Abstract
Papillomaviruses are small DNA viruses which have the capacity to establish a persistent infection in mammalian epithelial cells. The papillomavirus E2 protein is a central coordinator of viral gene expression, genome replication, and maintenance. We have investigated the distribution of bovine papillomavirus E2 protein in nuclei of proliferating cells and found that E2 is associated with cellular chromatin. This distribution does not change during the entire cell cycle. The N-terminal transactivation domain, but not the C-terminal DNA-binding domain, of the E2 protein is responsible for this association. The majority of the full-length E2 protein can only be detected in chromatin-enriched fractions but not as a free protein in the nucleus. Limited micrococcal nuclease digestion revealed that the E2 protein partitioned to different chromatin regions. A fraction of the E2 protein was located at nuclear sites that are resistant against nuclease attack, whereas the remaining E2 resided on compact chromatin accessible to micrococcal nuclease. These data suggest that there are two pools of E2 in the cell nucleus: one that localizes on transcriptionally inactive compact chromatin and the other, which compartmentalizes to transcriptionally active nuclear structures of the cell. Our data also suggest that E2 associates with chromatin through cellular protein(s), which in turn is released from chromatin at 0.4 M salt.
Human and animal papillomaviruses are widely spread pathogens; these are small, double-stranded DNA viruses causing proliferative lesions of the skin and mucosa. Papillomaviruses have the capacity to establish a persistent infection in mammalian epithelial cells. According to a current model, the virus enters a proliferating cell of the basal epithelial cell layer, where the circular viral genome is amplified. Following the establishment stage, viral persistence is set up through the maintenance of a constant low copy number of extrachromosomal viral genomes in the nuclei of dividing host cells.
Bovine papillomavirus type 1 (BPV1) has become the prototype virus for the molecular analysis of papillomaviruses because of its ability to transform rodent cells in tissue culture and to exist as an autonomously replicating plasmid in these cells (21). The plasmids containing upstream regulatory regions (URR) of BPV1 can also be stably maintained as extrachromosomal elements in certain cell lines, where the URR-containing plasmids replicate in synchrony with cellular DNA (35). The long-term episomal maintenance of BPV1 URR-containing plasmid requires only two virally encoded proteins, the viral helicase E1 and the regulatory protein E2 (35). Recent studies have shown that BPV1 genomes, as well as URR-containing plasmids, are localized to host cell chromosomes during mitosis to ensure their accurate segregation into daughter cells (14, 24, 43) and that the E2 protein alone is sufficient for the chromatin attachment of URR-containing plasmids (14).
The E2 protein of BPV1 is a regulatory protein playing a crucial role during the viral life cycle. First, it acts as a regulator of the transcription; it activates transcription from different BPV1 promoters as well as different heterologous promoters in mammalian and yeast cells (19, 31, 44, 45). Several cellular partners have been identified that interact with E2 proteins, including Sp1 (27), TBP (46), TFIIB (7, 36), AMF1 (9), p300/CBP (23, 32), p/CAF (22), and hNAP1 (37). Second, the papillomavirus E2 protein is the primary viral origin-recognition protein, which helps to recruit the viral helicase E1 to the origin in the initiation of viral DNA replication (40). E2 can also relieve nucleosome-mediated repression of papillomavirus DNA replication in vitro (25). Recently it was shown that BPV1 E2 interaction with Brd4 tethers the viral DNA to host mitotic chromosomes (54).
The E2 proteins are relatively well conserved among the papillomaviruses in two domains: an N-terminal part forming a transactivation domain and a C-terminal part forming a dimerization and sequence-specific DNA binding domain (DBD). The E2 protein binds as a homodimer to consensus sequence ACCN6GGT and can regulate transcription from promoters containing these E2-binding sites (E2BS) (4). The N-terminal activation domain is required for the replication, transactivation, and maintenance function of the protein (1, 5, 30, 49).
The eukaryotic cell nucleus has a highly compartmentalized structure. It is the control center of the eukaryotic cell, where the genomic processes of DNA replication, transcription, and RNA processing occur and are precisely coordinated in space and time. There is a growing realization that the coordination and regulation of different nuclear activities is mediated by the spatial compartmentalization of structural components and enzymatic and regulatory activities (8, 20). Small DNA viruses, like papillomaviruses, utilize the cellular regulatory machineries to support their gene expression and DNA replication. The viral E2 protein is a multifunctional protein, and in order to fulfill its role in papillomavirus life cycle, it must interact with different nuclear compartments. Immunofluorescence studies have shown that the bovine papillomavirus type 1 (BPV1) E2 protein is concentrated in numerous small domains, scattered throughout the nucleus (3, 42). In this study, we have used biochemical approaches to characterize the BPV1 E2 protein subnuclear distribution and compartmentalization. We show that the BPV1 E2 protein associates with cellular chromatin during all stages of the cell cycle. We have also performed a detailed biochemical characterization of the in vivo interaction of the E2 protein with chromatin using independent procedures like limited micrococcal nuclease digestion and differential salt extraction. These studies showed that there are at least two pools of E2 in the cell nucleus and these different pools of E2 partition to different nuclear substructures.
MATERIALS AND METHODS
Cell culture and plasmids.
Chinese hamster ovary cell line (CHO) and its derivatives CHO49 (expressing BPV1 E2 protein) and CHOBgl40 (expressing BPV1 E1 and E2 proteins and BPV1 URR containing plasmid pNeoBgl40) (35) were maintained in Ham′s F12 medium supplemented with 10% fetal calf serum. For cell cycle synchronization, cells were synchronized in metaphase by culturing them with 50 ng/ml nocodazole for 4 h and then releasing the mitotic cells obtained by mechanical shake-off into cell cycles.
Transfections into CHO cells were performed as described previously (49). Bovine papillomavirus E2 expression vector pCGE2, pCG-E2R37A, pCG-E2E39A, and expression vectors for E2 deletion mutants and chimeric proteins in pCG vector have been described previously (2, 18, 49).
C127 cells were grown in Iscove's modified Dulbecco's medium containing 10% fetal bovine serum. For BPV1 transformed C127 cell lines, the C127 cells were transfected with BPV1 genome and 4 weeks later transformed foci were isolated and were dispersed in a separate culture dish. The transformed cells grown out from the isolated foci were subcloned. The cell line D9:D3 used in this study contains 160 copies of episomally replicating BPV1 genomes per tetraploid cell.
Cell cycle distribution analysis (FACS).
For flow cytometric analysis, cells were washed with phosphate-buffered saline (PBS) and fixed by adding equal volume of ice-cold ethanol and incubated at 4°C at least 1 h. Cells were then pelleted, resuspended in 400 μl of PBS with 1 mM MgCl2 and 100 μg/ml of RNase, and incubated at 37°C for 45 min to digest cellular RNA. Propidium iodide was added to a final concentration of 50 μg/ml to stain the nuclear DNA. Signals from 30,000 cells were collected using a FACSCalibur flow cytometer (BD Biosciences), and samples were analyzed using the standard software provided by the manufacturer.
Subcellular fractionation.
Chromatin fractionation with Triton X-100 was performed as described previously (33) with modifications. CHOBgl40 cells or CHO cells transfected with expression plasmids were treated with PBS-3 mM EDTA, collected by centrifugation, washed with ice-cold PBS, and centrifuged again. Cells (2 × 106) were resuspended in 100 μl of cytoskeleton buffer (CSK), which contained 10 mM piperazine-N,N′-bis(2-ethanesulfonic acid), pH 6.8, 100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, and 1 mM EGTA supplemented with leupeptin, aprotinin (5 ng/μl), 0.5 mM phenylmethylsulfonyl fluoride, 1 mM dithiothreitol (DTT). Triton X-100 (0.5%, vol/vol) was added and incubated for 15 min at 0°C. Alternatively, the first extraction was performed with 0.1% Triton X-100 for 5 min, and then cells were washed with PBS and extracted again with 0.5% Triton X-100 for 15 min. Soluble and insoluble fractions were separated in a microcentrifuge (7,500 rpm, 3 min), and the pellet was washed at 0°C with CSK buffer. The insoluble fraction was further incubated with CSK buffer containing either 0.2, 0.3, or 0.4 M NaCl for 20 min. The insoluble fraction was resuspended in CSK buffer, the sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer was added to soluble as well as insoluble fractions, and these fractions were subjected to immunoblotting. The concentration of total protein was determined by Bio-Rad Protein assay (Bio-Rad) using bovine serum albumin as a standard. For cross-linking of proteins and DNA, formaldehyde was added to corresponding fractions (final concentration, 1%), and after incubation of 15 min at room temperature the reaction was stopped by adding 0.125 M glycine. The cross-linked samples were boiled in the SDS-PAGE loading buffer for 30 s before loading onto the gel.
In the case of C127-BPV and CHO49 cells, the E2 protein was immunoprecipitated from fractions with 3F12 antibody bound to magnetic beads (Dynabeads). For immunoprecipitation, soluble as well as insoluble fractions were diluted in radioimmunoprecipitation assay (RIPA; 50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% NP-40, 0.5% deoxycholate, 0.1% SDS, 0.1 mM DTT, protease inhibitors) buffer, and the fractions were incubated with 3F12 antibody beads for 3 h at 4°C. Beads were then washed twice with RIPA containing 0.5 M NaCl and twice with RIPA alone, resuspended in SDS loading buffer, and subjected to immunoblotting analysis with horseradish peroxidase-conjugated 5E11 (subclone of monoclonal antibody [MAb] 3F12) antibody.
Subcellular fractionation was also carried out with cells attached to the glass substrates as described previously (48). Briefly, cells were grown on coverslips, washed with PBS, and extracted to generate various subcellular fractions as described above. For chromatin solubilization, cells were incubated with CSK buffer containing DNase (25 μl/ml) (Amersham) for 15 min at room temperature and then extracted in the same solution adjusted to 0.25 M (NH4)2SO4 for 10 min at room temperature. For RNase treatment, RNase (1 mg/ml) was added to CSK buffer. Cells were washed with ice-cold PBS, fixed, and subjected to immunofluorescence analysis.
Micrococcal nuclease digestion and chromatin fractionation.
The micrococcal nuclease digestion and chromatin fractionation procedure was described previously (38). For isolation of cell nuclei, 107 cells were resuspended in 300 μl of HNB (0.5 M sucrose, 15 mM Tris-HCl, pH 7.6, 60 mM KCl, 0.25 mM EDTA, 0.125 mM EGTA, 0.5 mM spermidine, 1 mM DTT, 0.5 mM phenylmethylsulfonyl fluoride, 5 ng/μl aprotinin, 5 ng/μl leupeptin) and supplemented with 0.5% Triton X-100. After 5 min of incubation at 0°C, nuclei were sedimented by centrifugation at 7,500 rpm for 4 min in a microcentrifuge, and then 106 nuclei were resuspended in 50 μl of nuclear buffer (20 mM Tris-HCl, pH 7.6, 70 mM NaCl, 20 mM KCl, 5 mM MgCl2, and 3 mM CaCl2 supplemented with protease inhibitors). Nuclei suspension was incubated with 30 U of micrococcal nuclease (Fermentas) at room temperature during the indicated time. The digestion was terminated by the addition of EDTA and EGTA to 5 mM each, the mixture was centrifuged at 7,500 rpm for 4 min, and the supernatant was designated the S1 fraction. The nuclear pellet was lysed in 2 mM EDTA for 15 min at 0°C. This was followed by centrifugation, and the supernatant and pellet were designated the S2 and P fractions, respectively. Proteins were analyzed by SDS-PAGE; E2 was detected with specific antibodies and core histones were detected by staining with Coomassie blue. For DNA analysis of S1, S2, and P fractions, aliquots were treated with lysis buffer (50 mM Tris-HCl, pH 7.6, 100 mM NaCl, 5 mM EDTA, 0.5% SDS) supplemented with RNase (100 μg/ml) and incubated at 37°C for 20 min and then supplemented with proteinase K (200 μg/ml) and incubated at 37°C for 4 h. DNA was precipitated with ethanol after phenol-chloroform extraction. DNA was transferred onto a 0.45-μm nylon transfer membrane (AppliChem) and incubated with 32P-labeled BPV1 URR probe (base pairs [bp] 6946 to 7900 of BPV1) under standard conditions.
For chromatin immunoprecipitation, chromatin from 2 × 107 CHO and CHOBgl40 cells was fractionated as depicted above, and the fractions were cross-linked with formaldehyde (final concentration, 1%) for 15 min at room temperature. The reaction was stopped by adding 0.125 M glycine. After sonication, the fractions were cleared by centrifugation at 13,000 rpm for 10 min in a microcentrifuge and the supernatant was incubated with 500 ng of E2-specific MAb 3E8 (18) bound to magnetic beads at room temperature for 2 h in immunoprecipitation (IP) buffer (20 mM Tris, pH 7.5, 150 mM NaCl, 150 mM sucrose, 1 mM EDTA, and 1 mM DTT supplemented with protease inhibitors). The beads were washed three times with 1 ml IP buffer but with 400 mM NaCl, and the immunoprecipitated material was removed by boiling the beads in SDS loading buffer for 20 min. After phenol-chloroform extraction and ethanol precipitation, DNA was resuspended in 15 μl of Tris-EDTA and analyzed by PCR using primers specifically amplifying bp 7338 to 7654 of the BPV1 genome. PCR was performed using 2 μl of a 1:100 dilution of IP fraction for 30 cycles.
Immunofluorescence and immunoblotting analysis.
For immunofluorescence analysis, CHOBgl40 cells were grown on coverslips and fixed in cold (−20°C) acetone/methanol (1:1) for 15 min. Coverslips were washed with PBS and blocked with bovine serum albumin (1 mg/ml in PBS) for 30 min. Mouse monoclonal antibodies against the E2 proteins 5H4, 1H10, and 1E4 (final concentration, 5 ng/μl), RNAPol II CTD repeat 4H8 (used at 1/2,000) (Abcam) (17), and CD43 protein 3A1 (1/5) (41) and rabbit polyclonal antibody against the Brd4 C terminus (1/50) (Abgent) were used as primary antibodies. As a secondary antibody, fluorescein isothiocyanate-conjugated goat anti-mouse or anti-rabbit immunoglobulin G (LabAs Ltd.) was used at the concentration recommended by the manufacturer. Images were captured using the Olympus DP70 CCD camera mounted on an Olympus BX-41 microscope.
For immunoblotting analysis, proteins were separated by SDS-polyacrylamide gel electrophoresis and transferred by a semidry blotting method to a polyvinylidene difluoride membrane (Millipore Corp.). Membranes were incubated with anti-E2 antibodies 3F12, 1E4, 1H10, and 1E2 (1 ng/μl) (18), anti-TBP antibody SL35 (1/4,000) (29), or anti-RNAPol II antibody 4H8 (1/10,000) (Abcam) and with a secondary horseradish peroxidase-conjugated antibody (LabAs Ltd., Tartu, Estonia) according to the manufacturers' recommendations. Detection was performed using an ECL detection kit (Amersham Pharmacia Biotech) following the manufacturers' manuals. Digital images were collected by scanning the film, and band intensities were measured using UVP GelWorks 1D Intermediate 3.0 software (Nonlinear Dynamics Ltd.).
Glycerol density-gradient centrifugation.
CHOBgl40 cells were first extracted with CSK supplemented with 0.5% Triton X-100 and after that with CSK buffer containing 0.4 M NaCl. The 0.4 M salt supernatant fraction of CHOBgl40 cells was either directly loaded onto the gradient (extract of 1.5 × 107 cells) or treated with 1% formaldehyde (extract of 3 × 107 cells) for 15 min at room temperature and stopped by adding 0.125 M glycine. Protein extracts (300 μl) were centrifuged through 5-ml linear glycerol gradient (10 to 35%; 10 mM Tris-HCl, pH 7.6, 0.1 mM EDTA, 100 mM NaCl) at 45,000 rpm for 5 h at 4°C in a Beckman SW55 rotor. Marker proteins, apoferritin (440 kDa), and alcohol dehydrogenase (150 kDa) (Sigma) were run in parallel gradients. The gradients were dripped into 15 fractions, and the bottom of the tube was washed with 100 μl of SDS loading buffer. Collected fractions were subjected to immunoblotting analysis.
RESULTS
Subnuclear distribution of the BPV1 E2 protein.
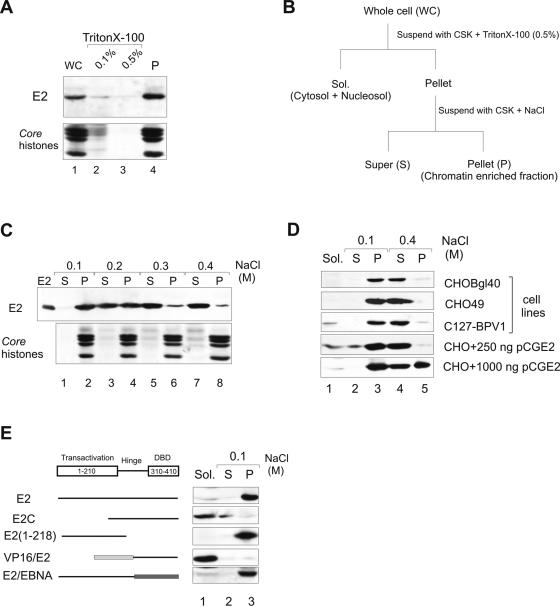
In this study, we have used, if not mentioned otherwise, the CHOBgl40 cell line that mimics the stable replication of papillomavirus genome in dividing cells. CHOBgl40 cells express the BPV1 E1 and E2 proteins from integrated cassettes and maintain extrachromosomally the BPV1 URR containing plasmid pNeoBgl40 (14, 35). BPV1 E2 is a nuclear protein (3, 42) as determined by immunofluorescence microscopy and biochemical analysis (13). To investigate the subnuclear distribution of the BPV1 E2 protein, we first fractionated the CHOBgl40 cells into soluble cytoplasmic and nucleoplasmic and insoluble chromatin and nuclear matrix-associated fractions (Fig. 1B). First, cells were lysed in a Triton X-100-containing buffer, soluble proteins were separated from the insoluble pellet by centrifugation, and all the fractions were analyzed by immunoblotting with E2-specific antibodies (18). As shown in Fig. 1A, more than 90% of the E2 protein was detected in the nonionic detergent-insoluble fraction (lane 4). We estimated that approximately 60 to 65% of total cellular proteins were extracted after treatment with 0.5% Triton X-100, and the remaining fraction (∼35% total protein) contained core histones used here as a marker of chromatin. Thus, the majority of the BPV1 E2 protein was associated with insoluble chromatin-nuclear matrix fraction. Under the same conditions, the other viral protein, E1, was easily extractable with Triton X-100 (data not shown). Next, the nonionic detergent-insoluble E2 fraction was further incubated with increasing concentrations of NaCl, and the solubilized proteins (∼20% total protein in the case of 0.4 M NaCl) were separated from insoluble fractions by centrifugation. The E2 protein, which remained in the chromatin-nuclear matrix fraction at 0.1 M NaCl (Fig. 1C, lane 2), was removed from the fraction by increasing the salt concentration to 0.3 M (lane 6). Intermediate salt concentrations gave an intermediate degree of extraction. A lot of chromatin-associated proteins are released from chromatin by moderate salt concentrations (10, 33), and salt solubility has been sometimes used to monitor binding of proteins to the chromatin compartment. So, from these experiments we suggested that the E2 protein associates with cellular chromatin.
FIG. 1.
Subcellular localization of the BPV1 E2 protein. (A) Association of E2 with a chromatin-containing fraction. CHOBgl40 cells were extracted with Triton X-100 and fractions were analyzed by immunoblotting with E2-specific antibodies. The lower part of the gel was analyzed for the presence of histones (Coomassie blue staining). (B) Protocol for cellular extraction. (C) Salt extractability of E2 expressed by the CHOBgl40 cell line. The chromatin-enriched fraction from panel A was further incubated for 15 min with CSK buffer containing the indicated salt concentrations, separated into soluble (S) and insoluble (P) fractions, and the two fractions were subjected to immunoblotting as in panel A. Proteins from 106 cells were loaded in each lane. (D) Salt extractability of E2 expressed by various cell lines and in CHO cells transiently. For transient expression, two different plasmid concentrations were used and the cells were lysed 24 h after transfection. Cells were extracted as in panel B, and the E2 protein from all fractions of CHO49 and C127-BPV1 cells was immunoprecipitated with E2-specific antibody 3F12 coupled to magnetic beads. Proteins from 106 CHOBgl40 cells, 1.5 × 107 CHO49 cells and 1.5 × 107 C127-BPV1 cells were loaded on S and P lanes, and Sol. fractions contain half of the material of S and P fractions. All fractions were subjected to immunoblotting with E2-specific antibodies. (E) E2 mutant association with a chromatin-containing fraction. CHO cells were transfected with 250 ng of expression plasmid encoding the indicated protein, and cells were lysed 24 h after transfection extracted as in panel B and subjected to immunoblotting with E2-specific antibodies.
In order to test that this phenomena is not specific for the CHOBgl40 cell line only, the same experiments were carried out with the CHO49 cell line (expressing the E2 protein alone on low levels) and with the BPV1-transformed C127 cell line. BPV1-transformed C127 cells maintain and replicate the entire BPV1 genome. The CHO49 and C127-BPV cells were biochemically fractionated as shown in Fig. 1B, and the E2 protein was immunoprecipitated from all fractions and analyzed by immunoblotting. As expected, the E2 protein in CHO49 and C127-BPV1 cells was detected in detergent-resistant chromatin-nuclear matrix fraction (Fig. 1D, lane 3) and was extractable with 0.4 M salt (lane 4), similar to results obtained with the CHOBgl40 cell line. So the E2 protein in all cell lines tested associates with cellular chromatin.
We also studied the E2 protein distribution in transiently transfected cells. We transfected CHO cells with different concentrations of the expression plasmid for E2 protein, and 24 h after transfection the cells were collected and subjected to biochemical fractionation as depicted in Fig. 1B. As shown on Fig. 1D, transiently expressed E2 was found predominantly in the detergent-resistant chromatin-nuclear matrix fraction (lane 3). Extraction of this fraction with 0.4 M salt released either all E2 (250 ng of expression plasmid used for transfection) or some part of the E2 protein (1,000 ng of expression plasmid used for transfection) from chromatin (Fig. 1D). We suggest that the ratio of released and tightly bound E2 protein after extraction with 0.4 M salt is dependent on the E2 protein amount expressed in the cell; in the case of high E2 expression levels, part of the protein remained bound to insoluble nuclear substructures, and in the case of low E2 expression levels, the protein was extractable. These data show that overexpressed E2 protein localizes to some other nuclear compartment.
In order to determine the region of E2 responsible for chromatin association, the subnuclear distribution of various E2 mutants was determined. As shown in Fig. 1E, the E2 repressor protein E2C (containing amino acids [aa] 162 to 410) was extractable from the cell structures already with 0.5% Triton X-100 (Fig. 1E, lane 1). By contrast, E2 transactivation domain E2 (aa 1 to 218) was preferentially recovered in the chromatin-nuclear matrix fraction (Fig. 1E, lane 3). Chimeric protein VP16/E2, which consists of the transactivation domain of another viral transactivator VP16 and DNA-binding domain of E2, was recovered in the soluble fraction (lane 1), and E2/EBNA (containing transactivation domain of E2 and DNA-binding domain of EBNA1 of Epstein-Barr virus) in the insoluble chromatin-nuclear matrix fraction (lane 3). E2 (aa 1 to 218) and E2/EBNA proteins associated with chromatin-nuclear matrix fraction were extractable from this fraction with 0.4 M salt (data not shown). At the same time the full-length EBNA protein is extractable from nuclear structures already with 0.5% Triton X-100 (28). These data suggest that the N-terminal transactivation domain but not the C-terminal DNA-binding-dimerization domain of E2 is responsible for its association with cellular chromatin. Therefore, the interaction of E2 with chromatin cannot be mediated by binding to cellular DNA; it is obviously mediated by chromatin proteins.
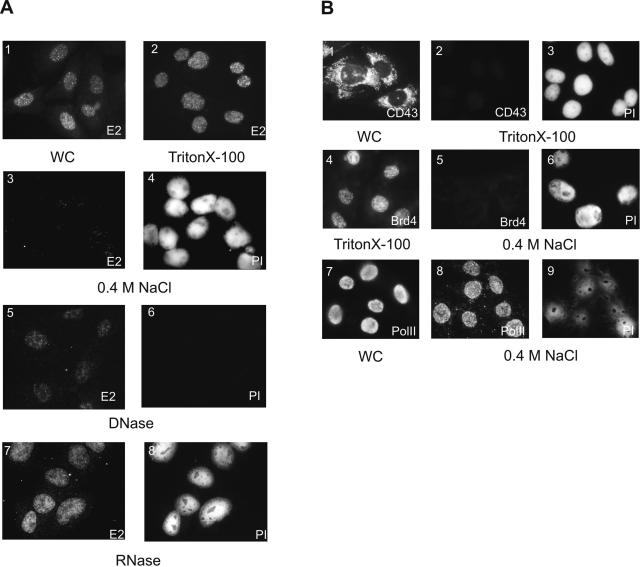
The results, obtained by biochemical fractionation, were further supported by indirect immunofluorescence analysis. The CHOBgl40 cells were fractionated while attached to the glass coverslips, and the E2 protein was visualized with specific antibodies while DNA in the nuclei was revealed by propidium iodide (PI) contained in the mounting medium. Treatment of cells with 0.5% Triton X-100, which removed the cytoplasm, nuclear membrane, and soluble nucleoplasm, had no effect on E2 staining (Fig. 2A, image 2). Incubation of remaining nuclei structures with 0.4 M NaCl released the E2 protein from nuclei (images 3 and 4). However, some dots of E2 remained sometimes even after extensive washing of cells. To verify the fractionation of cells on coverslips, some cellular controls were used. The cytoplasmic CD43 protein, which compartmentalizes into endoplasmatic reticulum of cells (41), was readily extracted with 0.5% Triton X-100 (Fig. 2B, images 1 to 3). A bromodomain protein, Brd4, which binds to acetylated chromatin and was shown to be extractable from chromatin at 150 to 200 mM salt (10), was in our case also detectable in the CHOBgl40 cell nucleus after Triton X-100 treatment and was extracted at 0.4 M NaCl (Fig. 2B, images 4 to 6). At the same time RNA Pol II was not extractable from the cell nucleus at 0.4 M salt (Fig. 2B, images 7 to 9), which is consistent with previous results that it is a nuclear matrix protein (34, 38). In order to study the association of E2 with RNA and DNA, the Triton X-100-treated nuclei were further incubated with RNase or DNase. As shown on Fig. 2A, the RNase treatment had no effect on E2 staining (Fig. 2A, images 7 and 8). In contrast, digestion of isolated nuclei with DNase led to the clearly diminished E2 protein staining (Fig. 2A, images 5 and 6). However, some E2 signal always retained after DNase treatment of cells (Fig. 2A, image 5). This raised the possibility that only some part but not all of the E2 protein associates with cellular chromatin.
FIG. 2.
(A) Subnuclear distribution of the E2 protein by immunofluorescence analysis. CHOBgl40 cells (image 1) grown on coverslips were treated with 0.5% Triton X-100 (image 2), and further detergent-treated cells were incubated with 0.4 M NaCl (images 3 and 4), DNase (images 5 and 6), or RNase (images 7 and 8). Cells were fixed in acetone/methanol, and the E2 protein was detected with specific antibodies. In panels 1, 2, 3, 5, and 7 E2 is fluorescein isothiocyanate labeled, and panels 4, 6, and 8 show cellular DNA stained with propidium iodide in the same field of cells. (B) Subnuclear distribution of cellular proteins. CHOBgl40 cells grown on coverslips were either directly or after treatment with 0.5% Triton X-100 (image 2-4) and 0.4 M NaCl (images 5, 6, 8, and 9) incubated with antibodies against CD43 (images 1 and 2), Brd4 (images 4 and 5) or RNA Pol II (images 7 and 8). PI, propidium iodide staining of cellular DNA (images 3, 6, and 9).WC, whole cells.
The E2 protein associates with cellular chromatin during the cell cycle.
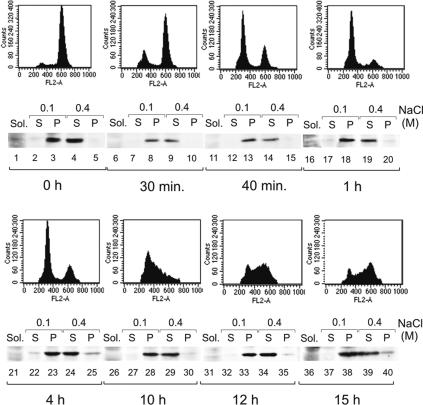
The experiments described in the previous section were performed with asynchronously proliferating cells. To study the E2 compartmentalization during the cell cycle, the CHOBgl40 cells were first synchronized in metaphase by brief treatment with nocodazole followed by mitotic shake-off. Cells were then either collected as mitotic populations or released into the cell cycle and collected at various time points after release from mitotic block. Cell cycle was determined by FACS analysis, and the CHOBgl40 cells were fractionated as depicted on Fig. 1B. As shown on Fig. 3A, the E2 protein remained bound to chromatin during all stages of the cell cycle (lanes 19, 24, 29, 34, and 39) including mitosis (lanes 4, 9, and 14). However, the E2 protein level, being high in G1, S, G2, and prometaphase cells, was reduced during progression of mitosis (Fig. 3A, compare lanes 3 and 4 with lanes 8 and 9).
FIG. 3.
Chromatin association of the E2 protein during the cell cycle. (A) CHOBgl40 cells were synchronized in metaphase, collected by mitotic shake-off and then released into the cell cycle. At the indicated time, cells were collected and subjected to biochemical fractionation described for Fig. 1B, and cell synchronization was confirmed by flow cytometry analysis. Equivalent amounts of each extract (extract of 106 cells for S and P lanes and of 2 × 105 cells for Sol. fraction) were subjected for immunoblotting with E2-specific antibodies. Sol., soluble.
In summary, these experiments suggest that the BPV1 E2 protein, or at least some part of it, is associated with cellular chromatin during all stages of the cell cycle. E2 remained bound to chromatin and was not detected free in nucleoplasm at any stage of the cell cycle. These results also support and confirm biochemically the immunofluorescence colocalization studies of our group and others, which have shown that the E2 protein associates with chromosomes during mitosis and remains associated with condensed chromosomes at every stage of mitosis (1, 5, 14, 43, 51).
E2 associates with nuclear substructures via cellular proteins.
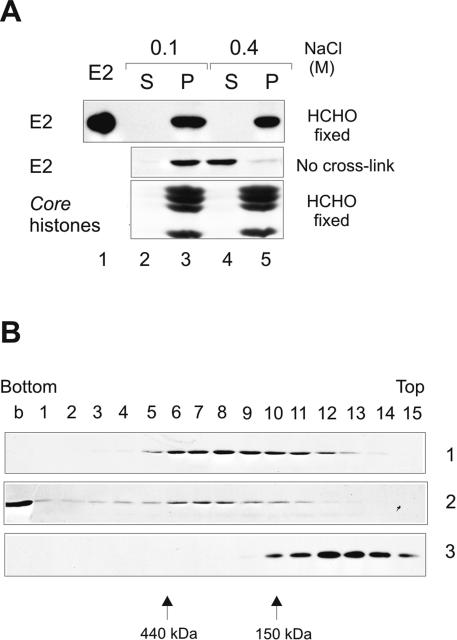
Biochemical fractionation of CHOBgl40 cells showed that E2 was extractable from chromatin-nuclear matrix fraction with the buffer containing 0.4 M salt. Next we fractionated CHOBgl40 cells as depicted in Fig. 1B, and the CSK plus 0.5% Triton X-100 pellet fraction (chromatin-enriched fraction) was cross-linked with 1% formaldehyde. Formaldehyde generates protein-protein and protein-DNA cross-links providing a snapshot of protein-protein and protein-DNA interactions within the cell. After cross-linking, the chromatin-enriched fraction was further incubated with buffer containing either 0.1 M or 0.4 M NaCl and separated by centrifugation into super (S) and pellet (P) fractions. As a result, the E2 protein became resistant to salt extraction (Fig. 4A, lanes 4 and 5), indicating that it was covalently cross-linked to DNA and/or salt-insoluble nuclear protein(s).
FIG. 4.
Biochemical analysis of localization of E2 after formaldehyde cross-link. CHOBgl40 cells were fractionated as depicted in Fig. 1B, and the CSK plus 0.5% Triton X-100 pellet fraction was cross-linked (upper panel) using 1% formaldehyde. After cross-linking, the pellet fraction was further incubated with buffer containing either 0.1 M (lanes 2 and 3) or 0.4 M (lanes 4 and 5) NaCl and separated by centrifugation into super (S) and pellet (P) fractions. The lower part of the gel was analyzed for the presence of histones (Coomassie blue staining). (B) Glycerol gradient centrifugation of the E2 protein released from chromatin by 0.4 M salt. Gel 1, a total of 1.5 × 107 CHOBgl40 cells were extracted as depicted in Fig. 1B, and the S fraction of 0.4 M salt treatment was centrifuged through glycerol as detailed in Materials and Methods. Gel 2, a total of 3 × 107 CHOBgl40 cells were extracted as depicted in Fig. 1B, and the S fraction of 0.4 M salt treatment was cross-linked with formaldehyde before loading onto the gradient. Glycerol gradient fractions were precipitated with trichloroacetic acid and subjected to SDS-PAGE and immunoblotted with E2-specific antibodies. Gel 3, the E2 protein (2 μg) purified from Escherichia coli was run in parallel gradient.
We also analyzed the glycerol gradient sedimentation profile of the E2 protein released from chromatin at 0.4 M NaCl. For this, the CHOBgl40 cells were fractionated as depicted in Fig. 1B and the proteins solubilized with 0.4 M NaCl were sedimented through 10 to 40% glycerol gradient. After centrifugation, 5-ml gradients were divided into 15 fractions and E2 was analyzed by immunoblotting. The E2 protein is a dimer with the size of 96 kDa in solution and the bacterially purified E2 protein sedimented in the gradient in the position of the dimeric protein (Fig. 4B, panel 3). The E2 protein released from chromatin by salt sedimented in the gradient in a broad peak mostly with a size bigger than expected for a free dimeric protein (Fig. 4B, panel 1). It was detected in fractions 6 to 10, which corresponds to ∼200- to 400-kDa complex (panel 1). We also performed the sedimentation analysis of a formaldehyde cross-linked E2-containing 0.4 M salt supernatant fraction. For this, we first fractionated the CHOBgl40 cells as depicted in Fig. 1B, and then the 0.4 M salt supernatant fraction was cross-linked with 1% formaldehyde before loading onto the glycerol gradient. As a result, the E2 protein was detected in the bottom of the gradient as well as in fractions 6 to 8 (Fig. 4B, panel 2). A weak signal of E2 was also detected in fractions 1 to 5. Probably the E2 protein-containing complex detected in panel 1 was cross-linked to other chromatin proteins and the resulted complex was big enough to sediment into the bottom of the gradient. Taken together, these data show that E2 associates with chromatin through cellular protein(s), which in turn is released from chromatin at 0.4 M salt. These data also suggest that there is very little free uncomplexed dimeric E2 in the nucleus of the cell; most of the protein is associated with some cellular proteins.
The E2 protein localizes into two different chromatin compartments.
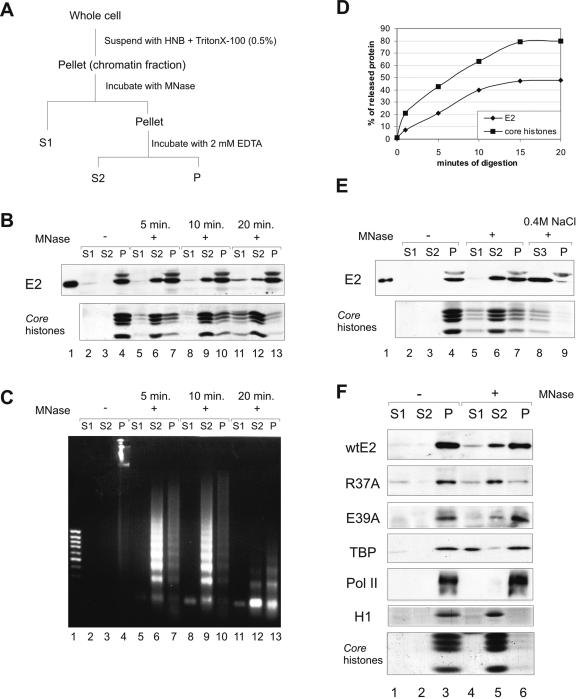
In order to investigate further the E2 association with cellular chromatin, we performed biochemical subfractionation of chromatin. For this, the CHOBgl40 cells were subjected to partial digestion with micrococcal nuclease and subsequent extraction with EDTA as depicted previously (39). Micrococcal nuclease is an enzyme that preferentially cleaves the linker DNA between adjacent nucleosomes, and the size of chromatin fragments depends on the extent of digestion. After nuclease treatment of CHOBgl40 cells for the indicated times and centrifugation and separation of the supernatant fraction S1, the pellet was resuspended in 2 mM EDTA, recentrifuged, and separated into a second supernatant fraction, S2, and a pellet, P (Fig. 5A). As shown by DNA analysis (Fig. 5C), fraction S1 was mainly composed of mononucleosomal-sized DNA fragments, whereas the S2 fraction contained a nucleosomal ladder of DNA fragments. The fraction P contained heterogenous-sized DNA that remained bound to the nuclear scaffold after the fractionation. The fractions S1, S2, and P were analyzed by immunoblot analysis, and the E2 protein was detected in fractions S2 and P (Fig. 5B lanes 6, 7, 9, and 10). Only after long incubation times, when the S2 fraction also contained mostly mononucleosomal DNA, the E2 protein was also detectable in fraction S1 (lane 11). Higher nuclease concentrations or prolonged incubation times failed to decrease the fraction of nuclease-resistant E2 protein; nearly 50% of E2 remained in fraction P even when more than 80% of the chromatin was detached by nucleases (Fig. 5D). Fraction S1 has been described to consist of open genetically active chromatin that is deficient in histone H1. Fraction S2 consists of more compact chromatin that contains histone H1 and the remaining insoluble fraction P includes the nuclear matrix but also nuclease-resistant chromatin including actively transcribed gene sequences (39). Their resistance against nuclease may be at least partially because of associated large protein complexes such as the RNA polymerase (15, 39) and the SWI/SNF chromatin remodeling complex (38) or the Orc1/Orc2 containing complex (16), which has been detected in fraction P. Our experiments showed that E2 occurs in two fractions: in the nuclease sensitive but compact chromatin fraction S2 and in the nuclease-resistant nuclear structures (P). Nuclease-resistant nuclear structures include nuclease-resistant chromatin and the nuclear matrix. Accordingly E2 may be associated either with chromatin or with the nuclear matrix or with both of them. The first explanation is that E2 is associated with chromatin that remained bound to nuclear pellet after nuclease digestion. However, because fraction P contained small amounts of DNA relative to E2 remained in this fraction, it is unlikely that E2 associated with chromatin only. The immunofluorescence analysis showed that part of the E2 protein remained bound to nuclear substructures after DNase I digestion (Fig. 2A, image 5), which also raised the possibility that part of E2 may associate with nuclear matrix. On the other hand, as shown on Fig. 5E, lane 8, the nuclease-resistant E2 was readily extracted with the buffer containing 0.4 M salt, which shows that E2 is not a classical nuclear matrix protein. Some proteins that are associated with the matrix in vivo may be stripped by salt extraction, and we suggest that some part of E2 may associate salt sensitively with nuclear matrix proteins or with transcription or other regulatory complexes instead of chromatin.
FIG. 5.
The E2 protein on chromatin. (A) Protocol for chromatin fractionation. (B) Nuclei from 2 × 107 CHOBgl40 cells were subjected to partial digestion with 30 U of micrococcal nuclease for the indicated times and separated into supernatant (S1) and pellet. The pellet was extracted with EDTA and centrifuged to yield supernatant (S2) and pellet (P) chromatin fractions. An equal volume of each fraction (extract of 106 cells) was subjected to SDS-PAGE and immunoblotted with E2-specific antibodies; the lower part of the gel was analyzed for the presence of histones (Coomassie blue staining). (C) DNA from each fraction was analyzed by agarose gel electrophoreses and stained with ethidium bromide. Lane 1, 1-kb DNA standard. (D) Mobilization of chromatin bound E2 by MNase. E2 and core histone bands obtained by treatment with 30 U of micrococcal nuclease were quantified by scanning and blotted together as a function of digestion times. The values of two independent experiments are expressed as percentages of total released material. (E) The pellet fraction from panel A was further incubated for 15 min with 0.4 M NaCl and separated into supernatant (S3) and pellet (P). All fractions were subjected to immunoblotting with E2-specific antibodies, and the lower part of the gel was analyzed for the presence of histones. (F) CHO cells were transfected with 200 ng of expression plasmids for E2 and E2 mutants R37A and E39A, and nuclei from 5 × 106 cells were subjected to digestion with 30 U of micrococcal nuclease for 10 min. An equal volume of each fraction was subjected to SDS-PAGE and immunoblotted with E2-specific antibodies and with anti-TBP and anti-Pol II antibodies to detect endogenous proteins; the lower part of the gel was analyzed for the presence of histones (Coomassie blue staining).
Limited nuclease digestion has been used to distinguish between transcriptionally active and inactive chromatin (38). Next we followed the distribution of E2 transcription-deficient mutant R37A (2) in this assay. For this, the CHO cells were transfected with 200 ng of expression plasmids for wild-type E2 (wtE2) or E2 mutants R37A and E39A and were subjected to digestion with micrococcal nuclease and subsequent extraction with EDTA. We tried to keep the E2 protein level as low as possible in order to avoid artifacts caused by overexpression of E2 (Fig. 1D). Our results showed that in transiently transfected cells, 65% of wtE2 protein retained the nuclease-resistant nuclear structures (P) (Fig. 5F, lane 6). At the same time, transcription-deficient E2 mutant R37A was detected mostly in fraction S2 and only 15% of the protein retained in fraction P (Fig. 5F, lanes 5 and 6). The transcription-competent E2 mutant E39A behaved similar to wtE2 (Fig. 5F). These results suggested that the nuclease-resistant nuclear structures (fraction P) contained the transcription-competent E2 but not the transcription-deficient E2 protein. The Coomassie blue staining of histones of the lower part of the same gel was used to follow the fractionation. As shown on Fig. 5F, the core histones as well as H1 were detected mostly in fraction S2. For comparison, the same filter was also incubated with antibodies against the general transcription factor TBP (TATA-binding protein) and RNA Pol II. Previous works have shown that some part of TBP is associated with nuclease-sensitive chromatin, and some part is present in transcription complexes associated with the nuclear substructure (15). Indeed, TBP signal was detected mostly in fractions S1 and P (Fig. 5F) that is in regions highly sensitive to micrococcal nuclease and in the fractions of the transcriptionally active chromatin that remain bound to the nuclear pellet after digestion. RNA Pol II was detected only in fraction P (Fig. 5F) as shown previously (15, 38). These data together confirm that the transcriptional activity is engaged with fractions S1 and P.
In summary, our data suggest that there are two pools of the E2 protein in the nucleus of the cell: one that localizes on transcriptionally inactive compact chromatin and the other, which compartmentalizes to transcriptionally active nuclear structures of the cell.
The URR-containing plasmid cofractionates with E2 into two different chromatin fractions.
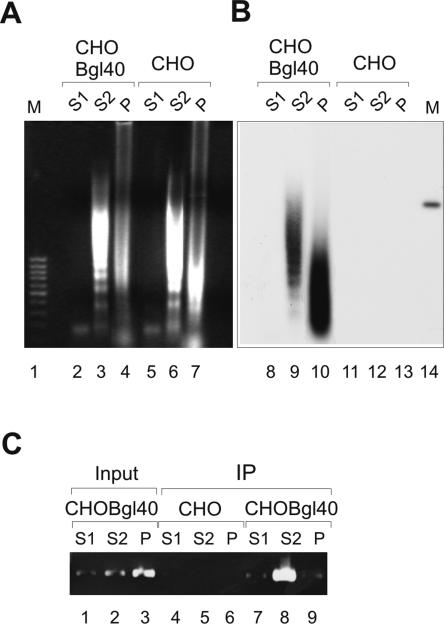
We also examined the distribution of URR-containing plasmid after chromatin fractionation with MNase. The CHOBgl40 cells were treated with MNase as depicted above (Fig. 5A), and the DNA was isolated from each fraction (Fig. 6A). DNA was then transferred onto a membrane and hybridized with a radiolabeled BPV1 URR-specific probe. As shown in Fig. 6B, lanes 9 and 10, the URR-specific signal was detected in fraction S2 as well as in fraction P. The URR containing plasmid Bgl40 is stably replicated in CHOBgl40 cells, and it is also packed into nucleosomes, therefore the MNase treatment reveals a specific ladder of DNA (Fig. 6B); linearized Bgl40 plasmid is shown on line 14. The CHO cells, which do not contain the Bgl40 plasmid, are negative for URR-specific signal (Fig. 6A, lanes 5 to 7, and B, lanes 12 and 13). These data show that the URR-containing plasmid fractionates into two different chromatin fractions: in the nuclease-sensitive but compact chromatin fraction S2 and in the nuclease-resistant nuclear structures (P).
FIG. 6.
The chromatin fractionation of URR-containing plasmid. (A) The nuclei from 107 CHO and CHOBgl40 cells were treated with 30 U of MNase as depicted above (Fig. 5A), and the DNA isolated from each fraction was transferred onto a membrane and hybridized with radiolabeled BPV1 URR-specific probe. Lane 1, 1-kb DNA standard. Lane 14, 100 pg of linearized plasmid Bgl40. (B) After MNase treatment of CHO and CHOBgl40 cells (Fig. 5A), the S1, S2, and P fractions were separated and cross-linked with 1% formaldehyde. The E2/DNA complexes were immunoprecipitated with E2-specific MAb 3E8, and the presence of URR-containing plasmid in the DNA/protein complexes recovered was analyzed by PCR using primers that specifically amplify a region of 7338 to 7654 bp of BPV1 genome. Lanes 1 to 3, the input chromatin from each fraction was tested in parallel with the immunoprecipitated DNA. The image of an agarose gel stained with ethidium bromide is shown.
To find out whether the URR-containing plasmid was tethered to cellular chromatin via the viral E2 protein, we performed the chromatin immunoprecipitation analysis. For this, after MNase treatment of CHO and CHOBgl40 cells (Fig. 5A), the S1, S2, and P fractions were separated and cross-linked with 1% formaldehyde followed by sonication. The E2/DNA complexes were immunoprecipitated with E2-specific MAb 3E8 recognizing the conformational epitope in the C-terminal DNA-binding domain (18), and the presence of URR-containing plasmid in the DNA/protein complexes recovered was analyzed by PCR using primers that specifically amplify a URR region of BPV1 genome. As shown in Fig. 6C, URR DNA could be detected in complex with E2 in compact chromatin fraction S2 (lane 8). The PCR product was not seen in CHO cells, which do not harbor URR-containing plasmid, indicating that the signal is specific for Bgl40 plasmid. In fractions S1 and P, the PCR product was detected only on background level (lanes 7 and 9). Fraction P contained Bgl40 plasmid (Fig. 6B, lane 10) as well as 50% of E2 signal after MNase treatment (Fig. 5), but we were not able to immunoprecipitate the E2 protein from this fraction because most of the protein remained insoluble after sonication.
DISCUSSION
The papillomavirus E2 proteins localize in the nucleus of the cell, where they are concentrated in numerous small dots, scattered throughout the nucleus as determined by immunofluorescence analysis. In this study we show that the BPV1 E2 protein, or at least some part of it, associates with cellular chromatin during the entire cell cycle. The N-terminal transactivation domain, but not the C-terminal DNA-binding domain, of the E2 protein, is responsible for this association. The majority of the full-length E2 protein can only be detected in chromatin-enriched fraction but not as a free protein in the nucleus. Our conclusions are based on different observations. First, the E2 protein fractionated biochemically into nonionic detergent resistant chromatin-nuclear matrix fraction. The E2 protein association with this fraction was salt sensitive. It was released from the fraction by 0.3 M salt, under conditions where chromatin components are extractable. Second, E2 was released from chromatin by DNase and MNase but not by RNase treatment.
Our data presented here show that the BPV1 E2 protein associates with cellular chromatin in interphase cells. In our study, the E2 protein was extractable from chromatin at 0.3 M salt; under these conditions the histone H1 was not yet released from the chromatin, so E2 could be released without disruption of the chromatin higher structure. We speculate that E2 associates preferably with compact, highly packed chromatin. The association of E2 with chromatin was mediated by cellular protein(s), which in turn were released from chromatin by salt as shown by in vivo cross-linking analysis. Furthermore, after mobilizing E2 from chromatin either with MNase or salt, E2 still remains associated with some cellular protein(s) as the E2-containing complex sedimented in the glycerol gradient with the size bigger than expected for a free uncomplexed dimeric protein.
The chromatin association was observed in the CHOBgl40 cell line, which expresses the E2 protein from heterologous promoters (35) as well as in BPV1-transformed C127 cells. These data are consistent with earlier work where the subcellular localization of E2 proteins in BPV1-transformed cells was determined. Hubbert et al. have shown that the full-length E2 protein associates with the nonionic detergent-insoluble fraction (13). However, they did not separate this fraction further and the exact compartment remained unknown. The E2 protein of human papillomavirus type 11 (HPV11) has been shown to associate with nuclear matrix (48). Nuclear matrix is a fibrogranular structure which remains after the nucleus has been treated with DNase and high salt (8). We also observed that 5 to 10% of E2 always retained attached to nuclear substructures after elution with 0.4 M salt (Fig. 1B), but the majority of E2 was extractable. It is possible that the E2 proteins of BPV1 and HPV11 compartmentalize differently within the cell nucleus; BPV1 E2 associates preferentially with chromatin and HPV11 E2 with nuclear matrix. This hypothesis is supported by the fact that these two viruses use different targets to enable viral DNA partitioning (50). On the other hand, the biochemical methods used have also some limitations. Some proteins that are associated with nuclear substructures in vivo may be stripped by salt extraction, and some proteins may coagulate under high-salt conditions and be thereby artificially detected as matrix elements. We suggest that the salt solubility does not reflect the association of E2 with chromatin only. It is clear that some part of E2 is associated with cellular chromatin, but this part of the E2 protein, which was not released by DNase or Mnase treatment, may localize to some other nuclear compartment. One possibility is that this part of E2 may compartmentalize into transcription initiation sites localizing in the nuclear-resistant chromatin as E2 interactions with TFIID and RNA Pol II have been shown to be salt sensitive (53).
In transiently transfected cells the E2 protein occurs in two fractions: one that associates with chromatin similar to E2 expressed by stable cell line and a second fraction that remains bound to nuclear substructures after extraction with moderate salt concentrations (Fig. 1D). We suggest that in case of overexpression the E2 protein tends to aggregate and therefore accumulate in some other foci. The other explanation is that the amount of E2 molecules that can bind to cellular chromatin is limited and is probably dependent on the potent binding sites on chromatin. When E2 binding sites on chromatin are occupied, the protein is concentrated in some other, probably storage sites within the cell nucleus.
Limited micrococcal nuclease digestion revealed that the E2 protein partitioned to different chromatin regions. A considerable fraction of the E2 protein was located at nuclear sites that are resistant against nuclease attack, whereas the remaining E2 resided on compact chromatin accessible to micrococcal nuclease. Thus, there are two pools of the E2 protein in the nucleus of the cell: one that localizes on transcriptionally inactive, compact chromatin and the other, which compartmentalizes to transcriptionally active nuclear structures of the cell. The URR-containing plasmid Bgl40 cofractionated with E2 into the same chromatin fractions: into compact chromatin fraction S2 and transcriptionally active chromatin fraction P. The URR-containing plasmid Bgl40 coimmunoprecipitated with E2 from compact chromatin fraction S2, which shows that URR-containing plasmid is linked to heterochromatin via the E2 protein. We can hypothesize that separate pools of E2 locating differently within the cell nucleus are responsible for different activities, for example for segregation and transcription activity of protein. On the other hand, some of these sites may represent storage sites from which protein can be recruited if necessary.
Papillomaviruses are small viruses, and they have to utilize the cellular transcription and replication machinery to support their gene expression and viral DNA replication. E2 is a transcription factor, which interacts with multiple proteins of the transcription machinery and is able to recruit holoenzymes to promoters (53). Therefore, it was not surprising that E2 fractionated into transcriptionally active nuclease-resistant nuclear structures containing active Pol II complex (15, 38). Another important nuclear process, DNA replication, is also associated with nuclease-resistant nuclear structures (52). E1 and E2 are the only virally encoded proteins required for the initiation of papillomavirus DNA replication in mammalian cells (49), the other replication factors are provided by host cell. It is not exactly known where the papillomavirus replication takes place. The formation of HPV replication compartments in transiently transfected cells on the basis of colocalization of viral E1 and E2 proteins, the HPV ori plasmid, the host RPA, and BrdU incorporation has been proposed (47). However, it is not yet clear that these HPV replication compartments are distinct domains or that the E1 and E2 proteins are targeted to restricted host replication sites. E2 has been reported to associate with RPA (26), and E1 and E2 could be targeted to a limited number of domains where RPA is normally found, and the E2 protein can play a role in recruiting E1 to replication sites. We suggest that some part of E2 found in nuclease-resistant nuclear structures can be associated also with cellular replication compartments.
Part of the E2 protein in the nucleus was associated with compact, transcriptionally inactive chromatin. This pool of E2 is probably not involved in transcription, and its function remains unclear. One possibility is that this is some kind of storage site of E2. The other possibility is that this part of E2 is required for segregation of viral genomes. Recently it was shown that BPV1 E2 interacts with BET family protein Brd4, and this interaction tethers the viral DNA to host mitotic chromosomes (6, 54). Brd4 binds to acetylated chromatin during interphase and mitosis and is extracted from chromatin with low salt (10) (Fig. 2B). Thus, it is a potent candidate for mediating interaction of E2 with heterochromatin. Unfortunately, we were not able to follow the distribution of Brd4 after chromatin fractionation with MNase, because antibodies against the C terminus of Brd4 were not able to recognize denaturated protein in immunoblot analysis. However, in this study we could not detect any differences in E2 binding to mitotic versus interphase chromatin. In both cases the association was salt sensitive and resistant to RNase treatment. This allows us to speculate that the E2 binding cellular factor can be the same in interphase and in mitosis.
The cell nucleus is increasingly recognized a spatially organized structure. If spatiotemporal regulation occurs, it would require subnuclear organization of transcription factors. Recent findings have shown that transcription factors are not homogenously distributed within the nuclear volume but rather are enriched at steady-state levels in discrete nuclear structures. Interestingly, studies that have examined codistribution of transcription factor foci with sites of transcription or RNA polymerase II show only minor overlap (12). Furthermore, different transcription factors generally occupy separate sites (11). It has proposed that many of the transcription factor-rich nuclear domains are not actively involved in transcription; they may represent storage sites from which proteins can be recruited if necessary. Transcription factor foci compartments are dynamic, with transcription factors continually exchanging into and out of these domains (11). We hypothesize that E2 is also pretty mobile in the nucleus and interacts with chromatin with a rapid on and off mode of binding, similar to other nuclear proteins. We speculate that in living cells, the E2 protein associates temporally with different nuclear compartments and protein complexes required for its functioning.
Acknowledgments
We are indebted to Aare Abroi for valuable discussions and help during the course of this work and to N. Hernandez for providing TBP antibodies and A. Sikut for CD43 antibodies.
This study was supported in part by grant 5994 from the Estonian Science Foundation to R.K. and grant INTNL 55000339 from the Howard Hughes Medical Institute to M.U.
REFERENCES
- 1.Abroi, A., I. Ilves, S. Kivi, and M. Ustav. 2004. Analysis of chromatin attachment and partitioning functions of the bovine papillomavirus type 1 E2 protein. J. Virol. 78:2100-2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abroi, A., R. Kurg, and M. Ustav. 1996. Transcriptional and replicational activation functions in the bovine papillomavirus type 1 E2 protein are encoded by different structural determinants. J. Virol. 70:6169-6179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allikas, A., D. Ord, R. Kurg, S. Kivi, and M. Ustav. 2001. Roles of the hinge region and the DNA binding domain of the bovine papillomavirus type 1 E2 protein in initiation of DNA replication. Virus Res. 75:95-106. [DOI] [PubMed] [Google Scholar]
- 4.Androphy, E., D. Lowy, and J. Schiller. 1987. Bovine papillomavirus E2 trans-activating gene product binds to specific sites in papillomavirus DNA. Nature 325:70-73. [DOI] [PubMed] [Google Scholar]
- 5.Bastien, N., and A. McBride. 2000. Interaction of the papillomavirus E2 protein with mitotic chromosomes. Virology 270:124-134. [DOI] [PubMed] [Google Scholar]
- 6.Baxter, M. K., M. G. McPhillips, K. Ozato, and A. A. McBride. 2005. The mitotic chromosome binding activity of the papillomavirus E2 protein correlates with interaction with the cellular chromosomal protein, Brd4. J. Virol. 79:4806-4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benson, J., R. Lawande, and P. Howley. 1997. Conserved interaction of the papillomavirus E2 transcriptional activator proteins with human and yeast TFIIB proteins. J. Virol. 71:8041-8047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berezney, R. 2002. Regulating the mammalian genome: the role of nuclear architecture. Adv. Enzyme Reg. 42:39-52. [DOI] [PubMed] [Google Scholar]
- 9.Breiding, D. E., F. Sverdrup, M. J. Grossel, N. Moscufo, W. Boonchai, and E. J. Androphy. 1997. Functional interaction of a novel cellular protein with the papillomavirus E2 transactivation domain. Mol. Cell. Biol. 17:7208-7219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dey, A., F. Chitsaz, A. Abbasi, T. Misteli, and K. Ozato. 2003. The double bromodomain protein Brd4 binds to acetylated chromatin during interphase and mitosis. Proc. Natl. Acad. Sci. USA 100:8758-8763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grande, M., I. Van der Kraan, L. De Jong, and R. Van Driel. 1997. Nuclear distribution of transcription factors in relation to sites of transcription and RNA polymerase II. J. Cell Sci. 110:1781-1791. [DOI] [PubMed] [Google Scholar]
- 12.Henzel, M., M. Kruhlak, N. MacLean, F.-M. Boisvert, M. Lever, and D. Bazett-Jones. 2001. Compartmentalization of regulatory proteins in the cell nucleus. J. Steroid Biochem. Mol. Biol. 76:9-21. [DOI] [PubMed] [Google Scholar]
- 13.Hubbert, N., J. Schiller, D. Lowy, and E. Androphy. 1988. Bovine papilloma virus-transformed cells contain multiple E2 proteins. Proc. Natl. Acad. Sci. USA 85:5864-5868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ilves, I., S. Kivi, and M. Ustav. 1999. Long-term episomal maintenance of bovine papillomavirus type 1 plasmids is determined by attachment to host chromosomes, which is mediated by the viral E2 protein and its binding sites. J. Virol. 73:4404-4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kimura, H., Y. Tao, R. Roeder, and P. Cook. 1999. Quantitation of RNA polymerase II and its transcription factors in an HeLa cell: little soluble holoenzyme but significant amounts of polymerase attached to the nuclear substructure. Mol. Cell. Biol. 19:5383-5392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kreitz, S., M. Ritzi, M. Baack, and R. Knippers. 2001. The human recognition complex protein 1 dissociates from chromatin during S phase in HeLa cells. J. Biol. Chem. 276:6337-6342. [DOI] [PubMed] [Google Scholar]
- 17.Kristjuhan, A., J. Walker, N. Suka, M. Grunstein, D. Roberts, B. R. Cairns, and J. Q. Svejstrup. 2002. Transcriptional inhibition of genes with severe histone h3 hypoacetylation in the coding region. Mol. Cell 10:925-933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kurg, R., J. Parik, E. Juronen, T. Sedman, A. Abroi, I. Liiv, U. Langel, and M. Ustav. 1999. Effect of bovine papillomavirus E2 protein-specific monoclonal antibodies on papillomavirus DNA replication. J. Virol. 73:4670-4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lambert, P., N. Dostatni, A. McBride, M. Yaniv, P. Howley, and B. Arcangioli. 1989. Functional analysis of the papilloma virus E2 trans-activator in Saccharomyces cerevisiae. Genes Dev. 3:38-48. [DOI] [PubMed] [Google Scholar]
- 20.Lamond, A. I., and W. C. Earnshaw. 1998. Structure and function in the nucleus. Science 280:547-553. [DOI] [PubMed] [Google Scholar]
- 21.Law, M., D. Lowy, I. Dvorezky, and P. Howley. 1981. Mouse cells transformed by bovine papillomavirus contain only extra-chromosomal viral DNA sequences. Proc. Natl. Acad. Sci. USA 78:2727-2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee, D., S. Hwang, J. Kim, and J. Choe. 2002. Functional interaction between p/CAF and human papillomavirus E2 protein. J. Biol. Chem. 277:6483-6489. [DOI] [PubMed] [Google Scholar]
- 23.Lee, D., B. Lee, J. Kim, D. Kim, and J. Choe. 2000. cAMP response element-binding protein-binding protein binds to human papillomavirus E2 protein and activates E2-dependent transcription. J. Biol. Chem. 275:7045-7051. [DOI] [PubMed] [Google Scholar]
- 24.Lehman, C. W., and M. R. Botchan. 1998. Segregation of viral plasmids depends on tethering to chromosomes and is regulated by phosphorylation. Proc. Natl. Acad. Sci. USA 95:4338-4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li, R., and M. Botchan. 1994. Acidic transcription factors alleviate nucleosome-mediated repression of DNA replication of bovine papillomavirus type 1. Proc. Natl. Acad. Sci. USA 91:7051-7055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li, R., and M. Botchan. 1993. The acidic transcriptional activation domains of VP16 and p53 bind the cellular replication protein A and stimulate in vitro BPV-1 DNA replication. Cell 73:1207-1221. [DOI] [PubMed] [Google Scholar]
- 27.Li, R., J. Knight, S. Jackson, R. Tjian, and M. Botchan. 1991. Direct interaction between Sp1 and the BPV enhancer E2 protein mediates synergistic activation of transcription. Cell 65:493-505. [DOI] [PubMed] [Google Scholar]
- 28.Lim, C., C. Choi, and J. Choe. 2004. Mitotic chromosome-binding activity of latency-associated nuclear antigen 1 is required for DNA replication from terminal repeat sequence of Kaposi's Sarcoma-associated herpesvirus. J. Virol. 78:7248-7256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lobo, S. M., M. Tanaka, M. L. Sullivan, and N. Hernandez. 1992. A TBP complex essential for transcription from TATA-less but not TATA-containing RNA polymerase III promoters is part of the TFIIIB fraction. Cell 71:1029-1040. [DOI] [PubMed] [Google Scholar]
- 30.McBride, A., J. Byrne, and P. Howley. 1989. E2 polypeptides encoded by bovine papillomavirus type 1 form dimers through the common carboxyl-terminal domain: transactivation is mediated by the conserved amino-terminal domain. Proc. Natl. Acad. Sci. USA 86:510-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morrissey, L., J. Barsoum, and E. Androphy. 1989. Trans-activation by the bovine papillomavirus E2 protein in Saccharomyces cerevisiae. J. Virol. 63:4422-4425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Müller, A., A. Ritzkowsky, and G. Steger. 2002. Cooperative activation of human papillomavirus type 8 gene expression by the E2 protein and the cellular coactivator p300. J. Virol. 76:11042-11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Okuno, Y., A. McNairn, N. den Elzen, J. Pines, and D. Gilbert. 2001. Stability, chromatin association and functional activity of mammalian pre-replication complex proteins during the cell cycle. EMBO J. 20:4263-4277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Patturajan, M., X. Wei, R. Berezney, and J. L. Corden. 1998. A nuclear matrix protein interacts with the phosphorylated C-terminal domain of RNA polymerase II. Mol. Cell. Biol. 18:2406-2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Piirsoo, M., E. Ustav, T. Mandel, A. Stenlund, and M. Ustav. 1996. Cis and trans requirements for stable episomal maintenance of the BPV-1 replicator. EMBO J. 15:1-11. [PMC free article] [PubMed] [Google Scholar]
- 36.Rank, N., and P. Lambert. 1995. Bovine papillomavirus type 1 E2 transcriptional regulators directly bind two cellular transcription factors, TFIID and TFIIB. J. Virol. 69:6323-6334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rehtanz, M., H. Schmidt, U. Warthorst, and G. Steger. 2004. Direct interaction between nucleosome assembly protein 1 and the papillomavirus E2 proteins involved in activation of transcription. Mol. Cell. Biol. 24:2153-2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reyes, J., C. Muchardt, and M. Yaniv. 1997. Components of the human SWI/SNF complex are enriched in active chromatin and are associated with the nuclear matrix. J. Cell Biol. 137:263-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rose, S., and W. Garrard. 1984. Differentiation-dependent chromatin alterations precede and accompany transcription of immunoglobulin light chain genes. J. Biol. Chem. 259:8534-8544. [PubMed] [Google Scholar]
- 40.Sedman, J., and A. Stenlund. 1995. Co-operative interaction between the initiator E1 and the transcriptional activator E2 is required for replicator specific DNA replication of bovine papillomavirus in vivo and in vitro. EMBO J. 14:6218-6228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sikut, R., C. X. Andersson, A. Sikut, J. Fernandez-Rodriguez, N. G. Karlsson, and G. C. Hansson. 1999. Detection of CD43 (leukosialin) in colon adenoma and adenocarcinoma by novel monoclonal antibodies against its intracellular domain. Int. J. Cancer 82:52-58. [DOI] [PubMed] [Google Scholar]
- 42.Skiadopoulos, M., and A. McBride. 1996. The bovine papillomavirus type 1 E2 transactivator and repressor proteins use different nuclear localization signals. J. Virol. 70:1117-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Skiadopoulos, M., and A. McBride. 1998. Bovine papillomavirus type 1 genomes and the E2 transactivator protein are closely associated with mitotic chromatin. J. Virol. 72:2079-2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Spalholz, B., Y. Yang, and P. Howley. 1985. Transactivation of a bovine papilloma virus transcriptional regulatory element by the E2 gene product. Cell 42:183-191. [DOI] [PubMed] [Google Scholar]
- 45.Stanway, C., M. Sowden, L. Wilson, A. Kingsman, and S. Kingsman. 1989. Efficient activation of transcription in yeast by the BPV1 E2 protein. Nucleic Acids Res. 17:2187-2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Steger, G., J. Ham, O. Lefebvre, and M. Yaniv. 1995. The bovine papillomavirus 1 E2 protein contains two activation domains: one that interacts with TBP and another that functions after TBP binding. EMBO J. 14:329-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Swindle, C. S., N. Zou, B. A. Van Tine, G. M. Shaw, J. A. Engler, and L. T. Chow. 1999. Human papillomavirus DNA replication compartments in a transient DNA replication system. J. Virol. 73:1001-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zou, N., B. Lin, F. Duan, K.-Y. Lee, G. Jin, R. Guan, G. Yao, E. Lefkowitz, T. Broker, and L. Chow. 2000. The hinge of the human papillomavirus type 11 E2 protein contains major determinants for nuclear localization and nuclear matrix association. J. Virol. 74:3761-3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ustav, M., and A. Stenlund. 1991. Transient replication of BPV-1 requires two viral polypeptides encoded by the E1 and E2 open reading frames. EMBO J. 10:449-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Van Tine, B., L. Dao, S.-Y. Wu, T. Sonbuchner, B. Lin, N. Zou, C.-M. Chiang, T. Broker, and L. Chow. 2004. Human papillomavirus (HPV) origin-binding protein associates with mitotic spindles to enable viral DNA partitioning. Proc. Natl. Acad. Sci. USA 101:4030-4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Voitenleitner, C., and M. Botchan. 2002. E1 protein of bovine papillomavirus type 1 interferes with E2 protein-mediated tethering of the viral DNA to mitotic chromosomes. J. Virol. 76:3440-3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wei, X., J. Samarabandu, R. Devdhar, A. Siegel, R. Acharya, and R. Berezney. 1998. Segregation of transcription and replication sites into higher order domains. Science 281:1502-1505. [DOI] [PubMed] [Google Scholar]
- 53.Wu, S., and C. Chiang. 2001. TATA-binding protein-associated factors enhance the recruitment of RNA polymerase II by transcriptional activators. J. Biol. Chem. 276:34235-34243. [DOI] [PubMed] [Google Scholar]
- 54.You, J., J. Croyle, A. Nishimura, K. Ozato, and P. Howley. 2004. Interaction of the bovine papillomavirus E2 protein with brd4 tethers the viral DNA to host mitotic chromosomes. Cell 117:349-360. [DOI] [PubMed] [Google Scholar]