Abstract
Our previous results demonstrated that the DNAβ satellite (Y10β) associated with Tomato yellow leaf curl China virus Y10 isolate (TYLCCNV-Y10) is essential for induction of leaf curl symptoms in plants and that transgenic expression of its βC1 gene in Nicotiana plants induces virus-like symptoms. In the present study, in vitro DNA binding activity of the βC1 proteins of Y10β and DNAβ (Y35β) found in the Tobacco curly shoot virus Y35 isolate (TbCSV-Y35) were studied following their expression as six-His fusion proteins in Escherichia coli. Electrophoretic mobility shift assays and UV cross-linking experiments revealed that βC1 proteins could bind both single-stranded and double-stranded DNA without size or sequence specificity. Suppression of green fluorescent protein (GFP) transgene silencing was observed with the new leaves of GFP-expressing Nicotiana benthamiana plants coinoculated by TYLCCNV-Y10 plus Y10β or by TbCSV-Y35 plus Y35β. In a patch agroinfiltration assay, the transiently expressed βC1 gene of Y10β or Y35β was able to suppress host RNA silencing activities and permitted the accumulation of high levels of GFP mRNA in the infiltrated leaf patches of GFP transgenic N. benthamiana plants. The βC1 protein of Y10β accumulated primarily in the nuclei of plant and insect cells when fused with β-glucuronidase or GFP and immunogold labeling showed that the βC1 protein is present in the nuclei of infected N. benthamiana plants. A mutant version of Y10β carrying the mutations within the putative nuclear localization sequence of the Y10 βC1 protein failed to induce disease symptoms, suppress RNA silencing, or accumulate in the nucleus, suggesting that nuclear localization of the βC1 protein is a key requirement for symptom induction and silencing suppression.
RNA silencing is an evolutionarily conserved surveillance system that occurs in many eukaryotic organisms, including animals (RNA interference), fungi (quelling) and plants (posttranscriptional gene silencing [PTGS]) (2, 10, 19, 34). One of the key intermediary elements in initiation of the RNA silencing pathway is double-stranded RNA (dsRNA). The conserved mechanism of RNA silencing involves the processing of dsRNA into 21 to 25 nucleotides and the production of small interfering RNAs (siRNAs) by an RNase III-like enzyme, DICER (22). These siRNAs subsequently guide a nuclease complex, referred to as RNA-induced silencing complex (4), to degrade target RNAs in a sequence-specific manner. In plants, an unidentified systemic signal is also generated and amplified to elicit long-range RNA silencing in distant tissues and across graft unions (41). RNA silencing clearly plays an important antiviral role in plants, animals, and perhaps other eukaryotes, because viruses are both initiators and targets of RNA silencing (14). Consistent with its antiviral nature, some viruses have evolved or acquired functional proteins (suppressors) that suppress RNA silencing by targeting different steps of silencing pathways (46, 57, 58).
Geminiviruses within the genus Begomovirus are transmitted by the whitefly Bemisia tabaci and have either one or two circular single-stranded DNA (ssDNA) genomic components (18, 23). Some monopartite begomoviruses, such as Ageratum yellow vein virus (AYVV), Bhendi yellow vein mosaic virus (BYVMV), Cotton leaf curl Multan virus (CLCuMV), and Eupatorium yellow vein virus (EpYVV) are unable to induce disease symptoms in their natural hosts. A novel type of circular single-stranded satellite DNA, referred to as DNAβ, has recently been found to be associated with some of these viruses and to be essential for the induction of typical disease symptoms in Ageratum, bhendi, cotton, or Eupatorium, respectively (6, 28, 48, 49). DNAβ is approximately half of the size (1.3 to 1.4 kb) of the begomovirus DNA, on which it depends for replication, encapsidation, insect transmission, and movement in plants (6, 48). Sequence analyses have shown that the complementary-sense strand of all DNAβ molecules encodes a βC1 gene that is conserved in position and size. Typically, this gene has the capacity to encode a 13- to 14-kDa protein comprising 118 amino acids, although some DNAβ molecules have additional N-terminal amino acids (50, 60). The precise function of DNAβ and its βC1 protein in pathogenesis is unknown, although it has been proposed that DNAβ may play a direct or an indirect role in replication, facilitating movement, or countering host defense response (48).
We demonstrated previously that DNAβ (Y10β) associated with the Tomato yellow leaf curl China virus Y10 isolate (TYLCCNV-Y10) is required for production of leaf curl symptoms in tobacco, tomato, and petunia and that TYLCCNV-Y10 alone produced asymptomatic infections in these hosts (12). Y10β mutants unable to express the βC1 protein lost the ability to induce disease symptoms when coinoculated with TYLCCNV-Y10 (12, 60). Transgenic Nicotiana benthamiana and Nicotiana tabacum plants expressing the Y10 βC1 gene also produced virus-like leaf curl symptoms (12). In the present study, we report that the βC1 protein binds DNA in a size- and sequence-nonspecific manner. Furthermore, we demonstrate that the βC1 protein functions as a suppressor of RNA silencing and is targeted to the nuclei of insect and plant cells.
MATERIALS AND METHODS
Protein expression and purification.
For expression of the βC1 proteins in Escherichia coli, the Y10 βC1 gene and the βC1 gene of DNAβ (Y35β) associated with the Tobacco curly shoot virus Y35 isolate (TbCSV-Y35) were amplified by PCR. For this purpose, primer pairs Y10βC1-F/Y10βC1-R and Y35βC1-F/Y35βC1-R (Table 1) were used to amplify the βC1 genes from the plasmids pGEM-Y10β and pGEM-Y35β, which contain full-length inserts of Y10β and Y35β, respectively (60; Z. H. Li, Y. Xie, and X. P. Zhou, unpublished data). After digestion with BamHI and SalI, the PCR fragments were cloned into pET-30a (Novagen, Madison, WI) to produce pET-Y10βC1 and pET-Y35βC1. The integrity of these constructs and those described below was confirmed by nucleotide sequencing. Protein expression was obtained in super broth medium (32 g/liter polypeptone, 20 g/liter yeast extract, 5 g/liter NaCl, 0.17 M KH2PO4, and 0.72 M K2HPO4 [pH 7.0]) by adding isopropyl-β-D-thiogalactopyranoside (IPTG; Promega, Madison, WI) to bacterial cultures harboring pET-Y10βC1 and pET-Y35βC1 plasmids. The recombinant βC1 proteins were purified over Ni2+-nitrilotriacetic acid affinity columns under denaturing conditions, as instructed by the manufacturer (QIAGEN, Hilden, Germany). The purified proteins were refolded by dialysis against the binding buffer (10 mM Tris-HCl [pH 7.8], 50 mM NaCl, 5 mM MgCl2,1 mM EDTA, 1 mM dithiothreitol, 0.1 mM phenylmethylsulfonyl fluoride, and 10% glycerol) three times at 4°C. The sizes of the purified βC1 proteins were determined by analyzing protein samples in 15% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels. Protein concentration was estimated by the Bradford assay as described previously (5). The polyclonal antibody against the recombinant Y10 βC1 protein was produced in a rabbit as described previously (43).
TABLE 1.
Primers used for PCR amplification
| Purpose and primer | Sequence (5′→3′)a | Modification introduced |
|---|---|---|
| Construction of expression vectors | ||
| Y10βC1-F | acggatccATGTATCATCCACAAC | BamHI |
| Y10βC1-R | ccgtcgacTCATACATCTGAATTTG | SalI |
| Y35βC1-F | accggatccATGACTATCAAATACAAC | BamHI |
| Y35βC1-R | ccgtcgacTCATACATTAGCTATTGTC | SalI |
| GFP/Sal-R | ccgtcgacTTATTTGTATAGTTCAT | SalI |
| Production of βC1:GUS or βC1:GFP fusion gene | ||
| Y10βC1/Sac-F | acgagctcATGTATCATCCACAAC | SacI |
| Y10βC1/Eco-R | ccgaattcTACATCTGAATTTG | EcoRI |
| GUS/Eco-F | ccgaattcATGTTACGTCCTGTAG | EcoRI |
| GUS/Eco-R | ccgaattcTCATTGTTTGCCTCCC | EcoRI |
| GFP/Sal-F | gcgtcgacATGAGTAAAGGAGAAGA | SalI |
| GFP/Bam-R | ccggatccTTATTTGTATAGTTCAT | BamHI |
Introduced restriction endonuclease sites are underlined; modified nucleotides are in lowercase type.
DNA binding assays.
Purified βC1 recombinant proteins that had been extracted from bacteria and renatured as described above were used in binding assays. To prepare radioactive DNA fragments, seven recombinant plasmids, pGEM-Y10β, pGEM-Y35IR, pGEM-P1, pGEM-P4, pGEM-P5, pGEM-MP, and pGEM-SGT1, were digested with EcoRI to release a full-length insert of Y10β, an approximately 400-bp fragment of TbCSV-Y35 genome, the 250-bp (P1), 700-bp (P4), and 1-kb (P5) fragments of Y10β, 795 bp of the Tomato mosaic virus movement protein (MP) gene and a 500-bp fragment of the Lycopersicon esculentum SGT1 gene. The released DNA fragments were end-labeled in the presence of 50 μCi [α-32P]dATP (3,000 Ci/mmol) using the Klenow fragment (Promega). Unlabeled competitor ssDNAs or 32P-labeled ssDNA probes were prepared by denaturing dsDNA in boiling water for 5 min and quickly immersing it into ice water.
Electrophoretic mobility shift assays (EMSA) were performed essentially as described previously (3), except that the binding reactions commonly contained 2 μg of protein and 2.0 ng of labeled DNA in 20 μl of binding buffer. Reactions were incubated at 25°C for 30 to 45 min, and samples were analyzed by electrophoresis in nondenaturing 4% polyacrylamide gels in 1× Tris-borate-EDTA. In competition assays, a 50-fold molar excess of unlabeled DNA was added in the binding reactions.
For UV cross-linking assays, βC1 proteins (2 μg) were tested for binding with radiolabeled DNA probe (2.0 ng) in binding buffer at 25°C for 30 to 45 min. The protein-DNA mixture was then cross-linked with a UV light (4 W at 254 nm) for 10 min and treated with 1 U of RNase-free DNase Q (Promega) for 30 min at 37°C. The protein-DNA complex was analyzed by 15% SDS-PAGE as described for EMSA. To determine the effects of NaCl on stability of the protein-DNA complexes, a series of concentrations (100, 200, 400, 800, 1,200, and 1,800 mM) of NaCl were added into the binding buffer before UV cross-linking. All gels in DNA binding assays were exposed to a phosphor screen and scanned with a Typhoon 9200 Imager (Amersham Pharmacia, Piscataway, NJ).
Suppression assays of RNA silencing.
A test for silencing suppression was based on a previously described experimental system (7, 27). In this system, systemic green fluorescent protein (GFP) silencing in transgenic N. benthamiana plants (line 16c) carrying a highly expressed GFP transgene was induced by infiltration of lower leaves of transgenic seedlings with a strain of Agrobacterium tumefaciens harboring the same functional GFP in a binary Ti plasmid (35S:GFP), kindly provided by D. C. Baulcombe, John Innes Center, United Kingdom. By 20 to 25 days postinfiltration (dpi), silencing of the GFP was extensive in all vegetative tissues of the plants (7). In order to test whether DNAβ is able to suppress RNA silencing, the 16c seedlings having two to three leaves were agroinfiltrated with A. tumefaciens harboring 35S:GFP for inducing GFP transgene silencing. After 25 dpi, the GFP-silenced seedlings were agroinoculated with an infectious clone of TYLCCNV-Y10 (pBinplus-Y10A-1.7) or TbCSV-Y35 (pBinplus-Y35A-1.9) alone or together with its associated DNAβ (pBinplus-Y10β-dimer or pBinplus-Y35β-dimer) (60; Li et al., unpublished). Agroinoculation assays were also conducted in order to determine the effects of DNAβ on the onset of GFP silencing. For this purpose, 16c seedlings at the two- to three-leaf stage were agroinoculated with cloned viral DNA alone or together with its associated DNAβ. By 20 days postinoculation, the leaf curling symptoms were obvious and the seedlings were induced for GFP silencing via agroinfiltration with A. tumefaciens harboring 35S:GFP. To test whether the βC1 protein can suppress RNA silencing, the Y10 and Y35 βC1 genes were also cloned between the 35S promoter and the nopaline synthase terminator (nos) in the binary vector pBin438 (35) to generate the constructs 35S:Y10βC1 and 35S:Y35βC1, respectively (Fig. 1). A similar strategy was used to generate the 35S:Y10βC1T construct using pGEM-Y10βC1T (12) as a template. This strain contains a stop codon located 45 nucleotides downstream of the first start codon in the Y10 βC1 gene (12). Y10βC1/3A, in which the three basic amino acids 49KKK51 were replaced with AAA substitutions, was constructed by insertion of overlap extension PCR products into pBin438. The 35S:2b plasmid containing the Cucumber mosaic virus (CMV) 2b gene and the 35S:P1/HC-Pro plasmid containing the Tobacco etch virus (TEV) P1/HC-Pro gene, used as controls, were kindly provided by D. C. Baulcombe and J. C. Carrington (Oregon State University), respectively. The plasmid 35S:asGFP was constructed by cloning the mGFP5 gene (25) into pBin438 in an antisense orientation using the primers GFP/Sal-F and GFP/Bam-R. All constructs were introduced into the A. tumefaciens strain C58C1 by triparental mating or by direct transformation. For patch coinfiltration assays, the suppressor constructs in A. tumefaciens strain C58C1 were grown to an optical density at 600 nm of 1.0 and mixed with equal volumes of A. tumefaciens harboring 35S:GFP or 35S:asGFP prior to coinfiltration. Visual detection of GFP fluorescence in whole plants and leaf patches was performed using a handheld 100-W, long-wave UV lamp (UV Products, Upland, CA). Plants were photographed with a Nikon 5000 digital camera (Tokyo, Japan) mounted with UV and yellow filters. Exposures were 4 to 8 s long, depending on the fluorescence intensity and distance from the plant, and the images were processed by using Adobe Photoshop.
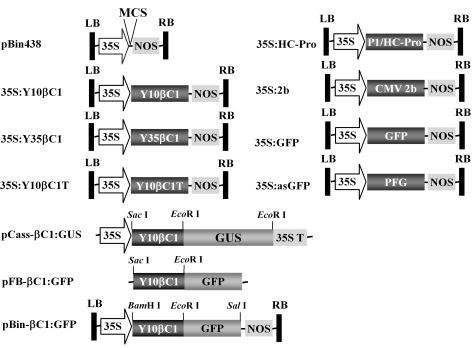
FIG. 1.
Schematic representation of constructs used for agroinfiltration and subcellular localization of the βC1 protein. LB, T-DNA left border; RB, T-DNA right border; 35S, CaMV 35S promoter; NOS, nopaline synthase terminator; Y10βC1, βC1 gene of Y10β associated with TYLCCNV-Y10; Y35βC1, βC1 gene of Y35β associated with TbCSV-Y35; 2b, CMV 2b gene; HC-Pro, TEV P1/HC-Pro gene; MCS, multiple cloning sites; 35ST, 35S terminator; PFG, antisense GFP gene.
Northern blot analysis.
Total RNA isolation and Northern blot analysis were conducted as described previously (12). Membranes were hybridized at 42°C with [α-32P]dCTP-labeled probes specific for the GFP gene. Hybridization signals were detected by phosphorimaging using a Typhoon 9200 imager (Amersham Pharmacia).
Localization of βC1 protein expression in onion, insect, or tobacco cells.
To obtain a βC1:β-glucuronidase (GUS) fusion cistron, the Y10 βC1 gene was amplified with the primers Y10βC1/Sac-F and Y10βC1/Eco-R (Table 1) to remove the stop codon of the Y10 βC1 gene. After digestion with SacI and EcoRI, the PCR fragments were cloned into a pCass II vector (51) to produce pCass-Y10βC1, which contains an in-frame fusion to the udiA reporter gene that codes for GUS (Fig. 1). A similar strategy was used to generate the Y10 βC1/3A:GUS fusion cistron. Monolayers of epidermal cells were removed from the inner peel of purple onion (Allium cepa) bulbs, placed on modified Murashige and Skoog (MS) medium (1× MS salts, 3% sucrose, and 2% agar [pH 5.8]), and bombarded with 1 mg of 1-μm gold particles in a mixture of 10 μg of the plasmid containing the βC1:GUS or βC1/3A:GUS plasmid. GUS activity was detected with the X-Gluc substrate at 12 to 16 h postbombardment.
To obtain a βC1:GFP fusion for insect cell expression, the vector pFB-GFP (33), which contains an enhanced green fluorescent protein (EGFP) gene, was digested with SacI/EcoRI and ligated with a SacI/EcoRI-digested Y10 βC1 gene to obtain the pFB-βC1:GFP plasmid, in which the Y10 βC1 coding sequence was fused in-frame to the 5′ end of the EGFP gene (Fig. 1). A similar strategy was used to generate the pFB-βC1/3A:GFP fusion cistron. The constructs and the control vector (pFB-GFP) were expressed in sf21 insect cells using the Bac-to-Bac baculovirus expression system as recommended by the supplier (Invitrogen, Carlsbad, CA). GFP-expressing cells were examined by confocal microscopy (FV500; Olympus, Tokyo, Japan) at 48 h posttransfection.
To obtain a βC1:GFP fusion for expression in tobacco cells, the Y10 βC1 gene was amplified with the primer pair Y10βC1-F and Y10βC1/Eco-R (Table 1) and cloned into the BamHI/EcoRI-digested pBin-mGFP5-ER (25) to produce pBin-βC1:GFP-ER. The βC1:GFP fusion gene was then amplified with Y10βC1-F and GFP/Sal-R from pBin-βC1:GFP-ER and cloned into pBin438 to generate pBin-βC1:GFP, in which the Y10 βC1 coding sequence was fused in-frame to the 5′ end of the mGFP5 ORF (Fig. 1). The plasmids pBin-mGFP5-ER and pBin-βC1:GFP were introduced into A. tumefaciens strain C58C1 as described above. The leaves of 4-week-old N. benthamiana plants agroinfiltrated with A. tumefaciens harboring pBin-mGFP5-ER and pBin-βC1:GFP, respectively. By 3 dpi, the GFP expression in the leaves was observed at 488 nm by using a confocal microscope (LSM 510; Zeiss, Germany). Images were captured using Zeiss LSM 510 software and converted to tagged image file format for export.
Fixation of plant material and immunogold labeling.
Leaf tissue of N. benthamiana plants infected with TYLCCNV-Y10 plus Y10β and healthy leaves were fixed in 50 mM phosphate buffered saline (PBS; pH 6.8) containing 2.5% (vol/vol) glutaraldehyde and 2% (vol/vol) polyformaldehyde overnight at 4°C. Thereafter, the samples were thoroughly rinsed with 50 mM PBS and postfixed with 1% (wt/vol) osmium tetroxide in the same buffer for 2 h at room temperature. All samples were then dehydrated in a graded ethanol series and embedded in LowicrylK4M resin (Electron Microscopy Sciences, Fort Washington, PA). Ultrathin sections (70 nm) were cut with a diamond knife and mounted onto nickel grids. After blocking for 30 min at room temperature with 50 mM PBS (pH 6.8) containing 1.0% bovine serum albumin (BSA) and 0.02% polyethylene glycol 2000 (blocking solution [BL]), the grids were incubated for 1 h in polyclonal antibody raised against the Y10 βC1 protein for 20 to 60 min at room temperature. The grids were washed with several changes of PBS and then incubated with Protein A-gold (Sigma, St. Louis, MO) diluted in BL for 1 h. After incubation, the grids were washed with PBS and double-distilled H2O, stained with uranyl acetate and lead citrate, and then examined by electron microscopy (JEM-1200EX; JEOL, Japan). The control experiment included replacing the polyclonal antibody against the Y10 βC1 protein with preimmune serum prior to incubation with Protein A-gold.
RESULTS
DNAβ-encoded βC1 protein binds DNA in a sequence-nonspecific manner.
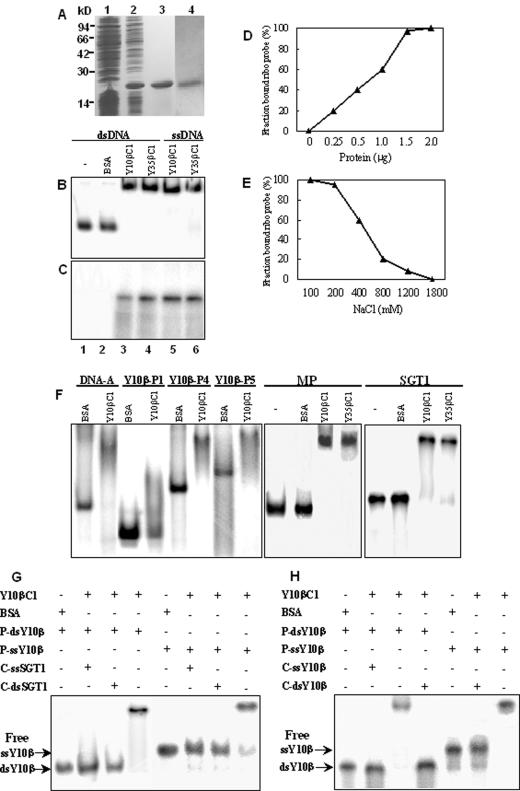
Cloning of the 381 bp Y10 βC1 and 357 bp Y35 βC1 genes into an E. coli expression vector pET-30a with an in-frame fusion with the six-His open reading frame (ORF), resulted in the production of significant amounts of His-βC1 fusion proteins after induction with IPTG (Fig. 2A). The fusion protein migrated in SDS-PAGE gels to a position corresponding to an Mr of ∼19 kDa, and Coomassie blue staining indicated that the His-Y10βC1 and His-Y35βC1 proteins were highly enriched and lacked detectable contaminants after purification over Ni2+-nitrilotriacetic acid affinity columns under denaturing conditions (Fig. 2A). The proteins were also detected in Western blots using an anti-six-His monoclonal antibody (data not shown). The purified His-Y10βC1 protein was then used for specific polyclonal antibody production, and the resulting antibody had a specific reaction with the His-Y10βC1 protein (Fig. 2A).
FIG. 2.
βC1 protein DNA binding assays. (A) βC1 proteins expressed in E. coli. Lane 1, Coomassie blue staining of an uninduced culture extract. Lane 2, Coomassie blue staining of culture extracts after IPTG induction and expression from the recombinant plasmid pET-Y10βC1. Lane 3, Coomassie blue staining of affinity-purified His-Y10βC1 recombinant protein. Lane 4, immunoblot using polyclonal antibodies raised against the purified Y10 βC1 protein. (B) EMSA binding assays of full-length Y10β probes. Lane 1, migration of unbound dsDNA probe. Lane 2, BSA control. Lane 3, Y10 βC1 protein binding to dsDNA probe. Lane 4, Y35 βC1 protein binding to dsDNA probe. Lane 5, Y10 βC1 protein binding to ssDNA probe. Lane 6, Y35 βC1 protein binding to ssDNA probe. (C) UV cross-linking assays of full-length Y10β probes. Lanes 1 to6 are as described for panel B. (D) Concentration effects of the Y10 βC1 protein on binding to dsDNA. The fraction of the bound protein was determined by EMSA analysis of labeled Y10β dsDNA (2 ng) binding in the presence of 0 to 2.0 μg of Y10 βC1 protein. (E) Effects of NaCl concentration on DNA binding by the Y10 βC1 protein. Labeled Y10β dsDNA (2 ng) was used in UV cross-linking analysis in the presence of 2.0 μg Y10 βC1 protein and 100 to 1,800 mM NaCl in the binding buffer. (F) EMSA assays to determine the effects of DNA size on binding activities. Labeled DNAs tested were a fragment of Y35 DNA-A (400 bp), P1 (250 bp), P4 (700 bp), and P5 (1 kb) from Y10β DNA fragments, the ToMV MP gene (795 bp), and a tomato SGT1 gene fragment (500 bp). (G) Competition of βC1 protein binding to labeled Y10β ssDNA or dsDNA in the presence of a 50-fold molar excess of unlabeled ssDNA or dsDNA from the SGT1 fragment. (H) Conditions were the same as for panel G except that unlabeled ssDNA or dsDNA from the Y10β fragment was used as the competitor. +, presence; −, absence; P, probe DNA; C, competitive DNA.
The purified Y10 and Y35 βC1 proteins were recovered from the column, renatured, and then tested for their abilities to bind full-length radiolabeled Y10β dsDNA or ssDNA probes in EMSA assays. Protein-DNA complexes with retarded electrophoretic mobilities were observed when either ssDNA or dsDNA probes were tested. Substitution of the βC1 proteins with the same amount of BSA in the assays did not result in retarded complexes (Fig. 2B). In addition, when the βC1 proteins were treated at 80°C for 15 min prior to the binding assay, the binding activity for dsDNA or ssDNA was destroyed (data not shown). These results suggest that the βC1 proteins are essential for complex formation and that the binding is affected by the conformation of the proteins. Protein-DNA binding was also confirmed with UV cross-linking assays. After DNase treatment, labeled bands were not observed with BSA controls or with reactions lacking the proteins, but complex formation was present in the presence of the βC1 proteins (Fig. 2C).
The dependence of the binding activity on βC1 protein concentration is shown in Fig. 2D. The proportion of binding increased linearly as the amount of the Y10 βC1 protein in the reaction increased, and it reached saturation when the molecular ratio of Y10 βC1 to dsDNA probe reached 1 × 103 (ng/ng). The requirement of such a high concentration for optimal binding probably is a result of incomplete renaturation or misfolding of the βC1 proteins after affinity purification. Binding assays to determine the effects of different concentrations of NaCl on complex formation demonstrated that the Y10 βC1 protein bound dsDNA strongly at 100 to 200 mM concentrations of NaCl; the binding activity declined precipitously as NaCl concentrations increased from 200 to 800 mM and was not detectable when the NaCl concentration reached 1.8 M (Fig. 2E). Thus, the Y10 βC1 proteins had a strong binding affinity for both ssDNA and dsDNA.
To determine whether there is a DNA size limitation for binding, three Y10β fragments with different lengths and a 400-bp fragment of TbCSV-Y35 DNA were used as probes for EMSA. The results indicated that the Y10 βC1 protein could bind each of these DNA fragments and produce retarded DNA bands (Fig. 2F). To determine whether there is a DNA sequence requirement for binding, additional DNA fragments from the ToMV MP and the tomato SGT1 genes were used in EMSA experiments. These derivatives, and both ssDNA and dsDNA fragments, were bound by the βC1 proteins, but BSA controls had no binding activity (Fig. 2F). Competitive binding experiments were also carried out to determine the possible DNA sequence and structural preferences for binding. Assays with labeled Y10β dsDNA or ssDNA were conducted in the presence of the Y10 βC1 protein and 50-fold molar excess of unlabeled SGT1 ssDNA or dsDNA (Fig. 2G). These results revealed that binding of the labeled Y10β dsDNA and the ssDNA probes was abolished in the presence of unlabeled SGT1 ssDNA or dsDNA. Homologous competition using the same probes in the presence of unlabeled Y10β ssDNA or dsDNA revealed no preferential binding for either structure (Fig. 2H). Hence, the βC1 protein binds both ssDNA and dsDNA independently of structure or sequence.
The βC1 protein functions as a suppressor of RNA silencing.
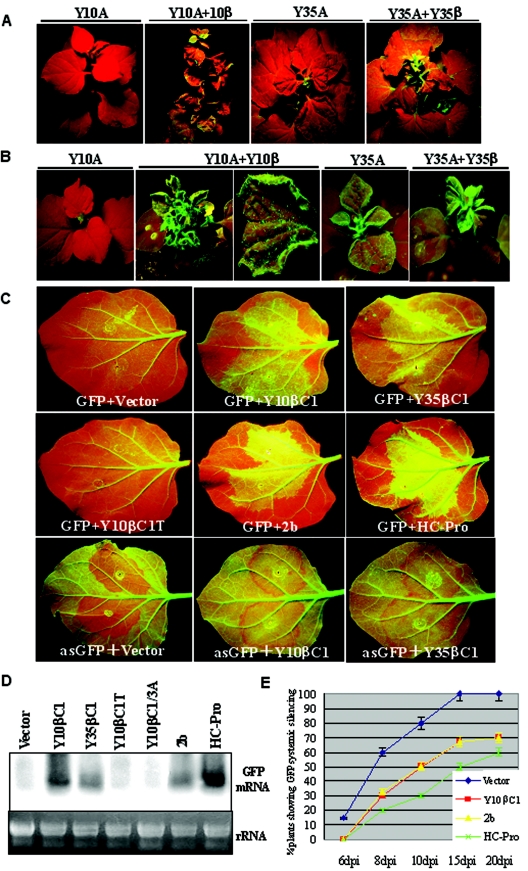
We have previously demonstrated that Y10β, when coinoculated with TYLCCNV-Y10, elicits severe leaf curling symptoms in N. benthamiana plants (60). Similarly, coinoculation of TbCSV-Y35 with its cognate Y35β resulted in severe downward leaf curling symptoms in N. benthamiana in comparison with upward leaf curling symptoms induced by TbCSV-Y35 alone (Li et al., unpublished). In order to determine whether virulence caused by DNAβ is correlated with its ability to suppress gene silencing, agroinoculation assays were performed with either the cloned viral DNA alone or the cloned viral DNA together with its cognate DNAβ after 16c seedlings were induced for GFP transgene silencing. By 25 dpi, GFP fluorescence in newly emerging leaves of GFP-silenced plants coinoculated with TYLCCNV-Y10 plus Y10β or TbCSV-Y35 plus Y35β could be observed (Fig. 3A). In contrast, the new leaves of seedlings infected by TYLCCNV-Y10 alone did not contain obvious GFP fluorescence but instead had a bright red color typical of chlorophyll autofluorescence, indicating that the established 35S:GFP silencing had not been suppressed. Leaves infected with TbCSV-Y35 exhibited a similar red color, and only minimal levels of green fluorescence were noted in emerging leaf tissue. These experiments show that DNAβ is a strong suppressor of established RNA silencing in infected plants.
FIG.3.
Suppression of RNA silencing by begomoviruses infection or transient expression of the βC1 protein. (A) Reversal of established GFP silencing by begomovirus infection. GFP silencing in the 16c plants was elicited by agroinfiltration of the lower leaves with 35S:GFP. After 25 days, the silenced plants showing red fluorescent leaves were agroinoculated with TYLCCNV-Y10 (Y10A) alone, TYLCCNV-Y10 plus the cognate Y10β, TbCSV-Y35 (Y35A), or TbCSV-Y35 plus the cognate Y35β and were photographed 25 days afterwards. (B) Prevention of the establishment of GFP silencing by begomoviruses infection. The 16c seedlings expressing GFP were agroinoculated with TYLCCNV-Y10 alone, TYLCCNV-Y10 plus Y10β, TbCSV-Y35 alone, or TbCSV-Y35 plus cognate Y35β. By 20 dpi, the lower leaves of the infected plants were infiltrated with 35S:GFP and evaluated for the ability to induce silencing 25 days later. (C) Ability of the βC1 protein to suppress local GFP silencing mediated by sense and antisense GFP in 16c leaf patches. Leaves expressing GFP were coinfiltrated with bacteria harboring 35S:GFP (GFP) or 35S:asGFP (asGFP), plus 35S promoter constructs suitable for expression of the Y10 βC1 protein, a loss-of-function Y10 βC1 protein (Y10βC1T), the Y35 βC1 protein, the CMV 2b protein (2b), the TEV P1/HC-Pro protein (HC-Pro) or the empty vector (pBin438, Vector), respectively. At 7 dpi, the photos were taken under UV light. (D) Northern blot analysis of GFP mRNA in the coinfiltrated leaf patches of 16c plants. The RNA samples tested were isolated from leaf patches coinfiltrated with 35S:GFP plus empty vector, Y10βC1, Y35βC1, Y10βC1T, Y10βC1/3A, 2b, and P1/HC-Pro driven by 35S promoter. The ethidium bromide-stained gel shown below the blot provides an RNA loading control. (E) Ability of the Y10 βC1, CMV 2b, and HC-Pro proteins to prevent 35S:GFP mediated systemic RNA silencing. Lower leaves of 16c seedlings were coinfiltrated with bacteria harboring 35S:GFP plus bacteria harboring the empty vector, the Y10βC1, 2b, or HC-Pro driven by the 35S promoter, respectively. Plants were subsequently monitored for the onset of systemic silencing at 6, 8, 10, 15, and 20 dpi. Triplicates of each experimental treatment contained a total of 10 individual seedlings.
To determine whether viral infection can interfere with the initiation of GFP silencing, GFP expressing 16c seedlings were agroinoculated with either cloned viral DNA alone or cloned viral DNA in combination with the associated DNAβ (Fig. 3B). By 20 dpi, the infected 16c seedlings were induced for GFP silencing via agroinfiltration. Plants coinoculated with TYLCCNV-Y10 plus Y10β or TbCSV-Y35 plus Y35β still exhibited a strong green fluorescence in the emerging leaves. Some suppression of GFP silencing occurred in the lower infiltrated leaves, and incomplete suppression appeared in the intermediate leaves. However, no green fluorescence or only minor flecks of fluorescence were observed with the seedlings inoculated with TYLCCNV-Y10 or TbCSV-Y35 alone (Fig. 3B). Therefore, DNAβ not only appears to interfere with established silencing but also prevents the establishment of silencing by 35S:GFP in infected plants.
In contrast to wild-type DNAβ, a Y10β mutant, pBinPLUS-C1M-Tβ carrying a loss-of-function βC1 gene (12) failed to reverse the GFP silencing or to block the onset of GFP silencing when coinoculated with TYLCCNV-Y10 (data not shown). To directly test the ability of the βC1 protein to suppress RNA silencing, a patch coinfiltration assay was performed with the leaves of the 16c plants. Examination of the infiltrated leaves under UV light at 7 dpi revealed that the patches receiving 35S:GFP plus pBin438 (referred to as Vector) showed greatly decreased green fluorescence and a deep red color (Fig. 3C), with a concomitant absence of GFP mRNA (Fig. 3D). In contrast, the patches receiving 35S:GFP plus 35S:Y10βC1 or 35S:GFP plus 35S:Y35βC1 exhibited a bright green fluorescence in the infiltrated leaf patches. Western blot analysis using the polyclonal antibody against the recombinant Y10 βC1 protein also confirmed the expression of the βC1 proteins in the patches receiving 35S:Y10βC1 and 35S:Y35βC1 (data not shown). Patches infiltrated with the CMV 2b construct were similar in GFP intensity to patches infiltrated with 35S:Y10βC1 and 35S:Y35βC1, but patches expressing 35S:P1/HC-pro had a brighter green fluorescence than those with the βC1 derivatives (Fig. 3C), and the HC-Pro patches also contained higher levels of GFP mRNA (Fig. 3D). However, the patches infiltrated with 35S:GFP plus 35S:Y10βC1T, containing a frame-shift version of Y10 βC1 gene, had a deep red color similar to that of tissue infiltrated with the vector (Fig. 3C), and these patches also lacked GFP mRNA (Fig. 3D). When patch coinfiltration assays were tested with the antisense GFP gene construct (35S:asGFP) plus either 35S:Y10βC1, 35S:Y35βC1, 35S:2b, or 35S:HC-Pro, bright green fluorescence in the infiltrated patches was also observed, whereas a red color was present when 35S:asGFP and either the empty vector or 35S:Y10βC1T were coinfiltrated (Fig. 3C and data not shown). Taken together, these findings show that the βC1 protein per se is a major suppressor of gene silencing that acts at the local level in cells to prevent silencing. Direct comparisons also indicate that the βC1 suppression is comparable to the level of suppressor activity exhibited by the CMV 2b protein, although it is somewhat weaker than that of HC-Pro.
GFP fluorescence persisted for 7 to 9 days in the patches that had received suppressor constructs plus 35S:GFP, and the upper leaves of most plants continued to fluoresce. In contrast, the patches that had received 35S:GFP plus Vector were undergoing silencing by 7 to 9 dpi, and the upper leaves were undergoing silencing by this time (data not shown). Systemic silencing in all of the plants that had received 35S:GFP plus Vector or 35S:Y10βC1T was observed at 20 dpi, whereas about 30% of the plants that received 35S:GFP plus either 35S:Y10βC1 or 35S:2b contained green fluorescence in the upper leaves by this time (Fig. 3E). Three independent experiments involving 10 plants for each infiltration were carried out, and they produced similar results. Thus, our observations indicate that the βC1 proteins have a persistent effect on systemic silencing in the coinfiltration assay, one similar to those of the 2b and P1/HC-Pro suppressor proteins.
βC1 protein targets the nucleus of insect and plant cells.
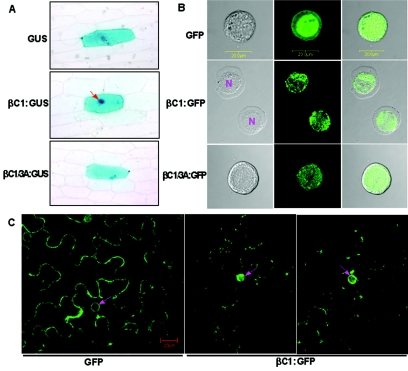
To determine the subcellular localization of the βC1 protein, the Y10 βC1 coding sequence was fused in-frame to the 5′ end of the GUS gene driven by a Cauliflower mosaic virus 35S promoter (Fig. 1), and the resultant construct was introduced into onion epidermal cells by particle bombardment. Following expression, GUS localization was detected histochemically. Free GUS expressed in onion cells was generally uniformly distributed throughout both the cell cytoplasm and the nuclear compartments (Fig. 4A). In contrast, there was a substantial nuclear enrichment of βC1:GUS fusion protein in most (over 90%) of the expressed cells, with a concomitant reduction of staining in the cell cytoplasm. To determine if the nuclear localization sequence (NLS) signals were recognized in cells of different taxa, the subcellular localization of the βC1 protein was investigated by using a baculovirus-insect cell expression system. As has been reported previously (54), GFP fluorescence is present throughout the cytoplasm and the nucleus when expressed from the GFP vector control. In contrast, fluorescence from the βC1:GFP fusion construct was confined primarily to the nucleus (Fig. 4B). The nuclear accumulation of βC1:GFP was also confirmed under bright-field illumination to identify the nucleus of sf21 insect cells.
FIG. 4.
Accumulation of the βC1 protein in the nuclei of plant and insect cells. (A) Localization of βC1:GUS fusion protein in onion epidermal cells. Arrows indicate the locations of nuclei. (B) Localization of the βC1 protein in the nuclei of sf21 insect cells. The panel shows insect cells expressing GFP (upper lane), βC1:GFP (middle lane), or βC1/3A:GFP (bottom lane) fusion protein, which were examined under bright-field illumination (left) to identify the nucleus, under fluorescent-field illumination (middle) to evaluate GFP fluorescence, and by confocal microscopy (right) for an overlay of bright and fluorescent illumination. N, cell nucleus. Bar, 20 nm. (C) Targeting of the βC1 protein to the nuclei of N. benthamiana plants. Images showing GFP fluorescence in N. benthamiana leaves expressing GFP (left) or βC1:GFP (middle and right) were examined using a confocal microscopy. Bar, 20 nm.
To investigate the subcellular localization of the βC1 protein in tobacco cells, the pBin-βC1:GFP construct and the pBin-mGFP5-ER (control) were introduced into N. benthamiana leaf cells via agroinfiltration. An examination of leaves expressing free GFP by confocal microscopy at 3 dpi showed that the fluorescence patterns were qualitatively the same in the cytoplasms and nuclei in nearly all of the cells (Fig. 4C). However, most of the fluorescence occurred in the nuclei of leaves expressing the βC1:GFP fusion protein (Fig. 4C) and, typically, the fluorescence in a large number of cells was much stronger in the nuclei than in the cytoplasm and around the cell wall.
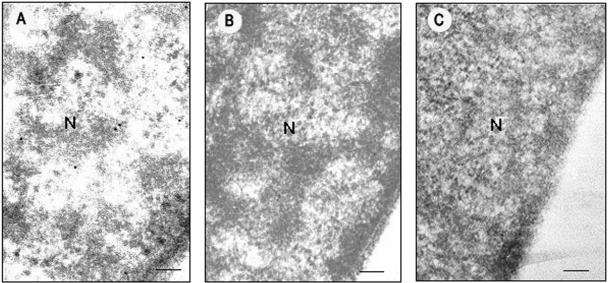
The subcellular location of Y10 βC1 protein was also studied by immunoelectron microscopy of leaf tissue of N. benthamiana plants infected with TYLCCNV-Y10 plus Y10β. Gold label was detected primarily in the nuclei of infected cells, but label was not concentrated in the nuclei of cells of healthy N. benthamiana plants (Fig. 5, Table 2). A small amount of label was also present in chloroplasts of infected cells but was not found in other organelles. Gold particles were not detected in sections when the polyclonal antibody raised against the Y10 βC1 protein was replaced with preimmune antiserum (Fig. 5), indicating that background labeling was minimal. An estimation of the number of gold particles per unit of surface area in the sections revealed that the βC1 protein labels primarily the nucleus, and these results are consistent with those obtained from transient expression of the βC1:GFP fusion protein in leaf cells of N. benthamiana plants. These results collectively provide persuasive evidence that the Y10 βC1 protein is localized primarily in the nucleus.
FIG. 5.
Immunogold labeling of the Y10 βC1 protein in N. benthamiana plant cells infected by TYLCCNV-Y10 plus Y10β. Label was detected in the nuclei of infected cells (A) but was not detected in the nuclei of healthy N. benthamiana plant cells (B) or in infected cells when the polyclonal antibody against βC1 protein was replaced with preimmune serum (C). Bar, 150 nm. N, cell nucleus.
TABLE 2.
Number of gold particles per μm2 of the sections of N. benthamiana cells
| Inoculum | No. of gold particles per μm2 of section indicated
|
||||
|---|---|---|---|---|---|
| Nucleus | Cytoplasm | Chloroplast | Mitochondrion | Cell wall | |
| TYLCCNV-Y10 + Y10β | 15 ± 1.89a | 0 | 3 ± 1.33 | 0 | 0 |
| TYLCCNV-Y10 + Y10βC1/3A | 0 | 0 | 4 ± 1.25 | 0 | 0 |
| Not inoculated | 0 | 0 | 0 | 0 | 0 |
Average result for 10 randomly selected fields plus or minus standard error.
Mutation analysis of the putative NLS of Y10 βC1 protein.
Many viral proteins targeted to the nucleus contain NLS. An inspection of the amino acid sequences of the DNAβ-encoded βC1 proteins reveals a conserved, basic amino acid-rich region between positions 41 and 56 of Y10 βC1. Online software predication (http://psort.nibb.ac.jp) indicated that this region of the Y10 βC1 protein contains a monopartite NLS consisting of seven amino acids, 45PALAKKK51. To determine whether the βC1 protein contains a functional NLS, the Y10β mutant pBin-Y10βC1/3A, in which the three basic amino acids 49KKK51 were replaced with AAA substitutions, was constructed. This mutant could be trans-replicated by TYLCCNV-Y10, but it failed to induce disease symptoms in N. benthamiana plants when coinoculated with TYLCCNV-Y10 (data not shown). Transient expression of the Y10 βC1 protein containing these mutations also failed to suppress local GFP silencing and target the nuclei of tobacco, onion, and insect cells (Fig. 3D and 4; Table 2). A Western blot using polyclonal antibody against the βC1 protein verified that Y10βC1/3A was stably produced in pBin-Y10βC1/3A-infiltrated N. benthamiana leaves, which eliminates the possibility that loss of these activities was a result of lack of expression of the mutation protein. These experiments thus suggest that the nuclear localization of the βC1 protein may be critical for eliciting disease symptoms and for inducing silencing suppression.
DISCUSSION
Numerous plant RNA viruses have associated satellite RNAs that are completely dependent on the helper virus for replication, movement, and encapsidation. Some satellite RNAs are known to exacerbate viral symptoms or induce symptoms distinct from those induced by the helper virus alone, but most attenuate symptoms, whether or not they contain an expressed ORF (11). Satellite DNAβ molecules associated with begomoviruses contain a functional βC1 gene and have been demonstrated to play a vital role in inducing yellow vein disease in Eupatorium and Ageratum, leaf curl disease in cotton, yellow vein mosaic disease in bhendi, and yellow leaf curl disease in tomato (6, 28, 48, 49, 60). Our previous studies have demonstrated that the TYLCCNV-Y10 βC1 protein functions as a pathogenicity determinant and induces virus-like leaf curl symptoms in transgenic Nicotiana plants in the absence of virus infection (12). In the present studies, we found that the βC1 protein binds DNA in a sequence-nonspecific manner, functions as a suppressor of RNA silencing, and targets the nucleus of insect and plant cells.
In the EMSA and UV cross-linking assays, purified βC1 proteins of Y10β and Y35β could bind dsDNA or ssDNA. However, database searches revealed no known proteins with discernible sequence relatedness to the βC1 protein. Zinc fingers or Cys-His-rich regions (24, 32) that might be involved in DNA binding were also absent, suggesting that the βC1 protein may contain a novel type of DNA-binding motif. The βC1 proteins bind dsDNA or ssDNA without binding preferences for DNA structure, sequence, or size. Sequence-nonspecific DNA binding activities have also been reported for geminivirus CP and C2/AC2 proteins. The CP binds dsDNA and ssDNA in a sequence-nonspecific manner in vitro (36, 40), and the C2/AC2 protein binds ssDNA but exhibits only weak binding affinity for dsDNA in vitro (38, 52, 56). However, the replication protein (Rep) of begomoviruses differs from CP and C2/AC2 by exhibiting sequence-specific binding of iterated DNA sequences (iterons) that function as essential elements for initiating rolling circle replication (23). DNA binding activities of these viral proteins may have important roles in the life cycle of geminiviruses. Systemic movement of monopartite begomovirus in plants also requires the CP, V2, and/or C4 proteins (29, 45). The CP of monopartite begomovirus is nuclear localized and shuttles the viral genome into and out of the host nucleus, whereas the V2 protein may function in cell-to-cell movement (20). In contrast, the movement of bipartite begomoviruses is mediated by the BV1 and BC1 proteins. BV1 is involved in shuttling the viral ssDNA both into and out of the nucleus and functions as the nuclear shuttle protein (NSP), and the BC1 protein functions in cell-to-cell movement of viral DNA and is associated with the plasma membrane and cell wall (39, 47). The BC1 was also postulated to be a symptom determinant for bipartite geminiviruses, because transgenic expression of the BC1 gene of Squash leaf curl virus in N. benthamiana (42), Tomato mottle virus in N. tabacum (16), and Bean dwarf mosaic virus in tomato produced virus-like symptoms (26). Similar symptoms were found with transgenic Nicotiana plants expressing the Y10 βC1 gene (12). However, DNAβ is not essential for systemic movement of monopartite begomoviruses, because a βC1 deletion mutant of Y10β can move systemically in plants and be replicated in trans and become encapsidated by TYLCCNV-Y10 (44). Thus, the βC1 protein appears not to parallel the BC1 cell-to-cell movement functions, and the present results raise a number of questions as to whether the βC1 protein acts cooperatively with the CP of monopartite begomovirus in shuttling viral DNA from the nucleus to the cytoplasm. These issues will be examined in subsequent experiments.
In plants, RNA silencing functions as a natural antiviral defense, and plant viruses have evolved suppressor proteins for counteracting host defenses (57). Compared with TYLCCNV-Y10 or TbCSV-Y35 infection alone, coinfection with cognate DNAβ could reverse established GFP silencing and could block the onset of GFP silencing in emerging leaves of 16c plants. In patch coinfiltration assays, the Y10 and Y35 βC1 proteins could also suppress local GFP silencing. In these experiments, the intensity of the silencing suppressor activity of the βC1 protein appears to be similar to that of CMV 2b. However, βC1 appears to be a weaker suppressor than HC-Pro, based on the accumulation of GFP mRNA transcripts in the coinfiltrated patches. A βC1 gene frame-shift mutant of Y10β failed to induce disease symptoms when coinoculated with TYLCCNV-Y10 and, consequently, did not play a role in silencing suppression (Fig. 3). Clearly, our data suggest that the βC1 protein functions as a suppressor of RNA silencing and that it is a pathogenicity protein that plays a vital role in symptom induction by suppression of the silencing defenses in plants. Thus, the βC1 protein appears to be similar to many silencing suppressor proteins, for example, potyvirus HC-Pro, CMV 2b, and tombusvirus P19, that function as pathogenicity determinants in RNA viruses (1, 7, 21, 30, 46, 56).
In begomoviruses, the AC2 protein encoded by African cassava mosaic virus and C2, a positional homologue of AC2 encoded by the TYLCCNV isolate from Guangxi province, have been identified as suppressors of RNA silencing (55, 58). Our results show that infections with TYLCCNV-Y10 alone failed to reverse established GFP silencing or to block the onset of GFP silencing in 16c plants. In contrast, infections with the bipartite African cassava mosaic virus lead to suppression of GFP silencing in newly emerging infected leaves and in fully expanded leaves at about 3 weeks postinoculation (58). This difference suggests that suppressors encoded by TYLCCNV-Y10 might be so weak that the suppressor genes could not overcome host RNA silencing and, consequently, TYLCCNV-Y10 infection failed to induce symptoms. Similarly, transient expression driven by the 35S promoter of the C2 protein of TYLCCNV-Y10 also suppressed local GFP silencing in 16c plants, but it had only a mild effect on systemic GFP silencing in the patch coinfiltration assay (13). TYLCCNV-Y10 has a functional C2 and C4 ORF, but it produced asymptomatic infections in tested host plants. In contrast, the TYLCCNV Guangxi isolate was previously reported to infect tobacco and tomato systemically and to induce leaf curl symptoms (15). The C4 gene of the TYLCCNV Guangxi isolate was also found to counter the Rep-induced hypersensitive response in N. benthamiana (53). Therefore, we propose that the different activities of the Guangxi and Y10 isolates of TYLCCNV in symptom induction and suppression of RNA silencing may be due to (i) the C2 and/or C4 proteins of the Guangxi isolate functioning as stronger suppressors of RNA silencing than the Y10 isolate or (ii) the accumulation of different amounts of the C2 and/or C4 proteins in infected plants.
Our localization studies showed that the βC1 protein localized to the nuclei of insect cells and onion epidermal cells. These results indicate that the nuclear enrichment of the βC1:GFP fusion protein occurs in a diverse range of cells derived from taxonomically distinct organisms. The fact that a βC1:GFP fusion construct also produced strong GFP fluorescence in the nuclei of N. benthamiana cells provides a strong argument that the observed nuclear localization of the βC1 protein is critical for interactions occurring during infection (Fig. 4C and Fig. 5). Additional support for this hypothesis was obtained in experiments indicating that an inactive mutant of the Y10 βC1 protein failed to accumulate in the nuclei of infected N. benthamiana cells. Taken together, our results demonstrate that the βC1 protein is targeted to the nucleus and that this targeting is essential for its functions during infection.
An NLS has not yet been characterized for the βC1 protein. However, mutations within a putative NLS of the Y10 βC1 protein abolished nuclear localization and suppression of RNA silencing as well as the ability to elicit symptom induction. Similar phenomena were also reported for the CMV 2b and TYLCCNV C2 suppressor proteins (15, 37). Both suppressors have a characterized NLS, and their nuclear localizations are absolutely indispensable for silencing suppression. Further experiments need to be performed to characterize the βC1 NLS and to determine the relationship between nuclear localization, symptom induction, and silencing suppression of the βC1 protein.
Which steps of the RNA silencing pathway targeted by the βC1 protein remain to be elucidated? This question may be answered by results from experiments with several suppressor proteins, such as HC-Pro, P19, and Turnip yellow mosaic virus P69, that have been demonstrated to affect the plant development by interference with micro-RNA (miRNA)-mediated silencing pathways that share components with the siRNA-mediated antiviral pathway (8, 9, 17, 31). Because transgenic expression of the Y10 βC1 gene in Nicotiana plants elicited virus-like symptoms in the absence of viral infection, our results suggest that the βC1 protein may also play a role in developmental regulation by interfering with miRNA pathways. In Arabidopsis, these pathways are affected by the DICER-like proteins (DCL1, DCL2, and DCL3) that are nuclear localized and are required for miRNA and siRNA biogenesis (59). Thus, the βC1 protein may affect the activity of the DICER-like proteins in plants during nuclear activities that function in silencing suppression. The other possibilities are that βC1 protein could down-regulate transcription of a host protein that acts in the PTGS pathway in the cytoplasm or that βC1 protein could activate transcription of a host PTGS inhibitor. The precise ways the biochemical function and subcellular localization events interfere with silencing pathways to affect symptom induction by the βC1 protein will be the focus of our upcoming research activities.
Acknowledgments
We thank Andrew Jackson (University of California—Berkeley) for critically reading the manuscript. We also thank David Baulcombe (John Innes Center, Norwich, United Kingdom) for providing N. benthamiana 16c plant seeds and the plasmids 35S:GFP and 35S:2b and James Carrington (Oregon State University) for providing the plasmid 35S:P1/HC-Pro.
This research was supported by the National Natural Science Foundation of China (grant 30270062), the National Key Basic Research and Development Program (G2000016204), and the National Outstanding Youth Foundation (grant 30125032).
REFERENCES
- 1.Anandalakshmi, R., G. J. Pruss, X. Ge, R. Marathe, A. C. Mallory, T. H. Smith, and V. B. Vance. 1998. A viral suppressor of gene silencing in plants. Proc. Natl. Acad. Sci. USA 95:13079-13084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baulcombe, D. C. 1999. Gene silencing: RNA makes RNA makes no protein. Curr. Biol. 9:599-601. [DOI] [PubMed] [Google Scholar]
- 3.Behjatnia, S. A. A., I. B. Dry, and M. A. Rezaian. 1998. Identification of the replication-associated protein binding domain within the intergenic region of tomato leaf curl geminivirus. Nucleic Acids Res. 26:925-931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernstein, E., A. A. Caudy, S. M. Hammond, and G. J. Hannon. 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409:363-366. [DOI] [PubMed] [Google Scholar]
- 5.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 6.Briddon, R. W., S. Mansoor, I. D. Bedford, M. S. Pinner, K. Saunders, J. Stanley, Y. Zafar, K. A. Malik, and P. G. Markham. 2001. Identification of DNA components required for induction of cotton leaf curl disease. Virology 285:234-243. [DOI] [PubMed] [Google Scholar]
- 7.Brigneti, G., O. Voinnet, W. X. Li, L. H. Ji, S. W. Ding, and D. C. Baulcombe. 1998. Viral pathogenicity determinants are suppressors of transgene silencing in Nicotiana benthamiana. EMBO J. 17:6739-6746. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 8.Chapman, E. J., A. I. Prokhnevsky, K. Gopinath, V. V. Dolja, and J. C. Carrington. 2004. Viral RNA silencing suppressors inhibit the microRNA pathway at an intermediate step. Genes Dev. 18:1179-1186. (Erratum, 18: 1510.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, J., W. X. Li, D. Xie, J. R. Peng, and S. W. Ding. 2004. Viral virulence protein suppresses RNA silencing-mediated defense but upregulates the role of microRNA in host gene expression. Plant Cell 16:1302-1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cogoni, C., and G. Macino. 2000. Post-transcriptional gene silencing across kingdoms. Curr. Opin. Genet. Dev. 10:638-643. [DOI] [PubMed] [Google Scholar]
- 11.Collmer, C. W., and S. H. Howell. 1992. Role of satellite RNA in expression of symptoms caused by plant viruses. Annu. Rev. Phytopathol. 30:419-442. [DOI] [PubMed] [Google Scholar]
- 12.Cui, X., X. Tao, Y. Xie, C. M. Clauquet, and X. Zhou. 2004. A DNAβ associated with Tomato yellow leaf curl China virus is required for symptom induction. J. Virol. 78:13966-13974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cui, X., X. Tao, and X. P. Zhou. 2004. AC2 and AC4 proteins of Tomato yellow leaf curl China virus and Tobacco curly shoot virus function as suppressors of RNA silencing. Chinese Sci. Bull. 49:2607-2612. [Google Scholar]
- 14.Ding, S. W., H. W. Li, R. Lu, F. Li, and W. X. Li. 2004. RNA silencing: a conserved antiviral immunity of plants and animals. Virus Res. 102:109-115. [DOI] [PubMed] [Google Scholar]
- 15.Dong, X. L., R. van Wezel, J. Stanley, and Y. G. Hong. 2003. Functional characterization of the nuclear localization signal for a suppressor of posttranscriptional gene silencing. J. Virol. 77:7026-7033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duan, Y. P., C. A. Powell, D. E. Purcifull, P. Broglio, and E. Hiebert. 1997. Phenotypic variation in transgenic tobacco expressing mutated geminivirus movement/pathogenicity (BC1) proteins. Mol. Plant-Microbe Interact. 10:1065-1074. [DOI] [PubMed] [Google Scholar]
- 17.Dunoyer, P., C. H. Lecellier, E. A. Parizotto, C. Himber, and O. Voinnet. 2004. Probing the microRNA and small interfering RNA pathways with virus-encoded suppressors of RNA silencing. Plant Cell 16:1235-1250. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Fauquet, C. M., D. M. Bisaro, R. W. Briddon, J. K. Brown, B. D. Harrison, E. P. Rybicki, D. C. Stenger, and J. Stanley. 2003. Revision of taxonomic criteria for species demarcation in the family Geminiviridae, and an updated list of begomovirus species. Arch. Virol. 148:405-421. [DOI] [PubMed] [Google Scholar]
- 19.Fire, A. 1999. RNA-triggered gene silencing. Trends Genet. 15:358-363. [DOI] [PubMed] [Google Scholar]
- 20.Gafni, Y., and B. L. Epel. 2002. The role of host and viral proteins in intra- and inter-cellular trafficking of geminiviruses. Physiol. Mol. Plant Pathol. 60:231-241. [Google Scholar]
- 21.Guo, H. S., and S. W. Ding. 2002. A viral protein inhibits the long range signaling activity of the gene silencing signal. EMBO J. 21:398-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hammond, S. M., E. Bernstein, D. Beach, and G. J. Hannon. 2000. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 404:293-296. [DOI] [PubMed] [Google Scholar]
- 23.Hanley-Bowdoin, L., S. B. Settlage, B. M. Orozco, S. Nagar, and D. Robertson. 1999. Geminiviruses: models for plant DNA replication, transcription, and cell cycle regulation. Crit. Rev. Plant Sci. 18:71-106. [PubMed] [Google Scholar]
- 24.Hartitz, M. D., G. Sunter, and D. M. Bisaro. 1999. The tomato golden mosaic virus transactivator (TrAP) is a single-stranded DNA and zinc-binding phosphoprotein with an acidic activation domain. Virology 263:1-14. [DOI] [PubMed] [Google Scholar]
- 25.Haseloff, J., K. R. Siemering, D. C. Prasher, and S. Hodge. 1997. Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc. Natl. Acad. Sci. USA 94:2122-2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hou, Y. M., R. Sanders, V. M. Ursin, and R. L. Gilberston. 2000. Transgenic plants expressing geminivirus movement proteins: Abnormal phenotypes and delayed infection by Tomato mottle virus in transgenic tomatoes expressing the Bean dwarf mosaic virus BV1 or BC1 proteins. Mol. Plant-Microbe Interact. 13:297-308. [DOI] [PubMed] [Google Scholar]
- 27.Johansen, L. K., and J. C. Carrington. 2001. Silencing on the spot. Induction and suppression of RNA silencing in the Agrobacterium-mediated transient expression system. Plant Physiol. 126:930-938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jose, J., and R. Usha. 2003. Bhendi yellow vein mosaic disease in India is caused by association of a DNAβ satellite with a begomovirus. Virology 305:310-317. [DOI] [PubMed] [Google Scholar]
- 29.Jupin, I., F. De Kouchkovsky, F. Jouanneau, and B. Gronenborn. 1994. Movement of tomato yellow leaf curl geminivirus (TYLCV): involvement of the protein encoded by ORF C4. Virology 204:82-90. [DOI] [PubMed] [Google Scholar]
- 30.Kasschau, K. D., and J. C. Carrington. 1998. A counterdefensive strategy of plant viruses: suppression of posttranscriptional gene silencing. Cell 95:461-470. [DOI] [PubMed] [Google Scholar]
- 31.Kasschau, K. D., Z. X. Xie, E. Allen, C. Llave, E. J. Chapman, K. A. Krizan, and J. C. Carrington. 2003. P1/HC-Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA function. Dev. Cell. 4:205-217. [DOI] [PubMed] [Google Scholar]
- 32.Kirthi, N., and H. S. Savithri. 2003. A conserved zinc finger motif in the coat protein of Tomato leaf curl Bangalore virus is responsible for binding to ssDNA. Arch. Virol. 148:2369-2380. [DOI] [PubMed] [Google Scholar]
- 33.Li, G. Y., K. F. Liu, S. A. Baldwin, and D. W. Wang. 2003. Equilibrative nucleoside transporters of Arabidopsis thaliana—cDNA cloning, expression pattern, and analysis of transport activities. J. Biol. Chem. 278:35732-35742. [DOI] [PubMed] [Google Scholar]
- 34.Li, H. W., W. X. Li, and S. W. Ding. 2002. Induction and suppression of RNA silencing by an animal virus. Science 296:1319-1321. [DOI] [PubMed] [Google Scholar]
- 35.Li, T. Y., Y. C. Tian, and X. F. Qin. 2004. Transgenic plants with efficient insect resistance. Sci. China B 37:1479-1489. [Google Scholar]
- 36.Liu, H., M. I. Boulton, and J. W. Davies. 1997. Maize streak virus coat protein binds single- and double-stranded DNA in vitro. J. Gen. Virol. 78:1265-1270. [DOI] [PubMed] [Google Scholar]
- 37.Lucy, A. P., H. S. Guo, W. X. Li, and S. W. Ding. 2000. Suppression of post-transcriptional gene silencing by a plant viral protein localized in the nucleus. EMBO J. 19:1672-1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Noris, E., I. Jupin, G. P. Accotto, and B. Gronenborn. 1996. DNA-binding activity of the C2 protein of tomato yellow leaf curl geminivirus. Virology 217:607-612. [DOI] [PubMed] [Google Scholar]
- 39.Noueiry, A. O., W. J. Lucas, and R. L. Gilbertson. 1994. Two proteins of a plant DNA virus coordinate nuclear and plasmodesmal transport. Cell 76:925-932. [DOI] [PubMed] [Google Scholar]
- 40.Palanichelvam, K., T. Kunik, V. Citovsky, and Y. Gafni. 1998. The capsid protein of tomato yellow leaf curl virus binds cooperatively to single-stranded DNA. J. Gen. Virol. 79:2829-2833. [DOI] [PubMed] [Google Scholar]
- 41.Palauqui, J. C., T. Elmayan, J. M. Pollien, and H. Vaucheret. 1997. Systemic acquired silencing: transgene-specific post-transcriptional silencing is transmitted by grafting from silenced stocks to non-silenced scions. EMBO J. 16:4738-4745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pascal, E., P. E. Goodlove, L. C. Wu, and S. G. Lazarowitz. 1993. Transgenic tobacco plants expressing the geminivirus BL1 protein exhibit symptoms of viral disease. Plant Cell 5:795-807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qi, Y. J., X. P. Zhou, X. Z. Huang, and G. X. Li. 2002. In vivo accumulation of Broad bean wilt virus 2 VP37 protein and its ability to bind single-stranded nucleic acid. Arch. Virol. 147:917-928. [DOI] [PubMed] [Google Scholar]
- 44.Qian, Y. J., and X. P. Zhou. 2005. Pathogenicity and stability of a truncated DNAβ associated with Tomato yellow leaf curl China virus. Virus Res. 109:159-163. [DOI] [PubMed] [Google Scholar]
- 45.Rojas, M. R., H. Jiang, R. Salati, B. Xoconostle-Cazares, M. R. Sudarshana, W. J. Lucas, and R. L. Gilbertson. 2001. Functional analysis of proteins involved in movement of the monopartite begomovirus, tomato yellow leaf curl virus. Virology 291:110-125. [DOI] [PubMed] [Google Scholar]
- 46.Roth, B. M., G. J. Pruss, and V. B. Vance. 2004. Plant viral suppressors of RNA silencing. Virus Res. 102:97-108. [DOI] [PubMed] [Google Scholar]
- 47.Sanderfoot, A. A., and S. G. Lazarowitz. 1995. Cooperation in viral movement: the geminivirus BL1 movement protein interacts with BR1 and redirects it from the nucleus to the cell periphery. Plant Cell 7:1185-1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saunders, K., I. D. Bedford, R. W. Briddon, P. G. Markham, S. M. Wong, and J. Stanley. 2000. A unique virus complex causes Ageratum yellow vein disease. Proc. Natl. Acad. Sci. USA 97:6890-6895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saunders, K., I. D. Bedford, T. Yahara, and J. Stanley. 2003. The earliest recorded plant virus disease. Nature 422:831. [DOI] [PubMed] [Google Scholar]
- 50.Saunders, K., A. Norman, S. Gucciardo, and J. Stanley. 2004. The DNAβ satellite component associated with ageratum yellow vein disease encodes an essential pathogenicity protein (βC1). Virology 324:37-47. [DOI] [PubMed] [Google Scholar]
- 51.Shi, B. J., S. W. Ding, and R. H. Symons. 1997. Plasmid vector for cloning infectious cDNAs from plant RNA viruses: high infectivity of cDNA clones of tomato aspermy cucumovirus. J. Gen. Virol. 78:1181-1185. [DOI] [PubMed] [Google Scholar]
- 52.Sung, Y. K., and R. H. Coutts. 1996. Potato yellow mosaic geminivirus AC2 protein is a sequence non-specific DNA binding protein. FEBS Lett. 383:51-54. [DOI] [PubMed] [Google Scholar]
- 53.van Wezel, R., X. L. Dong, P. Blake, J. Stanley, and Y. G. Hong. 2002. Differential roles of geminivirus Rep and AC4 (C4) in the induction of necrosis in Nicotiana benthamiana. Mol. Plant Pathol. 3:461-471. [DOI] [PubMed] [Google Scholar]
- 54.van Wezel, R., H. Liu, P. Tien, J. Stanley, and Y. Hong. 2001. Gene C2 of the monopartite geminivirus tomato yellow leaf curl virus-China encodes a pathogenicity determinant that is localized in the nucleus. Mol. Plant-Microbe Interact. 14:1125-1128. [DOI] [PubMed] [Google Scholar]
- 55.van Wezel, R., H. T. Liu, Z. R. Wu, J. Stanley, and Y. H. Hong. 2003. Contribution of the zinc finger to zinc and DNA binding by a suppressor of posttranscriptional gene silencing. J. Virol. 77:696-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vargason, J. M., G. Szittya, J. Burgyan, and T. M. T. Hall. 2003. Size selective recognition of siRNA by an RNA silencing suppressor. Cell 115:799-811. [DOI] [PubMed] [Google Scholar]
- 57.Voinnet, O. 2001. RNA silencing as a plant immune system against viruses. Trends Genet. 17:449-459. [DOI] [PubMed] [Google Scholar]
- 58.Voinnet, O., Y. M. Pinto, and D. C. Baulcombe. 1999. Suppression of gene silencing: a general strategy used by diverse DNA and RNA viruses of plants. Proc. Natl. Acad. Sci. USA 96:14147-14152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xie, Z., L. K. Johansen, A. M. Gustafson, K. D. Kasschau, A. D. Lellis, D. Zilberman, S. E. Jacobsen, and J. C. Carrington. 2004. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2:E104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhou, X. P., Y. Xie, X. R. Tao, Z. K. Zhang, Z. H. Li, and C. M. Fauquet. 2003. Characterization of DNAβ associated with begomoviruses in China and evidence for co-evolution with their cognate viral DNA-A. J. Gen. Virol. 84:237-247. [DOI] [PubMed] [Google Scholar]