Abstract
The expression of a membrane-anchored gp41-derived peptide (M87) has been shown to confer protection from infection through human immunodeficiency virus type 1 (HIV-1) (Hildinger et al., J. Virol. 75:3038-3042, 2001). In an effort to characterize the mechanism of action of this membrane-anchored peptide in comparison to the soluble peptide T-20, we selected resistant variants of HIV-1NL4-3 and HIV-1BaL by serial virus passage using PM1 cells stably expressing peptide M87. Sequence analysis of the resistant isolates showed different patterns of selected point mutations in heptad repeat regions 1 and 2 (HR1 and HR2, respectively) for the two viruses analyzed. For HIV-1NL4-3 a single amino acid change at position 33 in HR1 (L33S) was selected, whereas for HIV-1BaL the majority of the sequences obtained showed two amino acid changes, one in HR1 and one in HR2 (I48V/N126K). In both selections the most important contiguous 3-amino-acid sequence, GIV, within HR1, associated with resistance to soluble T-20, was not changed. Site-directed mutagenesis studies confirmed the importance of the characterized point mutations to confer resistance to M87 as well as to soluble T-20 and T-649. Replication capacity and dual-color competition assays revealed that the double mutation I48V/N126K in HIV-1BaL results in a strong reduction of viral fitness, whereas the L33S mutation in HIV-1NL4-3 did enhance viral fitness compared to the respective parental viruses. However, the selected point mutations did not confer resistance to the more recently described optimized membrane-anchored fusion inhibitor M87o (Egelhofer et al., J. Virol. 78:568-575, 2004), strengthening the importance of this novel antiviral concept for gene therapy approaches.
The development of highly active antiviral therapy has resulted in greater expected life spans and slower progression to AIDS for patient infected with human immunodeficiency virus (HIV) (25, 26). However, the emergence of drug-resistant viruses during treatment and extensive subsequent intraclass cross-resistance has been a limiting factor in the success of treatment (5, 6). Due to this limitation, antiretrovirals with a novel mechanism of action have been the focus of extensive drug discovery research.
Enfuvirtide is one novel HIV-1 inhibitor already in clinical use; it blocks fusion of viral and cellular membranes by binding to heptad repeat region 1 (HR1) of virus gp41 (10, 15). Enfuvirtide is homologous to a segment of the HR2 region of gp41 corresponding to amino acids 127 to 162 and binds to the HR1 region of gp41 (39, 40). Enfuvirtide interferes with the formation of the six-helix bundle, composed of an inner coiled-coil trimer of the HR1 and HR2 regions aligned in an antiparallel manner, thereby blocking the final step of virus entry, the fusion of the viral membrane with the target cell membrane (28). A slightly longer, 39-amino-acid peptide, T-1249, binds to a region of HR1 that overlaps the binding site of enfuvirtide and is active against HIV-1, HIV-2, and simian immunodeficiency virus (11). In addition, several other fusion inhibitors have been studied, for example, T-649 (corresponding to amino acids 117 to 152 of gp41) and C34 (corresponding to amino acid 117 to 150 of gp41). Both block the formation of the hairpin structure in a way similar to enfuvirtide (4, 34, 40).
A wide range of susceptibility to enfuvirtide has been described in virus isolates from naive patients (13, 16, 18). Early in vitro studies using enfuvirtide showed the development of resistance is associated with changes in a conserved amino acid triad (GIV) at positions 36 to 38 in the HR1 region of gp41 (34). These findings were confirmed by site-directed mutagenesis experiments and in vivo studies which expanded the core region of functional importance to amino acids 36 to 45 (16, 30, 35, 38). Single amino acid substitutions in this region are the most common and cause various degrees of susceptibility loss. Double amino acid substitutions have also been observed and these are associated with the highest levels of resistance, some combinations (G36S/V38M) exhibiting an approximately 100-fold reduction in enfuvirtide susceptibility (30, 34, 38). In addition, changes in HR2 may equally be involved in enfuvirtide resistance as well (20, 36). Most variants that are resistant to enfuvirtide maintain susceptibility to T-1249 and T-649 (12, 34).
The rapid emergence of enfuvirtide-resistant viruses and the lack of oral bioavailability are major obstacles that hinder widespread application of enfuvirtide as part of an extended therapy option (5). To overcome these problems, the enfuvirtide peptide was engineered for expression on the cell membrane, leading to a high local concentration of peptide at the site of action (14). Surface expression was achieved by fusing an N-terminal signal peptide and a C-terminal scaffold consisting of a hinge and a membrane anchor to the antiviral peptide M87. This membrane-anchored peptide was expressed from a retroviral vector (pM87) and had good antiviral activity in cell lines.
Recently, we developed a retroviral vector expressing a membrane-anchored antiviral peptide that was also highly effective in primary cells and had minimal potential immunogenicity and no detectable toxicity (8). This membrane-anchored peptide was expressed from an optimized retroviral vector (pM87o) and encodes a larger antiviral peptide 46 amino acids in length (M87o). This extended membrane-anchored antiviral peptide can be viewed as a combination of the previously described antiviral peptides T-20 and T-649 (or C34) and is almost similar to DP207, a soluble peptide 45 amino acids in length shown to be a potent fusion inhibitor (40).
In an effort to characterize the mechanism of action of this membrane-anchored peptide in comparison to the soluble peptide T-20, we selected resistant variants of HIV-1NL4-3 and HIV-1BaL by serial virus passage using PM1 cells stably expressing peptide M87. Sequence analysis of the resistant isolates showed different patterns of selected point mutations in heptad repeat regions 1 and 2 for the two viruses analyzed. Site-directed mutagenesis studies confirmed the importance of the characterized point mutations (L33S, I48V, and N126K) to confer resistance to M87 as well as to soluble T-20 and to T-649. In addition, replication capacity and dual-color competition assays revealed that the double mutation I48V/N126K in HIV-1BaL results in a strong reduction of viral fitness, whereas the L33S mutation in HIV-1NL4-3 did enhance viral fitness compared to the parental viruses.
MATERIALS AND METHODS
Cells and viruses.
The T-cell line PM1, a subclone of HuT78 expressing CD4, CXCR4, and CCR5, and the transduced cell lines PM1-M87 and PM1-M87o have been described (8, 14, 19). TZM-bl have been described (38) and were transduced using vesicular stomatitis virus G protein pseudotyped particles containing the transfer vectors M87 and M87o as described (8, 14), resulting in cell lines TZM-M87 and TZM-M87o, respectively. The HIV-1BaL isolate was propagated on PM1 cells as was the molecular clone HIV-1NL4-3. T-20-resistant gp41 variants (NL/GIA, NL/DTV, NL/DIM) were described previously (8, 38) and cloned into the HIV-1NL4-3-based viral vector expressing Renilla luciferase in the position of nef (TN7-NL) (23). T-20-insensitive gp41 variants have been characterized previously (TN7-R14, TN7-X10, and TN7-X23) (13, 23).
Determination of cell surface expression of membrane-anchored peptides by a quantitative flow cytometry assay.
Quantification of the antibody-binding capacity of the C peptide-expressing cell lines was performed as described before by Lee et al. (17) with minor modifications. Monoclonal 2F5 antibody was labeled with fluorescein isothiocyanate using the Fluorotag conjugation kit (Sigma Aldrich, Munich, Germany). Conjugation was carried out according to the manufacturer's protocol. Approximately 106 M87 peptide or M87o peptide-expressing cells from an exponentially growing culture were harvested by centrifugation. Supernatant was removed and cells were labeled with the antibody 2F5-fluorescein isothiocyanate for 30 min at 4°C. In addition, 50 μl Quantum Simply cellular beads (≈106 beads with defined immunoglobulin G-binding capacities, Bangs Laboratories Inc., Fishers, IN) were labeled with different amounts of 2F5-fluorescein isothiocyanate for 30 min at 4°C. After the incubation, cells and beads were washed twice with 2 ml phosphate-buffered saline-2% fetal calf serum. Samples were than resuspended in 300 μl phosphate-buffered saline-2% fetal calf serum. In each sample 2 × 104 living cells and the same amount of beads were analyzed by flow cytometry. The geometric mean fluorescence of each population was determined by flow cytometry and plotted against their individually immunoglobulin G-binding capacity to generate a linear regression curve. This curve was then used to convert the geometric mean fluorescence of each M87 peptide and M87o peptide-expressing cell line into antibody-binding capacity.
Reverse transcription-PCR and site-directed mutagenesis.
Viral genomic RNA was extracted using cell-free supernatant from PM1-M87 cell cultures infected with the passaged viruses HIVNL4-3 and HIVBaL (QIAamp viral RNA kit, QIAGEN, Germany). After reverse transcription using an oligo(dT) primer the entire envelope gene was amplified as described earlier (7, 23) and cloned into pCR4-TOPO (Invitrogen, Germany). Sequence analysis of the HR1 and HR2 regions of gp41 was performed as described (38). The observed point mutations selected due to the passage of HIVNL4-3 and HIVBaL on PM1-M87 cells were introduced into the MP11-env expression vectors using the QuikChange XL site-directed mutagenesis kit (Stratagene, Germany) and confirmed by sequence analysis. The MP11-env expression vector system has been described previously (33). These modified envelope genes were also cloned into the vector TN7, resulting in replication-competent virus variants (TN7-NL/S, TN7-BaL/V, TN7-BaL/K, and TN7-BaL/VK).
Production and titration of virus stocks.
To generate infectious virus stocks, 293T cells (106 cells) were transfected with 3 μg DNA and 9 μl Metafectene (Biontex, Germany) according to the manufacturer's instructions. Virus-containing supernatant was harvested 3 days later and after filtration stored at −80°C. For some experiments the supernatant was concentrated by ultracentrifugation through a 20% sucrose cushion. In addition, 293T cells were used to generate HIV-1 pseudotyped particles by cotransfection of TN7stopp, and MP11-derived env expression plasmids (MP11-NL, MP11-BaL, MP11-NL/S, MP11-BaL/V, MP11-BaL/K, and MP11-BaL/VK) or pM3, a vesicular stomatitis virus G protein expression vector, using Metafectene (Biontex, Germany). TN7stopp does not express gp160 due to the insertion of two nucleotides in the 5′ region of the signal peptide, resulting in a frameshift and premature termination of translation. Virus-containing supernatants were harvested 3 days later, filtered, concentrated, and stored at −80°C.
All virus stocks were titrated using the indicator cells TZM-bl, which were seeded at a density of 3 × 103 cells per well in a 96-well tray. The next day, fivefold dilutions of 293T-derived virus stocks were used to infect TZM-bl cells and 48 h later the cells were fixed with 3% paraformaldehyde and stained by addition of 5-bromo-4-chloro-3-indolyl-β-galactopyranoside (X-Gal; 0.5 mg/ml in phosphate-buffered saline containing 3 mM potassium ferricyanide, 3 mM potassium ferrocyanide, and 1 mM magnesium chloride) (38). Individual groups of blue-stained cells were counted as single foci of infection and virus infectivity was determined as focus-forming units.
Growth curve analysis.
Growth curve analysis was performed using the T-cell lines PM1, PM1-M87, and PM1-M87o. Using equal amounts of infectious particles, 106 PM1 cells were infected (multiplicity of infection of 0.01). After adsorption at 37°C for 4 h, the cells were washed three times with phosphate-buffered saline and observed for p24-CA production in the supernatant over time using an in-house enzyme-linked immunosorbent assay (7).
T-20 and T-649 inhibition studies with TZM-bl.
For the T-20 and T-649 inhibition studies on TZM-bl cells, equal amounts of infectious particles (1,000 IU/well) were added in the absence or presence of 20, 10, 2, 0.4, 0.08, and 0.004 μg of inhibitor/ml. Triplicate wells were analyzed for each drug concentration. The TZM-bl cells were lysed at 2 days postinfection and the firefly luciferase activity was measured using the Steady-Glo assay system (Promega, Germany) and a luminometer (Labsystems, Germany). Relative activity (% of the control) was calculated by dividing the mean number of relative light units at each drug concentration by the mean number of relative light units from wells containing no drug.
Inhibition of T-20-resistant-, T-20-insensitive-, and M87-selected HIV strains by TZM-M87 and TZM-M87o.
TZM-bl, TZM-M87, and TZM-M87o cells were used for infections with T-20-resistant (TN7-NL/GIA, TN7-NL/3DTV, and TN7-NL/DIM) and T-20-insensitive (TN7-R14, TN7-X10, and TN7-X23) (23) virus strains and the two M87-selected HIV isolates NL/sel and BaL/sel. In addition, the TZM-bl, TZM-M87, and TZM-M87o cells were used for single-round infections using the pseudotyped particles based on the TN7stopp vector (23). The TZM-bl-based cell lines were lysed at 2 days postinfection and the firefly luciferase activity was measured using the Steady-Glo assay system (Promega, Germany) and a luminometer (Labsystems, Germany). Relative infection (% of the control: TZM-bl cells) was calculated by dividing the mean number of relative light units measured for the infection of TZM-M87 and TZM-M87o in relation to TZM-bl cells not expressing the antiviral peptides. Single-round infection assays using PM1, PM1-M87, and PM1-M87o were analyzed 48 h postinfection after cell lysis and detection of Renilla luciferase using the Renilla Assay System (Promega, Germany).
Replication capacity.
To compare the replication capacity of viruses carrying the parental envelope proteins of HIV-1NL4-3 or HIV-1BaL to the M87-selected envelope proteins NL/S, BaL/V, BaL/K, and BaL/VK in the absence of drug selection, a modified single-cell phenotypic assay was used (27, 41). The efficiency of the 293T transfections to obtain the different pseudotyped viruses (TN7stopp and MP11-env) was determined by measuring Renilla luciferase as relative light units (RLU) and used to normalize the virus stocks. PM1 cells (104 cells per well) were challenged with the pseudotyped virus stocks and 2 days postinfection Renilla activity was determined. The quantitative values (as a ratio) obtained after infection of PM1 cells and after transfection of 293T cells for the viruses carrying the envelope proteins NL4-3 and BaL was set to 100%. Differences in replication capacity for the pseudotyped viruses carrying M87-selected envelope proteins were assayed at least three times in quadruplicate for each virus.
Dual-color competition assay.
The fitness of recombinant viruses carrying NL4-3 and BaL Env proteins in comparison to the NL/S and BaL/VK Env proteins was determined by dual infection of PM1 cells as described (23). In order to follow the spread of two virus variants, different only in their envelope proteins, the envelope genes NL/S and BaL/VK were cloned into an enhanced green fluorescent protein (EGFP)-encoding viral vector (TN6G) and a monomeric red fluorescent protein (mRFP)-encoding viral vector (TN8) yielding replication-competent viruses after transfection of 293T cells. Pairs of virus variants (e.g., TN6-NL and TN8-NL/S) were used to infect PM1 cells at a multiplicity of infection of 0.025 (determined using TZM-bl cells) in duplicate. Each well contained 106 PM1 cells in 1 ml RPMI medium. After 6 h adsorption at 37°C, the cells were washed once in medium to remove residual input virus.
To determine the relative replication capacity of one virus over the other, half of the cells were harvested 3 days postinfection, fixed with 3% paraformaldehyde, and analyzed using a multiwell plate fluorometer reader to quantify GFP and mRFP expression. The same analysis was performed with the remaining cells 7 days postinfection to confirm the first data set and to evaluate growth advantage. All growth competition assays were performed at least two times independently. The ratio between the relative EGFP fluorescence and the relative mRFP fluorescence indicates the growth difference for the recombinant virus pairs. Infections with two marker viruses expressing the same envelope (e.g., TN6G-NL and TN8-NL) were included in each competition experiment for standardization. Furthermore, the results of all growth competition experiments were confirmed by expressing the opposite envelope/reporter combination.
RESULTS
Selection and characterization of viruses-resistant to M87.
Initially we started to select M87o-resistant virus variants using the recently described optimized PM1 cells (PM1-M87o). These cells express M87o (46 amino acids in length) at the cell surface in comparison to PM1-M87 cells (only 36 amino acids in length) (see Fig. 1 for comparison to the soluble fusion inhibitors T-20 and T-649). Several attempts, including the use of different primary isolates from more diverse HIV-1 subtypes A and C, failed. Prolonged culture of challenged PM1-M87o cells (up to 40 days) did not result in the detection of replicating virus in the culture supernatant.
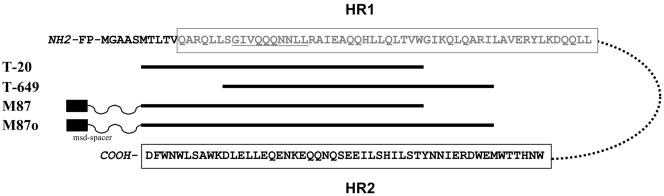
FIG. 1.
Schematic diagram of the fusion inhibitors used in this study. The N-terminal ectodomain of gp41, comprising the fusion peptide (FP), the two heptad regions (boxed, HR1 = gray, HR2 = black) linked by 28 amino acids (dotted line) is shown (HXB2 sequence). T-20 and T-649 are 36 amino acids in length and derived from HR2. M87 and M87o are 36 and 46 amino acids in length, respectively, and linked to a spacer and a membrane-spanning domain (msd-spacer) (8, 14).
As described before, the infection of PM1-M87 cells results in detectable HIV replication between 14 and 20 days postinfection (14). Therefore, these cells were challenged with HIV-1NL4-3 and HIV-1BaL (multiplicity of infection of 0.01) and virus-containing supernatant was harvested between days 17 and day 22. After titration of this first-round selected virus isolate (on TZM-bl cells), a second selection was initiated. After the third round of selection, the resulting virus isolates (NL/sel and BaL/sel) were analyzed for resistance to M87 using PM1 cells and PM1-M87 cells.
Monitoring the replication kinetics of the selected isolates in comparison to the parental viruses on PM1 cells reveals a similar growth curve for HIV-1NL4-3 and NL/sel, whereas the selected BaL variant (BaL/sel) displays a slow/low replication phenotype compared to HIV-1BaL (Fig. 2A, left). The challenge of PM1-M87 cells however shows a clear growth advantage for the selected isolates NL/sel and BaL/sel in comparison to the parental viruses HIV-1NL4-3 and HIV-1BaL (Fig. 2A, right). The detection of replicating parental virus indicates the emergence of resistant variants already at day 17 postinfection of PM1-M87 cells. Challenge experiments using the selected isolates (NL/sel and BaL/sel) and the optimized cell line PM1-M87o did not result in detectable HIV replication up to 25 days postinfection (data not shown).
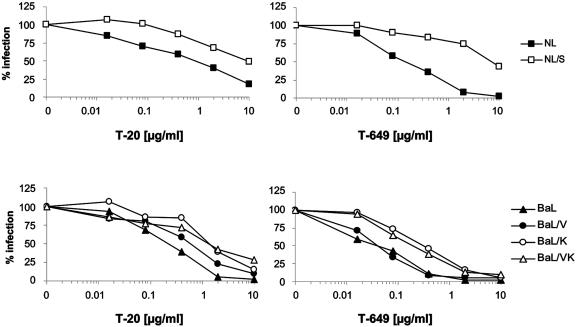
FIG. 2.
PM1-M87 selection results in M87-resistant isolates with increased IC50 values for both the T-20 and T-649 fusion inhibitors. (A) Growth of wild-type HIV-1NL4-3 and HIV-1BaL in comparison to the PM1-M87-selected isolates NL/sel and BaL/sel in PM1 cells (left) and PM1-M87 cells (right). Target cells were infected at a multiplicity of infection of 0.01 and monitored over time for p24 antigen release into the supernatant. (B) TZM-bl cells were infected with 1,000 IU/well of the parental viruses (HIV-1NL4-3 and HIV-1BaL) and selected isolates (NL/sel and BaL/sel) in the presence of increasing concentrations of T-20 or T-649 (0.02 to 10 μg/ml). Firefly luciferase activity was measured 48 h postinfection and is plotted on the y axis as percent infection relative to the control infection without fusion inhibitor. Shown are the means of triplicate infections for each inhibitor concentration. These data were used to calculate IC50 values and the fold change of IC50 (see Table 1).
Next, the susceptibility of the selected isolates to inhibition by soluble T-20 and soluble T-649 was analyzed. Using the indicator cell line TZM-bl (38) allows the quantification of infection (Fig. 2B). Increasing concentrations of fusion inhibitor were added before challenge of TZM-bl cells with 1,000 infectious units (IU) of the parental viruses and the selected isolates. The mean 50% inhibitory concentration (IC50) for the selected isolate NL/sel was 8 μg/ml for T-20 and 6 μg/ml for T-649, an increase of 8-fold and 40.2-fold, respectively, compared to the parental virus HIV-1NL4-3. The 40.2-fold change in susceptibility to T-649 was unexpected, since the peptide T-649 as a whole is not included in the membrane-anchored M87 peptide. For the selected isolate BaL/sel reduced levels of T-20 and T-649 sensitivity were also detected. A 16.2-fold increase in IC50 for T-20 and a modest 5.3-fold increase in IC50 for T-649 were determined (Table 1).
TABLE 1.
Evaluation of T-20 and T-649 sensitivity of PM1-M87-selected isolates (NL/sel and BaL/sel) and pseudotyped variants carrying envelope proteins with point mutations (NL/S [L33S], BaL/V [I48V], BaL/K [N126K], BaL/VK [I48V/N126K]) in TZM-bl indicator cellsa
| Isolate | Relative fusion inhibitor resistance
|
|
|---|---|---|
| T-20 | T-649 | |
| NL4-3 | 1 | 1 |
| NL/sel | 8.0 ± 1.0 | 40.2 ± 6.3 |
| NL/S | 10.1 ± 4.3 | 50.0 ± 5.7 |
| BaL | 1 | 1 |
| BaL/sel | 16.2 ± 3.3 | 5.3 ± 1.2 |
| BaL/V | 2.4 ± 0.9 | 1.0 ± 0.4 |
| BaL/K | 5.6 ± 1.3 | 5.3 ± 0.8 |
| BaL/VK | 5.6 ± 1.5 | 2.5 ± 0.5 |
The mean IC50 values were calculated from at least three independent TZM-bl infections. Relative fusion inhibitor resistance was calculated by dividing the IC50 value for the PM1-M87-selected isolates and each envelope protein variant by the IC50 value for HIV-1NL4-3 or HIV-1BaL.
Genetic analysis of HIV-1 env postselection on PM1-M87 cells.
Viral RNA was isolated from virus-containing cell culture supernatant and the entire envelope gene was amplified by reverse transcription PCR. Subsequently, the HR1/HR2 region was analyzed by sequencing individual clones. Figure 3 compares the amino acid sequences of the HR1 and HR2 regions of the parental viruses HIV-1NL4-3 and HIV-1BaL and 24 sequences analyzed for NL/sel and 20 sequences analyzed for BaL/sel. All NL/sel sequences obtained show one mutation N-terminal to the most important contiguous 3-amino-acid sequence (DIV, 36 to 38) within the HR1 motif. This single amino acid change is present in 18 out of 24 sequences obtained and present in the remaining seven sequences together with one additional mutation in either the HR1 or HR2 region. Thus, the major point mutation detected for the selected isolate NL/sel is L33S (codon change TTA→TCA). For the selected isolate BaL/sel the majority of the sequences analyzed (19 out of 20) encode two mutations, one in HR1 (I48V, codon change ATT→GTT), and one in HR2 (N126K, codon change AAT→AAA). Again, the HR1 region, described to contain mutations conferring resistance to T-20, was not changed. Note that the mutation N126K in HR2 has been described previously after selection in vivo (20) and recently been implicated in a T-20-dependent viral phenotype (2).
FIG. 3.
Alignment of the amino acid sequence of heptad repeats 1 and 2 of the parental viruses HIV-1NL4-3 and HIV-1BaL and the sequences obtained from PM1-M87-selected variants NL/sel and BaL/sel, respectively. Note that PM1-M87 selection did not change amino acids in the most important contiguous 3-amino-acid sequence (GIV/DIV, positions 36 to 38) of HR1.
Resistance determinants mapped by site-directed mutagenesis.
Next, the most common point mutations selected (L33S for NL4-3 and I48V, N126K, and I48V/N126K for BaL) were introduced into expression plasmids for the envelope proteins of HIV-1NL4-3 and HIV-1BaL, respectively. Pseudotyped particles were generated and used for single-round infection assays, again using the TZM-bl cell line as indicator cells and T-20 and T-649 to inhibit infection. As Fig. 4 shows, the single point mutation L33S confers a 50-fold increase in insensitivity to T-649 (IC50 = 7.5 μg/ml) and a 10-fold increase in insensitivity to T-20 (IC50 = 10 μg/ml) (Table 1). These results are in agreement with the previously described change in insensitivity due to selection with C34 (1) or T-20 (9) resulting in the same amino acid change at position 33 for HIV-1NL4-3.
FIG. 4.
Point mutations L33S (for HIV-1NL4-3) and N126K (for HIV-1BaL) are sufficient to confer resistance to T-20 and T-649. TZM-bl cells were infected with 1,000 IU/well of the pseudotyped viruses carrying the parental envelope proteins of HIV-1NL4-3 and HIV-1BaL and the point mutations indicated (NL/S [L33S], BaL/V [I48V], BaL/K [N126K], and BaL/VK [I48V/N126K]) in the presence of increasing concentrations of T-20 or T-649 (0.02 to 10 μg/ml). Firefly luciferase activity was measured 48 h postinfection and is plotted on the y axis as percent infection relative to the control infection (without fusion inhibitor). Shown are the means of triplicate infections for each inhibitor concentration. These data were used to calculate IC50 values and the fold change of IC50 (see Table 1).
The single-round infectivity assays performed using HIV-1BaL envelope expression plasmids (parental envelope BaL and the three point mutated envelope proteins) revealed a more differentiated pattern of sensitivity to T-20 and T-649. The mutation N126K in HR2 is sufficient to cause increased insensitivity to T-20 and T-649 (5.6-fold and 5.3-fold, respectively) similar to the observed increase in insensitivity for the selected isolate BaL/sel (Fig. 4 and Table 1), although not as pronounced. The mutation I48V in HR1 causes only a minor increase in insensitivity to T-20 and no increase in insensitivity to T-649. The combined mutation I48V/N126K, however, reveals a 5.6-fold increase in insensitivity to T-20 and a 2.5-fold increase in insensitivity to T-649 (Table 1). These results indicate that the N126K mutation is sufficient to confer partial resistance to T-20 and T-649 and that the I48V mutation is modulating this resistance phenotype.
Resistance to membrane-anchored fusion inhibitor M87.
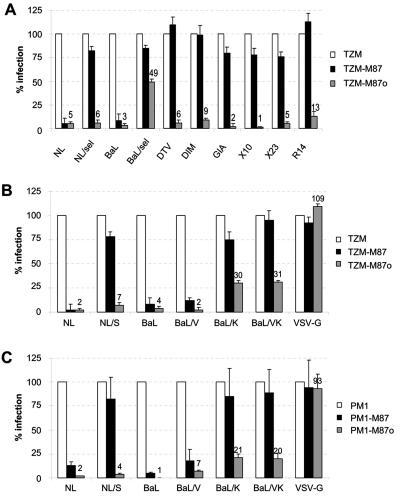
NL/sel and BaL/sel were selected using PM1-M87 cells. In order to analyze the resistance profile using another target cell, TZM-bl cells expressing either M87 or M87o, in a similar way to that described for PM1-M87 and PM1-M87o (8, 14) were generated. Using a quantitative flow cytometry analysis, the numbers of M87 and M87o molecules at the cell surface of TZM-bl cells in comparison to the two PM1-derived cell lines were determined. The newly established cell line, TZM-M87, expresses similar levels of membrane-anchored fusion inhibitor as described for the PM1 cell line (median number of molecules for TZM-M87: 315, and for PM1-M87: 533). In comparison, TZM-M87o cells show very high numbers of membrane-anchored fusion inhibitor (median number of molecules: 23,231), confirming the successful optimization strategy described by Egelhofer et al. (8).
These TZM-bl-derived cell lines were used for single-round infection assays using the pseudotyped particles described but also for infections using the selected isolates NL/sel and BaL/sel. In addition, these cell lines were challenged with replication-competent recombinant viruses carrying T-20-resistant envelope proteins and T-20-insensitive envelope proteins described previously (23, 34). As Fig. 5A shows, the amount of membrane-anchored M87 expressed by TZM-M87 cells is sufficient to cause a 95% reduction in infectivity for the parental viruses HIV-1NL4-3 and HIV-1BaL. As expected, the selected isolates NL/sel and BaL/sel displayed an M87-resistant phenotype and infected TZM-M87 cells almost as efficiently as TZM-bl cells (only 20% inhibition). No reduction in infectivity of TZM-M87 cells was measured for viruses carrying envelope proteins containing single point mutations in the HR1 region known to confer resistance to soluble T-20 (DTV, DIM, and GIA). Similarly, the three recombinant viruses carrying the envelope variants X10, X23, and R14 (13, 23) were able to infect the TZM-M87 cells with higher efficiency than HIV-1NL4-3 and HIV-1BaL, confirming their relative T-20 insensitivity.
FIG. 5.
Single-round infection assays with PM1-M87-selected viruses on TZM-bl cells and PM1 cells expressing M87 and M87o. TZM-bl, TZM-M87, and TZM-M87o were challenged with (A) different viruses carrying T-20-resistant (DTV, DIM, and GIA) and T-20-insensitive (X10, X23, and R14) and M87-resistant envelope proteins (NL/sel and BaL/sel) or (B) pseudotyped particles carrying envelope proteins with the point mutations indicated. In addition, PM1, PM1-M87, and PM1-M87o cells were challenged with pseudotyped particles carrying envelope proteins with the point mutations indicated (C). Forty-eight hours postinfection the cells were lysed and firefly luciferase activity (for A and B) and Renilla activity (for C) was determined. Luciferase activity obtained from TZM-bl and PM1 cells not expressing a membrane-anchored fusion inhibitor was set to 100%. Shown are the mean values of triplicate infections of a representative experiment.
Next, TZM-M87 cells were challenged with pseudotyped particles carrying the parental envelope proteins of HIV-1NL4-3 and HIV-1BaL and envelope proteins encoding the observed point mutations. The mutation L33S in the NL4-3 envelope confers resistance to M87, as did the mutations N126K and I48V/N126K in the BaL envelope protein (Fig. 5B). The single-round infection assay using PM1-M87 cells resulted in a similar pattern of resistance for NL/S, BaL/V, and BaL/VK (Fig. 5C). These results confirm the role of the selected point mutations for the M87-resistant phenotype.
In addition, we challenged TZM-bl cells and PM1 cells expressing the optimized M87o membrane-anchored fusion inhibitor (Fig. 5). This optimized fusion inhibitor exerts sufficient protection to infection by all the T-20-resistant, T-20-insensitive, and M87-resistant viruses analyzed, most likely due to the high expression of the membrane-anchored fusion inhibitor. Interestingly, the most effective infection was seen for the M87-resistant virus BaL/sel (49%, Fig. 5A) and the pseudotyped particles BaL/K and BaL/VK for both cell lines tested (BaL/K: 30% for TZM-M87o and 21% for PM1-M87o cells), although the selected variant BaL/sel does not establish a productive infection on PM1-M87o cells. The BaL/K and BaL/VK envelope proteins display only medium insensitivity against both soluble fusion inhibitors T-20 and T-649, whereas the other envelope proteins analyzed show either a high insensitivity against T-20 (DTV, DIM, GIA, and X10) or high insensitivity against T-649 (NL/sel and X23) (13, 23).
Replication capacity and viral fitness of HIV-1 recombinants carrying M87 resistance mutations.
Reduced replication capacity or reduced viral fitness contributes to the continued benefit of antiviral therapy despite the presence of drug resistance. For reverse transcriptase and protease inhibitors as well as for enfuvirtide, drug resistance is often associated with a reduction in viral fitness (18, 31, 32) but not in all cases (21, 41). Therefore a modified single-round infection assay was performed allowing the quantification of replication capacity for the pseudotyped viruses carrying the selected envelope proteins NL/S, BaL/V, Bal/K, and BaL/VK in comparison to the parental envelope proteins NL4-3 and BaL.
As Table 2 shows, the envelope protein NL/S displays a 1.5-fold higher replication capacity compared to the parental envelope NL4-3, despite a high resistance to soluble T-20 and T-649 and membrane-anchored M87. Thus, for NL/S we could not confirm a positive correlation between drug resistance and lower replication capacity. However, the selected BaL-derived envelope proteins BaL/V, BaL/K, and BaL/VK display a severely reduced replication capacity compared to the parental envelope BaL (44.2%, 15.2%, and 24.2%, respectively), indicating a positive correlation between drug resistance and impaired replication capacity.
TABLE 2.
Replication capacity of pseudotyped virus particles carrying the envelope proteins indicated (NL/S [L33S], BaL/V [I48V], BaL/K [N126K], BaL/VK [I48V/N126K])a
| Isolate | Replication capacity (% of control) |
|---|---|
| NL4-3 (control) | 100.0 |
| NL/S | 154.2 ± 13.1 |
| BaL (control) | 100.0 |
| BaL/V | 44.2 ± 18.1 |
| BaL/K | 15.2 ± 9.0 |
| BaL/VK | 24.2 ± 3.0 |
PM1 cells were infected with normalized virus stocks and Renilla activity determined 2 days postinfection. The ratio of relative light units obtained postinfection of PM1 cells to relative light units obtained after transfection of 293T cells was set to 100% for both pseudotype infections with particles carrying the parental envelope proteins NL4-3 and BaL. Values were calculated as the means of three independent experiments done in quadruplicate. Note that the L33S mutation does not cause reduced replication capacity despite being resistant to T-20, T-649, and M87.
Next, a viral fitness assay using replication-competent viruses was performed to confirm whether M87 resistance was correlated with a gain of fitness for NL/S and fitness impairment for the BaL-derived envelope proteins. Dual-color competition assays were performed in PM1 cells using the EGFP and mRFP marker viruses (TN6R and TN8) expressing M87-resistant Envs NL/S and BaL/VK or the envelope proteins of the parental viruses. As published previously, we established a proof of concept for the dual-color competition assay with marker viruses encoding the wild-type NL4-3 envelope protein and an envelope protein encoding a single amino acid change at position 38 of gp41 (V38A: GIA), known to confer resistance to T-20 in vitro and in vivo (23).
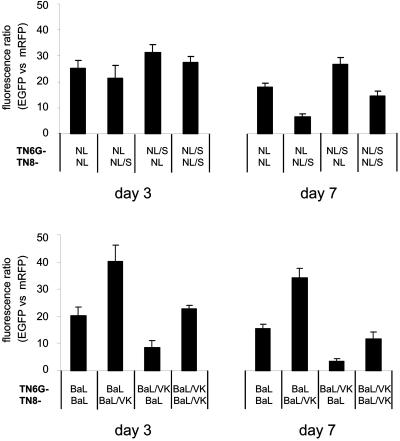
Here, PM1 cells were infected with equal infectious doses of either TN6G-NL and TN8-NL/S or TN6G-BaL and TN8-BaL/VK (as titrated on TZM-bl cells). Cells were harvested at day 3 and day 7 postinfection and total EGFP and mRFP fluorescence (at 508 nm and 609 nm, respectively) was determined using a fluorometer. Figure 6 illustrates the viral fitness data obtained. For the dual infection with the HIV-1BaL-derived viruses a reduced viral fitness for the M87-resistant envelope BaL/VK was observed (lower panel in Fig. 6), indicated by the higher fluorescence ratio (GFP versus mRFP) of the [TN6G-BaL]/[TN8-BaL/VK] infection compared to the dual infection carried out using the two fluorescent marker viruses expressing the same envelope protein ([TN6G-BaL]/[TN8-BaL] and [TN6G-BaL/VK]/[TN8-BaL/VK]). The opposite virus pair, where the M87-resistant envelope BaL/VK was expressed by the GFP-encoding marker virus ([TN6G-BaL/VK]/[TN8-BaL]), showed a reduced fluorescence ratio compared to control infections with the two marker viruses expressing the same envelope protein, confirming the growth advantage of the virus expressing the parental BaL envelope over the marker virus expressing the M87-resistant envelope protein. This difference in viral fitness was observed at both time points (day 3 and day 7) and therefore considered valid.
FIG. 6.
Relative fitness of viruses expressing the parental envelopes NL4-3 and BaL and the characterized point mutation NL/S [L33S] and BaL/VK [I48V/N126K]. A dual-color competition assay was performed on PM1 cells (multiplicity of infection of 0.025) with competing viruses expressing EGFP (TN6G-env) or mRFP (TN8-env) (23). Representative results from two independent infections (done in duplicate) are shown. The ratio between the relative EGFP fluorescence and the relative mRFP fluorescence indicates the growth difference for the respective recombinant virus pairs. Infections with two marker viruses expressing the same envelope (e.g., TN6G-NL and TN8-NL) were included in each competition experiment for standardization. Note that the M87-resistant envelope NL/S does not display reduced fitness compared to the parental virus HIV-1NL4-3.
The dual infection of PM1 cells with the two marker viruses TN6G-NL and TN8-NL/S resulted in a green-to-red fluorescence ratio below the fluorescence ratio obtained after the infection with the marker viruses expressing the same envelope proteins ([TN6G-NL]/[TN8-NL] and [TN6G-NL/S]/[TN8-NL/S]) at day 3 (Fig. 6, upper panel), indicating a growth advantage of the resistant virus variant over the parental virus TN6G-NL. At day 7 postinfection, this growth advantage was even more evident. Again, the opposite virus pair ([TN6G-NL/S]/[TN8-NL]) confirmed this result for both time points. Although the M87-selected point mutation L33S confers resistance to T-20, T-649, and M87, the replication-competent marker viruses TN6G-NL/S and TN8-NL/S do not display reduced viral fitness. The results of the dual-color competition assay therefore confirm the results obtained by the replication capacity assay.
DISCUSSION
The expression of a membrane-anchored gp41-derived peptide (M87) has been shown to confer protection from infection by HIV-1 of different subtypes (14). In an effort to characterize the mechanism of action of this membrane-anchored peptide in comparison to the soluble peptides T-20 and T-649, we selected resistant variants of HIV-1NL4-3 and HIV-1BaL by serial virus passage using PM1 cells stably expressing peptide M87. The selected virus variants display resistance to membrane-anchored M87 as well as to the soluble fusion inhibitors T-20 and T-649 (Fig. 2 and Table 1). Experiments performed using the newly developed TZM-bl-based cell lines TZM-M87 and TZM-M87o confirmed the resistance of NL/sel and BaL/sel to membrane-anchored M87 (Fig. 5A). In addition, these experiments clearly showed that T-20-resistant virus variants (DTV, DIM, and GIA) and T-20-insensitive variants (R14, X10, and X23) are able to infect TZM-bl cells expressing M87 at the cell surface as efficiently as the parental cell line TZM-bl, supporting the results obtained after challenging PM1-M87 cells (8). However, the selected virus variants did not infect (NL/sel) or only partially infected (BaL/sel) TZM-bl cells expressing the more recently described optimized membrane-anchored fusion inhibitor M87o (8).
Several challenge experiments using the two selected virus variants and PM1-M87o cells (expressing a very high concentration of M87o on the cell surface compared to TZM-M87o cells) did not result in detectable virus replication, strengthening the importance of this novel antiviral concept for gene therapy approaches. It is important to note that compared to M87 (and T-20) the optimized M87o construct encompasses 10 additional amino acids, now additionally targeting the deep pocket in HR1. It remains to be elucidated whether these additional amino acids or the high expression profile of M87o confers protection from infection through T-20- and M87-resistant HIV variants.
Site-directed mutagenesis studies confirmed the importance of the selected point mutations (L33S in NL-4.3 and I48V and N126K in BaL) to confer resistance to M87 as well as to soluble T-20 and T-646 (Fig. 4 and Table 1). It has been shown previously that replication of HIV-1NL4-3 in the presence of increasing concentrations of T-20 (9) or C34 (1) results in the selection of the L33S mutation. Here we show that the same mutation can be selected in the presence of M87. Amino acid 33 in HR1 marks the amino terminus of the potential T-20 (and therefore M87) binding site LLSGIV (37) and the change to serine at this position might reduce the binding of the soluble peptide as well as the membrane-anchored M87.
It is important to note that the HR1 of HIVNL-4.3 encodes aspartic acid at position 36 (36D) and not glycine (36G) as in most viral envelope proteins and that the mutation G36D alone confers 10-fold resistance to T-20. This could indicate that the combined mutation L33S/G36D is responsible for the observed phenotype. However, we introduced the L33S mutation into the BaL envelope protein (BaL/S, encoding glycine at position 36) and again observed a strong T-20 resistance (data not shown), arguing against L33S being a secondary mutation. Interestingly, the L33S mutation confers high resistance to soluble T-649, although this peptide was not present during selection. An explanation for this observation could be that the carboxy terminus of T-649 (and C34) interacts with the lysine at position 33 and facilitates efficient binding, but further experiments are needed to address this hypothesis.
Unexpectedly, the determination of the replication capacity and viral fitness of viruses encoding the L33S mutation indicates that the selected virus is fitter than the parental virus HIV-1NL4-3 (Table 2 and Fig. 6). It has been described previously by us (23) and others (21, 41) that the correlation of drug resistance or drug insensitivity and reduced fitness is not absolute. In support of our result for NL/S, Nameki et al. (22) analyzed the influence of C34 selected mutations for the Rev-responsive element partially overlapping the HR1 region of gp41. They point out that the nucleotides encoding lysine at position 33 are located in a single-strand bulge region of the stem IIc loop top and therefore the triplet change from TTA to TCA (now encoding serine at position 33) would only minimally interfere with the Rev-responsive element structure and function. As long as the interplay between HR1 and HR2 is not severely impaired, a drug-resistant virus could therefore replicate as efficiently as the parental virus (or better), as shown here for the L33S mutation in the envelope protein of NL4-3. Further studies are needed to address this hypothesis and to clarify a possible role of the existing mutation G36D in the NL/S envelope protein with regard to viral fitness. That L33S is a compensatory mutation to restore the fitness of this variant seems unlikely, since we have not observed differences in viral fitness for HIVNL4-3 versus HIVNL4-3(D36G) (data not shown).
The PM1-M87 selection of HIV-1BaL resulted in two point mutations being present in the majority of the analyzed sequences. Whereas I48V is located in the HR1 region, the N126K mutation is located in the HR2 region and causes a decrease in sensitivity to T-20 and to T-649 (Table 1). Recently, Nameki and colleagues (22) described the in vitro selection of N126K in the context of HIV-1NL4-3 using the fusion inhibitor C34, and Baldwin et al. (2) described the emergence of N126K after therapy failure. These reports and others (20, 29) support our observation that fusion inhibitor resistance can be obtained or modulated through changes in the HR2 region.
Resistance due to changes in HR2 might be highly context dependent (HIVNL4-3 variants encoding N126K could not be selected in our experiments) and difficult to detect in clinical trials, since this variant displays a considerable fitness defect. Therefore, the detection of N126K after prolonged T-20 therapy could also be the result of compensatory mutations being introduced into the early-emerging resistant variant N126K. More clinical data including a more detailed analysis of minor mutations emerging early after treatment initiation are needed to substantiate this hypothesis. Elegant studies of binding between an N126K-containing C34 peptide and wild-type N36 peptide showed a higher binding affinity than the wild-type C34 and N36 peptides, indicating a faster intramolecular HR1-HR2 interaction (22).
A stronger HR1-HR2 interaction might result in a severely reduced replication capacity for pseudotyped particles, as shown here for the viruses carrying the envelope proteins BaL/K and BaL/VK (Table 2, Fig. 6, in the background of the HIV-1BaL envelope protein) and by Nameki et al. for the HIV-1NL4-3 envelope protein (22). Recently, it has been described that the mutation N126K in combination with G38A results in a T-20-dependent virus (2). In the case of the M87-selected BaL variant (I48V/N126K), however, we could obtain infectious pseudotyped particles as well as replication-competent viruses in the absence of T-20 (although with a lower infectious titer compared to the parental virus HIV-1BaL; data not shown). The infectious titer for these particles could not be increased in the presence of T-20 or T-649. However, it is possible the patient-specific envelope described by Baldwin et al. (2) differs with respect to the intramolecular interaction of HR1 and HR2 from the BaL envelope studied here.
Taken together, these results provide further insight into the mechanisms underlying the development of resistance to entry inhibitors. Membrane-anchored M87-selected virus variants show distinct point mutations conferring resistance to M87 but also to the soluble fusion inhibitors T-20 and T-649. Increasing the concentration of the fusion inhibitor, as realized using the optimized M87o (8), however, results in a complete inhibition of virus replication for the M87-resistant variants described here as well as for T-20-resistant and T-20-insensitive variants, strengthening the importance of this novel antiviral concept for gene therapy approaches.
We conclude that two different ways to circumvent inhibition by fusion inhibitors seem to prevail: first, the mutation of HR1 results in reduced binding of the fusion inhibitor, mainly due to changes in the T-20 binding region LLSGIV and the neighboring amino acids (16, 30, 35, 38). Depending on the influence these point mutations have on the overall functionality of the Rev-responsive element and the intramolecular folding of the HR1-HR2 region, the selected virus is not impaired (L33S) or shows a reduced viral fitness (in most cases) (18, 21). Second, mutations in HR1 and/or HR2 (as shown here for BaL) favor the intramolecular folding of HR1-HR2 to prevent binding of soluble peptides or membrane-anchored M87. Such mutations most likely result in a severe reduction of viral fitness and therefore might have a greater implication for clinical practice (3, 24, 31).
Acknowledgments
We thank current and previous members of the lab for helpful suggestions and critical comments. PM1 cells and TZM-bl cells were provided by the EU program EVA/MRC centralized facility for AIDS reagents, NIBSC, United Kingdom. HIV-1 envelope genes encoding the T-20-resistant envelopes NL/DTV and NL/DIM were a gift from F. Kirchhoff (University of Ulm, Ulm, Germany).
This work was supported by a grant from Landesstiftung Baden-Württemberg to M.T.D.
This work counts as partial fulfillment of the Ph.D. requirements for S.L. at the University of Heidelberg and for F.H. at the University of Frankfurt.
REFERENCES
- 1.Armand-Ugon, M., A. Gutierrez, B. Clotet, and J. A. Este. 2003. HIV-1 resistance to the gp41-dependent fusion inhibitor C-34. Antiviral Res. 59:137-142. [DOI] [PubMed] [Google Scholar]
- 2.Baldwin, C. E., R. W. Sanders, Y. Deng, S. Jurriaans, J. M. Lange, M. Lu, and B. Berkhout. 2004. Emergence of a drug-dependent human immunodeficiency virus type 1 variant during therapy with the T20 fusion inhibitor. J. Virol. 78:12428-12437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bates, M., T. Wrin, W. Huang, C. Petropoulos, and N. Hellmann. 2003. Practical applications of viral fitness in clinical practice. Curr. Opin. Infect. Dis. 16:11-18. [DOI] [PubMed] [Google Scholar]
- 4.Chan, D. C., D. Fass, J. M. Berger, and P. S. Kim. 1997. Core structure of gp41 from the HIV envelope glycoprotein. Cell 89:263-273. [DOI] [PubMed] [Google Scholar]
- 5.Clavel, F., and A. J. Hance. 2004. HIV drug resistance. N. Engl. J. Med. 350:1023-1035. [DOI] [PubMed] [Google Scholar]
- 6.DeGruttola, V., L. Dix, R. D'Aquila, D. Holder, A. Phillips, M. Ait-Khaled, J. Baxter, P. Clevenbergh, S. Hammer, R. Harrigan, D. Katzenstein, R. Lanier, M. Miller, M. Para, S. Yerly, A. Zolopa, J. Murray, A. Patick, V. Miller, S. Castillo, L. Pedneault, and J. Mellors. 2000. The relation between baseline HIV drug resistance and response to antiretroviral therapy: re-analysis of retrospective and prospective studies using a standardized data analysis plan. Antivir. Ther. 5:41-48. [DOI] [PubMed] [Google Scholar]
- 7.Dittmar, M. T., S. Eichler, S. Reinberger, L. Henning, and H. G. Krausslich. 2001. A recombinant virus assay using full-length envelope sequences to detect changes in HIV-1 co-receptor usage. Virus Genes 23:281-290. [DOI] [PubMed] [Google Scholar]
- 8.Egelhofer, M., G. Brandenburg, H. Martinius, P. Schult-Dietrich, G. Melikyan, R. Kunert, C. Baum, I. Choi, A. Alexandrov, and D. von Laer. 2004. Inhibition of human immunodeficiency virus type 1 entry in cells expressing gp41-derived peptides. J. Virol. 78:568-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fikkert, V., P. Cherepanov, K. Van Laethem, A. Hantson, B. Van Remoortel, C. Pannecouque, E. De Clercq, Z. Debyser, A. M. Vandamme, and M. Witvrouw. 2002. env chimeric virus technology for evaluating human immunodeficiency virus susceptibility to entry inhibitors. Antimicrob. Agents Chemother. 46:3954-3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Greenberg, M. L., and N. Cammack. 2004. Resistance to enfuvirtide, the first HIV fusion inhibitor. J. Antimicrob. Chemother. 1:1. [DOI] [PubMed] [Google Scholar]
- 11.Greenberg, M. L., D. Davison, L. Jin, S. Mosier, T. Melby, P. Sista, R. DeMasi, D. Miralles, N. Cammack, and T. J. Matthews. 2002. In vitro antiviral activity of T-1249, a second generation fusion inhibitor. Antiviral Ther. 7:S14. [Google Scholar]
- 12.Greenberg, M. L., P. Sista, G. D. Miralles, T. Melby, D. Davison, L. Jin, S. Mosier, M. Mink, E. Nelson, R. DeMasi, L. Fang, N. Cammack, M. Salgo, F. Duff, and T. J. Matthews. 2002. Enfuvirtide (T-20) and T-1249 resistance: observations from phase II clinical trials of enfuvirtide in combination with oral antiretrovirals and a phase I/II dose-ranging mono-therapy trial with T-1249. Antiviral Ther. 7:S140. [Google Scholar]
- 13.Heil, M. L., J. M. Decker, J. N. Sfakianos, G. M. Shaw, E. Hunter, and C. A. Derdeyn. 2004. Determinants of human immunodeficiency virus type 1 baseline susceptibility to the fusion inhibitors enfuvirtide and T-649 reside outside the peptide interaction site. J. Virol. 78:7582-7589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hildinger, M., M. T. Dittmar, P. Schult-Dietrich, B. Fehse, B. S. Schnierle, S. Thaler, G. Stiegler, R. Welker, and D. von Laer. 2001. Membrane-anchored peptide inhibits human immunodeficiency virus entry. J. Virol. 75:3038-3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kilby, J. M., and J. J. Eron. 2003. Novel therapies based on mechanisms of HIV-1 cell entry. N. Engl. J. Med. 348:2228-2238. [DOI] [PubMed] [Google Scholar]
- 16.Labrosse, B., J. L. Labernardiere, E. Dam, V. Trouplin, K. Skrabal, F. Clavel, and F. Mammano. 2003. Baseline susceptibility of primary human immunodeficiency virus type 1 to entry inhibitors. J. Virol. 77:1610-1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee, B., M. Sharron, L. J. Montaner, D. Weissman, and R. W. Doms. 1999. Quantification of CD4, CCR5, and CXCR4 levels on lymphocyte subsets, dendritic cells, and differentially conditioned monocyte-derived macrophages. Proc. Natl. Acad. Sci. USA 96:5215-5220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu, J., P. Sista, F. Giguel, M. Greenberg, and D. R. Kuritzkes. 2004. Relative replicative fitness of human immunodeficiency virus type 1 mutants resistant to enfuvirtide (T-20). J. Virol. 78:4628-4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lusso, P., F. Cocchi, C. Balotta, P. D. Markham, A. Louie, P. Farci, R. Pal, R. C. Gallo, and M. S. Reitz, Jr. 1995. Growth of macrophage-tropic and primary human immunodeficiency virus type 1 (HIV-1) isolates in a unique CD4+ T-cell clone (PM1): failure to downregulate CD4 and to interfere with cell-line-tropic HIV-1. J. Virol. 69:3712-3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Melby, T., R. DeMasi, D. R. Kuritzkes, G. Heilek-Snyder, M. P. Salgo, N. Cammack, T. J. Metthews, and M. L. Greenberg. 2004. Analyses of virological response and enfuvirtide resistance through 48 weeks in the TORO 1 and TORO 2 studies. Antivir. Ther. 9:S181. [Google Scholar]
- 21.Menzo, S., A. Castagna, A. Monachetti, H. Hasson, A. Danise, E. Carini, P. Bagnarelli, A. Lazzarin, and M. Clementi. 2004. Genotype and phenotype patterns of human immunodeficiency virus type 1 resistance to enfuvirtide during long-term treatment. Antimicrob. Agents Chemother. 48:3253-3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nameki, D., E. Kodama, M. Ikeuchi, N. Mabuchi, A. Otaka, H. Tamamura, M. Ohno, N. Fujii, and M. Matsuoka. 2005. Mutations conferring resistance to human immunodeficiency virus type 1 fusion inhibitors are restricted by gp41 and rev-responsive element functions. J. Virol. 79:764-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neumann, T., I. Hagmann, S. Lohrengel, M. L. Heil, C. A. Derdeyn, H.-G. Kräusslich, and M. T. Dittmar. 2005. T20-insensitive HIV-1 from naive patients exhibits high viral fitness in a novel dual colour competition assay on primary cells. Virology 332:251-262. [DOI] [PubMed] [Google Scholar]
- 24.Nijhuis, M., S. Deeks, and C. Boucher. 2001. Implications of antiretroviral resistance on viral fitness. Curr. Opin. Infect. Dis. 14:23-28. [DOI] [PubMed] [Google Scholar]
- 25.Palella, F. J., Jr., K. M. Delaney, A. C. Moorman, M. O. Loveless, J. Fuhrer, G. A. Satten, D. J. Aschman, and S. D. Holmberg. 1998. Declining morbidity and mortality among patients with advanced human immunodeficiency virus infection. HIV Outpatient Study Investigators. N. Engl. J. Med. 338:853-860. [DOI] [PubMed] [Google Scholar]
- 26.Palella, F. J., Jr., M. Deloria-Knoll, J. S. Chmiel, A. C. Moorman, K. C. Wood, A. E. Greenberg, and S. D. Holmberg. 2003. Survival benefit of initiating antiretroviral therapy in HIV-infected persons in different CD4+ cell strata. Ann. Intern. Med. 138:620-626. [DOI] [PubMed] [Google Scholar]
- 27.Petropoulos, C. J., N. T. Parkin, K. L. Limoli, Y. S. Lie, T. Wrin, W. Huang, H. Tian, D. Smith, G. A. Winslow, D. J. Capon, and J. M. Whitcomb. 2000. A novel phenotypic drug susceptibility assay for human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 44:920-928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pierson, T. C., and R. W. Doms. 2003. HIV-1 entry and its inhibition. Curr. Top. Microbiol. Immunol. 281:1-27. [DOI] [PubMed] [Google Scholar]
- 29.Poveda, E., B. Rodes, C. Toro, L. Martin-Carbonero, J. Gonzalez-Lahoz, and V. Soriano. 2002. Evolution of the gp41 env region in HIV-infected patients receiving T-20, a fusion inhibitor. AIDS 16:1959-1961. [DOI] [PubMed] [Google Scholar]
- 30.Poveda, E., B. Rodes, C. Toro, and V. Soriano. 2004. Are fusion inhibitors active against all HIV variants? AIDS Res. Hum. Retroviruses 20:347-348. [DOI] [PubMed] [Google Scholar]
- 31.Quinones-Mateu, M. E., and E. J. Arts. 2002. Fitness of drug resistant HIV-1: methodology and clinical implications. Drug Resist. Update 5:224-233. [DOI] [PubMed] [Google Scholar]
- 32.Rangel, H. R., J. Weber, B. Chakraborty, A. Gutierrez, M. L. Marotta, M. Mirza, P. Kiser, M. A. Martinez, J. A. Este, and M. E. Quinones-Mateu. 2003. Role of the human immunodeficiency virus type 1 envelope gene in viral fitness. J. Virol. 77:9069-9073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reuter, S., P. Kaumanns, S. B. Buschhorn, and M. T. Dittmar. 2005. Role of HIV-2 envelope in Lv2-mediated restriction. Virology 332:347-358. [DOI] [PubMed] [Google Scholar]
- 34.Rimsky, L. T., D. C. Shugars, and T. J. Matthews. 1998. Determinants of human immunodeficiency virus type 1 resistance to gp41-derived inhibitory peptides. J. Virol. 72:986-993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sista, P. R., T. Melby, D. Davison, L. Jin, S. Mosier, M. Mink, E. L. Nelson, R. DeMasi, N. Cammack, M. P. Salgo, T. J. Matthews, and M. L. Greenberg. 2004. Characterization of determinants of genotypic and phenotypic resistance to enfuvirtide in baseline and on-treatment HIV-1 isolates. AIDS 18:1787-1794. [DOI] [PubMed] [Google Scholar]
- 36.Stanfield-Oakley, S. A., J. Jeffrey, C. B. McDanal, S. Mosier, L. Talton, L. Jin, P. Sista, N. Cammack, T. J. Matthews, and M. L. Greenberg. 2003. Determinants of susceptibility to enfuvirtide map to gp41 in enfuvirtide-naive HIV-1. Antiviral Ther. 8:S25. [Google Scholar]
- 37.Trivedi, V. D., S. F. Cheng, C. W. Wu, R. Karthikeyan, C. J. Chen, and D. K. Chang. 2003. The LLSGIV stretch of the N-terminal region of HIV-1 gp41 is critical for binding to a model peptide, T20. Protein Eng. 16:311-317. [DOI] [PubMed] [Google Scholar]
- 38.Wei, X., J. M. Decker, H. Liu, Z. Zhang, R. B. Arani, J. M. Kilby, M. S. Saag, X. Wu, G. M. Shaw, and J. C. Kappes. 2002. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob. Agents Chemother. 46:1896-1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wild, C., T. Greenwell, and T. Matthews. 1993. A synthetic peptide from HIV-1 gp41 is a potent inhibitor of virus-mediated cell-cell fusion. AIDS Res. Hum. Retroviruses 9:1051-1053. [DOI] [PubMed] [Google Scholar]
- 40.Wild, C. T., D. C. Shugars, T. K. Greenwell, C. B. McDanal, and T. J. Matthews. 1994. Peptides corresponding to a predictive alpha-helical domain of human immunodeficiency virus type 1 gp41 are potent inhibitors of virus infection. Proc. Natl. Acad. Sci. USA 91:9770-9774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang, H., Y. Zhou, C. Alcock, T. Kiefer, D. Monie, J. Siliciano, Q. Li, P. Pham, J. Cofrancesco, D. Persaud, and R. F. Siliciano. 2004. Novel single-cell-level phenotypic assay for residual drug susceptibility and reduced replication capacity of drug-resistant human immunodeficiency virus type 1. J. Virol. 78:1718-1729. [DOI] [PMC free article] [PubMed] [Google Scholar]