire great benefit in terms of growth and development in the presence of BL.
INTRODUCTION
Investigations concerning brassinosteroid (BR)-deficient mutants such as deetiolated2 (det2) (Li et al., 1996; Fujioka et al., 1997), dwarf4 (dwf4) (Choe et al., 1998), and constitutive photomorphogenesis and dwarfism (cpd) (Szekeres et al., 1996) in Arabidopsis thaliana, dwarf in tomato (Lycopersicon esculentum; Bishop et al., 1999), and lkb in pea (Pisum sativum; Nomura et al., 1997, 1999) showed that BR deficiency causes reduced shoot elongation, reduced fertility, delayed senescence, and altered vasculature and photomorphogenesis. Pleiotropic abnormal development can be rescued only by application of BRs. BR-insensitive mutants such as brassinosteroid-insensitive1 (bri1) (Li and Chory, 1997), brassinosteroid-insensitive2 (bin2) (Li et al., 2001; Li and Nam, 2002), and bri1-associated receptor kinase (bak1) (Nam and Li, 2002) in Arabidopsis, lka in pea (Nomura et al., 1997, 2003), and curl-3 in tomato (Koka et al., 2000) also exhibited abnormal phenotypes similar to those found in BR-deficient mutants. However, the phenotype of these mutants could not be restored to that of the wild type by application of BRs. Consequently, BRs are now considered as essential chemical signals and plant hormones, endogenous levels of which must be properly maintained in plants for normal growth and development.
To date, >50 BRs have been identified from the entire plant kingdom (reviewed in Fujioka, 1999; reviewed in Bajguz and Tretyn, 2003). These have been classified as C27-, C28-, and C29-BRs based on the nature of the alkyl groups at the C-24 position in the side chain of the 5α-cholestane carbon skeleton. Among these, C28-BRs such as castasterone (CS) and brassinolide (BL), which possess a C-24 methyl group, have been frequently identified in plant materials. Given their strong biological activity, CS and BL are considered to be the most important BRs in the plant kingdom. The biosynthesis of these BRs in plants has been extensively investigated by employing feeding experiments using isotope-labeled substrates, in addition to molecular genetic analyses of BR-deficient mutants (reviewed in Sakurai, 1999; reviewed in Bishop and Yokota, 2001; reviewed in Fujioka and Yokota, 2003). As a result, two parallel pathways, namely, the early and late C-6 oxidation pathway for C28-BRs, have been fully established (Figure 1). Campesterol is initially converted to campestanol (CN). In the early C-6 oxidation pathway, CN is initially oxidized to 6-oxoCN, which then undergoes successive oxidation to cathasterone (CT), teasterone (TE), 3-dehydroteasterone (3-DHT), typhasterol (TY), and finally CS. In the late C-6 oxidation pathway, CN is initially oxidized at C-22 to yield 6-deoxocathasterone, which then undergoes successive oxidation to 6-deoxoteasterone (6-deoxoTE), 6-deoxo-3-dehydroteasterone (6-deoxo-3-DHT), 6-deoxotyphasterol (6-deoxoTY), 6-deoxocastasterone (6-deoxoCS), and CS. Finally, CS is converted to BL via 7-oxalactonization.
Figure 1.
Biosynthetic Pathways to Generate BL in Plants.
Representative genes that catalyze BR biosynthesis are indicated. DET2, CYP85A1, CYP85A2, CYP90A1, and CYP90B1 are found in Arabidopsis. CYP90D2 (Hong et al., 2003) and CYP92A6 indicated with an asterisk are present in rice and pea, respectively.
The conversion of CS to BL has been demonstrated in cultured cells of Catharanthus roseus and Marchantia polymorpha and in the seedlings of C. roseus and Arabidopsis (Suzuki et al., 1993, 1995; Noguchi et al., 2000; Kim et al., 2003). Consequently, CS is now considered to be a direct biosynthetic precursor of BL. In certain plants, however, although a large amount of CS is present, not even trace levels of BL can be detected (reviewed in Yokota, 1997; reviewed in Fujioka, 1999). Additionally, the conversion rate of CS to BL is extremely low, even in plants where the conversion has been confirmed (Suzuki et al., 1995; reviewed in Sakurai, 1999; Noguchi et al., 2000). These findings suggested the possibility that CS is not only a biosynthetic precursor of BL, but also exerts its own biological activity. Although a higher concentration of CS was needed compared with BL, when exogenously applied to plants, CS displayed the same biological activity as that induced by BL and was capable of rescuing abnormal pleiotropic phenotypes in BR-deficient mutants. Curiously enough, no mutant of BL synthase (CS 6-oxidase), which catalyzes the conversion of CS to BL, has been found from Arabidopsis, tomato, pea, or rice (Oryza sativa), from which a variety of BR biosynthesis mutants has already been isolated. One reason for this could be that CS is biologically active on its own, as previously mentioned, so that any BL synthase mutant would not possess a significantly altered phenotype. This idea seems to be in accord with the recent finding that CS interacts with BRI1, the BL receptor protein, and competitively with BL (Wang et al., 2001). The conversion of CS to BL represents an interesting chemical reaction whereby an oxygen atom is inserted at a C-C bond, referred to as a Baeyer-Villiger oxidation. In some bacteria and fungi, the oxidation is mediated by Baeyer-Villiger monooxygenases (BVMOs), which are a class of flavoproteins, FAD-dependent monooxygenases (FMO) (Roberts and Wan, 1998). The presence of 26 FMOs in Arabidopsis (Fraaije et al., 2002) suggested that one of these might act as BL synthase, catalyzing the Baeyer-Villiger oxidation of CS to BL in the plant. However, we recently found that in Phaseolus vulgaris, BL synthase is located in the endoplasmic reticulum membrane, requires molecular oxygen and NADPH, and is inhibited by cytochrome P450 monooxygenase (Cyt P450) inhibitors, including carbon monoxide, the effect of which was reversed by irradiation with blue light (Kim et al., 2004a). This indicated that a Cyt P450 catalyzes the Baeyer-Villiger oxidation to generate BL.
We demonstrate here that an Arabidopsis Cyt P450, CYP85A2, a known BR C-6 oxidase, also possesses BL synthase activity responsible for the conversion of CS to BL. This demonstrates that a Cyt P450 mediates Baeyer-Villiger oxidation in plants. In this study, we also present the phenotype of Arabidopsis CYP85A2 knockout and overexpression lines. Molecular genetic and biochemical analyses of these mutants confirmed that CS, in addition to BL, should be a bioactive BR controlling the normal growth of plants and provided useful insights into the precise physiological role of CS and BL, which are the most important BRs in plants.
RESULTS
Arabidopsis CYP85A1 and CYP85A2 Possess Different Substrate Specificities
Arabidopsis contains two homologs (CYP85A1 and CYP85A2) of tomato CYP85 that mediate the conversion of 6-deoxoCS to CS, 6-deoxoTY to TY, 6-deoxo-3DHT to 3-DHT, and 6-deoxoTE to TE (Bishop et al., 1999; Shimada et al., 2001). Heterologously expressed Arabidopsis CYP85A1 and CYP85A2 in yeast also mediated the aforementioned C-6 oxidation reactions of BRs and demonstrated that both CYP85A1 and CYP85A2 possess BR C-6 oxidase activity (Shimada et al., 2001, 2003). However, why Arabidopsis contains two sets of genes encoding enzymes that possess the same function remains unknown. Recently, we demonstrated that CS is biosynthesized from cholesterol (CHR) via cholestanol and C27-BRs that have the same carbon skeleton as that of 28-norcastasterone (28-norCS) in young tomato plants (Kim et al., 2004b). Additionally, we demonstrated that 28-norCS could be converted to CS by S-adenosyl-l-Met–dependent sterolmethyltransferase in tomato plants. In Arabidopsis, the conversion of CHR to cholestanol has been demonstrated (Nakajima et al., 2002), and we recently determined that the conversion of 28-norCS to CS also occurs in Arabidopsis (T.-W. Kim and S.-K. Kim, unpublished data). This suggested that the C27-BR biosynthetic pathway from CHR to generate CS is also operative in the plant (Figure 1).
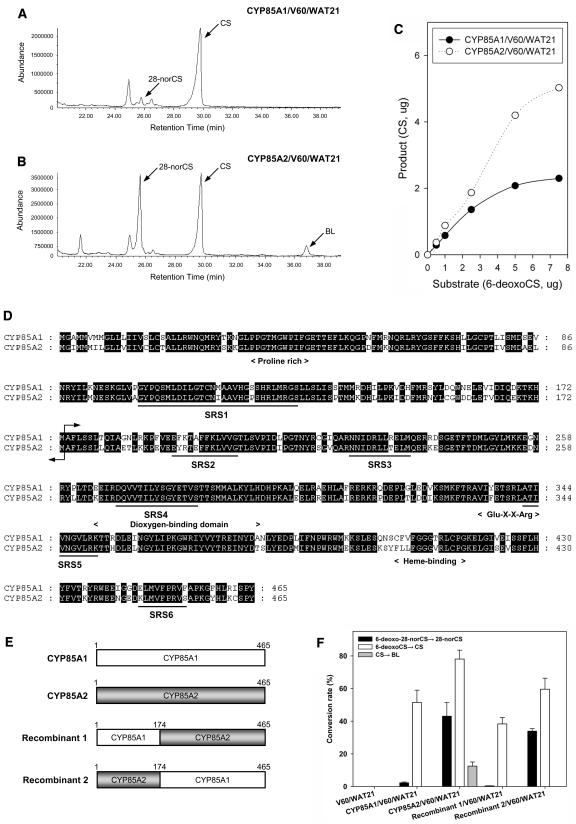
In the biosynthesis of C27-BRs to generate CS, heterologously expressed CYP85A1 also mediated the conversion of 6-deoxo-28-norcastasterone (6-deoxo-28-norCS) to 28-norCS (Kim et al., 2004b), although the conversion rate was clearly lower than that of 6-deoxoCS to CS. This suggested that CYP85A1 possesses higher substrate affinity for C28-BR and that the C-6 oxidation of C27-BRs might represent the predominant function of CYP85A2. In an effort to examine this possibility, CYP85A1 and CYP85A2 were overexpressed in the yeast strain WAT21, which carries Arabidopsis NADPH-Cyt P450 reductase (Urban et al., 1997). Enzyme activity was reexamined after feeding a mixture of 6-deoxo-28-norCS and 6-deoxoCS as sources of C27-BR and C28-BR to the yeast strains CYP85A1/V60/WAT21 and CYP85A2/V60/WAT21. The products were purified and analyzed by full-scan gas chromatography–mass spectrometry (GC-MS). As shown in Figure 2A, overexpressed CYP85A1 catalyzed the conversion of 6-deoxo-28-norCS to 28-norCS and of 6-deoxoCS to CS; however, the conversion rate of the former (2.0%) was much lower than that of the latter (43.9%). This suggested that the predominant function of CYP85A1 is to mediate the C-6 oxidation of a C28-BR. Overexpressed CYP85A2 also mediated the C-6 oxidation of 6-deoxo-28-norCS and 6-deoxoCS (Figure 2B). The conversion rate of 6-deoxo-28-norCS to 28-norCS (54.0%) and 6-deoxoCS to CS (83.5%) by CYP85A2 was significantly higher than that mediated by CYP85A1. Additionally, the results of in vivo kinetics that reflected the difference in C-6 oxidase activity between the two strains and determined by feeding experiments with 6-deoxoCS indicated that the activity of CYP85A2 was much higher than that of CYP85A1 (Figure 2C). These findings indicated that CYP85A2 is a stronger BR C-6 oxidase than CYP85A1 and that the C-6 oxidation of a C27-BR is predominantly catalyzed by CYP85A2. Taken together, it is clear that CYP85A1 and CYP85A2 possess different affinity for C27-BRs and C28-BRs as substrates, although they both possess BR C-6 oxidase activity.
Figure 2.
Biochemical Analysis of CYP85A1 and CYP85A2 Enzyme Activity.
(A) and (B) Total ion chromatograms of the products of CYP85A1 (A) and CYP85A2 (B) heterologously expressed in the transformed yeast strains CYP85A1/V60/WAT21 and CYP85A2/V60/WAT21. The same amount of 6-deoxo-28-norCS and 6-deoxoCS was simultaneously fed to each yeast strain.
(C) Comparison of BR C-6 oxidase activity in the CYP85A1/V60/WAT21 and CYP85A2/V60/WAT21 strains. Varying concentrations (0.5, 1, 2.5, 5, and 7.5 μg) of 6-deoxoCS as substrate were fed to the two yeast strains.
(D) Alignment of amino acid sequences of CYP85A1 and CYP85A2. The conserved domains (Pro-rich, dioxygen binding, Glu-X-X-Arg motif, and Heme binding) of most Cyt P450 enzymes are indicated below the aligned sequence. Black lines indicate the six substrate recognition sites (SRSs) in Cyt P450 (Gotoh, 1992; Werk-Reichhart and Feyereisen, 2000; Williams et al., 2004). The arrow refers to the recombination site of CYP85A1 and CYP85A2.
(E) Schematic illustration of chimeric enzymes generated by exchanging the N-terminal region of CYP85A1 and CYP85A2.
(F) Change in the substrate specificity of the CYP85A1 and CYP85A2 chimeric enzymes. No metabolite was detected in the V60/WAT21 strain (control).
Both CYP85A1 and CYP85A2 consist of 465 amino acids. They share 83% identity and 92% similarity (Figure 2D). In spite of this, CYP85A1 and CYP85A2 possess different substrate specificity for C27-BRs and C28-BRs. Consequently, a small number of amino acids that differ between CYP85A1 and CYP85A2 may determine the differential substrate specificity of CYP85A1 and CYP85A2. The CYP85A1 and CYP85A2 genes possess completely identical exon and intron structures. Part of the CYP85A1 and CYP85A2 genes (522 bp) encoding the N-terminal 174 amino acids was digested with the same restriction enzyme, and recombinant genes were constructed by the exchange of gene fragments (Figure 2E). The recombinant genes were overexpressed in the WAT21 yeast strain, and the respective enzyme activities were compared with that of CYP85A1 and CYP85A2. As shown in Figure 2F, recombinant 1, in which the N-terminal 174 amino acids of CYP85A2 were replaced by that of CYP85A1, mediated the C-6 oxidation of 6-deoxoCS to CS but possessed very weak activity for the conversion of 6-deoxo-28-norCS to 28-norCS, catalyzed by CYP85A2. Recombinant 2, in which the N-terminal 174 amino acids of CYP85A1 were substituted with the N terminus of CYP85A2, not only mediated the conversion of 6-deoxoCS to CS but also the conversion of 6-deoxo-28-norCS to 28-norCS, which is weakly catalyzed by CYP85A1. These findings indicated that the N terminus of CYP85A1 reduces the substrate specificity for C27-BRs and that the N terminus of CYP85A2 is important for the recognition of C27-BRs as substrates.
Arabidopsis CYP85A2 also Possesses BL Synthase Activity Responsible for the Conversion of CS to BL
In the aforementioned GC-MS analysis of CYP85A2-catalyzed products from 6-deoxoCS, an unexpected peak (36.78 min) was detected on the total ion chromatogram (Figure 2B). The product gave a molecular ion at a mass-to-charge ratio (m/z) of 528 and prominent ions at m/z 374, 344, 332, 177, 163, and 155 (base peak), which are characteristic ions of BL bismethaneboronate. By direct comparison of the mass spectrum and GC retention time with that of authentic BL bismenthaneboronate, the product was unambiguously identified as BL. The identified BL was thought to be derived from 6-deoxoCS via CS. To confirm this, 6-deoxoCS, CS, and deuterium-labeled CS (26, 28-[2H6]CS) were added to the culture media of CYP85A2/V60/WAT21, and the enzyme-catalyzed products were analyzed by GC-MS. When 6-deoxoCS was used as a substrate, two products, CS and BL, were identified. When CS and [2H6]CS were added, BL and [2H6]BL were identified (Table 1). Therefore, we confirmed that CYP85A2 mediates the conversion of not only 6-deoxoCS to CS but also of CS to BL. The possible involvement of CYP85A1 in similar conversions was also examined using the CYP85A1/V60/WAT21 yeast strain system. As expected, CS was identified as a product, although BL and [2H6]BL were not, strongly suggesting that CYP85A1 only possesses BR C-6 oxidase activity. Thus, we confirmed that the conversion of CS to BL is only mediated by CYP85A2.
Table 1.
GC-MS Analysis of CYP85A1 and CYP85A2 Enzyme Products
| Yeast Strains | Substrates | Productsa | RRt on GCb | Prominent Ions (m/z, Relative Intensity %) |
|---|---|---|---|---|
| CYP85A1/V60/WAT21 | 6-DeoxoCS | CS | 1.000 | 512 (M+, 72), 399 (7), 358 (29), 327 (10), 287 (27), 155 (100) |
| CS | NDc | – | – | |
| [26,28-2H6]CS | ND | – | – | |
| CYP85A2/V60/WAT21 | 6-DeoxoCS | CS | 1.000 | 512 (M+, 76), 399 (8), 358 (31), 327 (7), 287 (28), 155 (100) |
| BL | 1.242 | 528 (M+, 6), 374 (39), 344 (22), 332 (40), 177 (61), 163 (26), 155 (100) | ||
| CS | BL | 1.242 | 528 (M+, 6), 374 (40), 344 (21), 332 (38), 177 (60), 163 (25), 155 (100) | |
| [26,28-2H6]CS | [26,28-2H6]BL | 1.236 | 534 (M+, 6), 374 (39), 344 (23), 338 (44), 177 (64), 163 (28), 161 (100) |
Analyzed by its bismethaneboronate.
Relative retention time (RRt) with respect to CS (29.83 min).
Not detected.
Arabidopsis CYP85A2 Loss-of-Function Mutants Show Very Mild Abnormalities
Although the biochemical function of CYP85A2 was examined in some detail, the physiological role of CYP85A2 in plants remains unknown. In an effort to delineate the physiological role of CYP85A2, Arabidopsis CYP85A2 loss-of-function mutants cyp85a2-1 and cyp85a2-2 were selected from SIGnAL (http://siganl.salk.edu/cgi-bin/tdnaexpress) mutant pools. Sequence analysis of the genomic DNA flanking the T-DNA insertion site revealed that cyp85a2-1 possesses a T-DNA insertion at the 267th base pair position, and cyp85a2-2 possesses a single T-DNA insertion accompanying a deletion of 43 bp at the 2542th base pair position (Figure 3A). RT-PCR analysis using RNA isolated from the homozygous mutants and wild-type plants (ecotype Columbia-0 [Col-0]) showed that cyp85a2-1 and cyp85a2-2 were null alleles (Figure 3B).
Figure 3.
Phenotype of Arabidopsis CYP85A2-Deficient Mutants cyp85a2-1 and cyp85a2-2.
(A) Schematic diagram of T-DNA insertion in cyp85a2-1 and cyp85a2-2 mutants. Exons and introns are indicated by boxes and lines, respectively.
(B) RT-PCR analysis of CYP85A2 and CYP85A1 gene expression in the cyp85a2 mutant. Total RNA was isolated from the rosette leaves of 40-d-old plants. UBQ5 was used as a control.
(C) Comparison of rosette leaves of cyp85a2-2 (left) and cyp85a2-1 (right) with the wild type. Photographs were taken at 6 and 5 weeks, respectively, after germination. Bars = 1 cm.
(D) Ten-day-old seedlings of cyp85a2-2 and the wild type. Bar = 1 cm.
(E) Adult phenotype of the cyp85a2-2 mutant and the wild type. The photograph was taken 7 weeks after germination. Bar = 1 cm.
(F) Abnormal number of cauline leaves in the cyp85a2-2 mutant. 1, 2, and 3 indicate number of cauline leaves. Bar = 1 cm.
(G) Failure in silique development in an early inflorescence stem of the cyp85a2-2 mutant. Bar = 1 cm.
(H) Formation of siliques in the mature cyp85a2-2 mutant. Bar = 1 cm.
(I) and (J) Microscopy observation of a wild-type (I) and cyp85a2-2 mutant (J) flower. The cyp85a2-2 mutant possesses stamen filaments of short length.
Given that cyp85a2-1 and cyp85a2-2 displayed identical morphological defects (Figure 3C), further characterization focused on cyp85a2-2. Although young seedlings of cyp85a2-2 were a little smaller than the wild type, differences in growth were very subtle (Figure 3D). Although the life cycle is a little longer than that of the wild type, only slightly reduced stem elongation was observed in adult plants of cyp85a2-2 (Figure 3E). However, cyp85a2-2 showed dark green, rounded, and curled leaves and reduced petioles compared with those of wild-type plants (Figure 3C). After bolting, cyp85a2-2 showed abnormal cauline leaf development (Figure 3F) and silique formation (Figures 3G and 3H). Microscopy observations revealed that reduced silique formation was due to incomplete growth of stamens, leading to reduced self-pollination of stigmas and stamens (Figures 3I and 3J). Altogether, the overall phenotypic abnormalities in cyp85a2-2 were much milder than those in BR-deficient mutants and showed that Arabidopsis plants can grow normally to some extent in the absence of CYP85A2.
Abnormalities in the cyp85a2 Mutant Are Restored by Application of BL or CS
GC-MS analysis demonstrated that the cyp85a2 mutant was unable to generate detectable levels of BL like wild-type plants, but accumulated 6-deoxoCS (Table 2). This suggested that the abnormal phenotype of the cyp85a2 mutant might be caused by the accumulation of 6-deoxoCS. To confirm the possibility, 1 μM of 6-deoxoCS was applied to wild-type Arabidopsis for 1 week, and phenotypic changes of the wild type were examined. The phenotype of the wild type remained unchanged after application of 6-deoxoCS (data not shown) and demonstrated that the phenotype of the cyp85a2 mutant does not result from the accumulation of 6-deoxoCS.
Table 2.
Endogenous BR Levels in Wild-Type, cyp85a2, 35S-CYP85A2, cyp85a1, and 35S-CYP85A1 Plants
| Amount (ng/g Fresh Weight) |
||||||
|---|---|---|---|---|---|---|
| BR | Col-0 | cyp85a2-1 | cyp85a2-2 | 35S-CYP85A2 | cyp85a1 | 35S-CYP85A1 |
| 28-NorCSa | 0.24 | 0.07 | Trace | 0.32 | 0.22 | 0.21 |
| 6-DeoxoCS | 4.00 | 16.12 | 18.06 | 3.79 | 5.60 | 4.32 |
| CS | 2.01 | 1.54 | 1.71 | 2.51 | 1.92 | 2.18 |
| BL | NDb | ND | ND | 0.05 | ND | ND |
The endogenous amount was estimated by GC-selected ion monitoring using a calibration curve of authentic 28-norCS.
No detectable amount.
The endogenous level of CS in the cyp85a2 mutant decreased to 80% relative to wild-type Arabidopsis. This slight decrease suggested that a defect in BR C-6 oxidase in the cyp85a2 mutant might be complemented in part by overexpression of CYP85A1 in the mutants (Figure 3B). Therefore, use of the cyp85a2 mutant, which contains sufficient CS but no detectable amount of BL, is considered valuable in terms of delineating the physiological function of CS and BL.
The rescue of abnormalities in the cyp85a2 mutant, especially abnormal leaf development and male sterility, was examined by application of CS and BL in an effort to determine the biological activity of CS and BL. After application of BL to the center of the rosette of cyp85a2 plants, the overall growth of the rosette leaves was restored (Figure 4A). This resulted in almost complete recovery of the short petiole and round leaves in cyp85a2 in comparison with those of the wild type (Figure 4B). After application of BL to incomplete flowers in the early stage of bolting cyp85a2 (Figure 4D), the incompletely developed siliques on the inflorescence stem were rescued, resulting in normal silique formation (Figure 4E). The rescue of abnormalities in the cyp85a2 mutant was then examined after application of CS using the same methods as described above. As shown in Figures 4A, 4C, and 4F, abnormal rosette plant growth, leaf development, and self-fertilization in cyp85a2 were successfully restored to that of the wild type to almost the same degree as that observed after the application of BL. Because cyp85a2 lacks the ability to convert CS to BL, these findings established that CS, in and of itself, is also a biologically active BR that regulates the normal growth and development of plants.
Figure 4.
Rescue of Abnormalities in the cyp85a2 Mutant by Application of CS and BL.
(A) Restoration of rosette leaves of the cyp85a2-2 mutant after application of 1 μM CS and BL.
(B) and (C) Representative rosette leaves of the cyp85a2-2 mutant after application of 1 μM BL (B) and CS (C). Bars = 2 cm.
(D) to (F) Formation of siliques after application of 1 μM CS and BL. Inflorescence stem of the cyp85a2-2 mutant (D) before BR treatment. Floral buds indicated by dashed white circles were dipped into the BR solution. Silique development was restored after the exogenous application of BL (E) and CS (F).
In ecotype Col-0 used as background of cyp85a2, although CYP85A2 was highly expressed, BL was not detected (Table 2, Figure 3B) and suggested that BL can be synthesized in planta but is not detectable due to its presumed rapid turnover. A large accumulation of 6-deoxoCS in cyp85a2 can be explained by the lack of CYP85A2 activity that converts 6-deoxoCS to BL. Therefore, the aberrant growth and development of the cyp85a2 mutant is thought to result from the suppressed synthesis of CS and BL.
Overexpression of CYP85A2 Leads to Enhanced Growth and Development
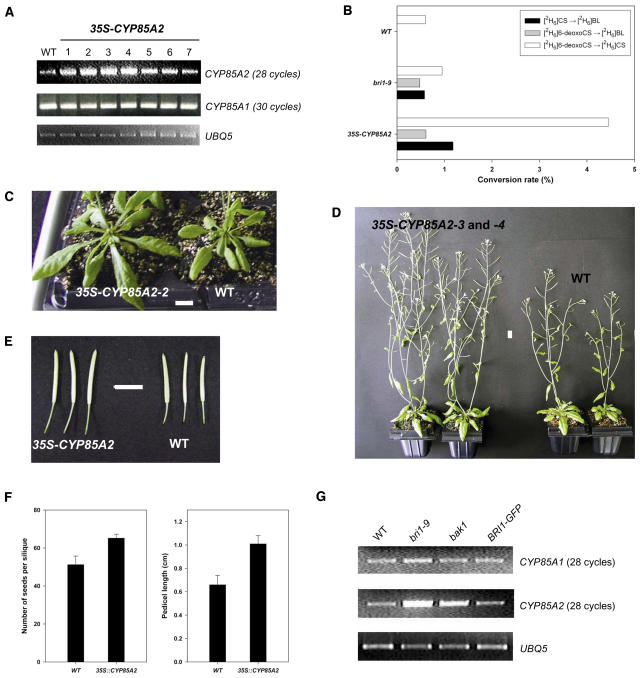
The Arabidopsis CYP85A2 overexpression lines (35S-CYP85A2) were generated in an effort to examine the physiological importance of CYP85A2 in plant growth and development. Seven independent transgenic plants that accumulated high levels of CYP85A2 transcript were screened by RT-PCR analysis (Figure 5A). In the 35S-CYP85A2 plants, the expression level of CYP85A1 was almost equal to that of wild-type plants.
Figure 5.
Biochemical Analysis and Phenotype of Arabidopsis CYP85A2 Overexpression Plants, 35S-CYP85A2.
(A) RT-PCR analysis of CYP85A2 and CYP85A1 mRNA levels in seven independent 35S-CYP85A2 lines.
(B) Result of feeding experiment on wild-type, bri1-9, and 35S-CYP85A2 plants using labeled BRs.
(C) Rosette leaves in 35-d-old 35S-CYP85A2 and wild-type (Col-0) plants. Bar = 1 cm.
(D) Inflorescence of 35S-CYP85A2 and wild-type plants. Bar = 1 cm.
(E) Siliques of 35S-CYP85A2 and wild-type plants. Bar = 1 cm.
(F) Comparison of the number of seeds per silique and pedicel length in 35S-CYP85A2 and wild-type plants. Error bars in the graph denote se (n > 30).
(G) RT-PCR analysis of CYP85A1 and CYP85A2 mRNA levels in wild-type, bri1-9, bak1, and BRI1-GFP plants. mRNA was isolated from 10-d-old seedlings. UBQ5 was used as a control.
Feeding experiments using [2H6]6-deoxoCS and [2H6]CS revealed that the conversion of [2H6]6-deoxoCS to [2H6]CS was significantly higher in 35S-CYP85A2 than in wild-type plants. Furthermore, 35S-CYP85A2 successfully mediated the conversion of [2H6]6-deoxoCS to [2H6]BL via [2H6]CS and [2H6]CS to [2H6]BL, which were not detected in the wild type (Figure 5B). Therefore, it was demonstrated that 35S-CYP85A2 is functionally operative for the overexpression of CYP85A2, resulting in a high level of BL in the mutants. As shown in Table 2, the endogenous level of 28-norCS and CS increased by ∼25 and 30%, respectively, compared with that of wild-type plants. Furthermore, BL was detected in 35S-CYP85A2 but not in the wild type.
As shown in Figure 5C, 35S-CYP85A2 possesses larger rosette leaves with longer petioles compared with wild-type plants. In adult plants, 35S-CYP85A2 possesses taller and multibranched stems compared with the wild type (Figure 5D). Additionally, larger siliques with longer pedicels were set on inflorescence stems in 35S-CYP85A2 compared with the wild type, resulting in an ∼30% increase in the number of seeds in a silique (Figures 5E and 5F). Consequently, the vegetative and reproductive growth of Arabidopsis was enhanced by overexpression of CYP85A2. In conclusion, BL produced by overexpression of CYP85A2 provided Arabidopsis plants with significant advantages in terms of growth and development.
Transcript Level of CYP85A2 Is Highly Maintained in BR Signaling Mutants
It was recently demonstrated that the endogenous level of CS and BL in the Arabidopsis bri1 mutant defective in a BR receptor kinase was higher than that in wild-type plants (Noguchi et al., 1999). This suggested that gene expression of CYP85A1 and/or CYP85A2 is highly maintained in BR signaling mutants. To verify this, the expression of CYP85A1 and CYP85A2 in three BR-signaling related plants, bri1, bak1, and BRI1-green fluorescent protein (GFP), was examined. As shown in Figure 5G, semiquantitative RT-PCR revealed that the expression of CYP85A1 was not significantly higher in the bri1-9 and bak1 mutants, whereas the expression of CYP85A2 was significantly higher in both mutants, especially in bri1-9. In contrast with bri1-9 and bak1, BRI1-GFP, which possesses an additional set of BRI1, did not show increased gene expression of CYP85A1 or CYP85A2. Taken together, the high level of CS and BL in bri1-9 (Noguchi et al., 1999) is considered to originate from enhanced gene expression of CYP85A2. In fact, [2H6]6-deoxoCS and [2H6]CS fed to bri1-9 were successfully converted into [2H6]CS and [2H6]BL (Figure 5B). This finding confirmed that the production of CS and BL is strongly associated with the high level of CYP85A2 expression in the mutants.
Overexpression and Knockout Mutation of CYP85A1 Do Not Cause Any Phenotypic Changes
The phenotypes of 35S-CYP85A1 and cyp85a1 null mutants were compared with that of wild-type plants in an effort to examine the function of CYP85A1, which also possesses BR C-6 oxidase activity. Four 35S-CYP85A1 lines that maintained higher levels of CYP85A1 than the wild type did not display any phenotypic changes compared with wild-type plants (Figures 6A and 6B). Similarly, the cyp85a1 null mutant possessing a T-DNA insertion at the 1819th base pair position did not display any phenotypic changes compared with wild-type plants (Figure 6C). These results were somewhat unexpected because in the 35S-CYP85A1 and null mutants, a higher and lower endogenous level of CS, respectively, was expected compared with wild-type plants. However, GC-MS analysis revealed that endogenous levels of CS in both the 35S-CYP85A1 and cyp85a1 mutants was almost the same as that in wild-type plants (Table 2). This indicates that the endogenous level of CS in 35S-CYP85A1 is not significantly affected by CYP85A1 activity. This result also suggests that the almost equal amount of CS in the cyp85a1 mutant compared with that in the wild type is maintained by the functional redundancy of CYP85A2 in Arabidopsis plants. Taken together, the physiological importance of CS in Arabidopsis growth and development has been reconfirmed.
Figure 6.
Phenotype of CYP85A1 Overexpression and cyp85a1 Mutant.
(A) RT-PCR analysis for overexpression of CYP85A1 in four independent transgenic lines.
(B) Inflorescence of 35S-CYP85A1 and wild-type (Col-0) plants. Bar = 2.5 cm.
(C) Rosette leaves in cyp85a1 and wild-type (Col-0) plants. Bar = 1 cm.
(D) Comparison of wild-type, cyp85a1, and cyp85a2-2 phenotypes.
DISCUSSION
We demonstrated here that Arabidopsis CYP85A1 and CYP85A2, two orthologs of tomato CYP85 known to mediate the same C-6 oxidation of BRs, are indeed functionally distinct. First, CYP85A1 and CYP85A2 display differential substrate specificity toward C27-BR and C28-BR substrates. CYP85A1 has a preference for C28-BR as substrate, whereas CYP85A2 possesses high affinity for both C28-BR and C27-BR. Chimeric enzymes constructed by the exchange of CYP85A1 and CYP85A2 gene fragments revealed that the SRS1 region in the N-terminal 174 amino acids of CYP85A2 might play an important role in the recognition of C27-BR substrates. As shown in Figure 2D, the N-terminal 174 amino acids contains a substrate recognition site (SRS1) of the six SRSs found in Cyt P450s. Given that alteration of SRSs may generate enzymes with differential substrate affinity, Ile 115 and Pro 121 in SRS1 of CYP85A2 might play a role in the recognition of C27-BRs. Tomato CYP85A1 only possesses BR C-6 oxidase activity. Examination of the amino acid sequence of tomato CYP85A1 shows that Ile 115 is the same as that in Arabidopsis CYP85A2, whereas Ser 121 is identical to that in CYP85A1. Investigation of the substrate specificity of tomato CYP85A1 for C27-BRs and C28-BRs could reveal the critical amino acid that determines the affinity for C27-BRs. Our results suggested that C-6 oxidation in C28-BR biosynthesis is mediated by both CYP85A1 and CYP85A2, whereas C-6 oxidation in C27-BR biosynthesis is predominantly catalyzed by CYP85A2. Therefore, CYP85A2 possesses versatile BR C-6 oxidase activity that can regulate both the biosynthesis of C27-BRs and C28-BRs. Secondly, unlike CYP85A1, CYP85A2 possesses both BL synthase and BR C-6 oxidase activity. In the 35S-CYP85A2 and a BR-insensitive mutant, bri1-9, which possesses significantly enhanced CYP85A2 expression, the conversion of 6-deoxoCS → CS → BL was demonstrated. This finding confirmed that CYP85A2 is a bifunctional enzyme that serves as a BR C-6 oxidase and BL synthase. The result that heterologously expressed CYP85A1 in yeast does not show activity for BL synthase strongly suggests that CYP85A1 possesses only BR C-6 oxidase activity. Nevertheless, the possibility for CYP85A1 as BL synthase is not completely excluded because heterologously expressed Cyt P450s involved in BRs synthesis, such as CYP90A1 and CYP90B1, have no biochemical function in yeast. In any case, our findings represent concrete evidence at the molecular level that a Cyt P450 catalyzes the conversion of CS to BL.
Cyt P450s have been shown to play crucial roles in the biosynthesis of BL. The hydroxylation of C-22 and C-23 was demonstrated to be catalyzed by Cyt P450s, namely DWF4 and CPD, respectively, as determined from biochemical and molecular genetic analyses of dwf4 and cpd Arabidopsis mutants. DWF4 and CPD were shown to possess significant homology to mammalian steroid hydroxylases (Szekeres et al., 1996; Choe et al., 1998). The C-6 oxidation of 6-deoxoCS to CS was shown to be mediated by Cyt P450, as determined from analysis of the tomato dwarf (d) gene, which is homologous to mammalian steroid hydroxylase genes (Bishop et al., 1999). Furthermore, two D orthologs, referred to as BR6OX1 and BR6OX2, have been isolated from Arabidopsis (Shimada et al., 2001, 2003). The proteins CPD, DWF4, BR6OX1, and BR6OX2 represent new members of the Cyt P450 group and have subsequently been referred to as CYP90A1, CYP90B1, CYP85A1, and CYP85A2, respectively. Recently, it was reported that the pea gene Dark induced DWF-like protein1 (CYP92A6) encodes a Cyt P450 that might catalyze the C-2 hydroxylation of TY to CS and of 6-deoxoTY to 6-deoxoCS and interact with a small GTP binding protein, Pra2 (Kang et al., 2001). In addition, involvement of a Cyt P450, referred to as CYP724B1, in BR biosynthesis was suggested in rice (Tanabe et al., 2005). In this study, we demonstrated that CYP85A2 also possesses BL synthase enzyme activity and mediates the conversion of CS to BL. Therefore, all oxygen insertion reactions involved in BL synthesis are catalyzed by Cyt P450s (Figure 1).
The Baeyer-Villiger oxidation reaction that forms part of the conversion of CS to BL involves insertion of an oxygen atom between the C-6 ketone and vicinal carbon at C-7 in CS, yielding a 7-oxa-lactonic B ring of BL. To date, this type of reaction was documented as being mediated by BVMOs, which are a class of FMO. BVMOs have been identified in bacteria and some fungi, which suggested that BVMOs play important roles in microbial oxidative metabolic pathways (Roberts and Wan, 1998). All prokaryotic BVMOs, such as steroid monooxygenase, cyclohexanone monooxygenase, cyclopentanone monooxygenase, and cyclododecanone monooxygenase, possess a specific domain carrying the unique sequence motif FXGXXXHXXXW(P/D) (Fraaije et al., 2002). To date, no BVMOs have been found in animals. However, the Baeyer-Villiger rearrangement involved in progesterone oxidation and lanosterol 14α-demethylation was shown to be mediated by CYP17 and CYP51, respectively (Fischer et al., 1991; Mak and Swinney, 1992; Swinney and Mak, 1994). In addition to BL, a steroidal plant hormone, plants produce various lactonic compounds, such as artemisinin, costunolide, nepetalactone, and digitoxigenin, as secondary metabolites (Croteau et al., 2000). However, characterization of the enzyme(s) responsible for mediating Baeyer-Villiger oxidation in plants has yet to be reported. In the Arabidopsis genome, as in animals, genes carrying BVMO sequence motifs have not been found. Nevertheless, the presence of 26 flavin-containing monooxygenases (FMOs) in the plant has suggested that they may act as enzyme(s) mediating Baeyer-Villiger oxidation (Fraaije et al., 2002). Lately, we demonstrated by biochemical analysis using Cyt P450 inhibitors that BL synthase, the enzyme mediating the conversion of CS to BL, is not an FMO but a Cyt P450 (Kim et al., 2004a). Subsequently, we demonstrated in this study that CYP85A2 possesses enzyme activity for the conversion of CS to BL. Recently, Nomura et al. (2005) reported that a Cyt P450 referred to as CYP85A3, which is preferentially expressed in tomato fruits, possesses BL synthase activity. They also demonstrated that CYP85A2 in Arabidopsis possesses the same BL synthase activity as that of tomato CYP85A3. Accordingly, tomato CYP85A3 and Arabidopsis CYP85A2 are Cyt P450s that mediate Baeyer-Villiger oxidation, suggesting that Baeyer-Villiger oxidation in plants, at least conversion of CS to BL, may be mediated by Cyt P450s.
Arabidopsis contains 272 Cyt P450s (http://www.p450.kvl.dk/p450.shtml). Among these, functions for only ∼30 Cyt P450s have been characterized (http://arabidopsis-p450.biotec.uiuc.edu/functions.shtml). Based on the finding that a Cyt P450 mediates Baeyer-Villiger oxidation, some unidentified Cyt P450s may be involved in the generation of useful lactonic compounds produced by plants. Considering that microbial BVMOs are used as biocatalysts to produce commercially valuable chiral lactones in synthetic chemistry (reviewed in Willetts, 1997; Burton, 2003), plant Cyt P450s mediating Baeyer-Villiger oxidation are also thought to be commercially promising enzymes for use as biocatalysts in green chemistry (Marko et al., 2002).
The precise physiological role of CS, whether CS is only a biosynthetic precursor of BL, and whether CS itself possesses the ability to control growth and development in plants remain unknown. The best way to unambiguously delineate the role of CS is through investigation of a BL synthase-deficient mutant and comparison of the phenotype with that of the wild type. If BL were the only biologically active BR, the mutant would show severe abnormalities found in other BR-deficient mutants such as det2, cpd, and dwf4. If CS were also a bioactive BR, the mutant would not display manifest abnormalities or show enhanced growth and development because of the accumulation of CS in the mutant. In this study, we demonstrated that CYP85A2 is a bifunctional enzyme that can regulate the endogenous level of both CS and BL. In fact, the cyp85a2 mutant contained no detectable BL and 20% less CS compared with wild-type plants. CYP85A1 transcript levels in cyp85a2 revealed that the CS level was maintained by functionally redundant CYP85A1 in the mutants. The cyp85a2 mutant did not display significant defects in growth, such as dwarfism, suggesting that normal stem elongation could be induced by endogenous CS. Silique formation was almost normal in completely matured mutants. In the early stage after bolting, however, the reduced growth of stamen filaments did not reach the head of the stigma, resulting in clearly reduced silique formation. The self-pollination was restored by application of CS as well as BL to the incomplete flowers of the cyp85a2 mutant. The abnormal development of cyp85a2 rosette leaves was also restored to that of the wild type by application of BL or CS. Application of CS almost completely restored the abnormalities in cyp85a2, indicating that CS must be a bioactive BR that can trigger the BR signal transduction pathway to express BR activity.
To maintain homeostatic levels, active BRs in plants must be deactivated or degraded after exerting their activity. The Arabidopsis CYP72B1 overexpression mutant, bas1-D (phyB activation-tagged suppressor 1-dominant), produced by activation tagging methods, facilitated the conversion of BL to 26-hydroxyBL, resulting in a reduced level of BL activity (Neff et al., 1999). We previously reported on the use of cell-free extracts prepared from P. vulgaris and M. polymorpha and determined that BL is biodegraded to 26-norbrassinolide, which also reduces BL activity (Kim et al., 2000a, 2000b). Therefore, 26-hydroxylation and 26-demethylation are thought to be deactivation or degradation reactions that maintain a steady state level of BL. It was recently reported that overexpression of Arabidopsis CYP72B1 in yeast also mediated the 26-hydroxylation of CS (Turk et al., 2003). Additionally, we demonstrated that 26-demethylation of CS occurs in Phaseolus and Marchantia cell-free extracts (Kim et al., 2004c). These data indicated that the endogenous level of CS is strictly controlled by oxidative deactivation as observed in the metabolism of BL.
Although abnormalities in the cyp85a2 mutant are much milder, the cyp85a2 mutant also displayed characteristic phenotypic alterations, such as delayed lifespan, short petiole, rounded leaves, and reduced self-pollination as shown in other BR-deficient mutants. The abnormal phenotype in cyp85a2 seems to be due to insufficient amounts of CS resulting from the absence of CYP85A2 in the mutant. Endogenous levels of CS in Arabidopsis are generated by the C-6 oxidation of 6-deoxoCS and the C-24 methylation of 28-norCS that was synthesized from 6-deoxo-28-norCS by C-6 oxidation (Figure 1). Deficiencies in CYP85A2 affect the C-6 oxidation of 6-deoxoCS and 6-deoxo-28-norCS, as well as the production of BL, despite the functional redundancy of CYP85A1 toward 6-deoxoCS. Consequently, it is thought that endogenous amounts of CS generated through the aforementioned two routes are not fully maintained in the cyp85a2 mutant compared with that of wild-type plants. Indeed, GC-MS analysis showed reduced levels of CS and 28-norCS in the cyp85a2 mutant (Table 2). On the other hand, 6-deoxoCS significantly accumulated in the cyp85a2 mutant. This probably resulted from the redundant higher levels of CYP85A1 in the mutant that are unable to sufficiently overcome the BR C-6 oxidase defect in cyp85a2. Taken together with the fact that CYP85A2 expressed in yeast showed stronger BR C-6 oxidase activity for a C28-BR than CYP85A1, this implied that CYP85A2 is a more efficient BR C-6 oxidase than CYP85A1. Consequently, CYP85A2 is a key enzyme that can regulate endogenous levels of CS by controlling both C28-BR and C27-BR biosynthetic pathways.
Real-time RT-PCR analysis revealed that CYP85A1 and CYP85A2 are expressed in an organ-specific manner in Arabidopsis plants (Shimada et al., 2003). Although CYP85A2 gene expression was most abundant in apical shoots, it is relatively ubiquitous. By contrast, CYP85A1 expression is predominant in apical shoots and siliques. Because apical shoot growth and silique formation require a high level of BR activity, it is thought that CYP85A1 supplies additional CS to the pool of CS and BL synthesized by CYP85A2. Therefore, CYP85A1 plays a supplementary role to CYP85A2 in terms of plant growth and development, at least in terms of apical shoot growth and seed formation.
In fact, the cyp85a1 null mutant showed no significant abnormalities in Arabidopsis development. In spite of the higher expression of CYP85A1, by contrast, the cyp85a2 mutant showed only mild abnormal phenotypes. This suggested that CYP85A2 is more important in the growth and differentiation of Arabidopsis, even though both CYP85A1 and CYP85A2 are functionally redundant in the plant. Based on the aforementioned relationship between phenotype and the functional redundancy of CYP85A1 and CYP85A2, cyp85a1 cyp85a2 double mutants may give rise to severe abnormalities during the growth and development of Arabidopsis, given that the double mutants have no CS because of a complete defect in BR C-6 oxidase. This hypothesis was recently verified by comparison of the phenotypes of Arabidopsis cyp85a1, cyp85a2, and cyp85a1 cyp85a2 (Nomura et al., 2005).
Endogenous levels of CS and 28-norCS in 35S-CYP85A2 were increased to only 25 and 33%, respectively, compared with those in Col-0. However, although there was no detectable amount of BL in Col-0, up to 0.05 ng/g fresh weight was found in 35S-CYP85A2, resulting in a phenotype with enhanced growth. Therefore, we conclude that the 25 and 33% change in CS and 28-norCS content, respectively, is physiologically important for generating BL in 35S-CYP85A2. In spite of the slight increase in endogenous CS in 35S-CYP85A1, the enhanced growth that was shown in 35S-CYP85A2 was not observed. This indicated that the enhanced growth of 35S-CYP85A2 might primarily occur as a result of increased levels of endogenous BL rather than CS in the mutant plant.
Despite reduced self-pollination in the early bolting stage of cyp85a2, completely mature adult cyp85a2 produced normal siliques. This could mean that most of the CS in cyp85a2 is primarily used for vegetative growth, such as stem elongation, and that the mutant uses CS to develop reproductive organs. In 35S-CYP85A2, pedicels grew longer than those of the wild type. Moreover, the number of seeds in a silique increased by as much as 30% compared with the wild type. BL has been identified from rape (Brassica napus) pollen (Grove et al., 1979), and in most cases, a high level of BL has been identified in the reproductive organs of plants (reviewed in Fujioka, 1999). In Arabidopsis Col-0 in particular, BL was not found in the shoot or leaf, but a high level of BL, which was higher than that of CS, was found in reproductive organs such as siliques and seeds (Fujioka et al., 1998; reviewed in Fujioka, 1999). This could suggest that Arabidopsis plants require BL at particular developmental stages, such as reproductive organ development, seed formation, and rapid cell elongation/division. Consequently, BL, the most active BR, is not always necessary for normal growth and development in Arabidopsis, although Arabidopsis requires BL when increased high BR activity is suddenly required. In other cases, CS functions as the bioactive BR that controls the overall growth and development of the plant.
METHODS
Plant Growth Conditions and Chemicals
Col-0 ecotype was used as the wild-type plant for phenotype comparisons. BR-related mutant (bri1-9, bak1, and BRI1-GFP) seeds were kindly provided by Jianming Li (University of Michigan, Ann Arbor, MI). Cold-treated plants were planted on soil or on 1× MS medium (Duchefa, Haarlem, The Netherlands) containing 0.8% (w/v) agar and 1% (w/v) sucrose and grown in an environmental growth chamber under a 22°C, 16-h-light/20°C, 8-h-dark cycle. When Arabidopsis seeds were planted on agar medium, seeds were surface-sterilized with 70% ethanol for 5 min and a 30% (v/v) bleach solution containing 0.025% (v/v) Triton X-100. All chemicals used in the biochemical analyses were obtained from Sigma-Aldrich Chemicals (St. Louis, MO).
Heterologous Expression of CYP85A1 and CYP85A2
Total RNA was extracted from Arabidopsis thaliana seedlings using TRI reagent (Sigma-Aldrich). cDNA was synthesized from 1 μg of total RNA using the MMLV-reverse transcriptase system (Promega, Madison, WI) according to the manufacturer's instructions. Specific primers used for the PCR amplification of CYP85A1 (At5g38970) and CYP85A2 (At3g30180) cDNA were designed to introduce a BamHI restriction site immediately upstream of the initiation codon and a KpnI site after the stop codon. Primers were as follows: CYP85A1 forward, 5′-ggatccATGGGAGCAATGATGGTGATGAT-3′; CYP85A1 reverse, 5′-ggtaccTTAGTAGGGTGAAATCCTAAGATG-3′; CYP85A2 forward, 5′-ggatccATGGGCATAATGATGATGATTTTG-3′; CYP85A2 reverse, 5′-ggtaccTCAGTAAGGTGAACACTTAAGATG-3′. cDNA was amplified using 32 thermal cycles (94°C 20 s, 61°C 30 s, and 72°C 2 min) with Ex Taq polymerase (Takara, Shuzo, Japan). CYP85A1 and CYP85A2 cDNA fragments were cloned separately into pGEM T easy vector (Promega). cDNAs were confirmed by DNA sequencing. The DNA fragments obtained from digestion using BamHI and KpnI were subcloned into the pYeDP60 vector (V60). The preparation of Cyt P450 overexpressed yeast strains (CYP85A1/V60/WAT21 and CYP85A2/V60/WAT21) and galactose induction were as previously described (Kim et al., 2004b). Galactose-induced cells were diluted in 20 mL of YPL to an OD550 of 0.4 to 0.6, and 5 μg of the appropriate substrates (6-deoxo-28-norCS, deoxoCS, CS, and [26,28-2H6]CS) were added to the cells. After 6 h, metabolites were extracted using 20 mL of ethyl acetate and then concentrated in vacuo. The extracts were passed through a silica column (SepPak SiO2 Plus; Waters, Milford, MA) and eluted with 8 mL of chloroform (CHCl3), 2% (v/v) methanol in CHCl3, and 8% (v/v) methanol in CHCl3. The 8% methanol in CHCl3 fraction was purified by passage through a C18 cartridge column (SepPak C18 Plus; Waters) and eluted with 10 mL of 50% (v/v) methanol in water and 5 mL of 90% methanol in water. The fraction eluted with 90% methanol in water was dried, dissolved in a small amount of methanol, and then subjected to reverse-phase HPLC (SenshuPak C18, 10 × 150 mm) with a flow rate of 2.5 mL min−1 using 45% acetonitrile (MeCN) in water. Fractions were collected every minute. Fractions corresponding to the retention times of authentic CS (19 to 21 min), 28-norCS (13 to 15 min), and BL (13 to 15 min) were collected and analyzed by GC-MS after bismethaneboronation. To calculate the amounts of 28-norCS and CS that were converted from 6-deoxo-28-norCS and 6-deoxoCS, respectively, the amount of CS was first calculated using [26,28-2H6]CS as an internal standard, and the amount of 28-norCS was estimated by the area ratio relative to CS on the total ion chromatogram.
Preparation of Chimeric Enzymes by Exchanging the N-Terminal Region of CYP85A1 and CYP85A2
In an effort to gain further insight into the structure-function relationship of the CYP85A1 and CYP85A2 enzymes, chimeric enzymes were generated by exchanging the N-terminal open reading frame of CYP85A1 with CYP85A2 and vice versa. In addition to possessing identical exon-intron structures, CYP85A1 and CYP85A2 possess a single restriction site for NdeI at the 516th base pair position. A unique restriction site for NdeI is located in the pGEM T easy vector (Promega). Thus, parent CYP85A1 and CYP85A2 cDNAs cloned into the pGEM T easy vector were digested with NdeI. After electrophoresis, the resulting four DNA bands (two 932-bp and two 3493-bp fragments) were isolated from the agarose gel using GENECLEAN Turbo (Q-BIO Gene, Montreal, Canada) and ligated to generate the chimeric cDNAs (Figure 2C). Newly cloned chimeric plasmids were analyzed by restriction enzymes and completely sequenced. Chimeric cDNAs were subcloned into the pYeDP60 shuttle vector and transformed into yeast (WAT21). Heterologous expression procedures for the two chimeric enzymes CYP85A1 and CYP85A2 were performed as described above.
35S-CYP85A1 and 35S-CYP85A2 Constructs and Transgenic Plants
Full-length CYP85A1 and CYP85A2 cDNA (1667 bp) including 5′ and 3′ untranslated regions were amplified by RT-PCR using Ex Taq polymerase. Specific primers used to introduce BamHI and SacI restriction sites were as follows: CYP85A1 forward primer, 5′-ggatccCTTTCTCTCTTTCTCTGGTCTGTT-3′; CYP85A1 reverse primer, 5′-gagctcGAGCTCGGGTTTAATATAGTAGAACATCATC-3′; CYP85A2 forward primer, 5′-ggatccAACCAAAGACTCTTAACCATCT-3′; CYP85A2 reverse primer, 5′-cgagctcACAACATTTCAAATATTTTATTT-3′. cDNA amplified by RT-PCR was cloned into the pGEM T easy vector and checked by DNA sequencing. The CYP85A1 and CYP85A2 cDNA/pGEM T easy constructs were digested with BamHI and SacI, and the resulting fragments subcloned into the pBI121 binary vector (CLONTECH, Palo Alto, CA) digested with BamHI and SacI. The CYP85A1 and CYP85A2 cDNA/pBI121 constructs were transformed into Agrobacterium tumefaciens (GV 3101) using electroporation. Agrobacterium-mediated transformation by floral dipping was used to generate transgenic Arabidopsis (Clough and Bent, 1998).
Isolation of cyp85a1 and cyp85a2 Mutants, and Rescue Experiments
The T-DNA insertional mutants cyp85a1 (SALK_148384), cyp85a2-1 (SALK_056270), and cyp85a2-2 (SALK_068754) were discovered on the Salk SIGnAL Web site (http://signal.salk.edu) and obtained from the ABRC (Columbus, OH). CYP85A1 forward primer 5′-ATGGGAGCAATGATGGTGATGAT-3′, CYP85A1 reverse primer 5′-TTAGTAGGGTGAAATCCTAAGATG-3′, CYP85A2 forward primer 5′-ATGGGCATAATGATGATGATTTTG-3′, CYP85A2 reverse primer 5′-TCAGTAAGGTGAACACTTAAGATG-3′, T-DNA left border primer 5′-CTTTGACGTTGGAGTCCACGTTCTTTAATA-3′, and T-DNA right border primer 5′-ATATTTGCTAGCTGATAGTGACCTTAG-3′ were used to verify the presence and location of the T-DNA insertion. Individual plants homozygous for a T-DNA insertion in the CYP85A2 gene were identified by PCR screening and segregation analysis.
To examine petiole elongation in cyp85a2 mutants by application of CS and BL, 1 μM of CS or BL (50 μL each) dissolved in 5% (v/v) ethanol was applied directly to the center of the rosette of plants where the inflorescence shoot was not elongated. This treatment was performed once a day for 5 d. Ten days after the first treatment, petiole length was examined using a digital camera. When the restoration of male sterility by BR was tested, floral buds of the mutants, which possessed inflorescence shoots 10 to 12 cm in length, were dipped in 1 μM of BR solution. This treatment was performed once a day for 3 d, and the formation of siliques was observed on the fifth day.
Expression Analysis of CYP85A1 and CYP85A2 by RT-PCR
First-strand cDNA was synthesized by MMLV-reverse transcriptase using 1 μg of total RNA extracted from the seedlings or leaves of plants with various genotypes. One-fifth and one-tenth of the RT products were used to amplify the cDNAs of CYP85A1 and CYP85A2, respectively. Semiquantitative RT-PCR was performed with Ex Taq polymerase using gene-specific primers (CYP85A1 forward, 5′-ATGGGAGCAATGATGGTGATGAT-3′; CYP85A1 reverse, 5′-TTAGTAGGGTGAAATCCTAAGATG-3′; CYP85A2 forward, 5′-ATGGGCATAATGATGATGATTTTG-3′; CYP85A2 reverse, 5′-TCAGTAAGGTGAACACTTAAGATG-3′). The concentration of UBQ5 (At3g62250) mRNA in each sample was determined so as to normalize any differences in the amount of total RNA present. Specific primers of UBQ5 used for RT-PCR (24 thermal cycles) were as follows: forward, 5′-GACCATAACCCTTGAGGTTGAATC-3′; reverse, 5′-AGAGAGAAAGAGAAGGATCGATC-3′.
Feeding Experiments
Col-0, 35S-CYP85A2, and the BR-insensitive mutant bri1-9 were used in this study. Arabidopsis seeds were surface-sterilized as described above and grown on 0.5× MS medium containing 0.8% (w/v) agar and 1% (w/v) sucrose in an environmental growth chamber under a 22°C, 16-h-light/20°C, 8-h-dark cycle. After 7 d, the seedlings (Col-0 and 35S-CYP85A2, 30 seedlings; bri1-9, 60 seedlings) were transferred into a 250-mL flask containing 30 mL of 0.5× MS medium supplemented with 1% sucrose. The seedlings were grown on a shaking incubator (120 rpm) at 22°C under a 16-h-light/8-h-dark cycle. After 5 d in culture, the appropriate deuterium-labeled substrates ([2H6]6-deoxoCS, 5 μg; [2H6]CS, 10 μg) were added to each flask and the cultures allowed to grow under the same conditions for 4 d. The seedlings were harvested and extracted with methanol. Seedling extracts were concentrated in vacuo and partitioned between water and CHCl3. The CHCl3 phases extracted from the culture media were combined with the seedling extracts and concentrated in vacuo. Because endogenous levels of BRs in the seedlings are negligible, nonlabeled CS and BL were added as internal standards for quantitative analysis of the residues. The purification of metabolites was performed as described above.
Quantitative Analysis of Endogenous BRs in Arabidopsis
Wild-type (45 g), cyp85a2-1 (30 g), cyp85a2-2 (30 g), cyp85a1 (40 g), 35S-CYP85A2 (45 g), and 35S-CYP85A1 (45 g) plants grown for 6 weeks on soil were harvested and extracted three times with 300 mL of 90% methanol. Deuterium-labeled 6-deoxoCS, CS, and BL were added as internal standards for quantitative analysis of the extracts (250 ng each). Evaporated extracts were partitioned three times between water and CHCl3. The CHCl3-soluble fractions were concentrated in vacuo and partitioned three times between 80% methanol and n-hexane. The concentrated 80% methanol extracts were repartitioned three times between ethyl acetate and phosphate buffer, pH 7.8. The ethyl acetate-soluble residues were subjected to silica gel chromatography. The column was eluted with 150 mL of CHCl3 containing 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 20, 50, and 100% (v/v) methanol. The 3 to 7% (v/v) methanol fractions were combined, concentrated in vacuo, and subsequently purified using a SepPak C18 silica cartridge column as described in the section detailing heterologous expression. The fraction obtained from the SepPak silica column was dried, dissolved in a small amount of methanol, and then subjected to reverse phase HPLC (SenshuPak C18, 10 × 150 mm). The column was eluted at a flow rate of 2.5 mL min−1 using MeCN-water gradients: 0 to 20 min, 45% MeCN; 20 to 40 min, 45 to 100% MeCN; 40 to 70 min, 100% MeCN. Fractions were collected every minute. Under the HPLC conditions eluted with the MeCN-water gradient, authentic 6-deoxoCS, CS, and BL/28-norCS were detected in fractions 41 to 43, 19 to 21, and 13 to 15, respectively. Therefore, these fractions were analyzed by capillary GC-MS after bismethaneboronation.
GC-MS
The GC-MS analyses were performed on a Hewlett-Packard 5973 mass spectrometer (electron impact ionization, 70 electron voltage; Palo Alto, CA) connected with 6890 gas chromatography fitted with a fused silica capillary column (HP-5, 0.25 mm × 30 m, 0.25-μm film thickness). The oven temperature was maintained at 175°C for 2 min, elevated to 280°C at a rate of 40°C min−1, and then maintained at 280°C. Helium was used as the carrier gas at a flow rate of 1 mL min−1, and samples were introduced using an on-column injection mode. Methaneboronation was performed by heating samples dissolved in pyridine containing methaneboronic acid (2 mg mL−1) at 80°C for 30 min.
Arabidopsis Genome Initiative locus identifiers for CYP85A1 and CYP85A2 are At5g38970 and At3g30180, respectively.
Acknowledgments
We thank D. Pompon and P. Urban for providing pYeDP60 and yeast strain WAT21. We are grateful to J. Li for supplying Arabidopsis seeds (bri1-9, bak1, and BRI1-GFP) and to T. Yokota for the syntheses of 6-deoxoCS and [2H6]6-deoxoCS. This research was supported by the Korean Science and Engineering Foundation (Grant R01-2002-000-00367-0 to S.-K.K. and S.C.C.) and by the Korea Research Foundation (Grant DP0483 to S.-K.K. and S.C.C).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Seong-Ki Kim (skkimbio@cau.ac.kr).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.033738.
References
- Bajguz, A., and Tretyn, A. (2003). The chemical characteristic and distribution of brassinosteroids in plants. Phytochemistry 62 1027–1046. [DOI] [PubMed] [Google Scholar]
- Bishop, G.J., Nomura, T., Yokota, T., Harrison, K., Noguchi, T., Fujioka, S., Takatsuto, S., Jones, J.D., and Kamiya, Y. (1999). The tomato DWARF enzyme catalyzes C-6 oxidation in brassinosteroid biosynthesis. Proc. Natl. Acad. Sci. USA 96 1761–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop, G.J., and Yokota, T. (2001). Plants steroid hormones, brassinosteroids: Current highlights of molecular aspects on their synthesis/metabolism, transport, perception and response. Plant Cell Physiol. 42 114–120. [DOI] [PubMed] [Google Scholar]
- Burton, S.G. (2003). Oxidizing enzymes as biocatalysts. Trends Biotechnol. 21 543–549. [DOI] [PubMed] [Google Scholar]
- Choe, S., Dilkes, B.P., Fujioka, S., Takatsuto, S., Sakurai, A., and Feldmann, K.A. (1998). The DWF4 gene of Arabidopsis encodes a cytochrome P450 that mediates multiple 22α-hydroxylation steps in brassinosteroid biosynthesis. Plant Cell 10 231–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Croteau, R., Kuichan, T.M., and Lewis, N.G. (2000). Natural products. In Biochemistry and Molecular Biology of Plants, B.B. Buchanan, W. Gruissem, and R.L. Jones, eds (Rockville, MD: American Society of Plant Biologists), pp. 1250–1318.
- Fischer, R.T., Trzaskos, J.M., Magolda, R.L., Ko, S.S., Brosz, C.S., and Larsen, B. (1991). Lanosterol 14α-methyl demethylase. J. Biol. Chem. 266 6124–6132. [PubMed] [Google Scholar]
- Fraaije, M.W., Kamerbeek, N.M., van Berkel, W.J., and Janssen, D.B. (2002). Identification of a Baeyer-Villiger monooxygenase sequence motif. FEBS Lett. 518 43–47. [DOI] [PubMed] [Google Scholar]
- Fujioka, S. (1999). Natural occurrence of brassinosteroids in the plant kingdom. In Brassinosteroids: Steroidal Plant Hormones, A. Sakurai, T. Yokota, and S.D. Clouse, eds (Tokyo: Springer-Verlag), pp. 21–45.
- Fujioka, S., Li, J., Choi, Y.-H., Seto, H., Takatsuto, S., Noguchi, T., Watanabe, T., Kuriyama, H., Yokota, T., Chory, J., and Sakurai, A. (1997). The Arabidopsis deetiolated2 mutant is blocked early in brassinosteroid biosynthesis. Plant Cell 9 1951–1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka, S., Noguchi, T., Yokota, T., Takatsuto, S., and Yoshida, S. (1998). Brassinosteroids in Arabidopsis thaliana. Phytochemistry 48 595–599. [DOI] [PubMed] [Google Scholar]
- Fujioka, S., and Yokota, T. (2003). Biosynthesis and metabolism of brassinosteroids. Annu. Rev. Plant Biol. 54 137–164. [DOI] [PubMed] [Google Scholar]
- Gotoh, O. (1992). Substrate recognition sites in cytochrome P450 family 2 (CYP2) proteins inferred from comparative analyses of amino acid and coding nucleotide sequences. J. Biol. Chem. 267 83–90. [PubMed] [Google Scholar]
- Grove, M.D., Spencer, G.F., Rohwededer, W.K., Mandava, N.B., Worley, J.F., Warthen, J.D., Jr., Steffens, G.L., Flippen-Anderson, J.L., and Cook, J.C., Jr. (1979). Brassinolide, a plant growth promoting steroid isolated from Brassica napus pollen. Nature 281 216–217.16066206 [Google Scholar]
- Hong, Z., Ueguchi-Tanaka, M., Umemura, K., Uozu, S., Fujioka, S., Takatsuto, S., Yoshida, S., Ashikari, M., Kitano, H., and Matsuoka, M. (2003). A rice brassinosteroid-deficient mutant, ebisu dwarf (d2), is caused by a loss of function of a new member of cytochrome P450. Plant Cell 15 2900–2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, J.G., Yun, J., Kim, D.H., Chung, K.S., Fujioka, S., Kim, J.I., Dae, H.W., Yoshida, S., Takatsuto, S., Song, P.S., and Park, C.M. (2001). Light and brassinosteroid signals are integrated via a dark-induced small G protein in etiolated seedling growth. Cell 105 625–636. [DOI] [PubMed] [Google Scholar]
- Kim, T.-W., Chang, S.C., Choo, J., Watanabe, T., Takatsuto, S., Yokota, T., Lee, J.S., Kim, S.Y., and Kim, S.-K. (2000. a). Brassinolide and [26,28-2H6] brassinolide are differently demethylated by loss of C-26 and C-28, respectively, in Marchantia polymorpha. Plant Cell Physiol. 41 1171–1174. [DOI] [PubMed] [Google Scholar]
- Kim, T.-W., Chang, S.C., Lee, J.S., Hwang, B., Takatsuto, S., Yokota, T., and Kim, S.-K. (2004. a). Cytochrome P450-catalyzed brassinosteroid pathway activation through synthesis of castasterone in Phaseolus vulgaris. Phytochemistry 65 679–689. [DOI] [PubMed] [Google Scholar]
- Kim, T.-W., Chang, S.C., Lee, J.S., Takatsuto, S., Yokota, T., and Kim, S.-K. (2004. b). Novel biosynthetic pathway of castasterone from cholesterol in tomato. Plant Physiol. 135 1231–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, T.-W., Han, K.-S., Joo, S.-H., Kang, M.-W., and Kim, S.-K. (2000. b). Metabolism of brassinolide in suspension cultured cells of Phaseolus vulgaris. Bull. Korean Chem. Soc. 21 1044–1046. [Google Scholar]
- Kim, T.-W., Park, H.-H., and Kim, S.-K. (2004. c). Cell-free conversion of castasterone in cultured cells of Phaseolus vulgaris and Marchantia polymorpha. Bull. Korean Chem. Soc. 25 955–956. [Google Scholar]
- Kim, Y.-S., Kim, T.-W., and Kim, S.-K. (2003). Conversion of 6-deoxocastasterone to brassinolide in a liverwort, Marchantia polymorpha. Bull. Korean Chem. Soc. 24 1385–1388. [Google Scholar]
- Koka, C.V., Cerny, R.E., Gardner, R.G., Noguchi, T., Fujioka, S., Takatsuto, S., Yoshida, S., and Clouse, S.D. (2000). A putative role for the tomato genes DUMPY and CURL-3 in brassinosteroid biosynthesis and response. Plant Physiol. 122 85–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J., and Chory, J. (1997). A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90 929–938. [DOI] [PubMed] [Google Scholar]
- Li, J., Nagpal, P., Vitart, V., McMorris, T.C., and Chory, J. (1996). A role for brassinosteroids in light-dependent development of Arabidopsis. Science 272 398–401. [DOI] [PubMed] [Google Scholar]
- Li, J., and Nam, K.H. (2002). Regulation of brassinosteroid signaling by a GSK3/SHAGGY-like kinase. Science 295 1299–1301. [DOI] [PubMed] [Google Scholar]
- Li, J., Nam, K.H., Vafeados, D., and Chory, J. (2001). BIN2, a new brassinosteroid-insensitive locus in Arabidopsis. Plant Physiol. 127 14–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak, A.Y., and Swinney, D.C. (1992). 17-O-Acetyltestosterone formation from progesterone in microsome from pig testes: Evidence for the Baeyer-Villiger rearrangement in androgen formation catalyzed by CYP17. J. Am. Chem. Soc. 114 8309–8310. [Google Scholar]
- Marko, D., Mihovilovic, M.D., Müller, B., and Stanetty, P. (2002). Monooxygenase-mediated Baeyer-Villiger oxidations. Eur. J. Org. Chem. 2002 3711–3730. [Google Scholar]
- Nakajima, N., Fujioka, S., Tanaka, T., Takatsuto, S., and Yoshida, S. (2002). Biosynthesis of cholestanol in higher plants. Phytochemistry 6 275–279. [DOI] [PubMed] [Google Scholar]
- Nam, K.H., and Li, J. (2002). BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 110 203–212. [DOI] [PubMed] [Google Scholar]
- Neff, M.M., Nguyen, S.M., Malancharuvil, E.J., Fujioka, S., Noguchi, T., Seto, H., Tsubuki, M., Honda, T., Takatsuto, S., Yoshida, S., and Chory, J. (1999). BAS1, a gene regulating brassinosteroid levels and light responsiveness in Arabidopsis. Proc. Natl. Acad. Sci. USA 96 15316–15323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi, T., Fujioka, S., Choe, S., Takatsuto, S., Tax, F.E., Yoshida, S., and Feldmann, K.A. (2000). Biosynthetic pathways of brassinolide in Arabidopsis. Plant Physiol. 124 201–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi, T., Fujioka, S., Choe, S., Takatsuto, S., Yoshida, S., Yuan, H., Feldmann, K.A., and Tax, F.E. (1999). Brassinosteroid-insensitive dwarf mutants of Arabidopsis accumulate brassinosteroids. Plant Physiol. 121 743–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura, T., Bishop, G.J., Kaneta, T., Reid, J.B., Chory, J., and Yokota, T. (2003). The LKA gene is a BRASSINOSTEROIDS INSENSITIVE 1 homolog of pea. Plant J. 36 291–300. [DOI] [PubMed] [Google Scholar]
- Nomura, T., Kitasaka, Y., Takatsuto, S., Reid, J.B., Fukami, M., and Yokota, T. (1999). Brassinosteroid/sterol synthesis and plant growth as affected by lka and lkb mutations of pea. Plant Physiol. 119 1517–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura, T., Kushiro, T., Yokota, T., Kamiya, Y., Bishop, G.J., and Yamaguchi, S. (2005). The last reaction producing brassinolide is catalyzed by cytochrome P-450s, CYP85A3 in tomato and CYP85A2 in Arabidopsis. J. Biol. Chem. 280 17873–17879. [DOI] [PubMed] [Google Scholar]
- Nomura, T., Nakayama, M., Reid, J.B., Takeuchi, Y., and Yokota, T. (1997). Blockage of brassinosteroid biosynthesis and sensitivity causes dwarfism in garden pea. Plant Physiol. 113 31–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts, S.M., and Wan, P.W.H. (1998). Enzyme-catalysed Baeyer-Villiger oxidations. J. Mol. Catal. 4 111–136. [Google Scholar]
- Sakurai, A. (1999). Biosynthesis. In Brassinosteroids: Steroidal Plant Hormones, A. Sakurai, T. Yokota, and S.D. Clouse, eds (Tokyo: Springer-Verlag), pp. 91–111.
- Shimada, Y., Fujioka, S., Miyauchi, N., Kushiro, M., Takatsuto, S., Nomura, T., Yokota, T., Kamiya, Y., Bishop, G.J., and Yoshida, S. (2001). Brassinosteroid 6-oxidases from Arabidopsis and tomato catalyze multiple C-6 oxidations in brassinosteroid biosynthesis. Plant Physiol. 126 770–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada, Y., Goda, H., Nakamura, A., Takatsuto, S., Fujioka, S., and Yoshida, S. (2003). Organ-specific expression of brassinosteroid-biosynthetic genes and distribution of endogenous brassinosteroids in Arabidopsis. Plant Physiol. 131 287–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, H., Fujioka, S., Takatsuto, S., Yokota, T., Murofushi, N., and Sakurai, A. (1993). Biosynthesis of brassinolide from castasterone in cultured cells of Catharanthus roseus. J. Plant Growth Regul. 12 101–106. [Google Scholar]
- Suzuki, H., Fujioka, S., Takatsuto, S., Yokota, T., Murofushi, N., and Sakurai, A. (1995). Biosynthesis of brassinosteroids in seedlings of Catharanthus roseus, Nicotiana tabacum, and Oryza sativa. Biosci. Biotechnol. Biochem. 59 168–172. [Google Scholar]
- Swinney, D.C., and Mak, A.Y. (1994). Androgen formation by cytochrome P450 CYP17. Solvent isotope effect and pL studies suggest a role for protons in regulation of oxene versus peroxide chemistry. Biochemistry 33 2185–2190. [DOI] [PubMed] [Google Scholar]
- Szekeres, M., Nemeth, K., Koncz-Kalman, Z., Mathur, J., Kauschmann, A., Altmann, T., Redei, G.P., Nagy, F., Schell, J., and Koncz, C. (1996). Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell 85 171–182. [DOI] [PubMed] [Google Scholar]
- Tanabe, S., Ashikari, M., Fujioka, S., Takatsuto, S., Yoshida, S., Yano, M., Yoshimura, A., Kitano, H., Matsuoka, M., Fujisawa, Y., Kato, H., and Iwasaki, Y. (2005). A novel cytochrome P450 is implicated in brassinosteroid biosynthesis via the characterization of a rice dwarf mutant, dwarf11, with reduced seed length. Plant Cell 17 776–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turk, E.M., Fujioka, S., Seto, H., Shimada, Y., Takatsuto, S., Yoshida, S., Denzel, M.A., Torres, Q.I., and Neff, M.M. (2003). CYP72B1 inactivates brassinosteroid hormones. An intersection between photomorphogenesis and plant steroid signal transduction. Plant Physiol. 133 1643–1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban, P., Mignotte, C., Kazmaier, M., Delorme, F., and Pompon, D. (1997). Cloning, yeast expression, and characterization of the coupling of two distantly related Arabidopsis thaliana NADPH-cytochrome P450 reductases with P450 CYP73A5. J. Biol. Chem. 272 19176–19186. [DOI] [PubMed] [Google Scholar]
- Yokota, T. (1997). The structure, biosynthesis and function of brassinosteroids. Trends Plant Sci. 2 137–143. [Google Scholar]
- Wang, Z.Y., Seto, H., Fujioka, S., Yoshida, S., and Chory, J. (2001). BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature 410 380–383. [DOI] [PubMed] [Google Scholar]
- Werk-Reichhart, D., and Feyereisen, R. (2000). Cytochrome P450: A success story. Genome Biol. 1 3000.1–3000.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willetts, A. (1997). Structural studies and synthetic applications of Baeyer-Villiger monooxygenases. Trends Biotechnol. 15 55–62. [DOI] [PubMed] [Google Scholar]
- Williams, P.A., Cosme, J., Vinkovic, D.M., Ward, A., Angove, H.C., Day, P.J., Vonrhein, C., Tickle, I.J., and Jhoti, H. (2004). Crystal structures of human cytochrome P450 3A4 Bound to metyrapone and progesterone. Science 305 683–686. [DOI] [PubMed] [Google Scholar]