Abstract
Sequences involved in the regulation of genetic and genomic processes are primarily located in noncoding regions. Identifying such cis-acting sequences from sequence data is difficult because their patterns are not readily apparent, and, to date, identification has concentrated on regions associated with gene-coding functions. Here, we used phylogenetic footprinting to look for gene-independent noncoding elements in the ribosomal RNA gene repeats from several Saccharomyces species. Similarity plots of ribosomal intergenic spacer alignments from six closely related Saccharomyces species allowed the identification of previously characterized functional elements, such as the origin-of-replication and replication-fork barrier sites, demonstrating that this method is a powerful predictor of noncoding functional elements. Seventeen previously uncharacterized elements, showing high levels of conservation, were also discovered. The conservation of these elements suggests that they are functional, and we demonstrate the functionality of two classes of these elements, a putative bidirectional noncoding promoter and a series of conserved peaks with matches to the origin-of-replication core consensus. Our results paint a comprehensive picture of the functionality of the Saccharomyces ribosomal intergenic region and demonstrate that functional elements not involved in gene-coding function can be identified by using comparative genomics based on sequence conservation.
Keywords: gene amplification, ribosomal RNA gene
Eukaryote genomes contain large amounts of noncoding DNA, and it is clear that a diversity of functional elements are embedded in this noncoding DNA (1). However, finding such functional elements has been difficult because rules for their sequence patterns have not been established, and they are usually short and can have relaxed sequence requirements. One of the most powerful methods is locating areas of sequence conservation in intergenic regions between related organisms (2, 3), a technique known as phylogenetic footprinting (4). The rationale is that functional elements will show a greater level of selective constraint than the background sequence, thus standing out as “footprints” of high sequence conservation (5). To date, discovery of noncoding functional elements by phylogenetic footprinting has been restricted largely to those elements involved in controlling gene expression, such as promoters and enhancers. In this study, we used phylogenetic footprinting to look for noncoding functional elements that are not involved in the regulation of gene expression, and we call these elements gene-independent noncoding elements (NOCs). We looked for NOCs in the well studied ribosomal RNA gene repeats (rDNA; Fig. 1A) that contain both coding (rRNA) and noncoding [intergenic spacer (IGS)] regions. The rDNA has previously been shown to be involved in several gene-independent noncoding functions, such as origin-of-replication activity [ribosomal autonomous replicating sequence (rARS)] (6, 7), replication-fork barrier (RFB) activity (8, 9), and gene silencing (10, 11). In particular, the rDNA is a highly repetitive region that is unstable because of recombination among the repeats. Therefore, we expect the rDNA to be a hotspot for the regulation of genomic instability through the action of NOCs.
Fig. 1.
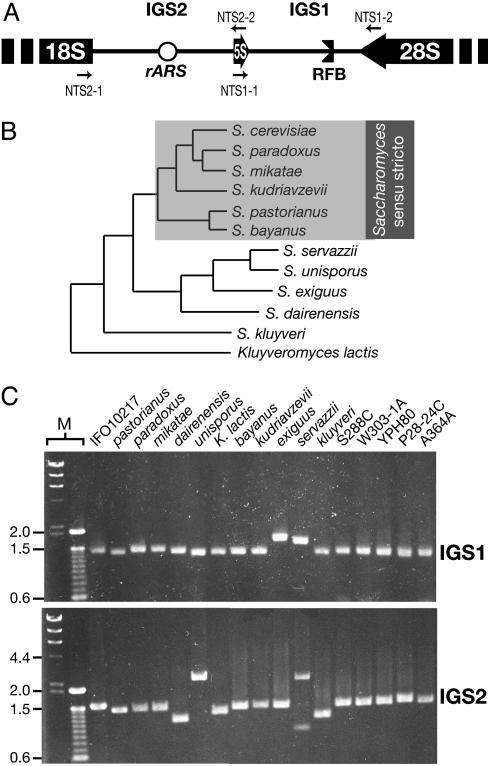
Sequencing the IGS regions from various Saccharomyces yeasts. (A) The rDNA IGS of S. cerevisiae. 18S, 28S, and 5S rRNA genes are shown as blocks and the IGS regions as lines. The rARS and RFB sites are indicated. Large arrows indicate directions of transcription. Primers used to amplify the IGS for sequencing are shown (small arrows). The diagram is not drawn to scale. (B) Phylogenetic tree, showing the relationships of the Saccharomyces yeasts that were sequenced [adapted from Kurtzman and Robnett (12)]. Species from the Saccharomyces sensu stricto clade that were used for the subsequent analyses are boxed. Branch lengths are not drawn to scale. (C) Gel, showing the PCR-amplified IGS1 and IGS2 regions from all 17 isolates. Names are as in B, except that the various S. cerevisiae strains are shown as strain names. M, size markers (λ HindIII, and 100-bp ladders). Size marks are shown to the left (kb).
We used species from the genus Saccharomyces (Ascomycota). Saccharomyces species have a long history of human association and a well established phylogeny, and the availability of many strains makes them excellent candidates for phylogenetic footprinting. Fig. 1B shows the phylogenetic relationships of the Saccharomyces species that were used in this study, including the group of Saccharomyces species most closely related to Saccharomyces cerevisiae, Saccharomyces sensu stricto species (12). Several studies have found that species within this phylogenetic range are useful for detecting noncoding functional regions (13–15), and, therefore, we sampled along similar lines.
To identify potential NOCs, the IGS regions of the rDNA were sequenced and compared. Most previously identified functional elements in the S. cerevisiae IGS could be recognized as phylogenetic footprints. We also identified a number of other conserved regions for which functions are not known and found evidence for functionality of two classes of these conserved elements. Our results suggest that comparative-genomics approaches are useful for identifying functional NOCs.
Methods
Strains. Twelve species from the “Saccharomyces complex,” including 6 isolates of S. cerevisiae (a total of 17 isolates; Table 1), were chosen to encompass a phylogenetic range appropriate for phylogenetic footprinting, and their phylogenetic relationships are shown in Fig. 1B.
Table 1. Yeast strains.
| Species | Strain | Reference no. |
|---|---|---|
| S. bayanus | – | BY4498 |
| S. cerevisiae | A364A | BY8233 |
| IFO10217 | BY21391 | |
| P28–24C | BY2986 | |
| S288C | BY611 | |
| W303–1A | BY4848 | |
| YPH80 | BY963 | |
| S. dairenensis | – | BY21386 |
| S. exiguus | – | BY21389 |
| S. kluyveri | – | BY20108 |
| S. kudriavzevii | – | BY20109 |
| S. mikatae | – | BY20110 |
| S. paradoxus | – | BY20111 |
| S. pastorianus | – | BY4497 |
| S. servazzii | – | BY21390 |
| S. unisporus | – | BY21387 |
| Kluyveromyces lactis | – | BY20597 |
All strains are from the Yeast Genetic Resource Center (YGRC; http://bio3.tokyo.jst.go.jp/jst/english/), Osaka University, Osaka, and reference no. reflect YGRC numbering. Strains in bold (10 isolates) are those used for the alignments in Fig. 2. S., Saccharomyces.
PCR and Sequencing. Standard techniques were performed as described in ref. 16. Primer combinations NTS1–1/NTS1–2 (GTTTGCGGCCATATCTACCAG and TGGGTGCTTGCTGGCGAATT, respectively) and NTS2–1/NTS2–2 (TAGGCAGATCTGACGATCAC and TACCAGCTTAACTACAGTTG, respectively) were used to amplify IGS1 and -2, respectively (Fig. 1C). Double-strand sequences of both IGS regions were obtained by primer-walking with the gel-purified PCR products. The sequences are deposited in the GenBank database under accession nos. DQ130071–DQ130103. The Kluyveromyces lactis IGS1 sequence was not determined.
Sequence Analysis. Sequences were aligned by using the program megalign (DNASTAR, Madison, WI) with the clustalw alignment function (17), and the alignments were adjusted by eye. Completed alignments were imported into the Seqlab (Göttingen, Germany) version of the Genetic Computer Group's software cluster wisconsin package (Version 10.3, Accelrys, San Diego), and the levels of DNA sequence conservation were visualized by using the plotsimilarity function. Similarity refers to the proportion of isolates in which a base is conserved, calculated across all of the bases within a single sliding window; e.g., for a 20-bp sliding window, if all 20 bases in that window are conserved across all 10 isolates, the score is 1.0.
Experimental Analysis of Ribosomal Conserved Noncoding Sequences (rCNS) 10 to 16. Deletion of the region covering rCNS 10 to 16, rDNA amplification assays, chromosomal plug preparation, contour-clamped homogeneous electric field (CHEF) electrophoresis, and Southern hybridization were all performed as described in ref. 18; CHEF conditions are as those shown in figure 4 of ref. 18.
Results
Constructing IGS Similarity Plots. We amplified the two rDNA IGS regions (IGS1 and -2) from all 17 Saccharomyces isolates by using PCR (Fig. 1C) and sequenced these PCR products directly (S. cerevisiae strain W303–1A was omitted from subsequent analyses because its IGS sequence was identical to S288C). The sequences were aligned, although only the 6 Saccharomyces sensu stricto species (Fig. 1B) (12) could be reliably aligned. Similarity plots, using a sliding window of comparison, were then used to look for phylogenetic footprints of highly conserved regions in the alignments of the Saccharomyces sensu stricto species/S. cerevisiae strains (10 isolates; see Table 1). The results are shown in Fig. 2, with the plots showing regions that are conserved above the average level of similarity for the whole alignment. The plots show a dynamic level of conservation across the IGS. IGS1 is characterized by a few broad regions of high conservation, whereas IGS2 is characterized by conserved regions that are more frequent but slightly less-highly conserved. IGS1 is more length-variable than IGS2, with several hotspots of length variation present. Previously identified length-variable regions (19) were present, along with additional length-variable regions. We also created similarity plots using various subsets of the species from Fig. 2. In general, we found that the phylogenetic range of the species used, rather than the number of isolates, is the most critical issue for creating informative similarity plots. A range that is too restricted results in the inability to reliably distinguish conserved regions from nonconserved regions (see Fig. 4, which is published as supporting information on the PNAS web site), whereas species that are evolutionarily too distant lose sequence conservation, as evidenced by the fact that the IGS sequences were unalignable outside of the Saccharomyces senso stricto species. Finally, the use of at least three species is necessary to avoid spurious sequence matches in regions that do not truly align.
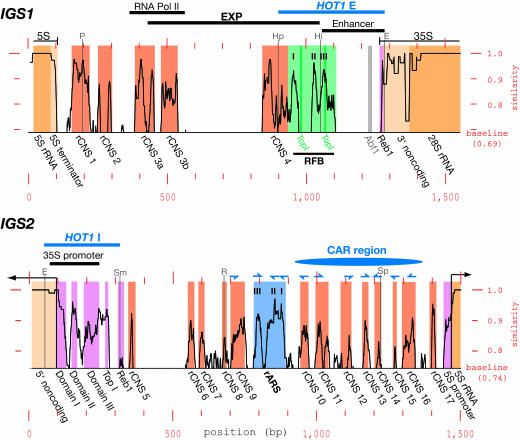
Fig. 2.
Similarity plots of IGS1 and IGS2. The level of similarity is given along the y axis by using sliding windows of 15 bp (IGS1, Top) or 20 bp (IGS2, Bottom) from the alignments of the 10 isolates (see Table 1). The “baseline” is the average level of similarity across the alignment for each region (actual values are also indicated). Elements of interest are boxed, with coding regions in yellow, transcribed regions upstream and downstream of the coding regions in pale yellow, rCNSs in orange, the RFB and rARS regions (each with three subelements indicated) in light green and blue, respectively, and other regions of interest in purple. The position of the nonconserved Abf1 site is shown in gray, and the positions of the two putative topoisomerase-I-binding sites (TopI) in the RFB are shown in dark green. Element names are given below the plots. Positions of other previously characterized, broad regions of interest are shown above the plots. Transcription-initiation and -termination sites are indicated by thin lines with arrowheads and block-ends, respectively. Conserved peaks matching the ARS consensus are marked with half arrowheads above the plots, and their strand locations and directions are indicated: alignment strand, upward-pointing half arrowhead; opposite strand, downward-pointing half arrowhead. Positions of familiar restriction enzyme sites are shown as thin gray lines (P, PvuII; Hp, HpaI; Hi, HindIII; E, EcoRI; Sm, SmaI; R, EcoRV; and Sp, SphI). The x axis is the position across each alignment.
Identification of Previously Identified Features. First, we looked for previously identified features in the IGS in these similarity plots. Many previously identified features could be easily identified as highly conserved peaks, as shown in Fig. 2.
rRNA Coding Regions. The rRNA coding regions are highly conserved, as expected, as are the 5′ and 3′ noncoding regions of the 35S rRNA transcript. The 35S rRNA promoter, located at one end of IGS2, shows peaks of conservation that correspond closely to the three promoter domains identified in ref. 20. Similarly, the putative 5S rRNA promoter (21) is conserved, as is the region immediately downstream of the 5S rRNA gene, a T-rich tract that overlaps with the transcription stop site (22).
rARS and RFB Sites. Two previously identified NOCs stood out as highly conserved regions in the similarity plots. One is the rARS present in IGS2. Its function depends on a region that contains three matches to the so-called ARS core-consensus sequence (ACS) (23, 24), and each of these three rARS core sequences corresponds to a highly conserved peak on the conservation profile. The other NOC is the RFB site in IGS1. Like the rARS, this is also made up of three core sequences (25, 26), and, as with the rARS, these three subregions each correspond to a peak of conservation. Interestingly, only one of the two topoisomerase I sites predicted in the RFB (27, 28) was found to be conserved; the topoisomerase I site near the 35S rRNA promoter (27) is also conserved.
Other Known Elements. The enhancer element for 35S rRNA transcription (190-bp EcoRI-HindIII IGS1 fragment) (29) is not conserved, aside from the ends of this element. These conserved ends correspond to a part of the RFB site and a Reb1-binding site, both of which have previously identified functions (see above and below). This lack of conservation is consistent with a report that the enhancer does not enhance endogenous 35S rRNA transcription (30). The HOT1 recombination hotspot consists of two regions, HOT1 E (in IGS1) and HOT1 I (located around the 35S rRNA promoter) (31). The conserved parts of HOT1 E and I correspond largely to conserved regions with previously identified functions (the RFB and Reb1 sites and the 35S rRNA promoter, respectively). Thus, it is likely that these elements provide HOT1 function, especially given the dependence of HOT1 activity on RNA polymerase I transcription, although the precise mechanism of HOT1-mediated recombination stimulation is still under investigation (32). Also conserved are two previously identified Reb1 sites (33, 34). The IGS1 Reb1 site is believed to be involved in transcription termination (35), and it is completely conserved in all 10 isolates, along with neighboring nucleotides and an adjacent T/A-rich region. Although the peak of conservation for the IGS 2 Reb1 site is not convincing, this small peak actually represents an 8-bp completely conserved motif embedded in a highly variable region. Because a 20-bp sliding window was used, the conservation at this site appears to be lower. Finally, an Abf1 (ARS-binding factor I) recognition sequence was predicted to be present in the enhancer (33), but this site is not conserved at all. This finding agrees with the recent observation that deletion of this Abf1 site has little effect on rARS activity (28).
Therefore, the majority of previously identified noncoding functional elements could be precisely located by the presence of highly conserved regions. This was true not only for noncoding elements involved in rRNA gene transcription (promoters and terminators) but also for NOCs involved in other functions. For the elements that did not exhibit appreciable sequence conservation, there was already evidence calling into question their functionality. Together, these results suggest that this phylogenetic-footprinting approach is a powerful predictor of functional elements with a range of biological functions.
Unknown Conserved Regions. We next turned our attention to the regions of high sequence conservation for which functions are not known. We report a total of 17 conserved elements (Fig. 2), 4 in IGS1 and 13 in IGS2, and name these rCNSs 1 to 17 (5). Sequence logos for these rCNSs are shown in Fig. 5, which is published as supporting information on the PNAS web site. The sequence conservation of these regions suggests that many of them are likely to be functionally important. Furthermore, given that they are embedded in the IGS and are not known to be involved in rRNA transcription, we believe that many will be functional NOCs. Therefore, we looked for clues to the function of some of these rCNSs.
Bidirectional Noncoding Promoter. The IGS1 in S. cerevisiae contains a region called EXP that is responsible for recombination-based amplification of rDNA and includes the RFB site (18). We reasoned that conserved regions in the EXP are likely to be cis-acting elements involved in amplification, with the RFB site being a known example. As can be seen in Fig. 2, there are two other rCNSs located in the EXP, rCNSs 3 and 4. Interestingly, rCNS 3 (composed of two adjacent sets of conserved peaks, rCNSs 3a and 3b) almost precisely corresponds to a previously identified bidirectional RNA polymerase II (pol II) promoter (36). No function has been attributed to this element, and it does not seem to promote transcription of any coding region. The conservation of this element suggested that it was a functional NOC involved in recombination-based amplification. By replacing this promoter with other inducible pol II promoters, we, indeed, showed that it is necessary for amplification and that it plays a key role in the regulation of recombination in the rDNA (T.K. and A.R.D.G., unpublished work). The role of rCNS 4 is, as yet, unknown. Unexpectedly, a large region between rCNSs 3 and 4 is devoid of conserved peaks, although a previous study indicated that this region is a functional part of the EXP (18). We suspect that, given the importance of transcription for EXP function, it is the presence of a transcriptional terminator in the construct used in the previous study that leads to the loss of EXP function, rather than the region actually being functional.
Series of Conserved ARS Sequence Matches. The conserved peaks in the IGS2 between the rARS and the 5S rRNA gene presented an intriguing pattern (Fig. 2). To look more closely at their possible function, we compared the conserved peaks in this region with each other. Extraordinarily, seven of the eight rCNSs between the rARS and the 5S rRNA gene (rCNSs 10 to 16) showed good matches to the ARS core-consensus sequence, as did rCNS 9 on the other side of the rARS (Fig. 2). Thus, including the rARS itself, there is a run of 11 conserved peaks that all show strong (at least 10 of 12-bp in the case of S. cerevisiae) matches to the ACS (Fig. 3A). Most matching nucleotides are conserved across all 10 isolates (69 of 96 total, with 16 of the nonconserved bases falling on ACS nucleotide sites, where redundancy is allowed) (23). For comparison, given the base and conservation composition of the region covering rCNS 10 to 16, one element with the average features of these rCNS ACS matches is expected every ≈1.7 kb per strand, by chance. Because this region is only ≈450 bp long, these matches are highly overrepresented. Furthermore, half of these matches face one direction, and half face the other direction; and half of the matches are on one strand, and half are on the other strand. This remarkable organization suggests that these conserved ACS matches are functional NOCs. The region of IGS2 covered by rCNS 10 to 16 forms a peak of cohesin association (called CAR) (37). There is evidence that cohesin localization is important in controlling recombination in the rDNA (38), thereby suggesting that this region may be involved in cohesin localization/recombination regulation.
Fig. 3.
Characterization of rCNSs in CAR. (A) Alignment of rCNSs that share similarity with the ACS. Sequences are from S. cerevisiae strain S288C, and the regions of similarity to the ACS are boxed. Nucleotides that match the ACS (shown above and below the alignments with accepted deviations) and that are conserved in all 10 isolates (Table 1) are black, those that match but are not completely conserved are blue, those that do not match but are completely conserved are green, and those that neither match nor are conserved are red. Flanking nucleotides are in lower case. The two alignments differ in their strand direction, as indicated by the arrows. rCNS names are shown to the right, along with the number of bases that match the ARS consensus. rv, the reverse complement of the alignment sequence was used. The three rARS ACSs are also included. (B) CHEF gel electrophoresis assaying rDNA amplification ability in the CAR replacement strain. The chromosome XII of two-copy rDNA strains with a replacement of CAR (ΔCAR) and the 5S rRNA gene (Δ5S) were resolved by CHEF electrophoresis. These strains were assayed after the introduction of FOB1 or an empty vector (+ and -, respectively). (Left) The ethidium bromide-strained gel. (Right) The gel probed with an rDNA probe, revealing chromosome XII (overlapping chromosomes VII and XV in the ΔCAR lanes of the left panel). A Hansenula wingei chromosome size marker (M; BioRad; missing from the Southern blot) and a parental two-rDNA-copy strain (C) were also included. Sizes of H. wingei chromosomes (Mb) and the positions of the other S. cerevisiae chromosomes are shown to the left.
Experimental Evidence that the ACS Matches Are Functional. To investigate further whether the region containing these ACS matches, which we will call CAR, is involved in rDNA recombination, we replaced CAR (encompassing rCNS 10 to 16) with a heterogeneous sequence (a URA3 marker) in a strain with only two copies of the rDNA (18). If these two rDNA copies are wild-type, they can amplify back up to wild-type copy number with the reintroduction of the FOB1 gene (which is deleted in the two-copy strain). This is the rDNA amplification assay (18). We observed the rDNA amplification ability of the two-copy strain containing the CAR replacement (ΔCAR), and a control strain in which the 5S rRNA gene (adjacent to CAR) is replaced by the URA3 marker (Δ5S), by using CHEF gel electrophoresis (Fig. 3B). After introduction of the FOB1 gene, the size of chromosome XII (the rDNA-containing chromosome) in the Δ5S strain increased dramatically and became smeared, indicating that rDNA amplification was occurring, as shown in ref. 18. In contrast, the ΔCAR strain showed no evidence of amplification after the introduction of the FOB1 gene. Therefore, we conclude that CAR, which contains the conserved ACS matches, is an important cis-acting sequence for rDNA amplification.
The rARS is currently defined as an ≈100-bp fragment that contains three ACS matches and is sufficient for full ARS function (24). CAR does not include this rARS, suggesting that the function of the majority of these ARS-like conserved peaks is not origin-of-replication activity. A recent study identified genetic interactions between cohesin and the origin-recognition complex (ORC) (39). Therefore, it is possible that these ARS-like rCNSs may associate with ORC and help localize cohesion to this region. However, the loss of amplification ability after CAR deletion suggests that this region has a positive role to play in rDNA recombination. Many of the peaks of conservation in this region are broader than the ACS match (see Fig. 5), therefore we cannot rule out that these other conserved motifs also help promote rDNA amplification. In any case, our results demonstrate that this region forms a functional NOC, thereby strengthening the conclusion that conserved peaks identified by this phylogenetic-footprinting approach are likely to be functional.
Features in Other rCNSs. The roles of the other rCNSs we identified remain unknown. We searched for matches of these remaining rCNSs to previously identified cis-acting sequences in the yeast genome by using the Transfac database through the Transcription Element Search System (TESS) web portal (www.cbil.u-penn.edu/tess/). Two potential matches were found. The first was a 12- of 13-bp match of the central part of rCNS 4 with the 5′ half of a TATA-binding protein (TBP) recognition site (40). TBP is involved in pol II transcription initiation, and therefore, its role, if any, is not clear. The second potential match is a 14- of 15-bp match of rCNS 17 to the 5′ half of a Rap1 (Grf1)-binding site (41). Rap1-binding sites have been implicated in a variety of roles, such as upstream activation and silencing, therefore the presence of this binding site is reasonable, but does not provide much of a clue to potential function. We also found an interesting expansion in the copy number of a 6-bp microsatellite-like sequence, of which rCNS 1 is composed, and of an adjacent 12-bp repeat in two S. cerevisiae strains. The conservation and proximity of these small repeats to the 35S promoter, together with a tendency for expansion, suggests a possible role in 35S transcription initiation (42).
Discussion
This phylogenetic-footprinting approach has given us a comprehensive picture of the functionality of the rDNA IGS in S. cerevisiae. Most previously identified functional regions do, indeed, show selective constraint, and we have reasons to doubt the functionality of those that do not (although it is important to note that some elements may be functional without showing DNA sequence conservation, such as spacing elements or simple-sequence motifs). Furthermore, this approach allowed us to identify a number of conserved regions for which functions have not been previously ascribed. The majority of these rCNSs are likely to be functional and are likely to function in roles independent of gene transcription. Supporting this conclusion, we have experimentally demonstrated that two classes of rCNSs are actually functional NOCs, and both of these appear to be involved in the maintenance of genomic stability. Therefore, phylogenetic footprinting is a powerful method for predicting functional NOCs from sequence data.
Previous phylogenetic-footprinting studies have identified a wide range of CNSs involved in the regulation of gene transcription. That we could identify functional NOCs by using the same approach suggests that the evolutionary dynamics of these two types of elements are similar, and, indeed, a comparison of similarity plots from randomly chosen noncoding regions from other parts of the Saccharomyces genome with the plots from this study showed broadly similar profiles of conserved peaks (data not shown). Such similarities are expected, because many NOCs probably function as cis-acting protein-binding sites. On the other hand, we were unable to align the IGS regions outside Saccharomyces sensu stricto species. The exception was the 35S rRNA promoter, which could be aligned across all 12 species (see Fig. 6, which is published as supporting information on the PNAS web site), implying some difference in the selective constraint of the 35S rRNA promoter, compared with other functional elements in the IGS. At least some rCNSs are expected to be present in these more distantly related species, based on the results of other Saccharomyces phylogenetic-footprinting studies (13, 43). Therefore, we searched for the presence of short, highly conserved subregions of all of the unknown rCNSs in Saccharomyces servazzii, Saccharomyces exiguus, and Saccharomyces dairenensis but were unable to find any convincing matches. We believe that these rCNSs are likely to be present but, for some reason, escape detection. One possibility is that the actual functional sequences are very short, sequence variation is permitted, and/or their position in the IGS varies widely, thus making them difficult to distinguish from background. Another possibility is that these rCNSs are more free to vary in sequence than are typical single-copy gene cis-acting elements. Many trans-acting factors recognize multiple cis-acting sites, acting to regulate suites of genes; thus, sequence changes in individual cis-acting sites are not easily tolerated. In the case of the rDNA, many NOCs may function there exclusively, thus allowing for rapid “one-on-one” coevolution between the NOC and its trans-acting factor. Finally, the rDNA is multicopy, and the evolutionary dynamics of the rDNA can act to quickly spread variants to all repeat copies (a process known as homogenization) (44, 45), thus, perhaps, aiding rapid evolutionary change. Indeed, it has even been suggested that this homogenization may promote pressure for evolutionary change on trans-acting factors (molecular drive) (46).
The advantages of identifying candidate NOCs is particularly clear in the case of higher eukaryotes, where only a small fraction of their large genomes are under selective constraint (1). It will be interesting to see what relationship, if any, yeast NOCs have to the recently discovered higher-eukaryote conserved nongenic sequences (47). Identification of potentially functional NOCs is just the first step in understanding their biological role. Once potential NOCs have been identified, the real challenge is elucidating their function. We expect that, as the number of available genome sequences increases, a combination of computer-based and experimental approaches will lead to rapid advances in our understanding of NOCs.
Supplementary Material
Acknowledgments
We thank Drs. Y. Kaneko and S. Harashima (Osaka University, Osaka) for kindly providing the yeast strains used in this study. This work was supported, in part, by Grants 13141205, 14380332, 17080010, and 17370065 from the Ministry of Education, Science and Culture, Japan, and a grant from the Human Frontier Science Program (to T.K.). A.R.D.G. was supported by a Japan Society for the Promotion of Science Postdoctoral Fellowship.
Author contributions: T.K. designed research; A.R.D.G., K.H., T.H., and T.K. performed research; A.R.D.G. and T.K. analyzed data; and A.R.D.G. and T.K. wrote the paper.
Abbreviations: ARS, autonomous replicating sequence; ACS, ARS core-consensus sequence; CAR, cohesin-association region; ΔCAR, CAR replacement; CHEF, contour-clamped homogeneous electric field; IGS, intergenic spacer; NOC, gene-independent noncoding element; rARS, ribosomal autonomous replicating sequence (origin of replication); rDNA, ribosomal RNA gene repeats; rCNS, ribosomal conserved noncoding sequence; RFB, replication-fork barrier.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. DQ130071–DQ130103).
References
- 1.Mouse Genome Sequencing Consortium (2003) Nature 420, 520-562. [DOI] [PubMed] [Google Scholar]
- 2.Pribnow, D. (1975) Proc. Natl. Acad. Sci. USA 72, 784-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schaller, H., Gray, C. & Herrmann, K. (1975) Proc. Natl. Acad. Sci. USA 72, 737-741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tagle, D. A., Koop, B. F., Goodman, M., Slightom, J. L., Hess, D. L. & Jones, R. T. (1988) J. Mol. Biol. 203, 439-455. [DOI] [PubMed] [Google Scholar]
- 5.Hardison, R. C. (2000) Trends Genet. 16, 369-372. [DOI] [PubMed] [Google Scholar]
- 6.Skryabin, K. G., Eldarov, M. A., Larionov, V. L., Bayev, A. A., Klootwijk, J., de Reg, V. C. H. F., Veldman, G. M., Planta, R. J., Georgiev, O. I. & Hadjiolov, A. A. (1984) Nucleic Acids Res. 12, 2955-2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brewer, B. J. & Fangman, W. L. (1991) BioEssays 13, 317-322. [DOI] [PubMed] [Google Scholar]
- 8.Kobayashi, T., Hidaka, M., Nishizawa, M. & Horiuchi, T. (1992) Mol. Gen. Genet. 233, 355-362. [DOI] [PubMed] [Google Scholar]
- 9.Brewer, B. J., Lockshon, D. & Fangman, W. L. (1992) Cell 71, 267-276. [DOI] [PubMed] [Google Scholar]
- 10.Bryk, M., Banerjee, M., Murphy, M., Knudsen, K. E., Garfinkel, D. J. & Curcio, M. J. (1997) Genes Dev. 11, 255-269. [DOI] [PubMed] [Google Scholar]
- 11.Smith, J. S. & Boeke, J. D. (1997) Genes Dev. 11, 241-254. [DOI] [PubMed] [Google Scholar]
- 12.Kurtzman, C. P. & Robnett, C. J. (2003) FEMS Yeast Res. 3, 417-432. [DOI] [PubMed] [Google Scholar]
- 13.Cliften, P. F., Hillier, L. W., Fulton, L., Graves, T., Miner, T., Gish, W. R., Waterson, R. H. & Johnston, M. (2001) Genome Res. 11, 1175-1186. [DOI] [PubMed] [Google Scholar]
- 14.Kellis, M., Patterson, N., Endrizzi, M., Birren, B. & Lander, E. S. (2003) Nature 423, 241-254. [DOI] [PubMed] [Google Scholar]
- 15.Liu, Y., Liu, X. S., Wei, L., Altman, R. B. & Batzoglou, S. (2004) Genome Res. 14, 451-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burke, D., Dawson, D. & Stearns, T. (2000) Methods in Yeast Genetics (Cold Spring Harbor Lab. Press, Woodbury, NY).
- 17.Thompson, J. D., Higgins, D. G. & Gibson, T. J. (1994) Nucleic Acids Res. 22, 4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kobayashi, T., Nomura, M. & Horiuchi, T. (2001) Mol. Cell. Biol. 21, 136-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jemtland, R., Maehlum, E., Gabrielsen, O. S. & Øyen, T. B. (1986) Nucleic Acids Res. 14, 5145-5158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Musters, W., Knol, J., Maas, P., Dekker, A. F., van Heerikhuizen, H. & Planta, R. J. (1989) Nucleic Acids Res. 17, 9661-9678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Challice, J. M. & Segall, J. (1989) J. Biol. Chem. 264, 20060-20067. [PubMed] [Google Scholar]
- 22.Brown, B. R., Bartholomew, B., Kassavetis, G. A. & Geiduschek, E. P. (1992) J. Mol. Biol. 228, 1063-1077. [DOI] [PubMed] [Google Scholar]
- 23.Van Houten, J. V. & Newlon, C. S. (1990) Mol. Cell. Biol. 10, 3917-3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller, C. A. & Kowalski, D. (1993) Mol. Cell. Biol. 13, 5360-5369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kobayashi, T. (2003) Mol. Cell. Biol. 23, 9178-9188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ward, T. R., Hoang, M. L., Prusty, R., Lau, C. K., Keil, R. L., Fangman, W. L. & Brewer, B. J. (2000) Mol. Cell. Biol. 20, 4948-4957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vogelauer, M. & Camilloni, G. (1999) J. Mol. Biol. 293, 19-28. [DOI] [PubMed] [Google Scholar]
- 28.Burkhalter, M. D. & Sogo, J. M. (2004) Mol. Cell 15, 409-421. [DOI] [PubMed] [Google Scholar]
- 29.Elion, E. A. & Warner, J. R. (1984) Cell 39, 663-673. [DOI] [PubMed] [Google Scholar]
- 30.Wai, H., Johzuka, K., Vu, L., Eliason, K., Kobayashi, T., Horiuchi, T. & Nomura, M. (2001) Mol. Cell. Biol. 21, 5541-5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Voelkel-Meiman, K., Keil, R. L. & Roeder, G. S. (1987) Cell 48, 1071-1079. [DOI] [PubMed] [Google Scholar]
- 32.Serizawa, N., Horiuchi, T. & Kobayashi, T. (2004) Genes Cells 9, 305-315. [DOI] [PubMed] [Google Scholar]
- 33.Morrow, B. E., Johnson, S. P. & Warner, J. R. (1989) J. Biol. Chem. 264, 9061-9068. [PubMed] [Google Scholar]
- 34.Kulkens, T., van Heerikhuizen, H., Klootwijk, J., Oliemans, J. & Planta, R. J. (1989) Curr. Genet. 16, 351-359. [DOI] [PubMed] [Google Scholar]
- 35.Lang, W. H. & Reeder, R. H. (1995) Proc. Natl. Acad. Sci. USA 92, 9781-9785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Santangelo, G. M., Tornow, J., McLaughlin, C. S. & Moldave, K. (1988) Mol. Cell. Biol. 8, 4217-4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laloraya, S., Guacci, V. & Koshland, D. (2000) J. Cell Biol. 151, 1047-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kobayashi, T., Horiuchi, T., Tongaonkar, P., Vu, L. & Nomura, M. (2004) Cell 117, 441-453. [DOI] [PubMed] [Google Scholar]
- 39.Suter, B., Tong, A., Chang, M., Yu, L., Brown, G. W., Boone, C. & Rine, J. (2004) Genetics 167, 579-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lue, N. F. & Kornberg, R. D. (1993) Proc. Natl. Acad. Sci. USA 90, 8018-8022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Buchman, A. R., Lue, N. F. & Kornberg, R. D. (1988) Mol. Cell. Biol. 8, 5086-5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reeder, R. H. (1984) Cell 38, 349-351. [DOI] [PubMed] [Google Scholar]
- 43.Cliften, P., Sudarsanam, P., Desikan, A., Fulton, L., Fulton, B., Majors, J., Waterson, R., Cohen, B. A. & Johnston, M. (2003) Science 301, 71-76. [DOI] [PubMed] [Google Scholar]
- 44.Ganley, A. R. D. & Scott, B. (1998) Genetics 150, 1625-1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hughes, K. W. & Peterson, R. H. (2001) Mol. Biol. Evol. 18, 94-96. [DOI] [PubMed] [Google Scholar]
- 46.Dover, G. (2002) Trends Genet. 18, 587-589. [DOI] [PubMed] [Google Scholar]
- 47.Dermitzakis, E. T., Reymond, A., Scamuffa, N., Ucla, C., Kirkness, E., Rossier, C. & Antonarakis, S. E. (2003) Science 302, 1033-1035. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.