Abstract
J virus (J-V) was isolated from feral mice (Mus musculus) trapped in Queensland, Australia, during the early 1970s. Although studies undertaken at the time revealed that J-V was a new paramyxovirus, it remained unclassified beyond the family level. The complete genome sequence of J-V has now been determined, revealing a genome structure unique within the family Paramyxoviridae. At 18,954 nucleotides (nt), the J-V genome is the largest paramyxovirus genome sequenced to date, containing eight genes in the order 3′-N-P/V/C-M-F-SH-TM-G-L-5′. The two genes located between the fusion (F) and attachment (G) protein genes, which have been named the small hydrophobic (SH) protein gene and the transmembrane (TM) protein gene, encode putative proteins of 69 and 258 amino acids, respectively. The 4,401-nt J-V G gene, much larger than other paramyxovirus attachment protein genes sequenced to date, encodes a putative attachment protein of 709 amino acids and distally contains a second open reading frame (ORF) of 2,115 nt, referred to as ORF-X. Taken together, these novel features represent the most significant divergence to date from the common six-gene genome structure of Paramyxovirinae. Although genome analysis has confirmed that J-V can be classified as a member of the subfamily Paramyxovirinae, it cannot be assigned to any of the five existing genera within this subfamily. Interestingly, a recently isolated paramyxovirus appears to be closely related to J-V, and preliminary phylogenetic analyses based on putative matrix protein sequences indicate that these two viruses will likely represent a new genus within the subfamily Paramyxovirinae.
Members of the family Paramyxoviridae are pleomorphic enveloped viruses possessing a nonsegmented negative-strand (NNS) RNA genome (29). They are divided into two subfamilies, Paramyxovirinae and Pneumovirinae, and until relatively recently, viruses within the subfamily Paramyxovirinae were classified into three genera: Respirovirus, Morbillivirus, and Rubulavirus. Since 1970 several novel paramyxoviruses have been isolated from a wide variety of aquatic and terrestrial animal species, including humans, and in many instances these isolations have been done in response to spectacular disease outbreaks. During the last decade, genome characterization of these novel viruses has illustrated the presence of much greater genetic diversity within the subfamily Paramyxovirinae than previously recognized. Although this has led to the creation of two new genera, Henipavirus and Avulavirus, several of the recently characterized viruses remain unclassified below the subfamily level (34, 61).
Since 1994 four new bat-associated paramyxoviruses have been isolated in Australia and Asia, three of which have caused zoonotic disease outbreaks. Hendra virus (HeV) and Nipah virus (NiV) were first isolated in 1994 and 1999, respectively (5, 40), and have caused several fatal disease outbreaks in both animals and humans. Menangle virus (MenV) was isolated in 1997 from stillborn piglets (43) and was subsequently associated with influenza-like illness in two humans; Tioman virus (TiV) was isolated in 2000 from the urine of island flying foxes (6) and has yet to be associated with disease. Genome characterization revealed that HeV and NiV are closely related, and they are now classified as the prototypic members of the new genus Henipavirus (16, 34, 60), while MenV and TiV were found to be closely related novel members of the genus Rubulavirus (2, 6, 7).
Several other recently characterized paramyxoviruses, isolated from a diverse range of animal species, have also contributed significantly to the genetic diversity of the subfamily Paramyxovirinae. Fer-de-lance virus (FDLV) was isolated from the lung of a fer-de-lance viper that died during an outbreak of disease on a Swiss snake farm in 1972 (8). Phylogenetic analyses did not consistently associate FDLV with any of the existing genera within the subfamily Paramyxovirinae, and it has been proposed that the snake paramyxoviruses should constitute a new genus (26). Tupaia paramyxovirus (TPMV) was isolated from the kidneys of an apparently healthy tree shrew of the species Tupaia belangeri captured in Thailand during the late 1970s (54, 62). Although protein sequence similarities and genome features indicate that TPMV is most closely related to members of the Henipavirus and Morbillivirus genera, it cannot be assigned to either of these genera (53, 54). Salem virus (SalV) was isolated from equine mononuclear cells collected during a disease outbreak that occurred simultaneously at three racetracks in New Hampshire and Massachusetts in 1992 (45). Epidemiological studies did not, however, identify a causal relationship between SalV and the outbreak. Genome characterization indicated that SalV is most closely related to the morbilliviruses and, to a lesser extent, to TPMV and HeV (45).
At least three novel paramyxoviruses have been isolated from rodents since the early 1960s. Nariva virus (NaV) was isolated on four occasions form the pooled organs of a forest rodent species trapped in Eastern Trinidad during 1962 and 1963 (55). It was classified as a paramyxovirus based on virion morphology and nucleocapsid structure (59) but has yet to be characterized molecularly and remains unclassified below the family level. Mossman virus (MoV) was isolated on two occasions during the early 1970s from the pooled organs of native rats trapped in Queensland, Australia (4). Recent characterization of the full-length MoV genome revealed it to be a novel member of the subfamily Paramyxovirinae and most closely related to TPMV (37). J virus (J-V) was isolated on four occasions by kidney autoculture from moribund mice (Mus musculus) trapped in 1972 in northern Queensland, Australia (23, 35). It was reported that all four mice from which the virus was isolated had extensive hemorrhagic lung lesions (23). Syncytium formation was observed in kidney autoculture monolayers, and electron microscopy revealed virion morphology and nucleocapsid structure typical of the paramyxoviruses. The name J virus was assigned in reference to M. H. Jun, the virologist responsible for much of the initial virus characterization. A serological study investigating the presence of antibodies against J-V in Australian mammals was carried out during the 1970s (23). Serum neutralizing antibodies were detected in 27/96 wild mice, 17/106 wild rats, 13/107 pigs, 1/157 cattle, and 2/91 humans. A smaller-scale serological survey carried out on North American mammals failed to detect antibodies against J-V in laboratory mice, humans, or hamsters. A number of J-V animal infection experiments were also carried out during the mid-1970s (22, 23). Intranasal and subcutaneous inoculation of J-V into weanling laboratory mice and rats was not associated with clinical signs of disease. Autopsies up to 3 weeks postinoculation revealed various degrees of hemorrhagic interstitial pneumonia, and virus was isolated from blood, lungs, and pooled liver, kidney, and spleen samples. Intranasal J-V inoculation of four pigs did not cause any clinical signs of disease or pathology. No virus was isolated from any of the four pigs, although two did develop significant serum neutralizing antibody levels.
Although the disease-causing potential of J-V remains somewhat unclear, we believed that characterization of the J-V genome could provide valuable information to aid in understanding of virus evolution within the family Paramyxoviridae. Here we describe the complete genome sequence of J-V, the largest paramyxovirus genome characterized to date. Containing eight genes, the J-V genome has a unique structure and represents a substantial contribution to the genetic diversity of the family Paramyxoviridae.
MATERIALS AND METHODS
Virus culture and RNA isolation.
J-V supplied in RK13 cells by the Queensland Department of Primary Industries (passage history unknown) was passaged once in Vero cells prior to three rounds of plaque purification in Vero cells. Vero cells were used for all subsequent J-V propagation. Monolayers were infected with J-V at a multiplicity of 0.005 50% tissue culture infective dose/cell, and at 3 days postinfection, J-V-infected cell cultures were processed by one of two methods. (i) For isolation of mRNA to be used in the cDNA subtraction protocol, infected cells were trypsinized and then pelleted at 1,500 × g for 10 min. Cytoplasmic RNA was extracted from infected and noninfected cells using the RNeasy Midi kit (QIAGEN, Germany). Quantities of cytoplasmic RNA extracted were measured by spectrophotometry using the GeneQuant II RNA/DNA calculator (Pharmacia). Poly(A)-containing mRNA was purified from cytoplasmic RNA using the Oligotex mRNA Midi kit (QIAGEN) and quantified as described above. (ii) For isolation of viral genomic RNA, the infected cell culture supernatant was decanted and centrifuged at 1,500 × g for 10 min to remove cellular debris, and the resulting supernatant was centrifuged at 206,000 × g for 25 min in a Beckman type 55.2 Ti rotor to pellet virus. Total RNA was extracted from crude virus pellets by using the RNeasy Mini kit (QIAGEN) and was quantified using the GeneQuant II RNA/DNA calculator. When J-V genomic RNA was prepared for use in the cDNA subtraction protocol, MoV genomic RNA was concurrently prepared from crude MoV pellets under similar conditions.
Isolation and characterization of viral cDNA using cDNA subtraction.
The Clontech PCR-Select cDNA subtraction kit (BD Biosciences) was used to select virus-specific fragments from (i) differentially expressed mRNA molecules in virus-infected cells (mRNA subtraction) and (ii) viral genomic RNA harvested from tissue culture supernatants (genomic subtraction), as described previously (2, 6, 37). Briefly, for mRNA subtraction, double-stranded cDNA was made using oligo(dT) primers and mRNA from J-V-infected cells (tester cDNA) and noninfected cells (driver cDNA), respectively. After digestion with RsaI, tester cDNA was ligated to kit adaptors 1 and 2R in separate reactions before being mixed with excess driver cDNA. Two hybridizations were performed, after which tester-specific cDNA fragments were amplified by primary and nested PCR. A slightly modified cDNA subtraction strategy was used for genomic subtraction. Double-stranded cDNA was made using random hexamer oligonucleotide primers and total RNA from pelleted J-V (tester cDNA) and MoV (driver cDNA), respectively. Digestion, adaptor ligation, hybridization, and PCRs were then carried out as described above. Nested PCR products from both subtractions were electrophoresed on 1% agarose gels and subsequently size purified in three fractions (0.2 to 0.6 kb, 0.6 to 1.0 kb, and 1.0 to 2.0 kb) using the QIAquick Gel Extraction kit (QIAGEN). Size-purified PCR products were digested with EagI and cloned into the pZErO-2 vector (Invitrogen) at the NotI site. Vector-insert ligation mixes were electroporated into Escherichia coli strain TOP10, and colonies were selected on Low Salt LB agar containing 50 μg/ml kanamycin. Colonies were randomly picked from each size-fractionated cloning experiment, and colony PCR was carried out to identify insert-containing clones. Insert sequences were obtained by DNA sequencing of either the purified colony PCR product (QIAquick Gel Extraction kit; QIAGEN) or purified plasmid DNA (QIAprep Spin Miniprep kit; QIAGEN).
Amplification of viral genomic RNA.
Total RNA was extracted from crude virus pellets as described above. cDNA was synthesized using the Omniscript RT kit (QIAGEN) with random hexamer primers at low concentrations (7.5 to 15 ng per μl reaction mix) to favor the production of relatively long cDNA molecules. Combinations of different types of oligonucleotide primers were used to synthesize PCR products covering the J-V genome (with the exception of 117 nucleotides [nt] at the genome 3′ end). These primers included (i) J-V-specific primers designed from known J-V sequence, (ii) J-V intergenic-region (IGR) primers designed based on highly conserved sequences at the known J-V gene borders (forward primer, 5′ CGCTAAGAAAAACTTAGGAGT; reverse primer, 5′ CGCACTCCTAAGTTTTTCTTA), (iii) a degenerate morbillivirus-specific genome terminus primer designed based on the 3′ and 5′ genome terminus sequences, highly conserved in all morbilliviruses (5′ CGCGGATCCACCARACAAAGTTGG), and (iv) a genome terminus primer based on the 3′ and 5′ genome terminus sequences of TPMV (5′ CGCGGATCCACCAGAAAAGG). All primers were synthesized by a commercial provider (GeneWorks, Australia). Platinum PCR SuperMix (Invitrogen) was used to perform PCR on the cDNA template synthesized as described above, and each 25-μl reaction mixture contained 20 pmol of each primer and 1 μl of the undiluted cDNA template. PCRs were carried out in a Gene Amp 2400 thermocycler using cycling parameters of 94°C for 100 s, followed by 35 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 1 to 2 min, and a final 2- to 5-min extension at 72°C. PCR products were visualized with ethidium bromide on 1 to 2% agarose gels and were purified using either the QIAquick PCR purification kit or the QIAquick Gel Extraction kit (both from QIAGEN). Purified PCR products were either sequenced directly or cloned into the pDrive vector (QIAGEN) for sequencing.
Characterization of genome termini.
A protocol adapted from a published 5′ rapid amplification of cDNA ends (5′ RACE) method (56) was used to obtain the sequence for 117 nt at the 3′ end of the J-V genome and the exact sequence of the 13 nt at the 5′ genome terminus. Total RNA was extracted from crude virus pellets (containing both genome and antigenome RNA) as described above. Single-strand cDNA synthesis, purification, and ligation of a single-strand anchor oligonucleotide (5′ GAAGAGAAGGTGGAAATGGCGTTTTGG, 5′ phosphorylated and 3′ blocked) was performed as described previously (37, 56). The ligation products were amplified by PCR using J-V-specific primers, nested with respect to the cDNA primer, and a 27-nt primer complementary in sequence to the anchor. When required, a heminested PCR, using the same anchor primer and an additional nested J-V-specific primer, was also performed. PCR products obtained were gel purified using the QIAquick Gel Extraction kit (QIAGEN) and were either sequenced directly or cloned into the pDrive vector (QIAGEN) for sequencing.
DNA sequencing.
Purified PCR products and plasmid DNA were sequenced using the BigDye Terminator v1.0 kit and an ABI PRISM 377 DNA sequencer (both from Applied Biosystems) in accordance with the manufacturer's instructions.
Analysis of nucleotide and deduced amino acid sequences.
Sequence data were routinely managed and processed using the Clone Manager 7 and Align Plus 5 programs in the Sci Ed Central software package (Scientific and Educational Software). Multiple nucleotide sequence alignments were performed using the Standard Linear scoring matrix, and the Blosum 62 matrix was used for multiple amino acid sequence alignments. Sequence similarity searches were conducted using the BLAST service at the National Center for Biotechnology Information (NCBI). Amino acid sequence identities of deduced J-V proteins with cognate proteins of other members of the Paramyxovirinae were determined by pairwise alignment (ClustalW [accurate] [52]) using MegAlign 4.00 in the Lasergene software package (DNASTAR Inc.).
Phylogenetic analyses were undertaken using programs available through the BioManager by ANGIS web interface (http://www.angis.org.au). Sequences were aligned by ClustalW (accurate) with published sequences from other members of the Paramyxovirinae, and evolutionary relationships between them were estimated using programs in the PHYLIP software package, version 3.2 (13). Modified data sets were generated by bootstrap resampling (100 replicates) using the Seqboot program, and both modified and unmodified data were analyzed by distance matrix (Protdist and Neighbor) programs. Consensus phylogeny trees were derived using Consense and drawn using DrawTree and TreeView (41).
Database accession numbers.
The full-length genome sequence of J-V has been deposited in GenBank under accession number AY900001. Accession numbers for other sequences used in this study are given below. For viruses where a full-length genome sequence was not available, individual gene sequences were used and are indicated by the abbreviated gene letter in parentheses following the accession number. Virus sequences used were as follows: avian paramyxovirus type 6 (APMV6), AY029299; bovine parainfluenza virus 3 (bPIV3), AF178654; bovine respiratory syncytial virus (bRSV), AF092942; canine distemper virus (CDV), AF014953; cetacean morbillivirus (CMV) strain dolphin morbillivirus (DMV), X75961 (N), Z47758 (P/V/C), Z30087 (M), Z30086 (F), and Z36978 (H); FDLV, AY141760; HeV, AF017149; human parainfluenza virus 1 (hPIV1), AF457102; hPIV2, X57559; hPIV3, AB012132; hPIV4a, M32982 (N), M55975 (P/V), D10241 (M), D49821 (F), and M34033 (HN); hPIV4b, M32983 (N), M55976 (P/V), D10242 (M), D49822 (F), and AB006958 (HN); human respiratory syncytial virus (hRSV), AF013254; measles virus (MeV), AB016162; MenV, AF326114 (N, P/V, M, F, HN); MoV, AY286409; mumps virus (MuV), AB040874; Newcastle disease virus (NDV) strain Beaudette C (for the 3′ leader sequence, see reference 27), AF064091 (N), X60599 (P/V), X04687 (M), X04719 (F), X04355 (HN), and X05399 (L); NiV, AF212302; peste-des-petits-ruminants virus (PPRV), X74443 (N), AJ298897 (P/V/C), Z47977 (M), Z37017 (F), and Z81358 (H); phocine distemper virus (PDV), X75717 (N), D10371 (P/V/C, M, F, H), and Y09630 (L); rinderpest virus (RPV), Z30697; SalV, AF237881 (N, P/V/C); Sendai virus (SeV), AB005795; simian virus 5 (SV5), AF052755; TiV, AF298895; TPMV, AF079780.
RESULTS
Characterization of the J-V genome.
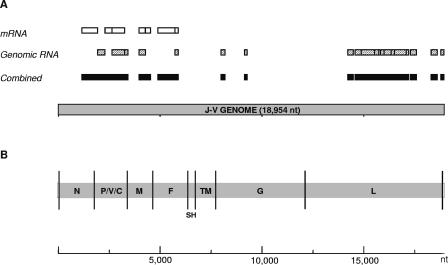
To obtain initial J-V-specific sequence, cDNA subtraction strategies were used. First, mRNA-based subtraction was used to isolate differentially expressed cDNA fragments synthesized from J-V-infected cells (2, 6). From the most successful cloning experiment, 30% of inserts sequenced were identified as virus specific by using BLASTx homology searches. All the virus “hits” were to paramyxovirus N, P, V, M, and F proteins, encoded by genes within the 3′ half of the genome (Fig. 1A). In an attempt to obtain sequence from the 5′ half of the J-V genome, genomic RNA-based subtraction was performed. The percentage of clones sequenced that were virus specific increased to 60%, and these were more evenly spread over the length of the genome, including numerous matches to the L protein (Fig. 1A). When the J-V-specific clones obtained by both subtraction strategies were combined, they covered a significant proportion of the genome (42%), from nt 1160 to nt 18934. PCR products were generated to fill in the gaps between cloned fragments and to cover the full genome length, using as a template randomly primed cDNA transcribed from J-V genomic RNA. A 5′ RACE method, which took advantage of the presence of both genome and antigenome RNA in crude J-V pellets, was used to elucidate the 5′ and 3′ genome termini. Every nucleotide in the J-V genome was sequenced with a minimum threefold redundancy, at least once in each direction and at least once directly from the PCR template without prior cloning.
FIG. 1.
Genome characterization and organization. (A) Schematic diagram of the J-V genome showing the genome fragments obtained by cDNA subtraction. Open boxes (top row), genome fragments obtained by mRNA-based subtraction; diagonally striped boxes (second row), fragments obtained by genomic RNA-based subtraction; solid boxes (at bottom), genome fragments obtained by both cDNA subtraction strategies combined. (B) J-V genome organization revealed by complete genome sequence analysis. The 3′ leader and 5′ trailer regions are boxed, genes are labeled, and the trinucleotide intergenic regions are represented by vertical black lines.
Genome features.
At 18,954 nt, the J-V genome is the largest in the family Paramyxoviridae sequenced to date, over 700 nucleotides longer than the next largest member, NiV (18,246 nt).
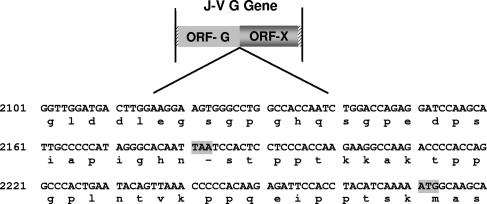
The J-V genome is composed of eight genes in the order 3′-N-P/V/C-M-F-SH-TM-G-L-5′ (Fig. 1B; Table 1). The two novel genes SH and TM, located between the F and G genes, encode putative proteins of 69 and 258 amino acids (aa), respectively. The J-V SH (small hydrophobic) protein gene was so named because it is in a position within the genome similar to that of the SH genes of other Paramyxovirinae, and the encoded SH protein shows significant similarity to the SH protein of MuV with respect to size and hydrophilicity profile. TM represents the “transmembrane” protein gene and refers to the predicted transmembrane association of the encoded 258-aa protein. No other member of the subfamily Paramyxovirinae has this genomic organization; the genome arrangement most similar to that of J-V is that of the rubulaviruses SV5 and MuV and the avulavirus APMV6 (3′-N-P/V-M-F-SH-HN-L-5′). In addition to the novel genes SH and TM, J-V also has an unusually large attachment protein gene. At 4,401 nt, the J-V G gene is 1,837 nt longer than the HeV G gene, previously the largest within the subfamily Paramyxovirinae. Although at 709 aa the putative J-V G protein is the largest attachment protein encoded by a member of the subfamily Paramyxovirinae, the dramatic increase in the size of the J-V G gene is principally due to the presence of an extensive second open reading frame (ORF), termed ORF-X. ORF-X has been arbitrarily defined as the 2,115-nt ORF commencing immediately after, and in frame with, the stop codon for the putative G protein. A graphical overview of this definition is shown in Fig. 2. ORF-X encodes a putative protein of 704 aa within which the first methionine is the 30th aa residue.
TABLE 1.
Predicted products of transcription and translation for each J-V gene
| Genea | mRNA size (nt)
|
Deduced protein
|
||||
|---|---|---|---|---|---|---|
| Full length | 5′ UTR | ORF | 3′ UTR | Size (aa) | Calculated molecular mass (Da) | |
| N | 1,696 | 46 | 1,569 | 81 | 522 | 58,374 |
| P/V/C (P) | 1,660 | 75 | 1,500 | 85 | 499 | 53,675 |
| P/V/C (C) | 1,660 | 118 | 459 | 1,083 | 152 | 17,715 |
| P/V/C (V) | 1,661 | 75 | 879 | 707 | 292 | 31,274 |
| P/V/C (W) | 1,662 | 75 | 933 | 654 | 310 | 33,742 |
| M | 1,214 | 14 | 1,023 | 177 | 340 | 37,693 |
| F | 1,750 | 66 | 1,635 | 49 | 544 | 59,474 |
| SH | 366 | 83 | 210 | 73 | 69 | 7,665 |
| TM | 996 | 104 | 777 | 115 | 258 | 29,097 |
| G | 4,401 | 53 | 2,130 | 2,218 | 709 | 78,135 |
| L | 6,759 | 68 | 6,615 | 76 | 2,204 | 254,458 |
The P/V/C gene of J-V potentially codes for four gene products by use of alternative reading frames and cotranscriptional mRNA editing. See the text for details.
FIG. 2.
Nucleotide and deduced amino acid sequences at the junction between the J-V G protein ORF (ORF-G) and ORF-X. The nucleotide sequence is presented as cDNA in the antigenome 5′→3′ sense. The single stop codon separating the two open reading frames and the first ATG codon within ORF-X are shaded.
The replication promoters of the Paramyxovirinae are bipartite in nature, consisting of 12 nt at the 3′ genome/antigenome terminus and an internal repeating element in the 13th, 14th, and 15th hexamers downstream (29). Conservation of the 12-nt terminal sequences is strong between members of the same genus, and consequently this feature is one of the criteria used for classifying viruses within the Paramyxovirinae. The downstream hexameric repeat is a 3′-GC dinucleotide for the avulaviruses and rubulaviruses, whereas for other members of the subfamily it is a single C (29). Complementarity between the J-V genome termini is imperfect, with differences at nt 4 and nt 12 (Fig. 3A). The J-V genome terminal sequences show the greatest similarity to those of MeV, and the associated downstream repeating element is like that of the morbilli-, respiro-, and henipaviruses. Consistent with the 3′ genome leader sequences of all Paramyxovirinae sequenced to date, the J-V leader sequence is 55 nt in length (Fig. 3 B). At 36 nt, the 5′ trailer of J-V is most similar in size to those of the henipaviruses (33 nt).
FIG. 3.
Alignment of genome terminal sequences. Dots indicate identical residues. See Materials and Methods for database accession numbers. (A) Alignment of the 3′ leader and 5′ trailer sequences of J-V. The 3′ leader is represented as antigenome written in the 5′→3′ sense, and the 5′ trailer is represented as genome written in the 5′→3′ sense. The first 12 nt of each terminus are shaded. (B) Alignment of 3′ leader sequences of selected members of the subfamily Paramyxovirinae. A gap has been introduced between nt 12 and nt 13, and the N gene transcription initiation site is shaded.
Conserved sequences at the gene borders of paramyxoviruses control the stop and reinitiation mechanism used by the viral RNA polymerase to transcribe individual genes (29). Like the henipaviruses, morbilliviruses, and respiroviruses, J-V has a conserved trinucleotide IGR sequence. The gene start, stop, and IGR sequences of J-V are shown in Table 2. At 18,954 nt, the J-V genome adheres to the “rule of six” (24). Most paramyxovirus genomes replicate efficiently only when they are a multiple of six nucleotides in length, presumably because each nucleocapsid protein associates with exactly six nucleotides (12) and it is the resulting helical nucleocapsid core that serves as the template for RNA synthesis. It has also been found that the hexamer phase position of the initiating nucleotide residue of each gene mRNA molecule is conserved between members of the same genus. J-V has the hexamer phase pattern “2,3,4,3,4,1,4,4.” If the phasing pattern of only the N, P, M, F, G, and L genes (2,3,4,3,4,4) is compared with those of other paramyxoviruses, the most similar to J-V are those of the henipaviruses (2,3,4,4,4,3) (61) and MoV (2,2,4,4,4,4).
TABLE 2.
Gene start and stop signals and IGRs of J-Va
| Virus and genes | Gene stop | IGR | Gene start |
|---|---|---|---|
| J-V | |||
| /N | TTT | AGGAGCAAAG | |
| N/P | TTAAGAAAAA | CTT | AGGAGTAAAG |
| P/M | TTAAGAAAAA | CTT | AGGAGTAAGG |
| M/F | ATAAGAAAAA | CTT | AGGAGTCAAG |
| F/SH | TAAATAAAAA | CTT | AGGACAAAAG |
| SH/TM | TTAGAAAAAA | CTT | AGGAGGAATG |
| TM/G | TTAAGAAAAA | CTT | AGGAGTGAAT |
| G/L | TTAAGAAAAA | CTT | AGGAGTGAAT |
| L/ | TTAAGAAAAA | CTT | |
| Consensus sequences | |||
| J-Vb | WWARDAAAAA | CTT | AGGASnVADK |
| MoVb | YWAHRAAAAA | CKT | AGGRBnMARG |
| TPMV | TTAVRRAAAA | CTT | AGGRnCMAAG |
| Henipaviruses | TWAHRAAAAA | CTT | AGGAnMCARG |
| Morbillivirusesc | HHWHDnAAAA | CKT | AGGRnBMARG |
| Respirovirusesc | DWAHVAAAAA | CYY | AGGRnnAAHG |
The coding strategy of the J-V P gene is most similar to that of the henipaviruses, morbilliviruses, and respiroviruses. The unedited mRNA is predicted to encode the phosphoprotein, and an edited mRNA molecule created by the addition of a single net nontemplated G residue at the editing site is predicted to encode the V protein. If two net nontemplated G residues were added at the editing site, the resulting mRNA molecule would encode the putative W protein. An alternative open reading frame present in both unedited and edited mRNA molecules encodes the putative C protein. The editing site of the J-V P gene (TTAAAAAAGGCA) is identical to that of TPMV and highly conserved with the henipavirus and morbillivirus editing sites. Like those of TPMV and MoV, the J-V editing site differs at one nucleotide from the core editing sequence AAAAAGGG, which is absolutely conserved among all henipaviruses, morbilliviruses, and respiroviruses.
Features of J-V genes common to all other members of the Paramyxovirinae.
As shown in Fig. 1 and Table 1, the majority of J-V genes (i.e., N, P, M, F, and L) have a genome location and molecular features similar to those of the cognate genes of other members of the subfamily Paramyxovirinae.
The J-V N gene is predicted to encode a 522-aa N protein (Table 1). A second open reading frame, beginning 1,213 nt downstream of the initiation codon for the N protein and encoding a putative 56-aa protein, was also identified. Open reading frames similar in size and position have been identified for NiV and MoV, encoding putative proteins of 72 aa and 61 aa, respectively. No significant sequence similarity between the 56-aa J-V protein and either of these proteins or other existing sequenced proteins was found. Like other paramyxovirus N proteins, the N protein of J-V can be divided into three regions on the basis of sequence variability. The central region (aa 171 to 383) is the most conserved, having 58% amino acid identity with MoV, and the C-terminal (aa 384 to 522) region is the least conserved, amino acid identities with other members of the Paramyxovirinae all being below 20%. The highly conserved amino acid sequence F-X4-Y-X3-Φ-S-Φ-A-M-G (where X represents any amino acid residue and Φ represents an aromatic amino acid residue), previously identified within the central region of N proteins of Paramyxovirinae and thought to be involved in N-N self-assembly contacts (29, 39, 63), was also identified in J-V.
As stated above, the J-V P/V/C gene has a coding strategy similar to that of the henipaviruses, morbilliviruses, and respiroviruses and has the capacity to encode up to four different proteins (Table 1). The putative 499-aa J-V P protein is most similar in size to the P proteins of the morbilliviruses (506 to 509 aa). The full-length protein showed limited similarity to the P proteins of other members of the Paramyxovirinae. Interestingly, the first 70 aa of the J-V and henipavirus P proteins showed high levels of similarity; the sequence F-X-Q-K-N-Q-X-X-I-Q-K-T-Y-G-R-S-X-I (aa 22 to 39) was particularly strongly conserved. In SeV, aa residues 33 to 41 of the P protein were found to be essential for genome replication. This 9-aa domain was demonstrated to be necessary for a stable N0-P interaction, apparently enabling the P protein to chaperone unassembled N protein (N0) during assembly of the nascent RNA genome (11). Although not significantly homologous to the 9-aa SeV P-protein domain, perhaps the highly conserved N-terminal region of the J-V and henipavirus P proteins could also play an important role in N0-P interactions. Like the P proteins of the henipa-, morbilli-, and respiroviruses, the J-V P protein has an acidic pI (4.5) and possesses many potential phosphorylation sites. The putative 152-aa J-V C protein is most similar in length to the C proteins of MoV (152 aa) and TPMV (153 aa) and, like most other C proteins, has a basic pI (10.2). Amino acid identities between the J-V C protein and other paramyxovirus C proteins were low. The V-specific region of the deduced 292-aa J-V V protein had significant amino acid identity with those of other members of the Paramyxovirinae; identity was maximal with hPIV4a (50%), bPIV3 (48%), and NiV (47%). Fifteen amino acid residues, including seven cysteine residues, perfectly conserved in all members of the subfamily were also conserved in J-V. The putative 310-aa W protein encoded by the J-V P/V/C gene shows no significant similarity to the W proteins of other paramyxoviruses, nor was any significant similarity identified between it and other known protein sequences in a BLASTp search.
Like matrix proteins of other paramyxoviruses, the deduced J-V matrix protein has a basic pI (9.4) and several hydrophobic domains, probably important factors contributing to ionic interactions with the acidic N proteins and association with cell membranes, respectively (29).
The 544-aa F protein of J-V is predicted to be a type I membrane protein with features similar to those of the F proteins of other paramyxoviruses. The first 18 aa of the J-V F protein are highly hydrophobic and so are predicted to contain the signal sequence, while the predicted transmembrane domain lies between aa 486 and 516, leaving a cytoplasmic tail of 28 aa. The predicted cleavage site of the J-V F protein, GVPGVR, is monobasic and does not conform to the consensus motif for cleavage by furin, R-X-K/R-R (20), conserved in the majority of the Paramyxovirinae. It is most similar to the cleavage sites of SeV (DVPQSR) and NiV (LVGDVR). The N-terminal 25-aa region of the J-V F1 subunit is highly hydrophobic and highly conserved with the fusion peptides of other Paramyxovirinae. Heptad repeats (HR), important for the formation of six-helix bundle structures which are necessary for membrane fusion in paramyxoviruses (47), were also identified in the region of the J-V F1 protein immediately adjacent to the fusion peptide (HRA), as well as in the region proximal to the transmembrane domain (HRB). Five potential sites for N-linked glycosylation of the J-V F protein were identified: one site within the F2 subunit (N64) and the remaining four within F1 (N357, N462, N466, N488). The fourth F1 site, N488, falls just within the putative transmembrane domain and is therefore unlikely to be used.
Alignment with the L proteins of representative members of the Paramyxovirinae demonstrated that the six strongly conserved linear domains described by Poch et al. (44) were also conserved in the J-V L protein. Poch et al. (44) also detected four sequence motifs within domain III of the NNS single-stranded RNA virus L proteins, which are conserved with other RNA-dependent polymerases and may represent elements of the active site for template recognition and/or phosphodiester bond formation. The four motifs are also highly conserved in the J-V L protein, including the functionally important GDNQ sequence within motif C. It is interesting that in this respect J-V contrasts with the other paramyxoviruses with relatively large genomes—the henipaviruses, TPMV, and MoV—which have a GDNE sequence at this site and are the only NNS single-stranded RNA viruses not to conserve the GDNQ sequence.
Genes encoded between the F and L genes of the J-V genome.
As shown in Fig. 1, the genome segment between the F and L genes of J-V displays significant divergence from the genomes of other members of the Paramyxovirinae. The features of the three genes encoded in this region are as follows.
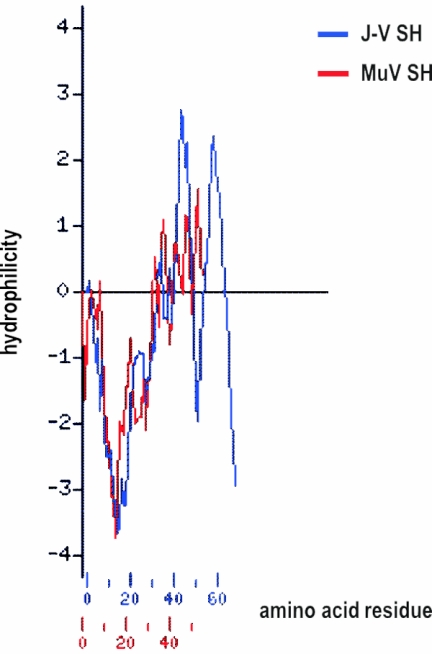
The SH gene is predicted to code for a 69-aa protein with a putative transmembrane domain between amino acid residues 6 and 28. Although no significant sequence similarity of the 69-aa SH protein to any sequenced protein was identified, the size and hydrophilicity profile of the J-V SH protein are very similar to those of the SH proteins of MuV (Fig. 4), bRSV, and hRSV (data not shown). As in the SH protein of MuV, no potential N-linked glycosylation sites were identified within the J-V SH protein sequence.
FIG. 4.
Hydrophilicity profile comparison of the J-V SH protein with the SH protein of MuV. The Kyte-Doolittle hydrophilicity profiles (28) were generated and aligned using the Protein Hydrophilicity/Hydrophobicity Search and Comparison Server provided by the Bioinformatics and Biological Computing Unit at the Weizmann Institute of Science, Rehovot, Israel (http://bioinformatics.weizmann.ac.il/hydroph/). Values above the axis line are hydrophilic; values below the axis line are hydrophobic. Each value is the average of the values of seven adjacent residues and is plotted at the middle residue.
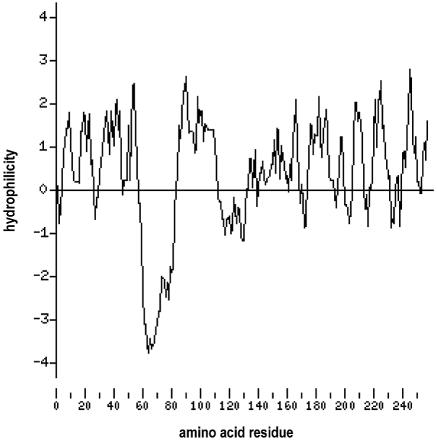
The TM gene, unique to J-V, is predicted to encode a 258-aa protein. No significant amino acid sequence homology was identified between the putative TM protein and any protein in sequence data banks. The hydrophilicity profile of the TM protein (Fig. 5) shows a highly hydrophobic putative transmembrane domain from aa 58 to 80. The HMMTOP server, an automatic server for predicting the topology of transmembrane proteins which is available on the Internet (http://www.enzim.hu/hmmtop/) (57, 58), predicted a transmembrane domain in TM from aa 58 to 81 and predicted that the 177-aa carboxy terminus of the protein would lie outside the cell. It should be noted, however, that this is merely a sequence-based prediction and that substantiating experiments have yet to be undertaken. The putative TM protein is basic, with a calculated pI of 10.0. Two potential N-linked glycosylation sites were identified, the first at aa 55, just N-terminal to the putative transmembrane domain, and the second at aa 167.
FIG. 5.
Hydrophilicity profile of the J-V TM protein generated by Kyte-Doolittle analysis (28) using the Protein Hydrophilicity/Hydrophobicity Search and Comparison Server provided by the Bioinformatics and Biological Computing Unit at the Weizmann Institute of Science, Rehovot, Israel (http://bioinformatics.weizmann.ac.il/hydroph/). Values above the axis line are hydrophilic; values below the axis line are hydrophobic. Each value is the average of the values of seven adjacent residues and is plotted at the middle residue.
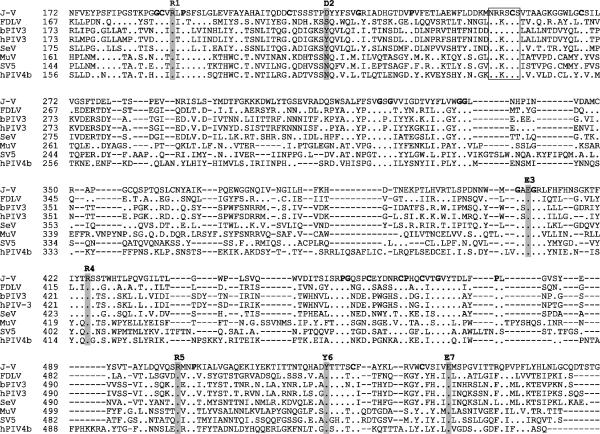
To date J-V has failed to display hemagglutinin or neuraminidase activity; therefore, in keeping with other members of the Paramyxovirinae for which these activities have not been identified, the J-V attachment protein gene has been designated G. The deduced 709-aa J-V G protein is the largest paramyxovirus attachment protein sequenced to date, the next largest being that of TPMV (665 aa). As inferred from the sequence alignment in Fig. 6, the increase in size is due mainly to the extension of the C-terminal region of the protein. Like the attachment proteins of other members of the Paramyxovirinae, the G protein of J-V is predicted to be a type II integral membrane protein, with a hydrophobic putative signal sequence/transmembrane domain lying between amino acid residues 31 and 59. Five potential sites for the addition of N-linked carbohydrate were identified: N51 (within the putative transmembrane domain and therefore unlikely to be used), N131, N619, N636, and N677. Conservation of structural determinants, such as the position of the hydrophobic transmembrane domain and cysteine, proline, and glycine residues, was strong between the J-V G protein and attachment proteins of other members of the Paramyxovirinae. Sequence and structural alignments of a wide range of viral, bacterial, protozoan, and eukaryotic neuraminidase and transneuraminidase proteins carried out by Langedijk et al. (30) identified seven common neuraminidase active-site residues. Six of these seven active-site residues are conserved in respirovirus and rubulavirus HN proteins, while four are conserved in the morbillivirus H proteins. The alignment of globular head regions of different paramyxovirus attachment proteins shown in Fig. 6 suggests that all seven active-site residues, including D2, which has no homologue in other paramyxovirus attachment proteins, may be present in the J-V G protein. The hexapeptide sequence NRKSCS, conserved in almost all respirovirus, rubulavirus, and avulavirus HN proteins and thought to form part of the sialic acid binding site (21, 38), has one conservative amino acid substitution in the J-V G protein (NRRSCS). Although the high level of conservation of neuraminidase active-site residues and the hexapeptide sialic acid binding site motif in the putative J-V G protein suggest that it is likely to possess neuraminidase activity, this was not observed experimentally on a fetuin substrate (data not shown). Conclusive definition of J-V neuraminidase activity awaits the outcome of alternative assays based on a range of different substrates. As stated earlier, the J-V G gene has an extraordinarily long 2,218-nt putative 3′ untranslated region (UTR) in which a 2,115-nt ORF, designated ORF-X, extends in frame immediately after the stop codon of the putative attachment protein ORF (Fig. 2). No significant level of amino acid identity was found between the encoded 704-aa X protein and any sequenced protein in a BLASTp search.
FIG. 6.
Sequence alignment of the globular head region of the J-V G protein with the HN proteins of FDLV and selected members of the Respirovirus and Rubulavirus genera. The seven proposed neuraminidase active-site residues are shaded and numbered. The putative sialic acid binding site hexapeptide is boxed. Proline, glycine, and cysteine residues conserved among all the attachment proteins in the alignment are boldfaced. Amino acid residue numbers for each protein are shown to the left of each sequence. Dots indicate identical residues, and dashes represent gaps introduced during sequence alignment. See Materials and Methods for database accession numbers.
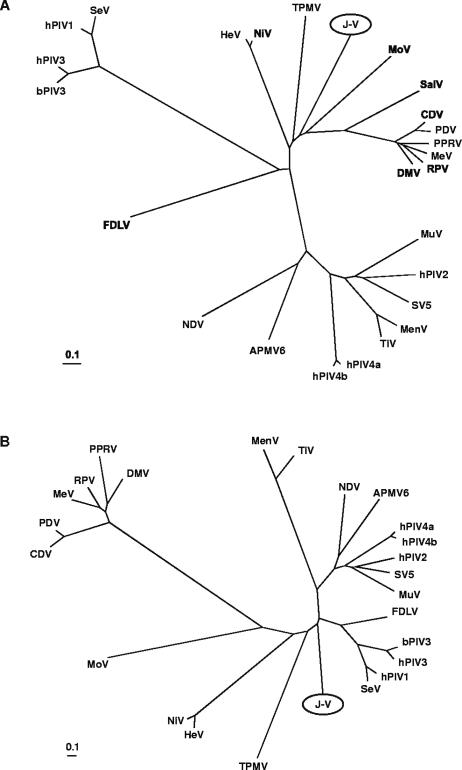
Phylogenetic analysis.
Phylogenetic analyses were carried out on full-length J-V N, P, M, F, G, and L protein sequences. Distance matrix methods were used to generate phylogenetic trees from alignments of J-V proteins with protein sequences of other members of the Paramyxovirinae. These trees clearly showed that J-V cannot be classified into any of the five existing genera within the subfamily. Along with MoV, TPMV, SalV, and the Henipavirus genus, J-V falls between the Respirovirus and Morbillivirus genera. Trees based on the N, P, M, and L proteins placed J-V between the henipavirus and morbillivirus clusters (Fig. 7A), whereas in the F and attachment protein trees, J-V was positioned between the henipavirus and respirovirus clusters. The tree based on the attachment protein sequences was unique in that J-V was estimated to be more closely related to the Respirovirus genus than to either the Henipavirus or the Morbillivirus genus (Fig. 7B). This suggests a somewhat different evolutionary pathway of the J-V attachment protein compared to the N, P, M, F, and L proteins. A similar observation has been made for the rubulaviruses MenV and TiV: while phylogenetic trees for the MenV and TiV N, P, M, and F protein sequences clearly place these viruses within the rubulavirus cluster, trees based on the HN protein sequences place them between the Avulavirus and Respirovirus genera (2, 7).
FIG. 7.
Unrooted phylogenetic trees based on complete nucleoprotein (A) and attachment protein (B) sequences of selected viruses within the subfamily Paramyxovirinae. The trees were generated from ClustalW (accurate) protein alignments using distance matrix programs (Protdist and Neighbor) within the PHYLIP software package and were drawn in TreeView. Branch lengths represent relative genetic distances. See Materials and Methods for database accession numbers.
DISCUSSION
Until recently, almost all viruses belonging to the subfamily Paramyxovirinae shared a uniform genome structure consisting of six genes in the order 3′-N-P-M-F-H/HN/G-L-5′. Only the rubulaviruses MuV and SV5 and the avulavirus APMV6 deviated from this arrangement, possessing between the attachment and large protein genes an additional gene coding for a small hydrophobic protein. In 2004, completion of the 15,378-nt genome sequence of the reptilian paramyxovirus FDLV revealed the presence of seven genes in the order 3′-N-U-P-M-F-HN-L-5′ (26). The gene designated U for unknown has 604 nt and encodes a putative 167-aa transmembrane protein. Like that of FDLV, the J-V genome has significantly increased the diversity of genome structure within the subfamily and provokes much thought regarding the evolution of the Paramyxovirinae. In particular, the large size and unique composition of the J-V genome suggest that it may represent an evolutionarily ancient paramyxovirus genome structure.
The position and size of the J-V SH gene are analogous to those of the SH genes of the rubulaviruses SV5 and MuV, which are also located between the F and attachment protein genes and which encode proteins of 44 and 57 aa, respectively. The avulavirus APMV6 and members of the Pneumovirinae also contain SH genes, and the proteins encoded by these genes range from 64 to 177 aa. At present the function of the paramyxovirus SH protein is largely unknown. It is not essential for growth of SV5 in cell culture (17), and recombinant hRSV in which the SH gene had been entirely deleted showed growth properties very similar to those of the wild-type virus both in vitro and in vivo (3). Recently, studies have shown that the SV5 SH protein is involved in the regulation of apoptosis in MDBK cells (18, 33). Although the sizes and hydrophilicity profiles of the SH proteins of MuV, bRSV, and hRSV and the putative SH protein of J-V are similar, no significant similarity in amino acid sequence between any of the paramyxovirus SH proteins and the J-V SH protein was found. This is not surprising considering that at the amino acid level the SH protein is poorly conserved between different paramyxoviruses and that the SH protein sequences of different MuV strains can differ from one another by as much as 23% (51). In contrast, the presence of an extensively hydrophobic region of approximately 30 aa with the potential to function as a transmembrane anchor is strongly conserved between the SH proteins of different paramyxoviruses. It has been determined that the SH proteins of SV5 and hRSV are type II integral membrane proteins which are packaged in virions (9, 10, 17, 19). Conversely, preliminary data suggest that the SH protein of MuV is orientated in cell membranes with its C terminus facing the cytoplasm, and it has not been detected in MuV virions (50).
In contrast to the J-V SH gene, the J-V TM gene does not have an obvious analogue in any of the paramyxoviruses sequenced to date. Although it shares some basic features with the G gene of members of the subfamily Pneumovirinae, the significance of these similarities is unclear. Like the pneumovirus and metapneumovirus G genes, the TM gene is positioned adjacent to and downstream from the SH gene. The putative 258-aa TM protein is similar in size to the G proteins of members of the Pneumovirinae, which range from 236 to 396 aa, and is also predicted to be a type II integral membrane protein. In addition to the full-length transmembrane protein form, the G protein of hRSV is synthesized as a secreted form that arises from translational initiation at the second in-frame ATG codon, located within the transmembrane domain (46). Similarly, the third in-frame ATG codon within the ORF for the TM protein, which is in a stronger context for initiation of translation than the preceding ATG codons (25), also falls within the putative transmembrane domain. However, unlike the ectodomains of most G proteins of Pneumovirinae, the J-V TM protein ectodomain does not have a high content of serine and threonine residues, potential acceptor sites for O-linked sugars.
It is interesting that the least-conserved J-V gene stop sequences, with differences at two nucleotide positions, occur at the F-SH and SH-TM boundaries. It has been shown that the Enders strain of MuV, which has a single nucleotide change in the F gene stop sequence, does not produce detectable levels of monocistronic SH or dicistronic SH-HN mRNA, and the SH protein cannot be detected in Enders-infected cells (50, 51). Perhaps the less-conserved J-V F and SH gene stop sequences could also lead to increased levels of polymerase read-through at the junctions, resulting in increased production of dicistronic and multicistronic mRNA molecules which in turn may affect the expression levels of the SH and TM proteins.
The presence of ORF-X within the J-V attachment protein gene has no counterpart in other paramyxovirus genome sequences, and no obvious mechanism for its translation has been identified. Conservation of a long open reading frame in an RNA virus implies that it is expressed and that in some way the function of its encoded protein is required; otherwise, one would expect mutations introducing stop codons within the sequence to have occurred. It is possible that ORF-X was created by a recent mutation, prior to which a 1,414-aa attachment protein was translated. Conversely, an unconventional mechanism may enable ORF-X to be expressed in its current context. The use of an internal ribosomal entry site (42), a ribosomal shunting mechanism (14), or ribosomal reinitiation of translation could enable ORF-X to be translated independently of the G protein. Alternatively, suppression of translational termination (15, 49) at the G protein ORF stop codon could enable translation of a 1,414-aa “G-X” protein.
Characterization of the full-length J-V genome has revealed that while J-V clearly belongs to the subfamily Paramyxovirinae, at present it cannot be classified below this level. Although protein-based phylogenetic analyses most frequently place SalV, TPMV, MoV, and J-V between the Henipavirus and Morbillivirus genera, these viruses do not show sufficient similarity to one another to be classified together in a new genus.
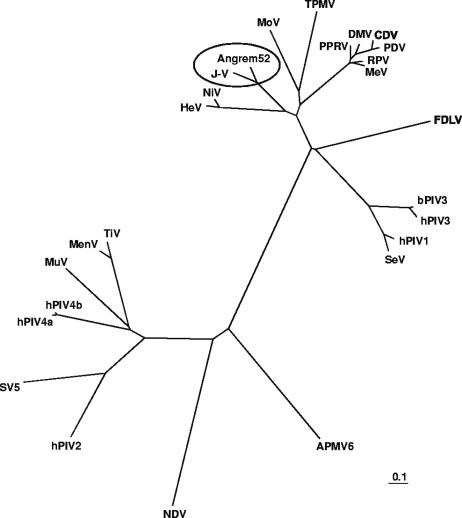
Interestingly, when J-V gene sequences were routinely submitted for BLASTx searches at the NCBI server, highly significant matches with protein sequences deduced from two putative novel human genes were identified (database accession numbers AAO91863 [matching fragments of the J-V P and V proteins], AAK76747 [matching the full-length J-V M protein], and AAL62340 [matching fragments of the J-V F protein]). These genes, designated Angrem104 (database accession number AF367870) and Angrem52 (AY040225), had been isolated by a subtractive hybridization experiment performed to screen and identify genes upregulated by angiotensin II in cultured human mesangial cells (31, 32). Analysis of the gene sequences revealed that Angrem104 appeared to be a paramyxovirus P gene mRNA sequence and that Angrem52 appeared to be a paramyxovirus dicistronic M-F mRNA sequence. This discovery was first documented in the Ph.D. thesis of an author of the present report (36). Similar findings have since been reported by two independent groups, both of which also concluded that the Angrem104 and Angrem52 sequences are likely derived from a novel paramyxovirus (1, 48). On alignment, the 340-aa J-V M protein showed 81% amino acid identity with the putative 340-aa M protein encoded by the Angrem52 sequence. In comparison, the next highest matches to the J-V M protein were the M proteins of NiV (50%) and HeV (49%). A phylogenetic tree based on an alignment of the J-V and Angrem52-derived M proteins with the M proteins of other members of the subfamily Paramyxovirinae placed J-V and the new virus in a cluster distinct from the Henipavirus and Morbillivirus genera and the unclassified viruses MoV and TPMV (Fig. 8). Because there appear to be sequencing errors in the putative Angrem P/V/C and F gene sequences, further phylogenetic analysis awaits definitive sequence data. In a collaborative effort between the Australian Animal Health Laboratory and the Renal Division of Peking University First Hospital, a novel paramyxovirus has recently been isolated from the mesangial cell line from which the Angrem52 and Angrem104 sequences were derived. Characterization of the full genome sequence of this virus is currently under way. The genome structure of the new virus will be of great interest, particularly with respect to the presence or absence of homologues of the J-V SH, TM, and G genes. Based on the close relationship between J-V and the new virus revealed by analysis of existing sequence information, it appears likely that they will constitute a new genus within the subfamily Paramyxovirinae.
FIG. 8.
Unrooted phylogenetic tree based on the putative matrix protein sequence encoded by Angrem52 and matrix protein sequences of selected viruses within the subfamily Paramyxovirinae. The tree was generated from a ClustalW (accurate) protein alignment using distance matrix programs (Protdist and Neighbor) within the PHYLIP software package and was drawn in TreeView. Branch lengths represent relative genetic distances. See Materials and Methods for database accession numbers.
Acknowledgments
We thank Gary Crameri for propagating and harvesting virus, Eric Hansson for help with the 5′ RACE, and Tony Pye and Maria Cardoso for providing the automated sequencing service.
REFERENCES
- 1.Basler, C. F., A. Garcia-Sastre, and P. Palese. 2005. A novel paramyxovirus? Emerg. Infect. Dis. 11:108-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bowden, T. R., M. Westenberg, L. F. Wang, B. T. Eaton, and D. B. Boyle. 2001. Molecular characterization of Menangle virus, a novel paramyxovirus which infects pigs, fruit bats, and humans. Virology 283:358-373. [DOI] [PubMed] [Google Scholar]
- 3.Bukreyev, A., S. S. Whitehead, B. R. Murphy, and P. L. Collins. 1997. Recombinant respiratory syncytial virus from which the entire SH gene has been deleted grows efficiently in cell culture and exhibits site-specific attenuation in the respiratory tract of the mouse. J. Virol. 71:8973-8982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campbell, R. W., J. G. Carley, R. L. Doherty, R. Domrow, C. Filippich, B. M. Gorman, and N. Karabatsos. 1977. Mossman virus, a paramyxovirus of rodents isolated in Queensland. Search 8:435-436. [Google Scholar]
- 5.Chua, K. B., W. J. Bellini, P. A. Rota, B. H. Harcourt, A. Tamin, S. K. Lam, T. G. Ksiazek, P. E. Rollin, S. R. Zaki, W. J. Shieh, C. S. Goldsmith, D. J. Gubler, J. T. Roehrig, B. Eaton, A. R. Gould, J. Olson, H. Field, P. Daniels, A. E. Ling, C. J. Peters, L. J. Anderson, and B. W. J. Mahy. 2000. Nipah virus: a recently emergent deadly paramyxovirus. Science 288:1432-1435. [DOI] [PubMed] [Google Scholar]
- 6.Chua, K. B., L. F. Wang, S. K. Lam, G. Crameri, M. Yu, T. Wise, D. Boyle, A. D. Hyatt, and B. T. Eaton. 2001. Tioman virus, a novel paramyxovirus isolated from fruit bats in Malaysia. Virology 283:215-229. [DOI] [PubMed] [Google Scholar]
- 7.Chua, K. B., L. F. Wang, S. K. Lam, and B. T. Eaton. 2002. Full length genome sequence of Tioman virus, a novel paramyxovirus in the genus Rubulavirus isolated from fruit bats in Malaysia. Arch. Virol. 147:1323-1348. [DOI] [PubMed] [Google Scholar]
- 8.Clark, H. F., F. S. Lief, P. D. Lunger, D. Waters, P. Leloup, D. W. Foelsch, and R. W. Wyler. 1979. Fer de Lance virus (FDLV): a probable paramyxovirus isolated from a reptile. J. Gen. Virol. 44:405-418. [DOI] [PubMed] [Google Scholar]
- 9.Collins, P. L., and G. Mottet. 1993. Membrane orientation and oligomerization of the small hydrophobic protein of human respiratory syncytial virus. J. Gen. Virol. 74:1445-1450. [DOI] [PubMed] [Google Scholar]
- 10.Collins, P. L., R. A. Olmsted, and P. R. Johnson. 1990. The small hydrophobic protein of human respiratory syncytial virus: comparison between antigenic subgroups A and B. J. Gen. Virol. 71:1571-1576. [DOI] [PubMed] [Google Scholar]
- 11.Curran, J., J. B. Marq, and D. Kolakofsky. 1995. An N-terminal domain of the Sendai paramyxovirus P protein acts as a chaperone for the NP protein during the nascent chain assembly step of genome replication. J. Virol. 69:849-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Egelman, E. H., S. S. Wu, M. Amrein, A. Portner, and G. Murti. 1989. The Sendai virus nucleocapsid exists in at least four different helical states. J. Virol. 63:2233-2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Felsenstein, J. 1989. PHYLIP—Phylogeny Inference Package (version 3.2). Cladistics 5:164-166. [Google Scholar]
- 14.Futterer, J., Z. Kiss-Laszlo, and T. Hohn. 1993. Nonlinear ribosome migration on cauliflower mosaic virus 35S RNA. Cell 73:789-802. [DOI] [PubMed] [Google Scholar]
- 15.Goff, S. P. 2001. Retroviridae: the retroviruses and their replication, p. 1871-1939. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 16.Harcourt, B. H., A. Tamin, K. Halpin, T. G. Ksiazek, P. E. Rollin, W. J. Bellini, and P. A. Rota. 2001. Molecular characterization of the polymerase gene and genomic termini of Nipah virus. Virology 287:192-201. [DOI] [PubMed] [Google Scholar]
- 17.He, B., G. P. Leser, R. G. Paterson, and R. A. Lamb. 1998. The paramyxovirus SV5 small hydrophobic (SH) protein is not essential for virus growth in tissue culture cells. Virology 250:30-40. [DOI] [PubMed] [Google Scholar]
- 18.He, B., G. Y. Lin, J. E. Durbin, R. K. Durbin, and R. A. Lamb. 2001. The SH integral membrane protein of the paramyxovirus simian virus 5 is required to block apoptosis in MDBK cells. J. Virol. 75:4068-4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hiebert, S. W., C. D. Richardson, and R. A. Lamb. 1988. Cell surface expression and orientation in membranes of the 44-amino-acid SH protein of simian virus 5. J. Virol. 62:2347-2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hosaka, M., M. Nagahama, W. S. Kim, T. Watanabe, K. Hatsuzawa, J. Ikemizu, K. Murakami, and K. Nakayama. 1991. Arg-X-Lys/Arg-Arg motif as a signal for precursor cleavage catalyzed by furin within the constitutive secretory pathway. J. Biol. Chem. 266:12127-12130. [PubMed] [Google Scholar]
- 21.Jorgensen, E. D., P. L. Collins, and P. T. Lomedico. 1987. Cloning and nucleotide sequence of Newcastle disease virus hemagglutinin-neuraminidase mRNA: identification of a putative sialic acid binding site. Virology 156:12-24. [DOI] [PubMed] [Google Scholar]
- 22.Jun, M. H. 1976. Studies on a virus isolated from wild mice (Mus musculus). M.Sc. thesis. James Cook University, Townsville, Queensland, Australia.
- 23.Jun, M. H., N. Karabatsos, and R. H. Johnson. 1977. A new mouse paramyxovirus (J virus). Aust. J. Exp. Biol. Med. Sci. 55:645-647. [DOI] [PubMed] [Google Scholar]
- 24.Kolakofsky, D., T. Pelet, D. Garcin, S. Hausmann, J. Curran, and L. Roux. 1998. Paramyxovirus RNA synthesis and the requirement for hexamer genome length: the rule of six revisited. J. Virol. 72:891-899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kozak, M. 1991. Structural features in eukaryotic mRNAs that modulate the initiation of translation. J. Biol. Chem. 266:19867-19870. [PubMed] [Google Scholar]
- 26.Kurath, G., W. N. Batts, W. Ahne, and J. R. Winton. 2004. Complete genome sequence of Fer-de-Lance virus reveals a novel gene in reptilian paramyxoviruses. J. Virol. 78:2045-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kurilla, M. G., H. O. Stone, and J. D. Keene. 1985. RNA sequence and transcriptional properties of the 3′ end of the Newcastle disease virus genome. Virology 145:203-212. [DOI] [PubMed] [Google Scholar]
- 28.Kyte, J., and R. F. Doolittle. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157:105-132. [DOI] [PubMed] [Google Scholar]
- 29.Lamb, R. A., and D. Kolakofsky. 2001. Paramyxoviridae: the viruses and their replication, p. 1305-1340. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 30.Langedijk, J. P., F. J. Daus, and J. T. van Oirschot. 1997. Sequence and structure alignment of Paramyxoviridae attachment proteins and discovery of enzymatic activity for a morbillivirus hemagglutinin. J. Virol. 71:6155-6167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liang, X., H. Zhang, A. Zhou, P. Hou, and H. Wang. 2003. Screening and identification of the up-regulated genes in human mesangial cells exposed to angiotensin II. Hypertens. Res. 26:225-235. [DOI] [PubMed] [Google Scholar]
- 32.Liang, X., H. Zhang, A. Zhou, and H. Wang. 2003. AngRem104, an angiotensin II-induced novel upregulated gene in human mesangial cells, is potentially involved in the regulation of fibronectin expression. J. Am. Soc. Nephrol. 14:1443-1451. [DOI] [PubMed] [Google Scholar]
- 33.Lin, Y., A. C. Bright, T. A. Rothermel, and B. A. He. 2003. Induction of apoptosis by paramyxovirus simian virus 5 lacking a small hydrophobic gene. J. Virol. 77:3371-3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mayo, M. A. 2002. A summary of taxonomic changes recently approved by ICTV. Arch. Virol. 147:1655-1656. [DOI] [PubMed] [Google Scholar]
- 35.Mesina, J. E., R. S. Campbell, J. S. Glazebrook, D. B. Copeman, and R. H. Johnson. 1974. The pathology of feral rodents in North Queensland. Tropenmed. Parasitol. 25:116-127. [PubMed] [Google Scholar]
- 36.Miller, P. J. 2004. The molecular and biological characterisation of two novel Australian paramyxoviruses. Ph.D. thesis. University of Melbourne, Parkville, Victoria, Australia.
- 37.Miller, P. J., D. B. Boyle, B. T. Eaton, and L. F. Wang. 2003. Full-length genome sequence of Mossman virus, a novel paramyxovirus isolated from rodents in Australia. Virology 317:330-344. [DOI] [PubMed] [Google Scholar]
- 38.Mirza, A. M., R. Deng, and R. M. Iorio. 1994. Site-directed mutagenesis of a conserved hexapeptide in the paramyxovirus hemagglutinin-neuraminidase glycoprotein: effects on antigenic structure and function. J. Virol. 68:5093-5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morgan, E. M. 1991. Evolutionary relationships of paramyxovirus nucleocapsid-associated proteins, p. 163-179. In D. W. Kingsbury (ed.), The paramyxoviruses. Plenum Press, New York, N.Y.
- 40.Murray, K., P. Selleck, P. Hooper, A. Hyatt, A. Gould, L. Gleeson, H. Westbury, L. Hiley, L. Selvey, and B. Rodwell. 1995. A morbillivirus that caused fatal disease in horses and humans. Science 268:94-97. [DOI] [PubMed] [Google Scholar]
- 41.Page, R. D. M. 1996. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 42.Pelletier, J., and N. Sonenberg. 1988. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature 334:320-325. [DOI] [PubMed] [Google Scholar]
- 43.Philbey, A. W., P. D. Kirkland, A. D. Ross, R. J. Davis, A. B. Gleeson, R. J. Love, P. W. Daniels, A. R. Gould, and A. D. Hyatt. 1998. An apparently new virus (family Paramyxoviridae) infectious for pigs, humans, and fruit bats. Emerg. Infect. Dis. 4:269-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Poch, O., B. M. Blumberg, L. Bougueleret, and N. Tordo. 1990. Sequence comparison of five polymerases (L proteins) of unsegmented negative-strand RNA viruses: theoretical assignment of functional domains. J. Gen. Virol. 71:1153-1162. [DOI] [PubMed] [Google Scholar]
- 45.Renshaw, R. W., A. L. Glaser, H. Vancampen, F. Weiland, and E. J. Dubovi. 2000. Identification and phylogenetic comparison of Salem virus, a novel paramyxovirus of horses. Virology 270:417-429. [DOI] [PubMed] [Google Scholar]
- 46.Roberts, S. R., D. Lichtenstein, L. A. Ball, and G. W. Wertz. 1994. The membrane-associated and secreted forms of the respiratory syncytial virus attachment glycoprotein G are synthesized from alternative initiation codons. J. Virol. 68:4538-4546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Russell, C. J., T. S. Jardetzky, and R. A. Lamb. 2001. Membrane fusion machines of paramyxoviruses: capture of intermediates of fusion. EMBO J. 20:4024-4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schomacker, H., P. L. Collins, and A. C. Schmidt. 2004. In silico identification of a putative new paramyxovirus related to the Henipavirus genus. Virology 330:178-185. [DOI] [PubMed] [Google Scholar]
- 49.Strauss, E. G., C. M. Rice, and J. H. Strauss. 1983. Sequence coding for the alphavirus nonstructural proteins is interrupted by an opal termination codon. Proc. Natl. Acad. Sci. USA 80:5271-5275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Takeuchi, K., K. Tanabayashi, M. Hishiyama, and A. Yamada. 1996. The mumps virus SH protein is a membrane protein and not essential for virus growth. Virology 225:156-162. [DOI] [PubMed] [Google Scholar]
- 51.Takeuchi, K., K. Tanabayashi, M. Hishiyama, A. Yamada, and A. Sugiura. 1991. Variations of nucleotide sequences and transcription of the SH gene among mumps virus strains. Virology 181:364-366. [DOI] [PubMed] [Google Scholar]
- 52.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tidona, C. A., and G. Darai. 2002. TPMV-like viruses (Paramyxoviridae, Paramyxovirinae), p. 660-662. In C. A. Tidona and G. Darai (ed.), The Springer index of viruses. Springer-Verlag, Berlin, Germany.
- 54.Tidona, C. A., H. W. Kurz, H. R. Gelderblom, and G. Darai. 1999. Isolation and molecular characterization of a novel cytopathogenic paramyxovirus from tree shrews. Virology 258:425-434. [DOI] [PubMed] [Google Scholar]
- 55.Tikasingh, E. S., A. H. Jonkers, L. Spence, and T. H. Aitken. 1966. Nariva virus, a hitherto undescribed agent isolated from the Trinidadian rat, Zygodontomys b. brevicauda (J. A. Allen & Chapman). Am. J. Trop. Med. Hyg. 15:235-238. [DOI] [PubMed] [Google Scholar]
- 56.Tillett, D., B. P. Burns, and B. A. Neilan. 2000. Optimized rapid amplification of cDNA ends (RACE) for mapping bacterial mRNA transcripts. BioTechniques 28:448-456. [DOI] [PubMed] [Google Scholar]
- 57.Tusnády, G. E., and I. Simon. 1998. Principles governing amino acid composition of integral membrane proteins: application to topology prediction. J. Mol. Biol. 283:489-506. [DOI] [PubMed] [Google Scholar]
- 58.Tusnády, G. E., and I. Simon. 2001. The HMMTOP transmembrane topology prediction server. Bioinformatics 17:849-850. [DOI] [PubMed] [Google Scholar]
- 59.Walder, R. 1971. Electron microscopic evidence of Nariva virus structure. J. Gen. Virol. 11:123-128. [DOI] [PubMed] [Google Scholar]
- 60.Wang, L., M. Yu, E. Hansson, L. I. Pritchard, B. Shiell, W. P. Michalski, and B. T. Eaton. 2000. The exceptionally large genome of Hendra virus: support for creation of a new genus within the family Paramyxoviridae. J. Virol. 74:9972-9979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang, L. F., K. B. Chua, M. Yu, and B. T. Eaton. 2003. Genome diversity of emerging paramyxoviruses. Curr. Genomics 4:263-273. [Google Scholar]
- 62.Wang, L. F., and B. T. Eaton. 2001. Emerging paramyxoviruses. Infect. Dis. Rev. 3:52-69. [Google Scholar]
- 63.Yu, M., E. Hansson, B. Shiell, W. Michalski, B. T. Eaton, and L. F. Wang. 1998. Sequence analysis of the Hendra virus nucleoprotein gene: comparison with other members of the subfamily Paramyxovirinae. J. Gen. Virol. 79:1775-1780. [DOI] [PubMed] [Google Scholar]