Abstract
To date, no molecular studies on group C viruses (Bunyaviridae, Orthobunyavirus) have been published. We determined the complete small RNA (SRNA) segment and partial medium RNA segment nucleotide sequences for 13 group C members. The full-length SRNA sequences ranged from 915 to 926 nucleotides in length, and revealed similar organization in comparison with other orthobunyaviruses. Based on the 705 nucleotides of the N gene, group C members were distributed into three major phylogenetic groups, with the exception of Madrid virus, which was placed outside of these three groups. Analysis of the Caraparu virus strain BeH 5546 revealed that it has an SRNA sequence nearly identical to that of Oriboca virus and is a natural reassortant virus. In addition, analysis of 345 nucleotides of the Gn gene for eight group C viruses and for strain BeH 5546 revealed a different phylogenetic topology, suggesting a reassortment pattern among them. These findings represent the first evidence for natural reassortment among the group C viruses, which include several human pathogens. Furthermore, our genetic data corroborate previous relationships determined using serologic assays (complement fixation, hemagglutination inhibition, and neutralization tests) and suggest that a combination of informative molecular, serological, and ecological data is a helpful tool to understand the molecular epidemiology of arboviruses.
Members of the family Bunyaviridae, genus Orthobunyavirus, have a genome consisting of three single-stranded, negative-sense RNA segments, designated large (LRNA), medium (MRNA), and small (SRNA) (9, 25). The L segment encodes a large protein that contains the RNA-dependent RNA polymerase activity for replication and transcription of genomic RNA segments. The M segment encodes a precursor polypeptide which gives rise to viral surface glycoproteins Gn and Gc and a nonstructural protein, NSm. The S segment encodes two proteins, the nucleocapsid (N) protein and a smaller nonstructural protein, NSs, in overlapping reading frames.
Group C viruses were first described in the Brazilian Amazon region during the 1950s (4, 5, 14, 27, 33). A total of 13 distinct group C viruses have been isolated from humans, wild animals (principally rodents, marsupials, and bats), and mosquitoes (16, 27). The current classification of group C is based on antigenic relationships determined using complement fixation (CF), neutralization (NT), and hemagglutination inhibition (HI) tests. According to this serological classification, the group C viruses can be divided into four antigenic complexes: the Caraparu complex, which includes Caraparu virus (CARV), Ossa virus (OSSAV), Apeu virus (APEUV), Vinces virus (VINV), and Bruconha virus (BRCV); the Madrid complex, which includes Madrid virus (MADV); the Marituba complex, which includes Marituba virus (MTBV), Murutucu virus (MURV), Restan virus (RESV), Nepuyo virus (NEPV), and Gumbo limbo virus (GLV); and the Oriboca complex, which includes Oriboca virus (ORIV) and Itaqui virus (ITQV) (5, 7, 14, 26, 28-30).
Geographically, group C viruses occur in tropical and subtropical areas of the Americas, including the United States, Mexico, Panama, Honduras, Guatemala, Trinidad, Brazil, Peru, Ecuador, Venezuela, and French Guiana (9, 14, 16, 27). Ten of the 13 registered viruses (CARV, ORIV, ITQV, NEPV, APEUV, MTBV, MURV, RESV, OSSAV, and MADV) have been associated with human disease, which generally presents as a self-limited, dengue-like illness consisting of fever, headache, myalgia, nausea, vomiting, weakness, etc., of 2 to 5 days in duration (17, 18, 32). Given the public and veterinary health importance of other viruses included in the genus Orthobunyavirus, surprisingly little attention has been paid to the molecular biology and genetics of the group C viruses.
To address this deficiency, we determined the complete SRNA sequence for group C viruses, as well as their SRNA genetic organization. We used nucleotide sequences for the nucleocapsid gene and partial nucleotide sequences (345 nucleotides [nt]) of the Gn glycoprotein of group C viruses, as well as homologous sequences for California encephalitis, Simbu, and Bunyamwera virus groups, to determine phylogenetic relationships among group C viruses and other orthobunyaviruses. We also attempted to correlate these data with serological relationships previously provided by Shope and Causey (28).
MATERIALS AND METHODS
Virus strains.
The group C viruses used in this study are listed in Table 1 and represent relatively low-passage isolates obtained from the World Reference Center for Emerging Viruses and Arboviruses at the University of Texas Medical Branch at Galveston and from the Department of Arbovirology and Hemorrhagic Fevers at the Evandro Chagas Institute, Belem, Pará State, Brazil.
TABLE 1.
Group C virus strains used in the current study
| Group C complex viruses | Strain | Country of origin | Host association | Isolation yr |
|---|---|---|---|---|
| Caraparu | ||||
| CARV | BeAn 3994 | Brazil | Cebus apella | 1956 |
| BeH 190863 | Brazil | Human | 1970 | |
| IQU 212 | Peru | Human | 1995 | |
| BeAr 299789 | Brazil | Culex portesi | 1975 | |
| BeH 5546 | Brazil | Human | 1956 | |
| APEUV | BeAn 848 | Brazil | Cebus apella | 1955 |
| BRCV | 77V-74814 | Brazil | Sentinel mouse | 1977 |
| OSSAV | BT 1820 | Panama | Human | 1961 |
| VINV | 75V-807 | Ecuador | Culex (Melanoconium) vomerifer | 1974 |
| Marituba | ||||
| MTBV | BeAn 15 | Brazil | Cebus apella | 1954 |
| BeAn 258481 | Brazil | Caluromys sp. | 1974 | |
| BeH 119190 | Brazil | Human | 1980 | |
| BeAr 186247 | Brazil | Culex melanoconium sp. | 1970 | |
| NEPV | TRVL 18462 | Trinidad | Culex (Aedinus) accelerans | 1957 |
| BeAn 10709 | Brazil | Sentinel mouse | 1959 | |
| BeAr 225808 | Brazil | Culex sp. | 1972 | |
| BeAn 275424 | Brazil | Nectomys squamipes | 1969 | |
| ITQV | BeAn 12797 | Brazil | Sentinel mouse | 1954 |
| BeAn 346113 | Brazil | Cebus apella | 1959 | |
| GLV | FE-3 71H | United States | Culex (Melanoconium) | 1963 |
| Madrid | ||||
| MADV | BT 4075 | Panama | Human | 1961 |
| Oriboca | ||||
| ORIV | BeAn 17 | Brazil | Cebus apella | 1954 |
| BeAn 263748 | Brazil | Oryzomys goeldi | 1974 | |
| BeAr 246614 | Brazil | Culex taeniopus | 1973 | |
| BeH 142102 | Brazil | Human | 1968 | |
| LCVP 217 | French Guyana | Human | 1977 | |
| MURV | BeAn 974 | Brazil | Cebus apella | 1955 |
| IQU 126 | Peru | Human | 1999 | |
| IQU 9891 | Peru | Human | 1999 | |
| BeH 257944 | Brazil | Human | 1974 | |
| RESV | TRVL 51144 | Trinidad | Human | 1963 |
Virus culture and RNA extraction.
Viruses were propagated in monolayer cultures of Vero cells. After 75% of cells exhibited cytopathic effects, the supernatants of infected cell cultures were collected, centrifuged at 3,000 rpm at 4°C for cell debris removal, and treated with 50% polyethylene glycol 8000 and 23% NaCl for viral RNA precipitation. After centrifugation, the virus pellets were eluted in 250 μl of RNase-free water. The RNA extraction was carried out using a commercial kit (QIAmp Viral RNA mini kit; QIAGEN, Valencia, CA).
SRNA and partial MRNA amplification.
A one-step reverse-transcription PCR (RT-PCR) protocol was used for amplification of the full-length SRNA, using primers that corresponded to the highly conserved termini of the SRNA segment of viruses included in the genus Orthobunyavirus (8). RT-PCRs were carried out in a 50-μl reaction mixture containing 10 μl (1 to 5 ng) of viral RNA, 10 pmol of a forward primer (AGTAGTGTGCTCCAC), 10 pmol of a reverse primer (AGTAGTGTGCTCCAC), 1× PCR buffer (50 mM Tris-HCl, pH 8.3, 75 mM KCl), 2.5 mM MgCl2, 2.5 mM dithiothreitol (DTT), 20 U of RNAsin RNase inhibitor (Invitrogen, Carlsbad, CA), 200 μM of deoxynucleoside triphosphates (dNTPs) (Invitrogen, Carlsbad, CA), 1.125 U of Platinum Taq DNA polymerase (Invitrogen, Carlsbad, CA), and 1 unit of Superscript II reverse transcriptase (Invitrogen). The RT reaction was first performed for 60 min at 42°C, followed by 35 PCR cycles, each consisting of 94°C for 40 s, 54°C for 40 s, and 72°C for 1 min.
For the amplification of the partial Gn glycoprotein gene, a standard two-step RT-PCR protocol was used. For the first-strand amplification, a 20-μl reaction mixture was used, consisting of 5 μl of virus RNA (1 ng to 5 μg) and 15 μl of the RT master mix including 1× first-strand buffer (250 mM Tris-HCl, pH 8.3, 375 mM KCl, 15 mM MgCl2, 0.1 M DDT), 20 U RNasin RNase inhibitor (Invitrogen), 200 μM of dNTPs, and 50 to 250 ng of random hexamer primers. The reactions were reverse transcribed for 60 min at 42°C. The PCR was performed using 2 ng of the RT products and a PCR mixture containing 1× PCR buffer, 2.5 mM MgCl2, 200 μM of dNTPs, 10 pmol of degenerate primer BUN-GnF (AC[T/A]AAG[C/T]TATA[C/T]AG[A/G]TA[T/C]AT) and 10 pmol of degenerate primer BUN-GnR (TGACATATG[C/T]TG[G/A]TT[A/G]AAGCA), with 1.125 U of Platinum Taq DNA polymerase adjusted for a final volume of 50 μl. The amplified products were visualized on a 1.2% agarose gel, purified using the GFX PCR DNA and Band purification kit (Amersham Biosciences, Piscataway, NJ), cloned, and sequenced.
cDNA cloning.
Cloning of the cDNA fragments was done with a plasmidial-bacterial system. Purified amplicons were ligated to the pGMT-Easy Vector (Invitrogen) at the lacZα peptide gene, and plasmid DNA from at least three recombinant bacterial colonies was recovered using the GFX microplasmid Prep kit (Amersham Biosciences, Piscataway, NJ). The restriction endonuclease EcoRI (Invitrogen) was used to release the ligated cDNA from the plasmid in order to verify the cloning efficiency.
Sequencing.
The nucleotide sequences of the cloned cDNAs were determined using the ABI PRISM Dye Terminator kit (Applied Biosystems, Foster City, CA) and an ABI 377 DNA sequencer. Universal T7 and SP6 primers were used to sequence the recombinant DNA in both directions. At least three plasmid clones were sequenced for each viral amplicon.
Sequence analyses.
The nucleotide sequences determined were aligned with homologous sequences from the GenBank library using ClustalX software. Phylogenetic trees were constructed using the neighbor-joining (NJ) (23), maximum-parsimony (MP), and maximum-likelihood (ML) methods, implemented with the PAUP 4.0 (31) and Mega 2.1 (15) software packages. For the likelihood analyses, the general time-reversible model was used and nucleotide frequencies were estimated empirically. For NJ analysis, a distance matrix was calculated from the aligned sequences using the Kimura two-parameter formula. Bootstrap analyses using 1,000 replicates were used to place confidence values on groupings (10) and the Kishino-Hasegawa test was used to evaluate competing topologies obtained from different viral RNA segments and using different methods (12).
RESULTS
The SRNA segment.
Full-length SRNA nucleotide sequences were obtained for the 13 prototype viruses in group C, as well as from 18 additional strains of ORIV (BeAn 263748, BeAr 246614, BeH 142102, and LCVP 217), MURV (IQU 9891, IQU 126, and BeH 257944), CARV (BeH 190863, IQU 212, BeAr 299789, and BeH 5546), MTBV (BeAn 258481, BeH 119190, BeAr 186247), NEPV (BeAn 10709, BeAr 225808, and BeAr 275424), and ITQV (BeAn 346113). The sequences showed similar organization to those reported for viruses in the Bunyamwera, California, and Simbu serogroups of the Orthobunyavirus genus, revealing two overlapping open reading frames (ORFs), N and NSs, predicted to encode the N and NSs proteins, respectively. The coding regions for all S segments were flanked by two terminal noncoding regions (NCRs) designated 5′ and 3′ NCRs (Table 2). The SRNA sequences for group C members showed nucleotide and deduced amino acid sequence identities ranging from 69.6% to 99.3% and 74% to 99.6%, respectively (Table 3).
TABLE 2.
Sequence characteristics of the SRNA of group C virusesa
| Virus | Sequence characteristic
|
|||||
|---|---|---|---|---|---|---|
| Nucleotides
|
Nucleotides/amino acids
|
|||||
| Total no. | 5′ NCR | 3′ NCR | A+U (%) | N | NSs | |
| Apeu | 916 | 76 | 135 | 57.5 | 705/234 | 297/98 |
| Nepuyo | 918 | 75 | 138 | 55.2 | 705/234 | 297/98 |
| Gumbo Limbo | 915 | 76 | 134 | 54.9 | 705/234 | 276/91 |
| Marituba | 917 | 75 | 137 | 56.9 | 705/234 | 297/98 |
| Oriboca | 920 | 75 | 140 | 53.3 | 705/234 | 297/98 |
| Murutucu | 924 | 75 | 144 | 53.6 | 705/234 | 297/98 |
| Restan | 919 | 73 | 141 | 54.0 | 705/234 | 276/91 |
| Caraparu | 922 | 76 | 141 | 54.1 | 705/234 | 297/98 |
| Bruconha | 922 | 76 | 141 | 54.1 | 705/234 | 297/98 |
| Ossa | 921 | 76 | 140 | 54.2 | 705/234 | 297/98 |
| Itaqui | 920 | 76 | 139 | 53.4 | 705/234 | 297/98 |
| Vinces | 923 | 74 | 145 | 53.5 | 705/234 | 297/98 |
| Madrid | 926 | 74 | 147 | 53.8 | 705/234 | 276/91 |
Shown are the sequence length, coding regions (N and NSs), 5′ and 3′ NCRs, and A+U composition.
TABLE 3.
N gene nucleotide and deduced amino acid sequence identity among group C viruses
| Virus | Nucleotide or amino acid sequence identity (%)a
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| APEUV | MTBV | MURV | ORIV | CARV | ITQV | BCRV | GLV | MADV | NEPV | OSSAV | RESV | VINV | |
| APEUV | 94.2 | 80.6 | 82.6 | 75.5 | 75.5 | 75.7 | 82.6 | 76.6 | 90.1 | 75.6 | 81.0 | 76.5 | |
| MTBV | 98.7 | 80.3 | 81.8 | 75.6 | 74.9 | 75.6 | 82.4 | 76.5 | 91.1 | 75.9 | 80.9 | 75.6 | |
| MURV | 88.9 | 89.8 | 95.5 | 84.3 | 82.6 | 84.1 | 77.6 | 72.6 | 82.1 | 83.4 | 91.1 | 81.8 | |
| ORIV | 90.2 | 89.4 | 97.9 | 82.7 | 81.4 | 82.7 | 78.6 | 73.5 | 84.0 | 82.6 | 90.9 | 81.6 | |
| CARV | 87.2 | 86.4 | 91.5 | 92.8 | 95.0 | 99.3 | 75.6 | 69.8 | 75.9 | 97.9 | 83 | 86.4 | |
| ITQV | 86.0 | 85.5 | 90.2 | 91.1 | 97.9 | 95.3 | 75.7 | 69.9 | 75.3 | 94.8 | 81.7 | 89.8 | |
| BRCV | 87.2 | 86.4 | 91.5 | 92.8 | 99.6 | 97.9 | 75.6 | 69.8 | 75.7 | 97.9 | 82.7 | 87.0 | |
| GLV | 89.4 | 88.5 | 85.5 | 86.8 | 84.3 | 83.4 | 84.3 | 78.6 | 86.1 | 76 | 79.4 | 76.2 | |
| MADV | 79.1 | 78.3 | 76.2 | 77.0 | 74.5 | 74.9 | 74.5 | 77.4 | 77.7 | 69.6 | 72.9 | 70.8 | |
| NEPV | 96.6 | 95.7 | 89.8 | 90.2 | 86.8 | 85.5 | 86.8 | 90.2 | 79.1 | 75.6 | 82.3 | 76.3 | |
| OSSAV | 86.8 | 86.0 | 91.1 | 92.3 | 99.1 | 97.4 | 99.1 | 84.3 | 74.0 | 86.4 | 83.4 | 86.8 | |
| RESV | 88.9 | 88.1 | 96.2 | 96.2 | 92.8 | 91.1 | 92.8 | 86.8 | 76.6 | 89.4 | 92.3 | 82.4 | |
| VINV | 83.8 | 83.8 | 88.1 | 88.5 | 93.2 | 93.6 | 93.2 | 82.1 | 74.9 | 84.3 | 93.6 | 87.7 | |
Values in the top right half of the data field represent nucleotide sequence identity, and values in the bottom left half represent amino acid sequence identity. Bold underlined values represent the genetic pair association established between the viruses APEUV-MTBV, MURV-ORIV, and CARV-ITQV. Bold italicized values correspond to the nucleotide and amino acid sequence identities between the viruses CARV and BRCV.
The ORIV strains isolated from different places and sources showed nucleotide sequence divergence of 0.6 to 1.4%. For MURV, 0.4 to 1.4% nucleotide sequence divergence was identified, whereas for NEPV isolates, nucleotide sequences diverged from 0.6% to 1.1%. One strain of ITQV (BeAn 346113) showed 98.9% nucleotide sequence identity with the prototype strain BeAn 12797. Furthermore, for CARV, all strains showed expected nucleotide sequence divergence levels of 0.3 to 1.0%, except for strain BeH 5546, which showed 96.5% identity with the N gene sequence of the prototype strain of ORIV, BeAn 17 (Table 4).
TABLE 4.
Nucleotide sequence divergence identity and among certain group C virusesa
| Nucleotide sequence identity (%)
|
||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CARV
|
ITQV
|
MTBV
|
MURV
|
NEPV
|
ORIV
|
|||||||||||||||||||
| H190 863 | IQU 212 | Ar 299789 | H 5546 | An 3994* | An 346113 | An 12797* | An 258481 | Ar 186247 | H 119190 | An 15* | ITQ 9891 | IQU 126 | H 257944 | An 974* | An 10709 | An 275424 | Ar 225808 | TRVL 18462* | An 263748 | Ar 246614 | H 142102 | LCVP 217 | An 17* | |
| BeH 190 863 | 99.7 | 99.3 | 81.7 | 99.4 | 95.2 | 94.9 | 75.5 | 75.6 | 76.2 | 75.5 | 84.1 | 83.8 | 83.8 | 84.1 | 75.9 | 76.2 | 76.3 | 75.7 | 82.1 | 82.4 | 81.8 | 81.8 | 82.4 | |
| IQU 212 | 0.3 | 99.0 | 81.6 | 99.3 | 95.0 | 94.8 | 75.6 | 75.7 | 76.3 | 75.6 | 84.0 | 83.7 | 83.7 | 84.0 | 75.9 | 76.2 | 76.3 | 75.7 | 82.0 | 82.3 | 81.7 | 81.7 | 82.3 | |
| BeAr 299789 | 0.7 | 1.0 | 81.6 | 99.3 | 94.8 | 94.9 | 75.7 | 75.9 | 76.3 | 75.9 | 84.1 | 83.8 | 83.8 | 84.1 | 76.2 | 76.2 | 76.3 | 76.0 | 82.3 | 82.6 | 82.0 | 82.0 | 82.6 | |
| BeH 5546 | 21.2 | 21.3 | 21.4 | 82.1 | 80.9 | 81.4 | 81.3 | 81.6 | 81.3 | 81.4 | 93.6 | 93.5 | 93.3 | 93.9 | 83.7 | 83.8 | 83.8 | 84.0 | 95.9 | 96.5 | 95.2 | 95.2 | 96.5 | |
| BeAn 3994* | 0.6 | 0.7 | 0.7 | 20.6 | 95.0 | 95.0 | 75.6 | 75.9 | 76.0 | 75.6 | 84.3 | 84.0 | 84.0 | 84.3 | 76.0 | 76.3 | 76.5 | 75.9 | 82.4 | 82.7 | 82.1 | 82.1 | 82.7 | |
| BeAn 346113 | 5.0 | 5.1 | 5.4 | 22.2 | 5.1 | 98.9 | 75.0 | 75.3 | 75.0 | 75.0 | 82.4 | 82.3 | 82.1 | 82.3 | 75.7 | 75.9 | 75.9 | 75.5 | 80.4 | 81.0 | 80.3 | 80.3 | 81.0 | |
| BeAn 12797* | 5.3 | 5.5 | 5.3 | 21.5 | 5.2 | 1.1 | 74.9 | 75.2 | 74.9 | 74.9 | 82.7 | 82.6 | 82.4 | 82.6 | 75.6 | 75.7 | 75.7 | 75.3 | 80.9 | 81.4 | 80.7 | 80.7 | 81.4 | |
| BeAn 258481 | 30.0 | 29.8 | 29.5 | 21.7 | 29.8 | 30.6 | 30.8 | 99.6 | 99.0 | 99.6 | 80.3 | 80.7 | 80.1 | 80.3 | 90.4 | 90.2 | 90.5 | 90.9 | 81.3 | 81.6 | 81.7 | 81.7 | 81.6 | |
| BeAr 186247 | 29.8 | 29.6 | 29.3 | 21.3 | 29.3 | 30.1 | 30.3 | 0.4 | 99.3 | 99.4 | 80.4 | 80.6 | 80.0 | 80.7 | 90.4 | 90.2 | 90.5 | 90.9 | 81.6 | 81.8 | 82.0 | 82.0 | 81.8 | |
| BeH 119190 | 28.9 | 28.7 | 28.7 | 21.7 | 29.1 | 30.5 | 30.8 | 1.0 | 0.7 | 99.0 | 80.3 | 80.4 | 79.9 | 80.6 | 89.6 | 89.5 | 89.8 | 90.2 | 81.3 | 81.6 | 81.7 | 81.7 | 81.6 | |
| BeAn 15* | 30.0 | 29.8 | 29.3 | 21.5 | 29.8 | 30.6 | 30.8 | 0.4 | 0.6 | 1.0 | 80.1 | 80.4 | 80.0 | 80.3 | 90.5 | 90.4 | 90.6 | 91.1 | 81.6 | 81.8 | 82.0 | 82.0 | 81.8 | |
| ITQ 9891 | 17.9 | 18.1 | 17.9 | 6.7 | 17.7 | 20.1 | 19.7 | 23.2 | 23.0 | 23.2 | 23.3 | 99.6 | 99.6 | 98.7 | 81.8 | 81.8 | 81.8 | 82.0 | 94.6 | 95.2 | 93.9 | 93.9 | 95.2 | |
| IQU 126 | 18.3 | 18.4 | 18.3 | 6.9 | 18.1 | 20.3 | 19.9 | 22.6 | 22.8 | 23.0 | 23.0 | 0.4 | 99.4 | 98.6 | 82.0 | 82.0 | 82.0 | 82.1 | 94.6 | 95.2 | 93.9 | 93.9 | 95.2 | |
| BeH 257944 | 18.3 | 18.4 | 18.3 | 7.0 | 18.1 | 20.4 | 20.1 | 23.3 | 23.5 | 23.7 | 23.5 | 0.4 | 0.6 | 98.7 | 81.7 | 81.7 | 81.7 | 81.8 | 94.3 | 94.9 | 93.6 | 93.6 | 94.9 | |
| BeAn 974* | 17.9 | 18.1 | 17.9 | 6.4 | 17.7 | 20.3 | 19.9 | 23.1 | 22.5 | 22.7 | 23.1 | 1.3 | 1.4 | 1.3 | 82.0 | 82.0 | 82.0 | 82.1 | 94.9 | 95.5 | 94.2 | 94.2 | 95.5 | |
| BeAn 10709 | 29.3 | 29.3 | 28.8 | 18.5 | 29.1 | 29.4 | 19.6 | 10.4 | 10.4 | 11.2 | 10.2 | 20.9 | 20.7 | 21.1 | 20.7 | 99.4 | 98.9 | 99.1 | 83.3 | 83.7 | 83.4 | 83.4 | 83.7 | |
| BeAn 275424 | 28.9 | 28.9 | 28.8 | 18.3 | 28.7 | 29.3 | 29.4 | 10.5 | 10.5 | 11.4 | 10.4 | 20.9 | 20.7 | 21.1 | 20.7 | 0.6 | 99.1 | 99.0 | 83.1 | 83.5 | 83.3 | 83.3 | 83.5 | |
| BeAr 225808 | 28.7 | 28.7 | 28.7 | 18.3 | 28.5 | 29.3 | 29.5 | 10.2 | 10.2 | 11.0 | 10.0 | 20.9 | 20.7 | 21.1 | 20.7 | 1.1 | 0.9 | 99.3 | 83.1 | 83.5 | 83.3 | 83.3 | 83.5 | |
| TRVL 18462* | 29.6 | 29.6 | 29.1 | 18.1 | 29.3 | 29.9 | 30.1 | 9.7 | 9.7 | 10.5 | 9.5 | 20.7 | 20.5 | 20.9 | 20.5 | 0.9 | 1.0 | 0.7 | 83.5 | 84.0 | 83.7 | 83.7 | 84.0 | |
| BeAn 263748 | 20.5 | 20.7 | 20.3 | 4.2 | 20.1 | 22.8 | 22.2 | 21.8 | 21.4 | 21.8 | 21.4 | 5.6 | 5.6 | 5.9 | 5.3 | 19.0 | 19.2 | 19.2 | 18.7 | 99.4 | 99.1 | 99.1 | 99.4 | |
| BeAr 246614 | 20.1 | 20.3 | 19.9 | 3.6 | 19.7 | 22.0 | 21.4 | 21.4 | 21.0 | 21.4 | 21.0 | 5.0 | 5.0 | 5.3 | 4.7 | 18.5 | 18.7 | 18.7 | 18.1 | 0.6 | 98.6 | 98.6 | 99.1 | |
| BeH 142102 | 20.9 | 21.1 | 20.7 | 5.0 | 20.5 | 23.0 | 22.4 | 21.2 | 20.8 | 21.2 | 20.8 | 6.4 | 6.4 | 6.7 | 6.1 | 18.8 | 19.0 | 19.0 | 18.5 | 0.9 | 1.4 | 99.4 | 98.6 | |
| LCVP 217 | 20.9 | 21.1 | 20.7 | 5.0 | 20.5 | 23.0 | 22.4 | 21.2 | 2.8 | 21.2 | 20.8 | 6.4 | 6.4 | 6.7 | 6.1 | 18.8 | 19.0 | 19.0 | 18.5 | 0.9 | 1.4 | 0.6 | 98.6 | |
| BeAn 17* | 20.1 | 20.3 | 19.9 | 3.6 | 19.7 | 22.0 | 21.4 | 21.4 | 21.0 | 21.4 | 21.0 | 5.0 | 5.0 | 5.3 | 4.7 | 18.5 | 18.7 | 18.7 | 18.1 | 0.6 | 0.6 | 1.4 | 1.4 | |
Values in the top right half of the data field represent nucleotide sequence identity, and values in the bottom left half represent nucleotide sequence divergence. *, prototype viruses. Underlined values indicate nucleotide sequence identities (top half) and italicized values represent amino acid sequence identities (bottom half) among each virus and its respective strains; bold underlined values represent homology among strain BeH 5546 and strains of CARV; bold italicized values represent identities among strain BeH 5546 and strains of ORIV.
Partial MRNA nucleotide sequences.
The partial sequence (345 nucleotides) of the MRNA segment was obtained for eight group C viruses (APEUV, MTBV, ORIV, MURV, RESV, CARV, and ITQV). The sequences showed considerable similarity to the partial nucleotide sequence of the Gn glycoprotein of other orthobunyaviruses, which is encoded in the first one-third of the MRNA segment. The sequences corresponded to the homologous nucleotide positions 193 to 573, 168 to 513, and 259 to 602 of representative members of the California (La Crosse virus), Simbu (Oropouche virus), and Bunyamwera (Bunyamwera virus) serogroups, respectively. The Gn partial nucleotide sequence comparisons among group C viruses are summarized in Table 5.
TABLE 5.
Partial Gn nucleotide and deduced amino acid sequence identity among group C viruses
| Virus | Nucleotide or amino acid sequence identity (%)a
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| MTBV | MURV | ITQV | ORIV | BeH 5546 | APEUV | CARV | RESV | VINV | |
| MTBV | 88.7 | 75.9 | 73.3 | 64.9 | 62.3 | 64.6 | 79.7 | 70.4 | |
| MURV | 95.7 | 71 | 69.6 | 66.1 | 69.3 | 66.1 | 85.2 | 67.5 | |
| ITQV | 70.4 | 70.4 | 86.7 | 69.9 | 70.4 | 70.7 | 71.6 | 78.3 | |
| ORIV | 70.4 | 68.7 | 96.5 | 71.3 | 72.5 | 71.6 | 69.9 | 78 | |
| BeH 5546 | 65.2 | 66.1 | 80 | 80.9 | 87 | 98.3 | 66.7 | 82.3 | |
| APEUV | 64.3 | 67 | 80.9 | 79.1 | 95.7 | 87.2 | 71 | 77.1 | |
| CARV | 65.2 | 66.1 | 80 | 80.9 | 100 | 95.7 | 67.2 | 83.2 | |
| RESV | 88.7 | 91.3 | 66.1 | 66.1 | 65.2 | 64.3 | 65.2 | 66.4 | |
| VINV | 69.6 | 71.3 | 82.6 | 83.5 | 89.6 | 87.8 | 89.6 | 67.8 | |
Values in the top right half of the data field represent nucleotide sequence identity, and values in the bottom left half represent amino acid sequence identity. Strains: APEUV, BeAn 848; CARV, BeAn 3994; ITQV, BeAn 12797; MTBV, BeAn 15; MURV, BeAn 974; ORIV, BeAn 17; RESV, TRVL 51144; VINV, 75V-807. Bold values represent the nucleotide and amino acid identities for the genetic pair association established between MTBV-MURV, ITQV-ORIV, and CARV-APEUV. Underlined and italicized values correspond to the nucleotide and amino acid sequence identities among strain BeH 5546 and CARV and ORIV, respectively.
Phylogenetic analyses.
To establish genetic relationships among group C and other orthobunyaviruses, phylogenetic trees were constructed using the complete N gene (705 nt) and partial Gn gene (345 nt) sequences using NJ, MP, and ML methods. Representative members of the California, Bunyamwera, and Simbu serogroups were used as outgroups to root the trees. Both NJ and MP methods generated trees with similar topology, although bootstrap support values were slightly lower for the MP construction.
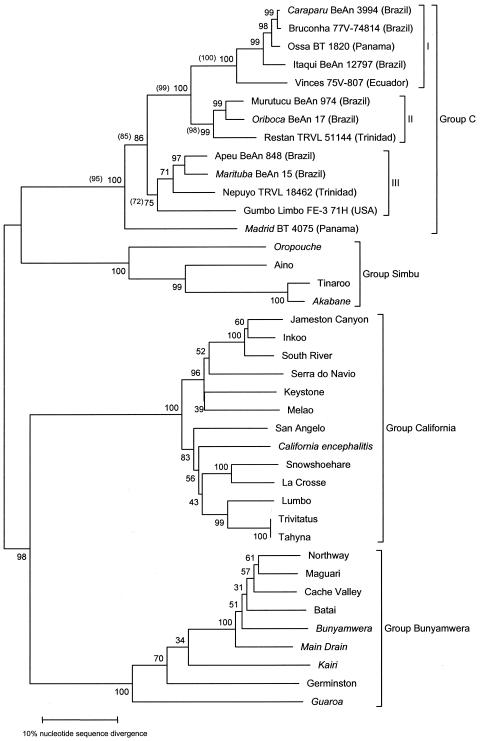
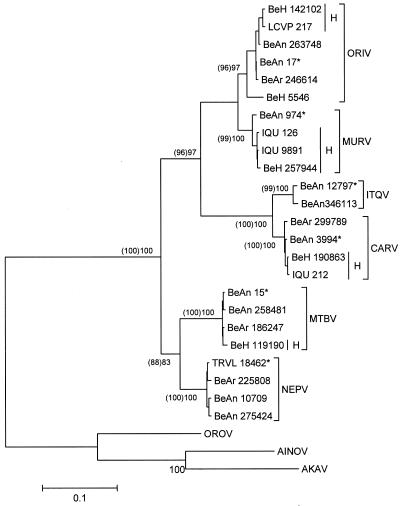
All trees generated from the S segment depicted the members of group C as a monophyletic group sharing a common ancestor, and group C appeared to be most closely related to viruses belonging to the Simbu serogroup (Fig. 1). In addition, group C viruses were distributed in three major lineages designated as group I (CARV, BRCV, OSSAV, ITQV, and VINV), group II (MURV, ORIV, and RESV), and group III (APEUV, MTBV, NEPV, and GLV). MADV represented a distinct lineage in the trees (Fig. 1). The phylogenetic analysis of additional strains of ORIV, CARV, ITQV, and MURV, as well as of additional strains of NEPV and MTBV, is shown in Fig. 2. Except for the CARV strain BeH 5546 that grouped with strains of ORIV, all other group C virus strains showed the expected positions in the trees (clades representing a given virus).
FIG. 1.
Phylogeny of group C virus members based on their N ORF complete nucleotide sequences. Phylogenetic analyses were carried out using both NJ and MP methods, yielding identical topologies, and the NJ tree is presented. Group C virus members were distributed into tree major lineages called groups I, II, and III. Numbers adjacent to each branch represent the percentage bootstrap support calculated for 1,000 replicates. Values inside and outside parentheses indicate bootstrap values obtained by the MP and NJ methods, respectively. Members of the California, Bunyamwera, and Simbu serogroups were used as outgroups to root the tree. The scale bar represent 10% nucleotide sequence divergence.
FIG. 2.
Phylogenetic analysis of additional group C virus strains of ORIV, MURV, ITQV, CARV, MTBV, and NEPV isolated from different geographic areas, times, and hosts. Vertical bars indicate strains isolated from humans. Bootstrap values were assigned for 1,000 replicates. Numbers inside the parentheses indicate percentage bootstrap support using MP, while numbers outside parentheses represent NJ values. Isolates labeled with stars represent group C virus prototype strains. Viruses belonging to the Simbu serogroup were used as an outgroup to root the tree. Scale bars represent 10% nucleotide sequence divergence. OROV, Oropouche virus; AINOV, Aino virus; AKAV, Akabane virus.
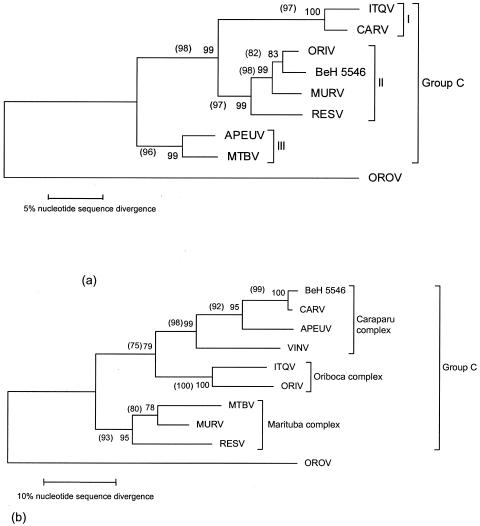
Trees generated from a 345-nt partial Gn sequences of eight group C virus species and for CARV strain BeH 5546 revealed a topology different from that generated using the S segment. Using all phylogenetic methods, several groupings determined using the N gene sequences (APEUV-MTBV, ORIV-MURV, and CARV-ITQV; see Fig. 1) changed (to APEUV-CARV, ORIV-ITQV, and MTBV-MURV) when the Gn partial sequences were analyzed. RESV, which was based on the SRNA analysis grouped with ORIV and MURV, grouped with MURV and MTBV in the MRNA analyses with strong bootstrap support. The CARV strain BeH 5546 also shifted its position in the MRNA trees; it grouped with ORIV in the S segment analysis, but grouped with the prototype CARV strain BeAn 3994 using M segment sequences (Fig. 3). Again, bootstrap support was strong using both genome segments and all phylogenetic methods. Maximum-likelihood analyses were used to test these competing S and M segment topologies using the Kishino-Hasegawa test. Sequence evolution models were optimized using both genome segments and using the competing topologies (Fig. 3). Regardless of which model was selected, each topology generated using MP and NJ with a given genome segment was significantly more likely than the competing topology generated using the other genome segment (P < 0.001) (Fig. 3). These results indicated that the topologies obtained with each genome segment were significantly different, suggesting that each RNA segment had a different evolutionary history.
FIG. 3.
Comparison between S and M phylogenetic tree topologies for group C viruses. (a) N gene (705 nt) tree showing the three major groups I, II, and III. (b) Gn glycoprotein gene (345 nt) tree. Analyses using NJ and MP methods yielded identical topologies. Numbers inside and outside parentheses indicate percentages of bootstrap support obtained by the NJ and MP analyses, respectively. Horizontal branches are proportional to the scale bar, which represents 5% and 10% nucleotide sequence divergence, respectively. OROV, Oropouche virus.
DISCUSSION
The group C viruses, together with members of the Guama serogroup, were some of the first arboviruses described in the Amazon Region during the early 1950s. Intense serological and ecological studies indicated that these viruses have a complex pattern of antigenic relationships yet circulate in a compact ecosystem that may favor coinfection of mosquito vectors and/or vertebrate hosts with two viruses (27)
Because of these observations, group C viruses were suggested as an ideal model to study reassortment as a natural evolutionary mechanism. The patterns of antigenic relationship were first established based on serological techniques reflecting different viral proteins (CF, HI, and NT). By CF, three related pairs were identified (APEUV-MTBV, CARV-ITQV, and ORIV-MURV); however, using HI and/or NT tests, different pairings were revealed (APEUV-ITQV, ORIV-CARV, and MTBV-MURV). These specific pair associations using multiple tests became a method for rapid identification of new group C viruses and for their grouping into four complexes (Caraparu, Oriboca, Marituba, and Madrid). This classification was determined by HI and/or neutralizing antibody properties that are predominantly encoded in the virus MRNA segment and that suggested natural reassortment (28).
To better understand the genetic relationships among group C viruses, we sequenced the S segment of 13 recognized members and the partial MRNA segments of eight prototype viruses and of strain BeH 5546 were determined. Significantly different phylogenetic tree topologies were obtained for APEUV, MTBV, ORIV, CARV, ITQV, MURV, VIN, and RESV and for strain BeH 5546 using S-versus-M segment sequences, with different pairings (Fig. 3).
The phylogenetic placement of group C viruses in three major groups (I, II, and III) based on nucleocapsid gene sequences is in agreement with serologic relationships determined using the CF test, which reflect antigens of the N protein, encoded in the SRNA segment. However, these groupings differ from the current classification based on the HI and NT test results. ITQV is currently included in the Oriboca complex, APEUV is antigenically related to other group C viruses of the Caraparu complex, and RESV, MURV, and strain BeH 5546 belong to the Marituba complex. However, using S segment sequences, these three viruses grouped with the Caraparu, Marituba, and Oriboca complexes, respectively (Fig. 3). These discrepancies, supported by strong bootstrap values and significantly different likelihood scores, suggest that the S and M segments have different evolutionary histories, reflecting natural reassortment.
The analysis of additional strains of ORIV (Table 1) revealed further evidence of reassortment among group C viruses. All strains analyzed showed high nucleotide sequence identity with their respective prototype strains, except for a single Brazilian strain (BeH 5546) of CARV. This 1956 human isolate was nearly identical (mean of 95.8% nucleotide sequence identity) to the ORIV S segment sequences (Table 4). In addition, the partial Gn nucleotide sequence of strain BeH 5546 shared 98.3% sequence identity with the prototype CARV (strain BeAn 3994) yet only 71.3% identity with the prototype ORIV (strain BeAn 17) (Table 5). These results are in agreement with previous findings of aberrant conflicting antigenic relationships for strain BeH 5546, namely, that it reacts by CF like ORIV and MURV and by HI like CARV and APEUV.
The BRCV strain 77V 14814 was isolated in Sao Paulo State, in the southeast of Brazil, and was identified as a new group C member (3). In our study it showed 99.3% nucleotide sequence identity with the N gene sequence of CARV. This suggests that BRCV obtained its S segment from CARV. Further studies on the BRCV M segment are needed to confirm the reassortant history of this strain.
Genetic and geographic data for the SRNA of group C viruses isolated from different localities in the Americas (Brazil, Peru, French Guiana, Panama, Trinidad, and United States) and from different hosts (humans, mosquitoes, and sentinel animals) suggests an intense traffic of these viruses through the Americas. Phylogenetic topologies also indicate that relationships are not correlated with geographic distributions. For example, closely related sister pairs including MURV and RESV were isolated in different localities, in Brazil and Trinidad; APEUV and GLV were isolated in Brazil and the United States; and CARV and OSSA were isolated in Brazil and Panama, respectively. Virus strains isolated from humans are generally more closely related to each other than to those isolated from arthropods and sentinel animals (Fig. 2).
Evolution of segmented viruses can occur by various mechanisms, such as mutation, genetic recombination, and genetic reassortment (1, 2, 6, 13, 19, 20, 21, 22, 24). However, little is known about the relative contributions of these mechanisms to generating virus biodiversity among orthobunyaviruses and other members of the family Bunyaviridae. Antigenic, ecological, and genetic characteristics of the group C viruses indicate that several of these agents represent natural reassortants. The ecosystems where group C viruses coexist, sometimes sharing the same arthropod vectors, and vertebrate hosts, probably facilitate natural reassortment.
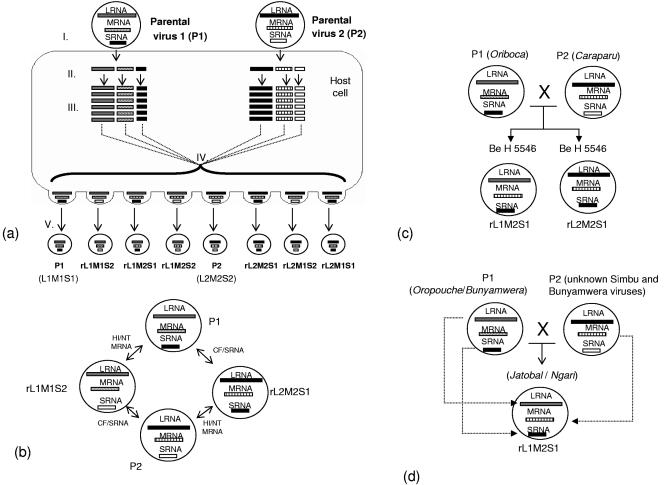
A hypothetic model for reassortment among group C viruses is presented in Fig. 4a. The antigenic and genetic relationships observed for group C viruses suggest a particular reassortment pattern where the reassortants rL1M1S2 and rL2M2S1 are most frequently generated (Fig. 4b). Two hypothetical origins of strain BeH 5546 are presented in Fig. 4c: (i) strain BeH 5546 received its MRNA from CARV and its S and L segments from ORIV; (ii) strain BeH 5546 received its SRNA from ORIV and its M and L segments from CARV. Nucleotide sequences for the LRNA segments of the three viruses involved (CARV, ORIV, and BeH 5546) are needed to test these hypotheses.
FIG. 4.
(a) Hypothetical model of orthobunyavirus reassortment pattern. (Part I) Coinfection of a host cell with parental viruses 1 (P1) and 2 (P2). (Part II) Uncoating of segmented virus genomes. (Part III) Replication of segmented genomes in the host cell. (Part IV) Reassortment of genome segments. (Part V) Progeny virus with independently reassorted genome segments. (b) Genetic relationships among hypothetical parental viruses (P1 and P2) and possible reassortant progeny viruses (rL1M1S2 and rL2M2S1) involved in the reassortment process of group C viruses. (c) Hypothetical reassortment patterns for strain BeH 5546. (d) Hypothetical pattern for Jatobal virus and Ngari virus.
Our group C virus reassortment model can also be applied to identify the reassortment pattern for Jatobal (Simbu group) and Ngari (Bunyamwera group) orthobunyaviruses. Our model suggests that both are rL1M2S1 reassortant progenies, since they received the S and L segments from Oropouche virus and Bunyamwera virus, and the M segment from unknown Simbu and Bunyamwera members, respectively (Fig. 4d) (11, 22).
Our findings represent the first genetic data for group C viruses and provide a better understand of the relationships among these viruses, as well as those of other orthobunyaviruses. They also show a good correlation between antigenic, ecological, and genetic data and should improve our understanding of the molecular epidemiology and evolution of arboviruses. However, a more complete understanding of the interactions between ecosystems, host associations, and antigenic and genetic relationships and their effects on arbovirus requires further study.
Acknowledgments
We are grateful to Ottis R. Causey, Calixta E. Causey, and Robert E. Shope (in memoriam), who led a team of researchers and technicians of the Instituto Evandro Chagas and Rockefeller Foundation Virus Laboratory during the early 1950s, for all available ecological and antigenic data about group C viruses. Their work inspired our genetic analyses. We also thank Evonnildo Gonçalves and Maria Paula Schneider from the DNA Polymorphism Laboratory of Federal University of Para, Department of Genetics, for the initial analysis of the group C virus sequences.
This work was supported in part by NIH contract N01-AI30027, CNPq grants 302770/02-0 and 550275/01-0, and a CAPES doctoral grant (MRTN).
REFERENCES
- 1.Archer, A. M., and R. Rico-Hesse. 2002. High specific genetic divergence and recombination in arenaviruses from the Americas. Virology 304:274-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bishop, D. H., and B. J. Beaty. 1988. Molecular and biochemical studies of the evolution, infection and transmission of insect bunyaviruses. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 311:326-338. [DOI] [PubMed] [Google Scholar]
- 3.Calisher, C. H., T. L. Coimbra, O. S. Lopes, D. J. Muth, L. E. A. Sacchetta, D. B. Francy, J. S. Lazuick, and C. B. Cropp. 1983. Identification of new Guama and group C serogroup bunyaviruses and an ungrouped virus from Southern Brazil. Am. J. Trop. Med. Hyg. 32:424-431. [DOI] [PubMed] [Google Scholar]
- 4.Casals, J., and L. Whitman. 1961. Group C, a new serological group of hitherto undescribed arthropod-borne viruses. Immunological studies. Am. J. Trop. Med. Hyg. 10:250-258. [DOI] [PubMed] [Google Scholar]
- 5.Causey, O. R., C. E. Causey, O. M. Maroja, and D. G. Macedo. 1961. The isolation of arthropod-born viruses including members of two hitherto undescribed serological groups, in the Amazon Region of Brazil. Am. J. Trop. Med. Hyg. 10:227-249. [DOI] [PubMed] [Google Scholar]
- 6.Charrel, R. N., H. Feldmann, C. F. Fulhorst, R. Khelifa, R. de Chesse, and X. de Lamballerie. 2002. Phylogeny of New World arenavirus based on the complete coding sequences of the small genomic segment identified an evolutionary lineage produced by intrasegmental recombination. Biochem. Biophys. Res. Commun. 296:1118-1124. [DOI] [PubMed] [Google Scholar]
- 7.Coimbra, T. L. M., I. M. Rocco, A. Susuki, L. E. Pereira, L. T. M. Souza, E. S. Nassar, I. B. Ferreira, and E. Chamelet. 1998. Arthropod and rodent-borne viruses detected in São Paulo state, Brazil, p. 168-176. In A. P. A. Travassos da Rosa, J. F. S. Travassos da Rosa, and P. F. C. Vasconcelos (ed.), An overview of arbovirology in Brazil and neighbouring countries. Instituto Evandro Chagas, Belém, Brazil.
- 8.Dunn, E. F., D. C. Pritlove, and R. M. Elliott. 1994. The SRNA genome segments of Batai, Cache Valley, Guaroa, Kairi, Lumbo, Main Drain and Northway Bunyaviruses: sequence determination and analysis. J. Gen. Virol. 70:597-608. [DOI] [PubMed] [Google Scholar]
- 9.Elliott, R. M., M. Bouloy, C. H. Calisher, R. Goldbach, J. T. Moyer, S. T. Nichol, R. Pettersson, A. Plyusnin, and C. S. Schmaljohn. 2000. Family Bunyaviridae, p. 599-621. In M. H. V. van Regenmortel, C. M. Fauguet, and D. H. L. Bishop (ed.), Virus taxonomy: seventh report of the International Committee on Taxonomy of Viruses. Academic Press, San Diego, Calif.
- 10.Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 11.Gerrard, S. R., L. Li, A. D. T. Barrett, and S. T. Nichol. 2004. Ngari virus is a Bunyamwera virus reassortant that can be associated with large outbreaks of hemorrhagic fever in Africa. J. Virol. 78:8922-8926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goldman, N., J. P. Anderson, and A. G. Rodrigo. 2000. Likelihood-based test of topologies in phylogenetics. Syst. Biol. 49:652-670. [DOI] [PubMed] [Google Scholar]
- 13.Holland, J., and E. Domingo. 1998. Origin and evolution of viruses. Virus Genes 16:13-21. [DOI] [PubMed] [Google Scholar]
- 14.Karabatsos, N. 1985. International catalogue of arbovirus including certain other viruses of vertebrates. American Society of Tropical Medicine and Hygiene, San Antonio, Tex.
- 15.Kumar, S., K. Tamura, and M. Nei. 2000. Molecular evolutionary genetic analysis. Version 1.01. The Pennsylvania State University, Philadelphia.
- 16.Pinheiro, F. P. 1981. Arboviral zoonoses in South America, p. 159. In J. H. Steele and G. W. Beron (ed.), CRC handbook series in zoonoses, section B: viral zoonoses. CRC Press, Inc., Boca Raton, Fla.
- 17.Pinheiro, F. P., A. P. A. Travassos da Rosa, R. B. Freitas, J. F. S. Travassos da Rosa, and P. F. C. Vasconcelos. 1986. Aspectos clínico—epidemiológicos dos arbovírus. Instituto Evandro Chagas: 50 anos de contribuição às ciências biológicas e à medicina tropical, Belém, Brazil, vol. 1, p. 375-408. Fundação Serviços de Saúde Publica, Belém, Brazil.
- 18.Pinheiro, F. P., and A. P. A. Travassos da Rosa. 1994. Arboviral zoonoses of Central and South America, p. 214-217. In G. Beran (ed.), Handbook of zoonoses. CRC Press, Inc., Boca Raton, Fla.
- 19.Pringle, C. R. 1996. Genetics and genome segment reassortment, p. 189-226. In R. M. Elliott (ed.), The Bunyaviridae. Plenum Press, New York, N.Y.
- 20.Pringle, C. R., J. F. Lees, W. Clark, and R. E. Elliott. 1984. Genome subunit reassortment among bunyaviruses analysed by dot hybridization using molecularly cloned complementary DNA probes. Virology 135:244-256. [DOI] [PubMed] [Google Scholar]
- 21.Rodriguez, L. L., J. H. Owens, C. J. Petters, and S. T. Nichol. 1998. Genetic reassortment among viruses causing hantavirus pulmonary syndrome. Virology 242:99-106. [DOI] [PubMed] [Google Scholar]
- 22.Saeed, M. F., H. Wang, M. Suderman, D. W. Beasley, A. P. A. Travassos da Rosa, L. Li, R. E. Shope, R. B. Tesh, and A. D. T. Barrett. 2001. Jatobal virus is a reassortant containing the small RNA of Oropouche virus. Virus Res. 77:25-30. [DOI] [PubMed] [Google Scholar]
- 23.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstruction phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 24.Sall, A., P. M. Zanotto, O. K. Sene, and H. G. Zeller. 1999. Genetic reassortment of Rift Valley fever virus in nature. J. Virol. 73:8196-8200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmaljohn, C. S., and J. W. Hooper. 2001. Bunyaviridae: the viruses and their replication, p. 1581-1602. In B. N. Fields, D. M. Knipe, P. M. Howley, and D. E. Griffin (ed.), Fields' virology. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 26.Shope, R. E., and L. Whitman. 1966. Nepuyo virus, a new group C agent isolated in Trinidad and Brazil. II Serological studies. Am. J. Trop. Med. Hyg. 15:772. [DOI] [PubMed] [Google Scholar]
- 27.Shope, R. E., J. P. Woodall, and A. P. A. Travassos da Rosa. 1988. The epidemiology of disease caused by viruses in group C and Guamá (Bunyaviridae), p. 37-52. In T. P. Monath (ed.), The arboviruses: epidemiology and ecology. CRC Press, Inc., Boca Raton, Fla.
- 28.Shope, R. E., and O. R. Causey. 1962. Further studies on the serological relationships of group C arthropod-borne viruses and the association of these relationships to rapid identification of types. Am. J. Trop. Med. Hyg. 11:283-290. [DOI] [PubMed] [Google Scholar]
- 29.Spence, L., C. R. Anderson, T. H. Aitken, and W. G. Downs. 1966. Nepuyo virus, a new group C agent isolated in Trinidad and Brazil. I. Isolation and properties of the Trinidadian strain. Am. J. Trop. Med. Hyg. 15:71-74. [DOI] [PubMed] [Google Scholar]
- 30.Spence, L., A. H. Jonkers, and L. S. Grant. 1968. Arboviruses in the Caribbean Islands. Prog. Med. Virol. 10:415-486. [PubMed] [Google Scholar]
- 31.Swofford, D. L. 1998. PAUP: Phylogenetic Analysis Using Parsimony (and other methods), version 4. Sinauer Associates, Sunderland, Mass.
- 32.Vasconcelos, P. F. C., A. P. A. Travassos da Rosa, F. P. Pinheiro, R. E. Shope, J. F. Travassos da Rosa, S. G. Rodrigues, N. Degallier, and E. S. Travassos da Rosa. 1998. Arboviruses pathogenic for man in Brazil, p. 72-99. In A. P. A. Travassos da Rosa, J. F. S. Travassos da Rosa, and P. F. C. Vasconcelos (ed.), An overview of arbovirology in Brazil and neighbouring countries. Instituto Evandro Chagas, Belém, Brazil.
- 33.Woodall, J. P. 1967. Vírus research in Amazônia. Atas Simpósio Sobre Biota Amazônica 6:31-63. [Google Scholar]