Abstract
d-GLYCERATE 3-KINASE (GLYK; EC 2.7.1.31) catalyzes the concluding reaction of the photorespiratory C2 cycle, an indispensable ancillary metabolic pathway to the photosynthetic C3 cycle that enables land plants to grow in an oxygen-containing atmosphere. Except for GLYK, all other enzymes that contribute to the C2 cycle are known by their primary structures, and the encoding genes have been identified. We have purified and partially sequenced this yet missing enzyme from Arabidopsis thaliana and identified it as a putative kinase-annotated single-copy gene At1g80380. The exclusive catalytic properties of the gene product were confirmed after heterologous expression in Escherichia coli. Arabidopsis T-DNA insertional knockout mutants show no GLYK activity and are not viable in normal air; however, they grow under elevated CO2, providing direct evidence of the obligatory nature of the ultimate step of the C2 cycle. The newly identified GLYK is both structurally and phylogenetically distinct from known glycerate kinases from bacteria and animals. Orthologous enzymes are present in other plants, fungi, and some cyanobacteria. The metabolic context of GLYK activity in fungi and cyanobacteria remains to be investigated.
INTRODUCTION
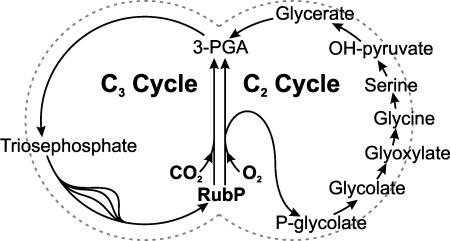
d-GLYCERATE 3-KINASE (GLYK; EC 2.7.1.31) is one of the core enzymes of plant photosynthetic carbon assimilation that is composed of two connected metabolic pathways: the photosynthetic Calvin or C3 cycle and the photorespiratory glycolate or C2 cycle (Figure 1). Both metabolic cycles are initiated by the same chloroplastic enzyme, ribulose 1,5-bisphosphate (RubP) carboxylase/oxygenase (Rubisco), whose dual catalytic activity reflects the evolutionary origin of photosynthesis in an anaerobic environment (Lorimer and Andrews, 1973). Under oxygen-free conditions, carboxylation of RubP yields 3-phosphoglycerate (3PGA) for further processing in the carbon reduction and assimilation reactions of the C3 cycle and its associated pathways. Later in earth history and plant evolution, the presence of atmospheric oxygen triggered the oxygenation of RubP, leading to the synthesis of equimolar amounts of 3PGA and 2-phosphoglycolate (2PG; Bowes et al., 1971; Ogren and Bowes, 1971). The ratio of 3PGA relative to 2PG synthesis is determined both by intrinsic properties of Rubisco and by the CO2:O2 ratio in the enzyme's microenvironment. In other words, a high CO2 concentration not only favors RubP carboxylation but reduces the rate of 2PG synthesis quite efficiently. It shall be mentioned that a minority of embryophytic plants (those with C4 or crassulacean acid metabolism) has exploited this opportunity and evolved appropriate mechanisms to increase their internal CO2:O2 ratio; however, this did not result in the elimination of photorespiratory metabolism (e.g., Osmond and Harris, 1971; Dever et al., 1996). Hence, under current atmospheric conditions and even more in combination with drought and high temperature, the high rate of RubP oxygenation with its inherent production of large amounts of 2PG represents a more or less heavy metabolic burden for all land plants.
Figure 1.
The C2 and C3 Cycles of Photosynthetic Carbon Metabolism Are Tied up with Each Other via the Oxygenation of RubP and the Phosphorylation of Glycerate.
The photorespiratory C2 cycle acts as an ancillary metabolic pathway that compensates for the futile withdrawal of RubP from the C3 cycle under aerobic conditions by serving as a carbon recovery system reconverting 2PG to 3PGA. In addition to Rubisco and GLYK, the complete C2 cycle needs at least eight more enzymes (Figure 1). The 2PG produced by oxygenation of RubP becomes dephosphorylated in the chloroplast followed by the peroxisomal synthesis of Gly from glycolate. In the course of the subsequent conversion of Gly to Ser, one-quarter of the bound carbon is released as photorespiratory CO2. Simultaneously, an equimolar amount of NH3 is lost at this step and needs to be reassimilated (Keys et al., 1978). Peroxisomal enzymes then convert Ser to glycerate, and the concluding chloroplastic phosphorylation of glycerate to 3PGA by GLYK completes the reactions of the C2 cycle (Husic et al., 1987). The described mass flow of carbon from 2PG through the C2 cycle to 3PGA has been supported by different approaches (discussed in Lorimer and Andrews, 1981), including 14C (Rabson et al., 1962) and 18O (Berry et al., 1978) labeling experiments.
Given this central position of GLYK in one of the major areas of primary plant metabolism, it is astonishing how poorly this enzyme is known. Except for some initial biochemical research (Kleczkowski and Randall, 1983; Schmitt and Edwards, 1983; Chaguturu, 1985; Kleczkowski et al., 1985; Kleczkowski and Randall, 1988), no further work has been published since the late 1980s, and neither the structure of the protein nor the encoding gene(s) are known. In other words, GLYK represents the last C2 cycle enzyme that is not known by its primary structure but only by its enzymatic activity and by its main metabolic function.
The conversion of d-glycerate to 3PGA is not unique to plants but occurs in other eukaryotes and in prokaryotic organisms as well. In fact, the enzyme was first isolated from bakers' yeast (Black and Wright, 1956) and from horse liver (Ichihara and Greenberg, 1957). Since then, primary structures of many nonplant glycerate kinases (GKs) were determined and their metabolic functions investigated. Malfunction of the human GK, for example, leads to the hereditary disease d-glyceric aciduria (Van Schaftingen, 1989). In bacteria and archaea, GK participates in several processes, including the metabolization of one-carbon compounds, glycolate, and glyoxylate via the Ser cycle and the d-glycerate pathway, respectively (Doughty et al., 1966; Ornston and Ornston, 1969; Hubbard et al., 1998; Cusa et al., 1999; Verhees et al., 2003). Using these known GK amino acid sequences, however, we could not find homologous proteins in current plant protein, genome, and EST databases. Similarly, putative homologous proteins were not found in the available yeast databases.
We therefore purified the enzyme from Arabidopsis thaliana leaves and identified the encoding single-copy gene At1g80380 (AtGLYK) with mass spectrometry. T-DNA insertional knockout mutants show no GLYK activity and accumulate very large amounts of glycerate. Similar to other photorespiratory mutants (e.g., Somerville, 1984), the mutants are not viable in normal air but, although slower, grow under elevated CO2, which provides direct and conclusive evidence of the obligatory nature of the supposed ultimate step of the C2 cycle. Notably, plant GLYK is structurally and phylogenetically distinct from known GKs, thus being prototypic for a novel protein family.
RESULTS AND DISCUSSION
Purification of GLYK and Identification of the Arabidopsis GLYK Gene
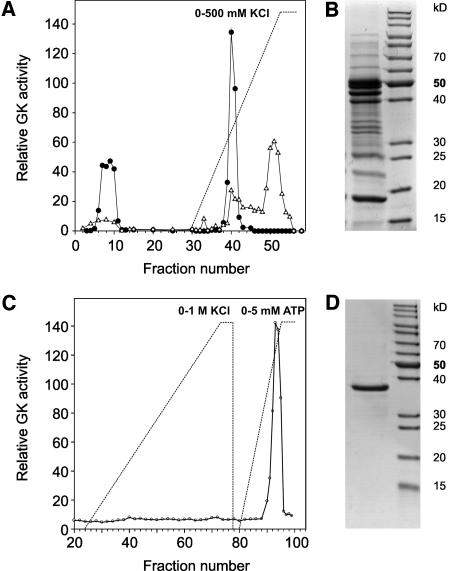
We first set out to obtain partial protein sequences for a plant GLYK and to identify the encoding gene(s). To this end, the enzyme was purified to homogeneity from Arabidopsis leaves using a combination of hydrophobic interaction, ion exchange, and affinity chromatography (Figure 2, Table 1). During DEAE ion-exchange chromatography, GLYK eluted as a single peak between 150 and 200 mM KCl and was further purified by two affinity chromatography steps first using Affi-Blue (Bio-Rad, Hercules, CA) and finally Matrex Green A (Millipore, Bedford, MA). The electrophoretically homogeneous protein had an approximate size of 39 kD by SDS-PAGE analysis and showed a specific activity of 54 μmol min−1 mg−1 (protein).
Figure 2.
Purification of GLYK from Arabidopsis Leaves.
(A) Elution profile during DEAE ion-exchange chromatography. Circles indicate GLYK activity, and triangles show protein concentration.
(B) SDS-PAGE of pooled fractions with GK activity.
(C) Final affinity chromatography on Matrex Green A affinity yields electrophoretically homogeneous enzyme.
(D) SDS-PAGE of 5.0 μg of purified enzyme.
Table 1.
Purification of Arabidopsis GLYK
| Purification Step | Volume (mL) | Protein (mg) | Total Activity (μmol min−1) | Specific Activity (μmol min−1 mg−1) | Purification (Fold) |
|---|---|---|---|---|---|
| Crude extract | 1560 | 2360.0000 | 147.70 | 0.0630 | 1 |
| (NH4)2SO4 | 225 | 2466.0000 | 26.90 | 0.0110 | n.c.a |
| Phenyl-Sepharose | 230 | 1040.0000 | 4.50 | 0.0043 | n.c. |
| DEAE-Fractogel | 16 | 5.0000 | 3.20 | 0.6430 | 10 |
| Affi-Gel Blue | 20 | 0.0700 | 127.00 | 24.3000 | 386 |
| Matrex Green A | 3 | 0.0079 | 0.43 | 54.6000 | 867 |
Factor was not calculated (n.c.) because the high ammonium ion concentration inhibits GLYK activity.
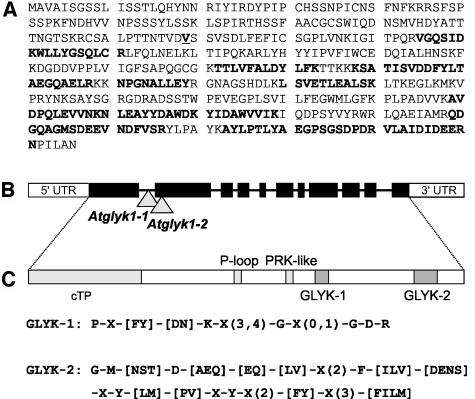
For identification, the protein band was digested with trypsin and analyzed using nanoflow liquid chromatography coupled to mass spectrometry (Wienkoop et al., 2004b). Eight amino acid sequences were obtained that all matched to only one Arabidopsis protein encoded by gene At1g80380 (AtGLYK; Figure 3A). AtGLYK comprises 11 exons, and the open reading frame of 1368 bp corresponds to a protein of 456 amino acids (Figure 3B). According to Target P (Emanuelsson et al., 2000) and ChloroP (Emanuelsson et al., 1999), the protein sequence includes a plastid-targeting sequence with three potential cleavage sites (after amino acids 63, 107, and 118). Sequence comparison with the homologous rice protein (BAD73764; Sasaki et al., 2002) points to a length most likely of 118 amino acids for the plastid-targeting peptide. This corresponds to a molecular mass of 38,160 D for the mature protein, which is very close to that found for the purified protein in our analyses. The protein contains an ATP/GTP binding site motif A (P-loop) domain typical for a family of nucleoside triphosphate hydrolases (SSF52540; amino acids 215 to 222) and short stretches of similarity to motifs of the phosphoribulokinase (PRK) family domain (IPR006083; Pfam 00485; amino acids 210 to 425). We therefore tested whether purified Arabidopsis GLYK exhibits PRK or uridine kinase (UK) activity and found that neither of the two reactions is catalyzed (Table 2).
Figure 3.
Arabidopsis GLYK Is Encoded by the Gene At1g80380.
(A) Complete amino acid sequence of Arabidopsis GLYK with experimentally determined partial amino acid sequences shown in bold.
(B) GLYK is localized at the very end of the right arm of chromosome 1 and comprises 11 exons (∼3500 bp). T-DNA insertion sites in the knockout lines are marked with triangles. UTR, untranslated region.
(C) The protein includes a 118–amino acid chloroplastic transit peptide (cTP), an ATP/GTP binding site motif A (P-loop), and the motifs GLYK-1 and GLYK-2, which are specific for the GLYK protein family.
Table 2.
Substrate Use of Purified and Recombinant Arabidopsis GLYK and Recombinant GLYK from S. cerevisiae
| GLYK Activity ±se (μmol min−1 mg−1) |
||||
|---|---|---|---|---|
| Substrate | Nucleotide | ArabidopsisNative | ArabidopsisRecombinant | S. cerevisiaeRecombinant |
| Glycerate | ATP | 53.6 ± 1.4 | 27.9 ± 6.2 | 21.3 ± 3.6 |
| Uridine | ATP | Not detectable | Not detectable | Not detectable |
| Ribulose 5-P | ATP | Not detectable | Not detectable | Not detectable |
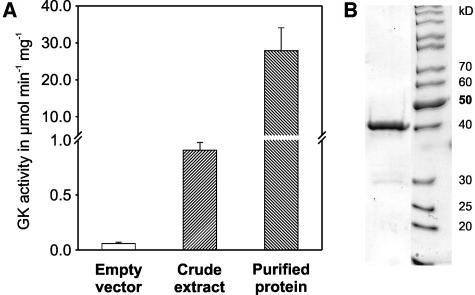
We next verified whether At1g80380 indeed encodes a functional GLYK. For heterologous expression, we cloned a 5′-terminally modified AtGLYK cDNA obtained by RT-PCR with appropriate primers into the Stratagene Escherichia coli expression vector pCALn. The N terminus of the encoded protein corresponds to amino acid position 65. After induction, the recombinant bacteria showed substantially greater GLYK activity in comparison with control cells bearing the empty vector (Figure 4). A large-scale isolation of the recombinant protein from E. coli lysate was performed using the leaf GLYK purification protocol. The purified enzyme migrated as a single band of the expected size on SDS-PAGE. To exclude possible contaminations by bacterial enzymes brought about by the affinity chromatography steps, its identity was reconfirmed by mass spectrometric analysis (data not shown). The specific activity of the purified recombinant enzyme (28 μmol min−1 mg−1) was lower than that of the native plant enzyme, which could be due to the extended N terminus or a missing posttranslational modification of the plant GLYK after expression in E. coli.
Figure 4.
The Recombinant Protein Encoded by At1g80380 Shows GLYK Activity.
(A) GLYK activities measured in E. coli BL-21 lysate harboring the empty and the recombinant vector, respectively, or with the purified recombinant protein.
(B) SDS-PAGE of recombinant GLYK purified by affinity chromatography on Matrex Green A.
GLYK Is the Exclusive Source of GK Activity in Arabidopsis
The Arabidopsis GLYK does not show any significant similarity to other proteins encoded by the Arabidopsis genome (Arabidopsis Genome Initiative, 2000) and is apparently encoded by a single gene. We next determined whether GLYK is the exclusive source of the respective enzymatic activity in Arabidopsis or, alternatively, whether any other Arabidopsis enzyme could exert GLYK activity. To this end, we isolated two allelic knockout mutants from T-DNA insertion lines (SALK lines SALK085479 and SALK036371; obtained via the Nottingham Arabidopsis Stock Centre). To verify the presence and position of the T-DNA insertions, we first sequenced PCR fragments obtained with primers specific for the T-DNA left border and right border, respectively, and for flanking genome sequences. The results confirmed the presence of a T-DNA insertion in the first intron (Atglyk1-1, SALK 085479) and in the second exon (Atglyk1-2, SALK 036371), respectively (Figure 3B).
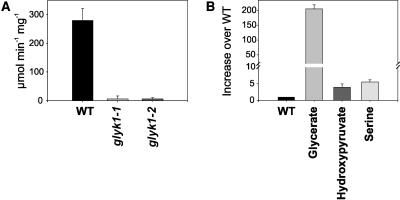
We then produced homozygous plants for both lines and found that the allelic mutations lead to the same morphological and biochemical phenotype with respect to all analyzed features. This confirmed that the observed mutant features are indeed due to the missing GLYK activity and not caused by any undiscovered background mutations. In ambient air, homozygous mutants germinate normally, but their growth becomes arrested at the early cotyledon stage. The seedlings die within approximately 2 weeks, unless being transferred to CO2-enriched air (Figure 5A). Such behavior has been observed with most but not all photorespiratory mutants (Somerville and Ogren, 1979; Somerville, 1984, 2001; Blackwell et al., 1988; Richter et al., 2005) and confirms the essential role of GLYK for the proper functioning of the photorespiratory C2 cycle already in very early stages of Arabidopsis development. Under the low-photorespiratory conditions of elevated CO2 (1200 μL L−1), homozygous Atglyk1-1 and Atglyk1-2 plants show reduced growth in comparison with wild-type plants, but they are fully viable and fertile. As examined by RT-PCR analysis, neither of the allelic homozygous knockout lines contains detectable amounts of AtGLYK transcripts (Figure 5B). Moreover, GLYK activity in leaves of these plants is undetectable (Figure 6A). A gas chromatography–time of flight–mass spectrometry (GC-TOF-MS)-based analysis of metabolites in leaves of wild-type and mutant plants grown under elevated CO2 reveals an up to ∼200-fold accumulation of glycerate in the mutants and an approximately fivefold increase in the downstream metabolites of the C2 cycle, hydroxypyruvate and Ser (Figure 6B). These data demonstrate that AtGLYK represents the only significant source of GLYK activity in Arabidopsis. They also show that the replenishment of the C3 cycle with salvaged C2 cycle carbon occurs exclusively via glycerate and 3PGA, thus providing direct and conclusive evidence on the nature of the final enzymatic step of the C2 cycle.
Figure 5.
GLYK Is Indispensable for Normal Plant Development and Growth, but This Absolute Requirement Is Relaxed under Elevated CO2.
(A) GLYK knockout mutants do not survive in normal air but recover at elevated CO2. From left to right: Arabidopsis wild type and Atglyk1-1 and Atglyk1-2 grown in normal air in comparison with Atglyk1-1 grown under 1200 μL L−1 CO2. All plants are 2 weeks after germination. The mutants do not develop primary leaves in normal air but do so within 4 to 5 d after transfer to elevated CO2.
(B) The homozygous knockout plants do not show residual GLYK mRNA. The 40S ribosomal S16 protein mRNA was used as internal control in the RT-PCR analysis.
Figure 6.
At1g80380 Is the Exclusive Source of GLYK Activity in Arabidopsis Leaves.
(A) GLYK activity in wild-type and knockout mutants. Residual activity is close to zero and within the range of experimental error in Atglyk1-1 and Atglyk1-2.
(B) Leaf glycerate content is greatly elevated, whereas the amounts of hydroxypyruvate and Ser are only approximately fivefold enhanced. Metabolite content is depicted in relative units with the respective wild-type values set to one. Bars represent standard errors of the mean.
GLYK Is Prototypic for a Novel GK Family
We already discussed the absence in the Arabidopsis proteome of homologs to known GK proteins from animals or bacteria. Vice versa, homologs to the Arabidopsis GLYK occur neither in animals nor in most bacteria but are present in plants, fungi, and some cyanobacteria. We therefore attempted to provide functional evidence for at least one of the nonplant GLYK homologous proteins. To this end, we expressed the homologous GLYK gene from Saccharomyces cerevisiae in E. coli, purified the recombinant protein to electrophoretic homogeneity (data not shown), and analyzed its enzymatic activity. As shown in Table 2, the yeast enzyme is a GLYK and does not accept other substrates, such as ribulose 5-phosphate or uridine.
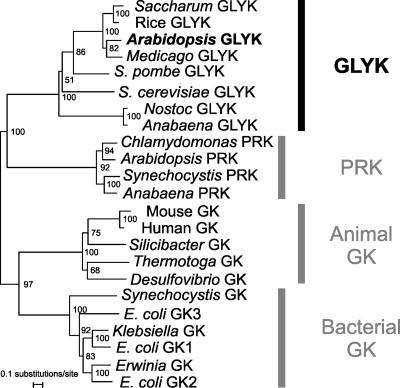
To analyze the evolutionary relationships between the different GKs, a phylogenetic tree was calculated with plant and nonplant GK amino acid sequences (Figure 7). PRK amino acid sequences were included in this analysis because of the presence in GLYK of short stretches of similarity to PRK domains. The tree branches into four clearly separated clades, three for the GKs and one for PRK. One of the GK clades comprises those bacterial GK that harbor a so-called GK domain (IPR004381). This domain occurs only in bacterial GK and, as the only known exception, in a homologous protein of the amoeba Dictyostelium discoideum. A second clade includes GK from animals, specialized bacteria like Thermotoga, Desulfovibrio, and Silicibacter, and archaea (data not shown). The respective proteins do not have a GK domain but a C-terminal multiorganism fragment rich with Leu (IPR007835), which is a rather unspecific domain of yet unknown function occurring in several nonplant reductases and kinases. The third clade is formed from AtGLYK homologous proteins. In addition to plants and green algae, such proteins are present only in fungi and in some filamentous, nitrogen-fixing, heterocyst-forming cyanobacteria with complex genomes.
Figure 7.
GLYK Represents a Novel Kinase Family Phylogenetically Distinct from Other GK Proteins.
The neighbor-joining tree was constructed from a ClustalX (Thompson et al., 1997) alignment of GLYK, GK, and PRK amino acid sequences with the program TREECON 1.3b (Van de Peer and De Wachter, 1994) using a Poisson correction for the calculation of the distance matrix. Bootstrap analysis with 1000 replicates (results shown as percentage values) confirmed the tree topology at all nodes and the differentiation into four unrelated classes. Sequences used for calculations are as follows: E. coli GK1 (E. coli glxK, P77364), E. coli GK2 (E. coli garK, P23524), E. coli GK3 (E. coli gcxK putative GK, AAL61903), Erwinia GK (E. carotovora garK, CAG76470), Klebsiella GK (K. pneumoniae glxK, BAD14998), Synechocystis GK (Synechocystis sp PCC 6803 hypothetical protein, P73408), mouse GK (Mus musculus transcript, AAH25834), human GK (Homo sapiens GLYCTK1, AAP41923), Silicibacter GK (Silicibacter sp TM 1040 putative GK, ZP_00336015), Desulfovibrio GK (D. desulfuricans putative GK, ZP00129227), Thermotoga GK (T. maritima putative GK, AAD36652), Anabaena GLYK (A. variabilis predicted kinase, ZP_00161317), Nostoc GLYK (Nostoc sp PCC 7120 hypothetical protein, NP_486913), S. cerevisiae GLYK (S. cerevisiae ATP binding protein, NP011721), S. pombe GLYK (S. pombe ATP binding protein, P78883), rice GLYK (Oryza sativa PRK/UK-like protein, BAD73764, Os01g48990), Arabidopsis GLYK (At1g80380, this article), Synechocystis PRK (Synechocystis sp PCC 6803 PRK, AAA27293), Anabaena PRK (A. variabilis PRK/UK, AD2321), Arabidopsis PRK (P25697), and Chlamydomonas PRK (C. reinhardtii PRK, P19824).
Green algae can excrete glycolate; however, they form the same C2 cycle metabolites as they are produced in higher plants (Stabenau and Winkler, 2005). Therefore, green algal GLYK is probably involved in photosynthetic-photorespiratory processes similar to land plants. By contrast, the complete knockout of Gly decarboxylase (which is an indispensable C2 cycle enzyme in higher plants) does not impair the performance of Synochocystis to any measurable extent (Hagemann et al., 2005), suggesting that the C2 cycle is of minor importance in this species and perhaps most cyanobacteria. Correspondingly, the Synechocystis genome does not harbor a plant-type GLYK gene, which seems to be restricted to complex cyanobacteria with larger genomes. Opposite to plants and cyanobacteria, fungi don't photosynthesize nor do they have a C2 cycle. This apparent relaxation (in cyanobacteria) or even complete absence (in fungi) of photorespiratory metabolism suggests a role of the nonplant GLYK in yet uninvestigated metabolic processes.
To possibly find domains that are distinctive for the GLYK protein family, we searched the available GLYK sequences from plants and nonplant microorganisms for characteristic amino acid patterns (PRATT 2.1; Jonassen et al., 1995) and identified two highly specific domains with the consensus sequences: P-X-[FY]-[DN]-K-X(3,4)-G-X(0,1)-G-D-R (named GLYK-1) and G-M-[NST]-D-[AEQ]-[EQ]-[LV]-X(2)-F-[ILV]-[DENS]-X-Y-[LM]-[PV]-X-Y-X(2)-[FY]-X(3)-[FILM] (named GLYK-2; Figure 3C). The GLYK-1 domain occurs in all analyzed GLYK-homologous proteins, whether of plant or nonplant origin. By contrast, the GLYK-2 domain is specific for the respective plant and yeast proteins only but does not occur in the homologous cyanobacterial proteins. Possible functions in catalysis or regulation of these highly conserved GLYK-specific domains remain to be analyzed. Notably, the crystal structure of the S. cerevisiae GLYK (still described as similar to E. coli pantothenate kinase) was recently determined (de La Sierra-Gallay et al., 2004). Our functional analysis, in combination with these new data on the enzyme's molecular structure, will initiate the analysis of structure-function relationships for this new class of enzymes and of their metabolic context in nonplant organisms.
METHODS
Plant Material and Growth Conditions
Arabidopsis thaliana, ecotype Columbia, was used for this study as the wild type. SALK lines SALK_085479 and SALK_036371 (Alonso et al., 2003) were obtained from the Nottingham Arabidopsis Stock Centre (http://nasc.nott.ac.uk). Seeds were incubated at 4°C for at least 2 d to break dormancy before germination. Seedlings and adult plants were grown on soil (Type Mini Tray; Einheitserdewerk, Uetersen, Germany) and vermiculite (1:5 mixture) and watered with 1× modified Hoagland solution. Unless otherwise stated, plants were grown under a 12-h-light/12-h-dark cycle (22/18°C) at 150 to 200 μE m−2 s−1 in growth chambers.
GK Purification
Approximately 1000 g of Arabidopsis leaves were homogenized in three liters of ice-cold extraction buffer (25 mM Hepes-KOH, 1 mM EDTA, 1 mM MgCl2, 1 mM KCl, 10 mM β-mercaptoethanol, 0.1 mM phenylmethanesulfonyl fluoride, and 10% glycerol, pH 7.6). All subsequent steps were performed at 2 to 6°C. Debris was removed by filtration through cheese cloth followed by centrifugation at 25,000g. The protein fraction obtained by (NH4)2SO4 precipitation (25 to 75%) was dissolved 0.1 M sodium phosphate buffer, pH 7.6, containing 1.3 M (NH4)2SO4, loaded onto a 1.6 × 10-cm HiLoad Phenyl-Sepharose column (Amersham, Buckinghamshire, UK), and eluted with a 500-mL linear (NH4)2SO4 gradient (1.3 to 0 M, 1.0 mL min−1) in 10-mL fractions. Fractions with adequate GLYK activity (Kleczkowski et al., 1985) were combined, rebuffered in 25 mM Tris-HCl (10 mM β-mercaptoethanol and 0.1 mM phenylmethanesulfonyl fluoride, pH 7.0), and applied to a 1.6 × 30-cm DEAE-Fractogel column (Merck, Rahway, NJ). During elution with a linearly increasing KCl concentration gradient (0 to 500 mM, 1 mL min−1), 5-mL fractions were collected and assayed for GLYK activity. Active fractions were combined, rebuffered in 20 mM Tris-HCl (28 mM NaCl, 0.1 mM phenylmethanesulfonyl fluoride, and 10 mM β-mercaptoethanol, pH 8.0), applied to an Affi-Gel Blue-Matrix prepacked 5-mL column (Bio-Rad, Hercules, CA), and eluted with a linear ATP concentration gradient (0 to 5 mM, 0.5 mL min−1). GLYK-containing fractions were rebuffered in 25 mM Tris-HCl (0.1 mM phenylmethanesulfonyl fluoride and 10 mM β-mercaptoethanol, pH 7.0) and loaded onto a 1.6 × 7.0-cm Matrex Green A affinity column (Millipore, Bedford, MA). Unspecifically bound protein was washed off with 1 M KCl. Finally, GLYK was eluted with a linear ATP concentration gradient (0 to 5 mM, 1 mL min−1, 3-mL fractions) and further analyzed.
Isolation of Atglyk1-1 and Atglyk1-2 Knockout Mutants
To verify the T-DNA insertion, leaf DNA was PCR amplified (1 min 94°C, 1 min 58°C, 1 min 30 s 72°C; 35 cycles) with primers specific for the left border (mLB1, 5′-AATCAGCTGTTGCCCGTCTCACTGGTGAA-3′) or right border (SALK-RB1, 5′-ATTAAACTCCAGAAACCCGCGGCTGAG-3′), respectively, and an AtGLYK-specific primer (AtGLYKC-S1, 5′-GAGTCAAGAGAGAAAAGGGGTCGAG-3′, or AtGLYKC-A1, 5′-CTAAAGGAGGTACATCATCTCCATC-3′). The resulting fragments were then sequenced to verify the insertion sites. Zygosity was examined using by PCR amplification of leaf DNA with primers AtGLYKC-S1 and mLB1. The knockout of AtGLYK in homozygous plants of both mutant lines was verified by RT-PCR using 2.5 μg of leaf RNA for cDNA synthesis and primers AtGLYKC-S1 (sense) and AtGLYKC-A1 (antisense), resulting in a 630-bp PCR fragment. Before PCR analysis, cDNA amounts were calibrated according to signals obtained from the constitutively expressed At2g09990 gene encoding the 40S ribosomal protein S16.
Heterologous Expression of AtGLYK
cDNA obtained from leaf mRNA (RevertAid cDNA synthesis kit; MBI Fermentas, Burlington, Canada) was PCR amplified with the two primers 5′-AAGGATCCTCTTCTTATTTATCCTCCAAGCTT-3′ and 5′-AACTCGAGGTTTGCGAGTATCGGGTTCCTTTC-3′ (underlined sequences indicate the BamHI and XhoI sites, respectively). The resulting fragment, encoding the whole GLYK, excluding a 63–amino acid N-terminal sequence, was first cloned in the pCR 2.1 vector (Invitrogen, Carlsbad, CA), excised, and then ligated via BamHI-XhoI into the expression vector pCALn (Stratagene, La Jolla, CA). Recombinant GLYK was isolated using the Affinity protein expression system (Stratagene) or, on a larger scale, purified according to the protocol for the nonrecombinant GLYK given above.
Heterologous Expression of Yeast GLYK
The GLYK gene was amplified by PCR using Saccharomyces cereviseae DNA as template and the two primers 5′-CTCGAGTGTGATAAGTCAAAAACGGTATTGGAC-3′ and 5′-GGTACCCTATTCAATACATCTCGTTTTCGTAG-3′ (underlined sequences indicate the XhoI and KpnI sites, respectively). The resulting fragment, encoding the complete GLYK, was first cloned in the pGEMT-T vector (Promega, Madison, WI), excised, and then ligated via XhoI-KpnI into the expression vector pBAD-HisA (Invitrogen). Recombinant yeast GLYK was purified using the affinity of the His-tagged fusion protein to a Ni-NTA resin (Pro Bond; Invitrogen).
Nano-Liquid Chromatography–MS/MS Analysis
The in-gel tryptic digest was analyzed using a monolithic RP nanocolumn (30 cm × 100 μm) (Wienkoop et al., 2004a) with a liquid chromatography–MS system comprising a Surveyor HPLC instrument with flow splitter and an LCQ Deca Xplus ion trap (Thermo Finnigan, Waltham, MA). The tryptic digest was first loaded onto a peptide trap with a flow rate of 2 μL min−1 and subsequently washed for 5 min with 100% solvent A (0.1% formic acid in water). After washing, elution of the peptides and gradient separation was done with a flow rate of 200 to 300 nL min−1 using a 90 min gradient with 0 to 80% solvent B (0.1% formic acid in acetonitrile). Eluting peptides were continuously analyzed by selecting the three most abundant signals of a survey scan (mass-to-charge ratio range of 500 to 2000) for sequential MS/MS fragmentation. The MS/MS spectra were searched against an Arabidopsis database (http://www.arabidopsis.org/) using Turbo-sequest implemented in Bioworks 3.1 (Thermo Finnigan). Matches were filtered with Xcorr versus charge filter of Bioworks 3.1 with minimum Xcorrs of 2.0, 2.0, and 3.3 for singly, doubly, and triply charged fully tryptic peptides, a minimum Δ correlation of 0.1, and a minimum of two peptides per locus. Using DTA Select (Tabb et al., 2002), all redundancy of scans, peptides, and proteins was sorted out. Additionally, spectra were evaluated manually using the DTA Select graphical user interface.
Metabolite Extraction and GC-TOF-MS Analysis
Leaves from two individually harvested plants were pooled, extracted, and analyzed in triplicate as described (Weckwerth et al., 2004a, 2004b). GC-TOF-MS analysis was performed on an HP 5890 gas chromatograph (Agilent Technologies, Palo Alto, CA) with deactivated standard split/splitless liners containing glass wool. A 1-μL sample was injected in the splitless mode at 230°C injector temperature. GC was operated on a MDN-35 capillary, 30 m × 0.32 mm inner diameter, 25 μm film (Supelco, Bellefonte, PA), at constant flow of 2 mL min−1 helium. The temperature program started with 2 min isocratic at 85°C, followed by temperature ramping at 15°C min−1 to a final temperature of 360°C, which was held for 8 min. Data acquisition was performed on a Pegasus II TOF mass spectrometer (Leco, St. Joseph, MI) with an acquisition rate of 20 scans s−1 in the mass-to-charge ratio range of 85 to 600. The obtained data were first analyzed by defining a reference chromatogram with the maximum number of detected peaks over a signal-to-noise threshold of 50. Afterwards, all chromatograms were matched against the reference with a minimum match factor of 800. Compounds were annotated by retention index and mass spectra comparison to a user-defined spectra library. Selected unique fragment ions specific for each individual metabolite were used for quantification.
Acknowledgments
We greatly appreciate technical assistance by Klaudia Michl and Ursula Bauwe and would like to further thank Sandra Messutat, Nadja Engel, and Sandra Schwarte for help in some experiments. This work was supported by a grant of the Deutsche Forschungsgemeinschaft.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Hermann Bauwe (hermann.bauwe@uni-rostock.de).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.033993.
References
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408 796–815. [DOI] [PubMed] [Google Scholar]
- Berry, J.A., Osmond, C.B., and Lorimer, G.H. (1978). Fixation of 18O2 during photorespiration. Kinetic and steady-state studies of the photorespiratory carbon oxidation cycle with intact leaves and isolated chloroplasts of C3 plants. Plant Physiol. 62 954–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black, S., and Wright, N.G. (1956). Enzymatic formation of glyceryl and phosphoglyceryl methylthiol esters. J. Biol. Chem. 221 171–180. [PubMed] [Google Scholar]
- Blackwell, R.D., Murray, A.J.S., Lea, P.J., Kendall, A., Hall, N.P., Turner, J.C., and Wallsgrove, R.M. (1988). The value of mutants unable to carry out photorespiration. Photosynth. Res. 16 155–176. [DOI] [PubMed] [Google Scholar]
- Bowes, G., Ogren, W.L., and Hageman, R.H. (1971). Phosphoglycolate production catalysed by ribulose diphosphate carboxylase. Biochem. Biophys. Res. Commun. 45 716–722. [DOI] [PubMed] [Google Scholar]
- Chaguturu, R. (1985). Glycerate kinase from spinach leaves: Partial purification, characterization and subcellular localization. Physiol. Plant. 63 19–24. [Google Scholar]
- Cusa, E., Obradors, N., Baldoma, L., Badia, J., and Aguilar, J. (1999). Genetic analysis of a chromosomal region containing genes required for assimilation of allantoin nitrogen and linked glyoxylate metabolism in Escherichia coli. J. Bacteriol. 181 7479–7484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de La Sierra-Gallay, I.L., et al. (2004). Crystal structure of the YGR205w protein from Saccharomyces cerevisiae: Close structural resemblance to E. coli pantothenate kinase. Proteins 54 776–783. [DOI] [PubMed] [Google Scholar]
- Dever, L.V., Bailey, K.J., Lacuesta, M., Leegood, R.C., and Lea, P.J. (1996). The isolation and characterization of mutants of the C4 plant Amaranthus edulis. C. R. Acad. Sci. Ser. III 319 951–959. [Google Scholar]
- Doughty, C.C., Hayashi, J.A., and Guenther, H.L. (1966). Purification and properties of D-glycerate 3-kinase from Escherichia coli. J. Biol. Chem. 241 568–572. [PubMed] [Google Scholar]
- Emanuelsson, O., Nielsen, H., Brunak, S., and von Heijne, G. (2000). Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300 1005–1016. [DOI] [PubMed] [Google Scholar]
- Emanuelsson, O., Nielsen, H., and von Heijne, G. (1999). ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 8 978–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemann, M., Vinnemeier, J., Oberpichler, I., Boldt, R., and Bauwe, H. (2005). The glycine decarboxylase complex is not essential for the cyanobacterium Synechocystis sp strain PCC 6803. Plant Biol. 7 15–22. [DOI] [PubMed] [Google Scholar]
- Hubbard, B.K., Koch, M., Palmer, D.R., Babbitt, P.C., and Gerlt, J.A. (1998). Evolution of enzymatic activities in the enolase superfamily: Characterization of the (D)-glucarate/galactarate catabolic pathway in Escherichia coli. Biochemistry 37 14369–14375. [DOI] [PubMed] [Google Scholar]
- Husic, D.W., Husic, H.D., and Tolbert, N.E. (1987). The oxidative photosynthetic carbon cycle or C2 cycle. Crit. Rev. Plant Sci. 5 45–100. [Google Scholar]
- Ichihara, A., and Greenberg, D.M. (1957). Studies on the purification and properties of D-glyceric acid kinase of liver. J. Biol. Chem. 225 949–958. [PubMed] [Google Scholar]
- Jonassen, I., Collins, J.F., and Higgins, D. (1995). Finding flexible patterns in unaligned protein sequences. Protein Sci. 4 1587–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keys, A.J., Bird, I.F., Cornelius, M.J., Lea, P.J., Wallsgrove, R.M., and Miflin, B.J. (1978). Photorespiratory nitrogen cycle. Nature 275 741–743.16069754 [Google Scholar]
- Kleczkowski, L.A., and Randall, D.D. (1983). Purification and partial characterization of spinach leaf glycerate kinase. FEBS Lett. 158 313–316. [Google Scholar]
- Kleczkowski, L.A., and Randall, D.D. (1988). Substrate stereospecificity of leaf glycerate kinase from C3 and C4 plants. Phytochemistry 27 1269–1273. [Google Scholar]
- Kleczkowski, L.A., Randall, D.D., and Zahler, W.L. (1985). The substrate specificity, kinetics, and mechanism of glycerate-3-kinase from spinach leaves. Arch. Biochem. Biophys. 236 185–194. [DOI] [PubMed] [Google Scholar]
- Lorimer, G.H., and Andrews, T.J. (1973). Plant photorespiration: An inevitable consequence of the existence of atmospheric oxygen. Nature 248 359–360. [Google Scholar]
- Lorimer, G.H., and Andrews, T.J. (1981). The C2 chemo- and photorespiratory carbon oxidation cycle. In The Biochemistry of Plants, Vol. 8, M.D. Hatch and N.K. Boardman, eds (New York: Academic Press), pp. 329–374.
- Ogren, W.L., and Bowes, G. (1971). Ribulose diphosphate carboxylase regulates soybean photorespiration. Nat. New Biol. 230 159–160. [DOI] [PubMed] [Google Scholar]
- Ornston, M.K., and Ornston, L.N. (1969). Two forms of D-glycerate kinase in Escherichia coli. J. Bacteriol. 97 1227–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmond, C.B., and Harris, B. (1971). Photorespiration during C4 photosynthesis. Biochim. Biophys. Acta 234 270–282. [DOI] [PubMed] [Google Scholar]
- Rabson, R., Kearney, P.C., and Tolbert, N.E. (1962). Formation of serine and glyceric acid by glycolate pathway. Arch. Biochem. Biophys. 98 154–163. [DOI] [PubMed] [Google Scholar]
- Richter, A., Hartwig, T., Boldt, R., van den Daele, K., Edner, C., Mangelsen, E., Bauwe, U., Michl, K., Kolukisaoglu, Ü., and Bauwe, H. (2005). Do current concepts of the C2 cycle need a revision? In Photosynthesis: Fundamental Aspects to Global Perspectives, A. van der Est and D. Bruce, eds (Lawrence, KS: Allen Press), in press.
- Sasaki, T., et al. (2002). The genome sequence and structure of rice chromosome 1. Nature 420 312–316. [DOI] [PubMed] [Google Scholar]
- Schmitt, M.R., and Edwards, G.E. (1983). Glycerate kinase from leaves of C3 plants. Arch. Biochem. Biophys. 224 332–341. [DOI] [PubMed] [Google Scholar]
- Somerville, C.R. (1984). The analysis of photosynthetic carbon dioxide fixation and photorespiration by mutant selection. Oxf. Surv. Plant Mol. Cell Biol. 1 103–131. [Google Scholar]
- Somerville, C.R. (2001). An early Arabidopsis demonstration resolving a few issues concerning photorespiration. Plant Physiol. 125 20–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville, C.R., and Ogren, W.L. (1979). A phosphoglycolate phosphatase-deficient mutant of Arabidopsis. Nature 280 833–836.16067381 [Google Scholar]
- Stabenau, H., and Winkler, U. (2005). Glycolate metabolism in green algae. Physiol. Plant. 123 235–245. [Google Scholar]
- Tabb, D.L., McDonald, W.H., and Yates III, J.R. (2002). DTASelect and Contrast: Tools for assembling and comparing protein identifications from shotgun proteomics. J. Proteome Res. 1 21–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F., and Higgins, D.G. (1997). The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Peer, Y., and De Wachter, R. (1994). TREECON for Windows: A software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput. Appl. Biosci. 10 569–570. [DOI] [PubMed] [Google Scholar]
- Van Schaftingen, E. (1989). D-glycerate kinase deficiency as a cause of D-glyceric aciduria. FEBS Lett. 243 127–131. [DOI] [PubMed] [Google Scholar]
- Verhees, C.H., Kengen, S.W., Tuininga, J.E., Schut, G.J., Adams, M.W., De Vos, W.M., and van der Oost, J. (2003). The unique features of glycolytic pathways in Archaea. Biochem. J. 375 231–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weckwerth, W., Loureiro, M.E., Wenzel, K., and Fiehn, O. (2004. b). Differential metabolic networks unravel the effects of silent plant phenotypes. Proc. Natl. Acad. Sci. USA 101 7809–7814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weckwerth, W., Wenzel, K., and Fiehn, O. (2004. a). Process for the integrated extraction, identification and quantification of metabolites, proteins and RNA to reveal their co-regulation in biochemical networks. Proteomics 4 78–83. [DOI] [PubMed] [Google Scholar]
- Wienkoop, S., Glinski, M., Tanaka, N., Tolstikov, V., Fiehn, O., and Weckwerth, W. (2004. a). Linking protein fractionation with multidimensional monolithic reversed-phase peptide chromatography/mass spectrometry enhances protein identification from complex mixtures even in the presence of abundant proteins. Rapid Commun. Mass Spectrom. 18 643–650. [DOI] [PubMed] [Google Scholar]
- Wienkoop, S., Zoeller, D., Ebert, B., Simon-Rosin, U., Fisahn, J., Glinski, M., and Weckwerth, W. (2004. b). Cell-specific protein profiling in Arabidopsis thaliana trichomes: Identification of trichome-located proteins involved in sulfur metabolism and detoxification. Phytochemistry 65 1641–1649. [DOI] [PubMed] [Google Scholar]