Abstract
Enhanced mutagenesis may result in RNA virus extinction, but the molecular events underlying this process are not well understood. Here we show that 5-fluorouracil (FU)-induced mutagenesis of the arenavirus lymphocytic choriomeningitis virus (LCMV) resulted in preextinction populations whose consensus genomic nucleotide sequence remained unaltered. Furthermore, fitness recovery passages in the absence of FU, or alternate virus passages in the presence and absence of FU, led to profound differences in the capacity of LCMV to produce progeny, without modification of the consensus genomic sequence. Molecular genetic analysis failed to produce evidence of hypermutated LCMV genomes. The results suggest that low-level mutagenesis to enrich the viral population with defector, interfering genomes harboring limited numbers of mutations may mediate the loss of infectivity that accompanies viral extinction.
Mutagenic agents administered alone or in combination with antiviral inhibitors can drive RNA virus populations to extinction (4, 18, 20, 21, 41, 42, 46, 48, 50, 51, 53, 65, 67, 72, 77, 79; reviewed in references 6, 27, 28, 31, and 39). Loss of infectivity has been interpreted as an irreversible transition that occurs when template copying fidelity falls below a critical value termed the error threshold. Such a transition has been termed virus entry into error catastrophe, or lethal mutagenesis (50), and its existence has been supported by several theoretical studies (5, 31, 60, 83). In agreement with this concept, analysis of the mutant spectra of virus populations on their way to extinction has shown 2- to 17-fold increases in mutation frequencies, calculated among components of the mutant spectra, as well as increases in Shannon entropy (a measure of the different types of sequences present in a mutant spectrum) to nearly maximal values (that is, each component of the mutant spectrum showed a unique sequence) (reviewed in references 27 and 28).
The prototypic arenavirus lymphocytic choriomeningitis virus (LCMV) (12, 63, 64) showed extreme sensitivity to 5-fluorouracil (FU)-induced mutagenesis (41, 72) compared with other RNA viruses subjected to comparable doses of the mutagen (79). The extreme sensitivity of LCMV to FU offered an opportunity to analyze the capacity of LCMV to regain infectivity following FU mutagenesis as well as the modification of genomic nucleotide sequence variations as the virus moves toward or away from the error threshold and to explore the possible dominance of hypermutated genomes in the transition to extinction. The results reveal a remarkable capacity of LCMV populations to modify their fitness level while maintaining an invariant consensus sequence. Multiple molecular clones were analyzed to define a sequence at nucleotides 470 to 550 within the intergenic region of genomic segment L. A number of standard and mutated oligonucleotide primers have failed to produce evidence of hypermutated viral sequences in the L polymerase gene. The results suggest that limited mutagenesis may be sufficient to drive LCMV close to the error threshold and that LCMV can rapidly regain fitness in the absence of a mutagen. The invariance of the consensus genomic sequence, associated with the movements toward and away from the error threshold, suggests a decisive participation of the mutant spectrum in determining infectivity levels and supports a lethal defection model for virus extinction through enhanced mutagenesis.
MATERIALS AND METHODS
Cells and virus.
Baby hamster kidney cells (BHK-21) were grown in Dulbecco's modified Eagle's medium (DMEM) with 5% fetal calf serum (FCS) as described previously (79). Vero cells were maintained in DMEM supplemented with 3% FCS and 2% l-glutamine. LCMV strain Armstrong (ARM) 53b is a triple plaque-purified clone of Armstrong CA 1371, passaged four times in BHK-21 cells (10, 23, 30, 62, 73, 80, 82). For the present experiments, a stock virus (LCMV p0) was prepared by infecting 1 × 107 BHK-21 cells with LCMV at a multiplicity of infection (MOI) of 0.01 PFU per cell (41) (LCMV ARM 53b was kindly supplied by P. Borrow).
Virus infections.
Semiconfluent (2.8 × 106 cells in 100-mm-diameter dishes [Falcon]) monolayers of BHK-21 were infected with 0.01 PFU of LCMV ARM 53b per cell in 5 ml of DMEM supplemented with 10% FCS, 10% tryptose phosphate broth, 2% l-glutamine, 0.52% glucose, and 50 μg/ml gentamicin. Viral supernatants were harvested 48 h postinfection, clarified by centrifugation at 2,500 rpm for 30 min at 4°C, and stored at −80°C. Supernatants were titrated on Vero cell monolayers under a semisolid agar medium, as described elsewhere (3). Viral yields usually were ≥108 PFU/ml.
Serial passages of LCMV ARM 53b were carried out by infecting control (D) or FU-treated (F) cell monolayers with the corresponding supernatants of the previous infection. For infections carried out with a virus obtained following a D passage, 2.8 × 106 BHK-21 cells were infected with about 3 × 104 PFU of LCMV, whereas for infections with a virus derived from an F passage virus, the input was ≤103 PFU of LCMV. Virus supernatants and cells were harvested 48 h postinfection. To obtain preextinction virus populations, semiconfluent BHK-21 monolayers (15 182-cm2 flasks with 1.6 × 107 cells per flask) were infected at an MOI of 0.01 PFU/cell and incubated for 48 h in 10 ml of infection medium (with 100 μg/ml of FU) per flask. No specific LCMV sequences could be amplified and no infectivity (<10 PFU/ml) obtained from cell culture supernatants after a second passage in 100 μg/ml FU, even after 25-fold concentration with a Centricon Plus-20 Biomax-100 (Millipore) filter. Virus titers are means of three titrations; standard deviations never exceeded 10% of the mean.
Drug treatment.
Preparation of FU stock solutions, BHK-21 cell viability assays, and infections in the presence of the mutagenic analogue were performed as previously described (41, 79). LCMV was considered extinguished when no reverse transcriptase PCR (RT-PCR)-amplifiable material and no infectivity (<10 PFU/ml) could be detected after three blind passages of the undiluted viral population in BHK-21 cells in standard culture medium in the absence of mutagens.
RNA extraction, RT-PCR, nucleotide sequencing, molecular cloning, and calculation of mutant-spectrum complexity.
RNA was extracted from supernatants of infected cell cultures by using Trizol (Invitrogen) according to the manufacturer's protocol. RNAs were amplified by two-step RT-PCR using ThermoScript reverse transcriptase (Invitrogen) and a reverse (antisense) primer (http://www.cbm.uam.es/mkfactory.esdomain/webs/CBMSO/plt_LineasInvestigacion.aspx?IdObjeto=17) at 60°C for 45 min, followed by PCR with Pfu DNA polymerase (Promega). As a control to confirm that an excess of template was being amplified, 1/10 of the initial amount of template RNA was processed in parallel and yielded a positive amplification band (4, 41). cDNAs either were purified with a Wizard PCR purification kit (Promega) or were subjected to agarose gel electrophoresis and the cDNA band extracted from the gel using a QIAEX II gel extraction kit (QIAGEN). Purified cDNA was sequenced using Big Dye chemistry (ABI, Perkin-Elmer), and the products obtained were analyzed using an ABI 377 automated sequencer (Perkin-Elmer). Molecular clones corresponding to the intergenic regions of L RNA and of part of the GP-C-coding region in the S segment (residues 247 to 759) were obtained by ligation of cDNA into the pGEM-T Easy vector (Promega) and transformation into Escherichia coli DH5α cells. DNA from positive colonies was amplified with a TempliPhi amplification kit (Amersham; based on rolling-circle amplification by the high-fidelity φ29 DNA polymerase) by following the manufacturer's protocol. Mutant-spectrum complexity was calculated by scoring mutations relative to the consensus sequence defined by all the clones analyzed. Mutation frequencies for mutant spectra of the GP-C-coding region were determined as the number of different mutations (repeated mutations were counted only once) divided by the total number of nucleotides sequenced (the length of the GP-C-coding region multiplied by the number of clones analyzed).
The entire LCMV genome was sequenced using the primers and the strategy given in the supplementary material (http://www.cbm.uam.es/mkfactory.esdomain/webs/CBMSO/plt_LineasInvestigacion.aspx?IdObjeto=17). For sampling of partial genomic sequences, three LCMV genomic regions were amplified by RT-PCR and sequenced, as follows: in the S genomic RNA, the glycoprotein-coding region between residues 268 and 738 was amplified using primers GP247F (forward) and GP759R (reverse); in the nucleoprotein-coding region, residues 2244 to 2723 were amplified with primers NP2223F (forward) and NP2743R (reverse); in the L RNA, the polymerase-coding region between nucleotides 2950 and 4240 was amplified with primers L2929F (forward) and L4260R (reverse) (this set amplified the region corresponding to amino acids 978 to 1422, which encompasses premotif A to motif E, including the catalytic domain of the viral polymerase [24, 52, 87]). Consensus nucleotide sequences were determined using entire viral RNA populations as templates, except for the L intergenic region, for which consensus sequences were derived from the alignment of sequences from multiple individual molecular clones. Most nucleotide sequences were determined two or more times using different sense and antisense primers (described at http://www.cbm.uam.es/mkfactory.esdomain/webs/CBMSO/plt_LineasInvestigacion.aspx?IdObjeto=17). Nucleotide positions are given in the viral (genomic) sense and refer to the consensus genomic sequence determined in the present study. LCMV genomic nucleotide sequences were retrieved from GenBank (74-76) (GenBank accession numbers BANKIT 691408 and AY 894816). Sequences were compared using SEQMAN from the DNASTAR software package (Lasergene), and alignments were done with the GeneDoc program (K. B. Nicholas and H. B. Nicholas, Jr., Pittsburgh Supercomputing Center, Pittsburgh, Pa.). Computer-based predictions of RNA secondary structure were carried out using the MFOLD program (92) with default folding.
Detection of hypermutated molecules.
Conditions for reverse transcription and PCR were set to increase the probability of amplification of putative hypermutated molecules. The RT reaction was done with avian myeloblastosis virus (AMV) reverse transcriptase (Promega) at 37°C for 45 min and then at 94°C for 5 min. This incubation temperature is that recommended for the synthesis of first-strand cDNA using random oligonucleotide primers. One microliter of RNA was thawed at 70°C for 3 min, and after cooling on ice, 8 pmol of reverse primer was added and the mixture incubated at 65°C for 5 min. A mixture of 2.5 U of AMV and 7.5 U of RNAsin RNA inhibitor (Promega) was added in a final reaction volume of 20 μl. Five microliters of the first-strand reaction was used in a touchdown PCR in a volume of 50 μl with 1.25 U of Pfu DNA polymerase and 16 pmol of reverse and forward primers. The reaction conditions were as follows: denaturation hold at 95°C for 2 min, followed by 20 cycles at 95°C for 1 min, 60°C for 30 s (starting at an annealing temperature of 60°C, with a decrease of 0.5°C per cycle), and 72°C for 2 min, and finally 10 additional cycles at an annealing temperature of 47°C. Assuming that the more-frequent mutations induced by FU are transitions of the types A→G and U→C (41, 65, 67, 72, 79), oligonucleotides with different degrees of degeneracy (which should favor hybridization with mutated templates) were synthesized. Oligonucleotides L3662F (forward) and L4260R (reverse) were modified to produce a battery of degenerate primers to amplify residues 3662 to 4260 of the L polymerase-coding region (Table 1). The cDNAs synthesized were purified with a Wizard PCR purification kit (Promega), ligated into the pGEM-T Easy vector (Promega), and used to transform E. coli DH5α; positive bacterial clones were amplified with the TempliPhi amplification kit (Amersham). Purified cDNA and molecular clones were subjected to cycle sequencing. Standard RNA consisted of an SP6 RNA polymerase runoff transcript of a molecular clone that contained the polymerase region amplified with primers L3662F and L4260R in the genomic sense. Quantification of RNAs of control and FU-treated virus populations was done by two-step RT-PCR with ThermoScript RT (Invitrogen) at 60°C for 45 min, followed by a hold at 85°C for 5 min; 2 μl of the first-strand reaction product was used in a 20-μl reaction mixture with the Fast Start DNA Master SYBR Green I kit (Roche) in a Light Cycler instrument (Roche). A standard curve employed serial 10-fold dilutions of the standard RNA from 1010 to 105 molecules. Mutation frequencies were calculated as described above.
TABLE 1.
Oligonucleotide primers with position degeneracies used in the present study
| Namea | Sequence (5′ to 3′) | Type of degeneracy | Degree of degeneracyb | No. of clonesc | Avg no. of mutations per clonec |
|---|---|---|---|---|---|
| L3662F | AGTTTAAGAACCCTTCCCGC | 0 | 0 | 15 (R)d | 2.06 ± 1.83 |
| L3662F R2 | ARTTTAARAACCCTTCCCGC | 2 A/G | 4 | NDe | ND |
| L3662F RY2 | AGTTYAARAACCCTTCCCGC | 1 A/G, 1 C/T | 4 | ND | ND |
| L3662F RY4 | ARTTTAARAAYCCTTCYCGC | 2 A/G, 2 C/T | 8 | 9 (RY4) | 1.77 ± 1.39 |
| L3662F Y5 | AGTTYAAGAAYCCYTCYCGY | 5 C/T | 10 | 5 (R) | 1.60 ± 1.14 |
| L3662F Y5Bis | AGYYYAAGAACCCYYCCCGC | 5 C/T | 10 | 10 (RY5) | 1.20 ± 1.03 |
| L3662F R5 | RGTTTRRGRRCCCTTCCCGC | 5 A/G | 10 | 9 (RR4) | 1.77 ± 0.97 |
| L3662F RY7 | ARTTYAARAAYCCYTCYCGY | 2 A/G, 5 C/T | 14 | NAf | |
| L3662F RY16 | RGYYYRRGRRYYYYYYYYGC | 5 A/G, 11 C/T | 32 | NA | |
| L4260R | TGTTGAGGGTTCCACAGAGC | 0 | 0 | 15 (R) | 2.06 ± 1.83 |
| L4260R YR2 | TGTTGARGGYTCCACAGAGC | 1 A/G, 1 C/T | 4 | ND | |
| L4260R R3 | TGTTGARGGTTCCACRGARC | 3 A/G | 6 | 9 (FRY4) | 1.66 ± 1.65 |
| L4260R R4 | TGTTGRGGGTTCCRCRGRGC | 4 A/G | 8 | 5 (FY5Bis) | 1.00 ± 0.70 |
| L4260R Y4 | YGTYGAGGGYTCYACAGAGC | 4 C/T | 8 | 15 (F) | 2.20 ± 2.33 |
| L4260R Y5 | YGYYGAGGGYYCCACAGAGC | 5 C/T | 10 | 13 (FR5) | 0.69 ± 0.94 |
| L4260R YR7 | YGTYGARGGYTCYACRGARC | 3 A/G, 4 C/T | 14 | NA | |
| L4260R RY16 | YRYYRRRRRYYCCRCRRRRC | 11 A/G, 5 C/T | 32 | NA |
Forward primer L3662F and reverse primers L4260R were modified to produce a battery of degenerate primers. The types (Y = C + T; R = A + G) and total number of degeneracies for each primer are indicated by boldfaced letters.
Calculated by multiplying the number of degenerate positions by the number of possible nucleotides (A, G, C, or T).
Results shown only for the FU-treated sample.
The paired oligonucleotide used in the PCR is given in parentheses.
ND, not determined. Amplifications using L3662F R2 and L3662F RY2 were positive using RNA from amplification of control samples (D) but negative with RNA from FU-treated virus (F).
NA, negative amplification (no DNA band observed) in control or FU-treated samples. All oligonucleotides yielded positive amplification of control and FU-treated virus samples, except those that had degeneracy over 10.
Nucleotide sequence accession numbers.
The consensus genomic sequence determined in the present study has been deposited in GenBank under accession numbers AY847350 for the S genomic segment and AY847351 for the L genomic segment.
RESULTS
Rapid FU-mediated decrease of LCMV infectivity and rapid recovery in the absence of mutagens.
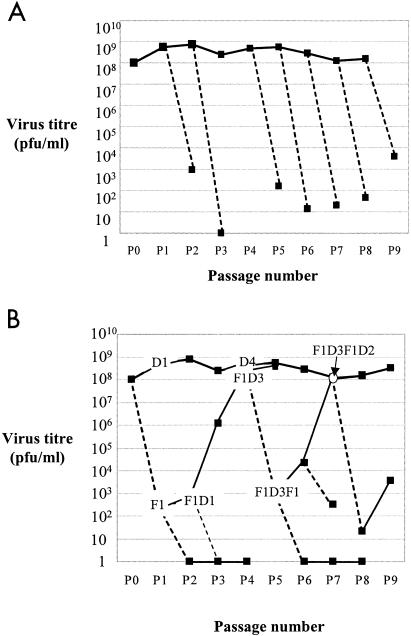
Serial population passages of RNA viruses under a defined set of stable conditions generally result in fitness gain (32, 58), and lethal mutagenesis is less effective with high-fitness virus (67, 79). To investigate whether loss of infectivity of LCMV upon replication in the presence of FU was dependent on the passage number of the virus in BHK-21 cells, LCMV was serially passaged in BHK-21 cells at an MOI of 0.01 PFU/cell in the absence of FU, as detailed in Materials and Methods. Using virus from passage 1, 2, 4, 5, 6, 7, 8, or 9, an infection in the presence of 100 μg/ml FU resulted in 105- to 108-fold decreases in infection progeny production, and the decrease did not show any trend with the passage number of LCMV in BHK-21 cells (Fig. 1A).
FIG. 1.
Effect of repeated mutagenesis-recovery cycles on LCMV infectivity during serial passages of LCMV ARM 53b in BHK-21 cells. LCMV (parental LCMV ARM 53b stock p0 with 1 × 108 PFU/ml) was serially passaged by infecting BHK-21 monolayers (2.8 × 106 cells) at an MOI of 0.01 PFU per cell in the absence (solid lines) or presence (dashed lines) of 100 μg/ml FU. Virus supernatants were harvested at 48 postinfection, clarified by centrifugation, and titrated on Vero cell monolayers. (A) Virus infectivity reduction following a single passage in the presence of FU. (B) Variation of infectivity of LCMV subjected to FU mutagenesis and recovery in the absence of FU. Passages in the absence or presence of FU are indicated by D and F, respectively (i.e., F1D3F1D2 means that the parental LCMV stock p0 was subjected successively to one passage in the presence of FU, three passages in the absence of FU, one passage in the presence of FU, and finally two passages in the absence of FU). Virus extinction at passages 2, 3, and 6 was confirmed by the absence of infectivity and the absence of virus-specific RT-PCR amplification after two further passages in the absence of drug. Procedures are detailed in Materials and Methods.
To study whether mutagenized populations could regain the capacity to produce progeny in the absence of FU, several FU-treated populations were allowed to replicate in the absence of the mutagen. In the first passage without FU, the capacity to produce progeny, which is taken as an estimate of relative viral fitness (33, 47), was impaired. However, two to three passages in the absence of mutagens allowed LCMV to regain the capacity to produce progeny at levels comparable to those of LCMV not subjected to mutagenic treatment (Fig. 1B). Two passages in the presence of FU led to virus extinction, as defined in Materials and Methods. In one case, extinction occurred despite an intervening recovery passage (Fig. 1B). A recovered preextinction population was driven to extinction by reiteration of the mutagenic process, and the preextinction population that resulted from this second round of mutagenesis could again attain the standard capacity to produce progeny (Fig. 1B). Thus, LCMV is very sensitive to FU-induced lethal mutagenesis, but preextinction populations maintain the potential to produce normal infectious progeny levels in BHK-21 cells upon replication in the absence of mutagen.
Fitness variations with an invariant consensus sequence.
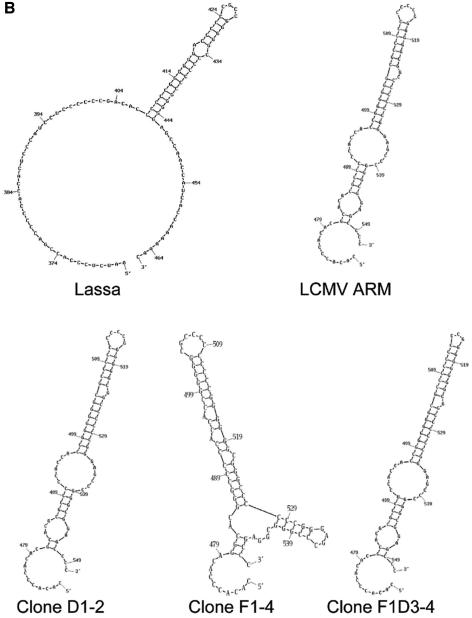
To study whether large fluctuations in replication capacity were associated with modification of the consensus LCMV genomic sequence, the entire genomic nucleotide sequences of the virus after one passage in the absence of drug (D1), the virus subjected to one passage in the presence of FU (F1), and the virus subjected to three subsequent recovery passages in the absence of FU (F1D3) were determined (the nomenclature of LCMV populations that defines the passage history in the presence or absence of FU is described in the legend for Fig. 1B). No differences in the consensus nucleotide sequence of the three viruses were observed. Although some differences were seen relative to the previously published LCMV ARM 53b sequence (73-76), our sequence is in good agreement with the LCMV genomic sequence deposited in GenBank with accession number AY894816. Under the standard amplification conditions, it was not possible to determine a defined nucleotide sequence (i.e., the peak pattern suggested the presence of insertions and/or deletions) for the intergenic region of segment L (nucleotides 470 to 550). RT-PCR at a higher temperature (25, 87) in 5% dimethyl sulfoxide was used to obtain cDNA that was cloned into the pGEM-T Easy vector (Promega). The nucleotide sequences of 13 clones from viral populations D1, F1, and F1D3 indicated a remarkable genetic heterogeneity with G-rich and C-rich sequences. Despite this heterogeneity, no evidence of dominance of different subpopulations was obtained for viruses D1, F1, and F1D3 (Fig. 2A). Two of the clones harbored large deletions which likely correspond to stem-loop structures deleted during the amplification or during the sequencing reaction by polymerase slippage (71, 89); however, heterogeneity in the viral RNA cannot be excluded. We have not subjected in vitro-transcribed RNA containing this region to RT-PCR to assess the contribution to the observed heterogeneity of errors during the amplification procedure. Previous studies have reported deletions in the intergenic regions of LCMV (75) and Lassa fever virus (87).
FIG. 2.
Nucleotide sequence and predicted secondary structure of an LCMV genomic region belonging to the L intergenic region. (A) Nucleotide sequence of residues 470 to 550, corresponding to the intergenic L region of clones from populations D1, F1, and F1D3. Sequences of clones are aligned under our consensus LCMV ARM 53b; at the bottom, the sequence previously published (termed here ARM 53b*) (76) is given. Our sequence is in agreement with a recent sequence (deposited in GenBank with accession number AY894816), determined independently of ours. Deletions introduced for best alignment of heteropolymeric stretches are indicated by dashes. (B) Predicted secondary structure of intergenic L residues 365 to 466 of Lassa fever virus and predicted secondary structures of residues 470 to 550 of LCMV ARM 53b, clones D1-2, F1-4, and F1D3-4 analyzed in the present study. Procedures for nucleotide sequence determination and secondary-structure prediction are described in Materials and Methods.
To further ascertain that variations in fitness were not accompanied by changes in the consensus genomic nucleotide sequence, three genomic regions (nucleotides 2950 to 4240 of the L segment, encoding amino acids 985 to 1415 of protein L; nucleotides 268 to 738 of the S segment, encoding amino acids 62 to 220 of GP-C; and nucleotides 2247 to 2723 of the S segment, encoding amino acids 198 to 357 of NP) were sequenced for viruses D1, F1, F1D1, D4, F1D3, F1D3F1, and F1D3F1D2 (passage history depicted in Fig. 1B). Again, no differences in consensus sequence were observed. These results show that extreme variations in fitness of an RNA virus, even the approach of population extinction, can occur without being reflected in any modification of the consensus genomic nucleotide sequence.
To rule out the possibility that the invariance of the consensus sequence reflected a genetically homogeneous RNA population, the amplified DNAs of the GP-C-coding region between residues 247 and 759 of the S segment of populations D1, F1, and FD3 were cloned into pGEM-T, and 8 to 13 molecular clones from each population were sequenced. The mutation frequencies for D1, F1, and FD3 were 1.4 × 10−4, 2.3 × 10−4, and 3.9 × 10−3, respectively (basal mutation frequency levels, determined with a runoff transcript, are included in Fig. 3), indicating that despite the invariance of the consensus sequence, the quasispecies contain highly heterogeneous mutant spectra (see also Discussion).
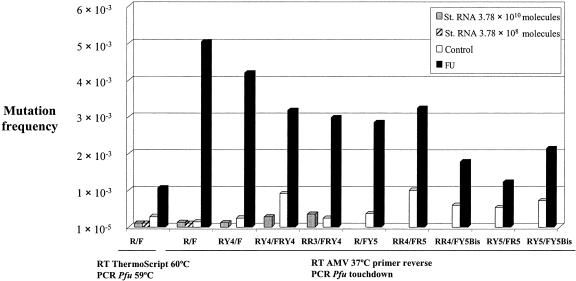
FIG. 3.
Mutation frequencies in control and FU-treated LCMV populations. RNAs from control (D) and FU-treated (F) virus populations were copied to cDNA (residues 3662 to 4260 of the L polymerase-coding region) under standard amplification conditions (RT reaction with ThermoScript RT at 60°C followed by Pfu DNA polymerase amplification at an annealing temperature of 59°C) or under less stringent amplification conditions (RT reaction with AMV RT at 37°C followed by Pfu amplification in a touchdown PCR) as indicated. Combinations of degenerate and wild-type primers are indicated (primers used are listed in Table 1, although the first letter, L, and the nucleotide position are omitted here for simplification). As an internal control, two dilutions of an LCMV L RNA runoff transcript (St. RNA, standard RNA) with similar numbers of molecules as the D and F virus populations were processed in parallel under both amplification conditions. Criteria for the design of degenerate primers and RT-PCR amplification conditions are detailed in Materials and Methods.
No evidence of dominance of hypermutated genomes.
We initially hypothesized that the transition into error catastrophe of RNA viruses could be accompanied by the synthesis of aberrant hypermutated, nonfunctional viral genomes and that despite being replication incompetent, such genomes could be detected by RT-PCR amplification (41). To search for such hypermutated genomes, we tested a collection of multiply substituted oligonucleotide primers (Table 1) by RT-PCR amplification assays using as a template RNA extracted from either control (D) or FU-treated (F) virus populations. Using 2.6 × 107 and 3.2 × 107 molecules for D and F populations, we observed positive amplification only with primers with a degree of degeneracy of 10 or lower (Table 1). The cDNA products of some of these amplifications were cloned and several molecular clones sequenced. No evidence of hypermutated molecules was obtained with any of the primer combinations tested. The mutant spectra of the F population showed higher genetic heterogeneity than those of the D population tested, but the average number of mutations per molecule was 2, with the exception of a single molecule whose number of mutations lies at the limit of viral genomes previously classified as hypermutated (56) (see Discussion).
For primers without degeneracy, lowering the temperature during first-strand synthesis, followed by touchdown amplification, allowed detection of genomes with a number of mutations that remained undetected under standard, more-stringent conditions of amplification. Under these new amplification conditions, the mutation frequency for the F population was 4.7-fold higher than the value obtained under standard amplification conditions whereas for the D population the mutation frequency was 2.0-fold lower (Fig. 3). Also, for the F population, the mutation frequencies scored were lower when any combination of degenerate oligonucleotides was used than with standard oligonucleotides representing the wild-type LCMV consensus sequence. Control amplifications employing similar numbers of molecules of L RNA runoff transcripts as those in the D and F populations indicate that the differences in the mutation frequency values were not due to the amplification conditions (Fig. 3). Thus, the results do not support our initial hypothesis that hypermutated molecules may be dominant during the transition into error catastrophe (41). Rather, the results suggest that mutant spectra, composed of genomes harboring modest numbers of fitness-decreasing mutations, dominate the molecular landscape in the LCMV transition into error catastrophe, supporting a lethal defection model for LCMV extinction (40).
DISCUSSION
Lethal mutagenesis has been approached with a number of RNA viruses (4, 18, 19, 21, 41, 42, 46, 48, 50, 51, 53, 65, 67, 72, 77, 79) and is currently being explored as a possible new antiviral strategy to provide an alternative to the use of inhibitors of viral replication (26, 35, 38). The molecular events that mediate the critical transition into viral extinction, also termed virus entry into error catastrophe (5, 31, 60, 83), are still poorly understood, but such understanding may be crucial for finding new virus-specific mutagenic agents that, in combination with inhibitors, could lead to a clinical application. The previous studies with LCMV documented a very high sensitivity of the virus to FU in cell culture (41, 72) and prevention by FU of the establishment of a persistent LCMV infection of RAG−/− (deficient in the adaptive immune response) mice (72). This latter study constituted a proof-of-principle of the potential of mutagenesis to prevent a viral infection in vivo. Moreover, there is evidence that the antiviral activity of the nucleoside analogue ribavirin, currently licensed for the treatment of several viral diseases (68, 81), might be mediated in some cases by its mutagenic action (4, 18-21, 39, 45, 53, 66, 88).
LCMV shows higher sensitivity to FU mutagenesis than foot-and-mouth disease virus (FMDV) in cell culture when the two viruses are subjected to similar FU doses (41, 65, 67, 72, 79). The basis for this difference is not known, but it may relate either to the size of the essential polymerase gene, which occupies 62.6% and 17.4% of the total genetic information of LCMV and FMDV, respectively; to a larger number of essential genomic sites susceptible to FU-induced mutations in LCMV than in FMDV; or to other replicative differences between the two viruses still to be elucidated (different affinities of the two polymerases for FU-triphosphate, etc.). The comparison of complete genomic LCMV sequences reported here has indicated that neither an abrupt decrease in infectivity associated with FU treatment nor the ensuing recovery of the capacity to produce infectious progeny upon replication in the absence of mutagen modified the consensus nucleotide sequence of the virus. Likewise, no variations were detected among LCMV populations subjected to one or several rounds of replication in the absence or presence of FU, or among populations that underwent alternations between the two passage regimens. In these cases the consensus sequences analyzed involved three genomic regions, one of which was a part of the polymerase (L) that included premotif A and motifs A to E (11, 24, 52, 70, 86, 87). The possibility that FU, in addition to its mutagenic activity documented by increases in mutant-spectrum complexity, could exert some mutagenesis-independent, inhibitory activity (i.e., directly on the viral polymerase or indirectly through alteration of nucleotide pools, etc.) on LCMV replication cannot be excluded. This would mean that FU could contribute to decreases in viral load, thereby favoring mutagenesis-induced extinction, as documented with FMDV (65-67).
It could be argued that fitness variations despite an invariant consensus sequence may occur without modification of the mutant spectrum, simply by an increase of dominance of a preexisting subset of genomes with the same consensus sequence. A static mutant spectrum is highly unlikely given the number of LCMV progeny per cell (Fig. 1), since it would imply mutation rates much lower than those that have been calculated for RNA viruses (29). Low mutation rates are not supported by the estimates of the frequencies of cytotoxic T-lymphocyte (CTL)- and antibody-escape mutants in LCMV populations (1, 15, 43, 49; reviewed in reference 78). Fitness variations without modification of consensus genomic sequences have been reported for vesicular stomatitis virus clones in the process of fitness recovery after bottleneck passages (59).
The L RNA intergenic region may function as a transcription termination signal, as previously suggested for LCMV (69, 74), other arenaviruses (25, 34, 44), and the stem-loop present in the S RNA intergenic region of LCMV (55), Pichinde virus (7), and Lassa fever virus (8, 16). RNA secondary-structure predictions suggest that the L RNA intergenic region of LCMV ARM 53b forms a stem-loop stabilized by 15 GC pairs (Fig. 3B), similar to a loop predicted for the L RNA intergenic regions of Tacaribe and Lassa fever virus (25, 34, 44). We have not distinguished whether the nucleotide sequence heterogeneity seen in the L intergenic region (Fig. 3A) corresponds to a heterogeneity present in the LCMV used (perhaps with functional implications) or is due to the production of altered copies during RT-PCR, or both.
Although it cannot be excluded that highly mutated or hypermutated RNA molecules derived from LCMV RNA may be present during the transition to error catastrophe, attempts using standard and multiply substituted oligonucleotide primers have failed to reveal the dominance of such genomes (Table 1 and Fig. 3). In the amplification with primers L3662F and L4260 RY4, one sequence was found that had nine mutations, of which six were U→C transitions and the others were A→G, C→A, and G→A. The mutation frequency for this molecule relative to the consensus sequence (1.6 × 10−2) would lie within the lower limit in the range of 3.3 × 10−2 to 1.0 × 10−1 mutations per nucleotide for the U→C (or A→G) hypermutation, previously characterized in genomes of measles virus, human parainfluenza virus 3, vesicular stomatitis virus, and Rous-associated virus, which has been related to posttranscriptional unwinding and mutagenic activities (13, 36, 54, 56, 57, 61). Hypermutated genomes have not been identified, either, among multiple molecular clones copied from preextinction populations of FMDV (4, 67, 79). In the FU-mediated transition to extinction of LCMV in the course of a persistent infection of BHK-21 cells, decreases in viral infectivity preceded decreases in viral RNA levels (40). The specific infectivity of LCMV reached values 102- to 103-fold lower than those for LCMV from parallel cultures not subjected to FU mutagenesis (40). Similar decreases in specific infectivity were noted for mutagenized FMDV during persistent or cytolytic infections (4, 37, 65, 66). These quantifications of specific infectivity, together with the complexity of mutant spectra of mutagenized LCMV and FMDV populations (4, 37, 40, 41, 65-67, 72, 79), suggest that rather than requiring extensive mutagenesis, virus extinction may be mediated by the presence of defective genomes (which have been termed “defectors” in several studies of RNA virus evolution [85, 90]). Therefore, the present evidence suggests that low-activity mutagens capable of being incorporated specifically by viral polymerases may be sufficient to enrich the defector population to a level at which it may interfere with standard genome replication. A likely molecular mechanism mediating this type of interference is the effect of multiple, low-frequency dominant-negative mutants generated in mutagenized virus populations, an extension to an intrapopulation level of the mechanisms of interference between different viruses, well documented in classical viral genetics (reviewed in references 2, 17, and 91). Because such interfering genomes—due to their partial or complete defectiveness—cannot dominate the population, no alteration of consensus sequences is observed despite profound fitness losses. Thus, the results reported here support a lethal defection model for the mutagenesis-induced genetic meltdown that results in viral extinction (40). Treatment with the mutagenic nucleoside analogue ribavirin of cell cultures infected with poliovirus or Hantaan virus resulted in decreases in infectivity that preceded decreases in viral RNA levels (19-21, 77), again suggesting a role of mutagenesis-induced defective genomes that maintain their replication competence prior to detectable losses of infectivity.
The results presented here underscore the relevance of mutant spectra in determining virus behavior, illustrated here by the capacity of LCMV to either decrease or increase its fitness without any alteration of the consensus nucleotide sequence. This observation must be added to results with several other virus-host systems showing suppressive or modulating effects of mutant spectra on specific types of variants with superior fitness (9, 14, 22, 37, 84).
Acknowledgments
We are indebted to J. C. de la Torre for pointing out to us the LCMV sequence (GenBank bankit 691408; accession number AY894816), for valuable comments, and for critical reading of the manuscript, and to P. Borrow for the generous gift of LCMV ARM 53b. We thank M. Dávila for expert technical assistance.
This work was supported by grants BMC 2001-1823-C02-01 and CAM 08.2/0015/2001.1 and by Fundación R. Areces. A.G.-P. was supported by a postdoctoral contract from CAM and a RyC contract from MCyT.
REFERENCES
- 1.Aebischer, T., D. Moskophidis, U. H. Rohrer, R. M. Zinkernagel, and H. Hengartner. 1991. In vitro selection of lymphocytic choriomeningitis virus escape mutants by cytotoxic T lymphocytes. Proc. Natl. Acad. Sci. USA 88:11047-11051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agol, V. I. 2002. Picornavirus genetics: an overview, p. 269-284. In B. L. Semler and E. Wimmer (ed.), Molecular biology of picornaviruses. American Society for Microbiology, Washington, D.C.
- 3.Ahmed, R., and M. B. Oldstone. 1988. Organ-specific selection of viral variants during chronic infection. J. Exp. Med. 167:1719-1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Airaksinen, A., N. Pariente, L. Menendez-Arias, and E. Domingo. 2003. Curing of foot-and-mouth disease virus from persistently infected cells by ribavirin involves enhanced mutagenesis. Virology 311:339-349. [DOI] [PubMed] [Google Scholar]
- 5.Alves, D., and J. F. Fontanari. 1998. Error threshold in finite populations. Phys. Rev. E 57:7008-7013. [Google Scholar]
- 6.Anderson, J. P., R. Daifuku, and L. A. Loeb. 2004. Viral error catastrophe by mutagenic nucleosides. Annu. Rev. Microbiol. 58:183-205. [DOI] [PubMed] [Google Scholar]
- 7.Auperin, D. D., V. Romanowski, M. Galinski, and D. H. Bishop. 1984. Sequencing studies of pichinde arenavirus S RNA indicate a novel coding strategy, an ambisense viral S RNA. J. Virol. 52:897-904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Auperin, D. D., D. R. Sasso, and J. B. McCormick. 1986. Nucleotide sequence of the glycoprotein gene and intergenic region of the Lassa virus S genome RNA. Virology 154:155-167. [DOI] [PubMed] [Google Scholar]
- 9.Borrego, B., I. S. Novella, E. Giralt, D. Andreu, and E. Domingo. 1993. Distinct repertoire of antigenic variants of foot-and-mouth disease virus in the presence or absence of immune selection. J. Virol. 67:6071-6079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Borrow, P., C. F. Evans, and M. B. Oldstone. 1995. Virus-induced immunosuppression: immune system-mediated destruction of virus-infected dendritic cells results in generalized immune suppression. J. Virol. 69:1059-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bruenn, J. A. 2003. A structural and primary sequence comparison of the viral RNA-dependent RNA polymerases. Nucleic Acids Res. 31:1821-1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buchmeier, M. J., M. D. Bowen, and C. J. Peters. 2001. Arenaviridae: the viruses and their replication, p. 1635-1668. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams and Wilkins, Philadelphia, Pa.
- 13.Cattaneo, R., and M. A. Billeter. 1992. Mutations and A/I hypermutations in measles virus persistent infections. Curr. Top. Microbiol. Immunol. 176:63-74. [DOI] [PubMed] [Google Scholar]
- 14.Chumakov, K. M., L. B. Powers, K. E. Noonan, I. B. Roninson, and I. S. Levenbook. 1991. Correlation between amount of virus with altered nucleotide sequence and the monkey test for acceptability of oral poliovirus vaccine. Proc. Natl. Acad. Sci. USA 88:199-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ciurea, A., P. Klenerman, L. Hunziker, E. Horvath, B. M. Senn, A. F. Ochsenbein, H. Hengartner, and R. M. Zinkernagel. 2000. Viral persistence in vivo through selection of neutralizing antibody-escape variants. Proc. Natl. Acad. Sci. USA 97:2749-2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clegg, J. C., S. M. Wilson, and J. D. Oram. 1991. Nucleotide sequence of the S RNA of Lassa virus (Nigerian strain) and comparative analysis of arenavirus gene products. Virus Res. 18:151-164. [DOI] [PubMed] [Google Scholar]
- 17.Condit, C. 2001. Principles of virology, p. 811-815. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed., vol. 2. Lippincott Williams and Wilkins, Philadelphia, Pa. [Google Scholar]
- 18.Contreras, A. M., Y. Hiasa, W. He, A. Terella, E. V. Schmidt, and R. T. Chung. 2002. Viral RNA mutations are region specific and increased by ribavirin in a full-length hepatitis C virus replication system. J. Virol. 76:8505-8517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Crotty, S., C. Cameron, and R. Andino. 2002. Ribavirin's antiviral mechanism of action: lethal mutagenesis? J. Mol. Med. 80:86-95. [DOI] [PubMed] [Google Scholar]
- 20.Crotty, S., C. E. Cameron, and R. Andino. 2001. RNA virus error catastrophe: direct molecular test by using ribavirin. Proc. Natl. Acad. Sci. USA 98:6895-6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crotty, S., D. Maag, J. J. Arnold, W. Zhong, J. Y. N. Lau, Z. Hong, R. Andino, and C. E. Cameron. 2000. The broad-spectrum antiviral ribonucleotide, ribavirin, is an RNA virus mutagen. Nat. Med. 6:1375-1379. [DOI] [PubMed] [Google Scholar]
- 22.de la Torre, J. C., and J. J. Holland. 1990. RNA virus quasispecies populations can suppress vastly superior mutant progeny. J. Virol. 64:6278-6281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de la Torre, J. C., G. Rall, C. Oldstone, P. P. Sanna, P. Borrow, and M. B. Oldstone. 1993. Replication of lymphocytic choriomeningitis virus is restricted in terminally differentiated neurons. J. Virol. 67:7350-7359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Djavani, M., I. S. Lukashevich, and M. S. Salvato. 1998. Sequence comparison of the large genomic RNA segments of two strains of lymphocytic choriomeningitis virus differing in pathogenic potential for guinea pigs. Virus Genes 17:151-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Djavani, M., I. S. Lukashevich, A. Sanchez, S. T. Nichol, and M. S. Salvato. 1997. Completion of the Lassa fever virus sequence and identification of a RING finger open reading frame at the L RNA 5′ end. Virology 235:414-418. [DOI] [PubMed] [Google Scholar]
- 26.Domingo, E. 2003. Quasispecies and the development of new antiviral strategies. Prog. Drug Res. 60:133-158. [DOI] [PubMed] [Google Scholar]
- 27.Domingo, E. 2005. Virus entry into error catastrophe as a new antiviral strategy. Virus Res. 107:115-228. [Google Scholar]
- 28.Domingo, E. (ed.). Viruses as quasispecies: biological implications. Curr. Top. Microbiol. Immunol., in press. [DOI] [PMC free article] [PubMed]
- 29.Drake, J. W., and J. J. Holland. 1999. Mutation rates among RNA viruses. Proc. Natl. Acad. Sci. USA 96:13910-13913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dutko, F. J., and M. B. A. Oldstone. 1983. Genomic and biologic variation among commonly used lymphocytic choriomeningitis virus strains. J. Gen. Virol. 64:1689-1694. [DOI] [PubMed] [Google Scholar]
- 31.Eigen, M. 2002. Error catastrophe and antiviral strategy. Proc. Natl. Acad. Sci. USA 99:13374-13376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Escarmís, C., M. Dávila, and E. Domingo. 1999. Multiple molecular pathways for fitness recovery of an RNA virus debilitated by operation of Muller's ratchet. J. Mol. Biol. 285:495-505. [DOI] [PubMed] [Google Scholar]
- 33.Escarmís, C., G. Gómez-Mariano, M. Dávila, E. Lázaro, and E. Domingo. 2002. Resistance to extinction of low fitness virus subjected to plaque-to-plaque transfers: diversification by mutation clustering. J. Mol. Biol. 315:647-661. [DOI] [PubMed] [Google Scholar]
- 34.Franze-Fernandez, M. T., S. Iapalucci, R. Lopez, and C. Rossi. 1993. Subgenomic RNAs of Tacaribe virus, p. 113-132. In M. S. Salvato (ed.), The Arenaviridae. Plenum Press, New York, N.Y.
- 35.Freistadt, M. S., G. D. Meades, and C. E. Cameron. 2004. Lethal mutagens: broad-spectrum antivirals with limited potential for development of resistance? Drug Resist. Updat. 7:19-24. [DOI] [PubMed] [Google Scholar]
- 36.Goff, S. P. 2003. Death by deamination: a novel host restriction system for HIV-1. Cell 114:281-283. [DOI] [PubMed] [Google Scholar]
- 37.González-López, C., A. Arias, N. Pariente, G. Gómez-Mariano, and E. Domingo. 2004. Preextinction viral RNA can interfere with infectivity. J. Virol. 78:3319-3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Graci, J. D., and C. E. Cameron. 2004. Challenges for the development of ribonucleoside analogues as inducers of error catastrophe. Antivir. Chem. Chemother. 15:1-13. [DOI] [PubMed] [Google Scholar]
- 39.Graci, J. D., and C. E. Cameron. 2002. Quasispecies, error catastrophe, and the antiviral activity of ribavirin. Virology 298:175-180. [DOI] [PubMed] [Google Scholar]
- 40.Grande-Pérez, A., E. Lázaro, P. Lowenstein, E. Domingo, and S. C. Manrubia. 2005. Suppression of viral infectivity through lethal defection. Proc. Natl. Acad. Sci. USA 102:4448-4452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grande-Pérez, A., S. Sierra, M. G. Castro, E. Domingo, and P. R. Lowenstein. 2002. Molecular indetermination in the transition to error catastrophe: systematic elimination of lymphocytic choriomeningitis virus through mutagenesis does not correlate linearly with large increases in mutant spectrum complexity. Proc. Natl. Acad. Sci. USA 99:12938-12943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Holland, J. J., E. Domingo, J. C. de la Torre, and D. A. Steinhauer. 1990. Mutation frequencies at defined single codon sites in vesicular stomatitis virus and poliovirus can be increased only slightly by chemical mutagenesis. J. Virol. 64:3960-3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hunziker, L., A. Ciurea, M. Recher, H. Hengartner, and R. M. Zinkernagel. 2003. Public versus personal serotypes of a viral quasispecies. Proc. Natl. Acad. Sci. USA 100:6015-6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Iapalucci, S., N. Lopez, O. Rey, M. M. Zakin, G. N. Cohen, and M. T. Franze-Fernandez. 1989. The 5′ region of Tacaribe virus L RNA encodes a protein with a potential metal binding domain. Virology 173:357-361. [DOI] [PubMed] [Google Scholar]
- 45.Jonsson, C. B., B. G. Milligan, and J. B. Arterburn. 2005. Potential importance of error catastrophe to the development of antiviral strategies for hantaviruses. Virus Res. 107:195-205. [DOI] [PubMed] [Google Scholar]
- 46.Lanford, R. E., D. Chavez, B. Guerra, J. Y. Lau, Z. Hong, K. M. Brasky, and B. Beames. 2001. Ribavirin induces error-prone replication of GB virus B in primary tamarin hepatocytes. J. Virol. 75:8074-8081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lazaro, E., C. Escarmis, J. Perez-Mercader, S. C. Manrubia, and E. Domingo. 2003. Resistance of virus to extinction on bottleneck passages: study of a decaying and fluctuating pattern of fitness loss. Proc. Natl. Acad. Sci. USA 100:10830-10835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee, C. H., D. L. Gilbertson, I. S. Novella, R. Huerta, E. Domingo, and J. J. Holland. 1997. Negative effects of chemical mutagenesis on the adaptive behavior of vesicular stomatitis virus. J. Virol. 71:3636-3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lewicki, H., A. Tishon, P. Borrow, C. F. Evans, J. E. Gairin, K. M. Hahn, D. A. Jewell, I. A. Wilson, and M. B. Oldstone. 1995. CTL escape viral variants. I. Generation and molecular characterization. Virology. 210:29-40. [DOI] [PubMed] [Google Scholar]
- 50.Loeb, L. A., J. M. Essigmann, F. Kazazi, J. Zhang, K. D. Rose, and J. I. Mullins. 1999. Lethal mutagenesis of HIV with mutagenic nucleoside analogs. Proc. Natl. Acad. Sci. USA 96:1492-1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Loeb, L. A., and J. I. Mullins. 2000. Lethal mutagenesis of HIV by mutagenic ribonucleoside analogs. AIDS Res. Hum. Retrovir. 13:1-3. [DOI] [PubMed] [Google Scholar]
- 52.Lukashevich, I. S., M. Djavani, K. Shapiro, A. Sanchez, E. Ravkov, S. T. Nichol, and M. S. Salvato. 1997. The Lassa fever virus L gene: nucleotide sequence, comparison, and precipitation of a predicted 250 kDa protein with monospecific antiserum. J. Gen. Virol. 78:547-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Maag, D., C. Castro, Z. Hong, and C. E. Cameron. 2001. Hepatitis C virus RNA-dependent RNA polymerase (NS5B) as a mediator of the antiviral activity of ribavirin. J. Biol. Chem. 276:46094-46098. [DOI] [PubMed] [Google Scholar]
- 54.Mangeat, B., P. Turelli, G. Caron, M. Friedli, L. Perrin, and D. Trono. 2003. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 424:99-103. [DOI] [PubMed] [Google Scholar]
- 55.Meyer, B. J., and P. J. Southern. 1993. Concurrent sequence analysis of 5′ and 3′ RNA termini by intramolecular circularization reveals 5′ nontemplated bases and 3′ terminal heterogeneity for lymphocytic choriomeningitis virus mRNAs. J. Virol. 67:2621-2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Meyerhans, A., and J.-P. Vartanian. 1999. The fidelity of cellular and viral polymerases and its manipulation for hypermutagenesis, p. 87-114. In E. Domingo, R. G. Webster, and J. J. Holland (ed.), Origin and evolution of viruses. Academic Press, San Diego, Calif.
- 57.Murphy, D. G., K. Dimock, and C. Y. Kang. 1991. Numerous transitions in human parainfluenza virus 3 RNA recovered from persistently infected cells. Virology 181:760-763. [DOI] [PubMed] [Google Scholar]
- 58.Novella, I. S., E. A. Duarte, S. F. Elena, A. Moya, E. Domingo, and J. J. Holland. 1995. Exponential increases of RNA virus fitness during large population transmissions. Proc. Natl. Acad. Sci. USA 92:5841-5844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Novella, I. S., and B. E. Ebendick-Corpus. 2004. Molecular basis of fitness loss and fitness recovery in vesicular stomatitis virus. J. Mol. Biol. 342:1423-1430. [DOI] [PubMed] [Google Scholar]
- 60.Nowak, M., and P. Schuster. 1989. Error thresholds of replication in finite populations mutation frequencies and the onset of Muller's ratchet. J. Theor. Biol. 137:375-395. [DOI] [PubMed] [Google Scholar]
- 61.O'Hara, P. J., S. T. Nichol, F. M. Horodyski, and J. J. Holland. 1984. Vesicular stomatitis virus defective interfering particles can contain extensive genomic sequence rearrangements and base substitutions. Cell 36:915-924. [DOI] [PubMed] [Google Scholar]
- 62.Oldstone, M. B., M. Salvato, A. Tishon, and H. Lewicki. 1988. Virus-lymphocyte interactions. III. Biologic parameters of a virus variant that fails to generate CTL and establishes persistent infection in immunocompetent hosts. Virology 164:507-516. [DOI] [PubMed] [Google Scholar]
- 63.Oldstone, M. B. A. (ed.). 2002. Current topics in microbiology and immunology, vol. 262 and 263. Arenaviruses I and III. Springer-Verlag, Berlin, Germany.
- 64.Oldstone, M. B. A. 2002. Biology and pathogenesis of lymphocytic choriomeningitis virus infection. Curr. Top. Microbiol. Immunol. 263:83-117. [DOI] [PubMed] [Google Scholar]
- 65.Pariente, N., A. Airaksinen, and E. Domingo. 2003. Mutagenesis versus inhibition in the efficiency of extinction of foot-and-mouth disease virus. J. Virol. 77:7131-7138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pariente, N., S. Sierra, and A. Airaksinen. 2005. Action of mutagenic agents and antiviral inhibitors on foot-and-mouth disease virus. Virus Res. 107:183-193. [DOI] [PubMed] [Google Scholar]
- 67.Pariente, N., S. Sierra, P. R. Lowenstein, and E. Domingo. 2001. Efficient virus extinction by combinations of a mutagen and antiviral inhibitors. J. Virol. 75:9723-9730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Parker, W. B. 2005. Metabolism and antiviral activity of ribavirin. Virus Res. 107:165-171. [DOI] [PubMed] [Google Scholar]
- 69.Pinschewer, D. D., M. Perez, and J. C. de la Torre. 2005. Dual role of the lymphocytic choriomeningitis virus intergenic region in transcription termination and virus propagation. J. Virol. 79:4519-4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Poch, O., B. M. Blumberg, L. Bougueleret, and N. Tordo. 1990. Sequence comparison of five polymerases (L proteins) of unsegmented negative-strand RNA viruses: theoretical assignment of functional domains. J. Gen. Virol. 71:1153-1162. [DOI] [PubMed] [Google Scholar]
- 71.Ripley, L. S. 1990. Frameshift mutation: determinants of specificity. Annu. Rev. Genet. 24:189-213. [DOI] [PubMed] [Google Scholar]
- 72.Ruiz-Jarabo, C. M., C. Ly, E. Domingo, and J. C. de la Torre. 2003. Lethal mutagenesis of the prototypic arenavirus lymphocytic choriomeningitis virus (LCMV). Virology 308:37-47. [DOI] [PubMed] [Google Scholar]
- 73.Salvato, M., P. Borrow, E. Shimomaye, and M. B. Oldstone. 1991. Molecular basis of viral persistence: a single amino acid change in the glycoprotein of lymphocytic choriomeningitis virus is associated with suppression of the antiviral cytotoxic T-lymphocyte response and establishment of persistence. J. Virol. 65:1863-1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Salvato, M., E. Shimomaye, and M. B. Oldstone. 1989. The primary structure of the lymphocytic choriomeningitis virus L gene encodes a putative RNA polymerase. Virology 169:377-384. [DOI] [PubMed] [Google Scholar]
- 75.Salvato, M., E. Shimomaye, P. Southern, and M. B. Oldstone. 1988. Virus-lymphocyte interactions. IV. Molecular characterization of LCMV Armstrong (CTL+) small genomic segment and that of its variant, Clone 13 (CTL−). Virology 164:517-522. [DOI] [PubMed] [Google Scholar]
- 76.Salvato, M. S., and E. M. Shimomaye. 1989. The completed sequence of lymphocytic choriomeningitis virus reveals a unique RNA structure and a gene for a zinc finger protein. Virology 173:1-10. [DOI] [PubMed] [Google Scholar]
- 77.Severson, W. E., C. S. Schmaljohn, A. Javadian, and C. B. Jonsson. 2003. Ribavirin causes error catastrophe during Hantaan virus replication. J. Virol. 77:481-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sevilla, N., E. Domingo, and J. C. de la Torre. 2002. Contribution of LCMV towards deciphering biology of quasispecies in vivo. Curr. Top. Microbiol. Immunol. 263:197-220. [DOI] [PubMed] [Google Scholar]
- 79.Sierra, S., M. Dávila, P. R. Lowenstein, and E. Domingo. 2000. Response of foot-and-mouth disease virus to increased mutagenesis. Influence of viral load and fitness in loss of infectivity. J. Virol. 74:8316-8323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Singh, M. K., F. V. Fuller-Pace, M. J. Buchmeier, and P. J. Southern. 1987. Analysis of the genomic L RNA segment from lymphocytic choriomeningitis virus. Virology 161:448-456. [DOI] [PubMed] [Google Scholar]
- 81.Snell, N. J. 2001. Ribavirin—current status of a broad spectrum antiviral agent. Expert Opin. Pharmacother. 2:1317-1324. [DOI] [PubMed] [Google Scholar]
- 82.Southern, P. J., M. K. Singh, Y. Riviere, D. R. Jacoby, M. J. Buchmeier, and M. B. Oldstone. 1987. Molecular characterization of the genomic S RNA segment from lymphocytic choriomeningitis virus. Virology 157:145-155. [DOI] [PubMed] [Google Scholar]
- 83.Swetina, J., and P. Schuster. 1982. Self-replication with errors. A model for polynucleotide replication. Biophys. Chem. 16:329-345. [DOI] [PubMed] [Google Scholar]
- 84.Teng, M. N., M. B. Oldstone, and J. C. de la Torre. 1996. Suppression of lymphocytic choriomeningitis virus-induced growth hormone deficiency syndrome by disease-negative virus variants. Virology 223:113-119. [DOI] [PubMed] [Google Scholar]
- 85.Turner, P. E., and L. Chao. 1999. Prisoner's dilemma in an RNA virus. Nature 398:441-443. [DOI] [PubMed] [Google Scholar]
- 86.van Poelwijk, F., M. Prins, and R. Goldbach. 1997. Completion of the impatiens necrotic spot virus genome sequence and genetic comparison of the L proteins within the family Bunyaviridae. J. Gen. Virol. 78:543-546. [DOI] [PubMed] [Google Scholar]
- 87.Vieth, S., A. E. Torda, M. Asper, H. Schmitz, and S. Gunther. 2004. Sequence analysis of L RNA of Lassa virus. Virology 318:153-168. [DOI] [PubMed] [Google Scholar]
- 88.Vignuzzi, M., J. K. Stone, and R. Andino. 2005. Ribavirin and lethal mutagenesis of poliovirus: molecular mechanisms, resistance and biological implications. Virus Res. 107:173-181. [DOI] [PubMed] [Google Scholar]
- 89.Viguera, E., D. Canceill, and S. D. Ehrlich. 2001. In vitro replication slippage by DNA polymerases from thermophilic organisms. J. Mol. Biol. 312:323-333. [DOI] [PubMed] [Google Scholar]
- 90.Wilke, C. O., D. D. Reissig, and I. S. Novella. 2004. Replication at periodically changing multiplicity of infection promotes stable coexistence of competing viral populations. Evol. Int. J. Org. Evol. 58:900-905. [DOI] [PubMed] [Google Scholar]
- 91.Youngner, J. S., and P. Whitaker-Dowling. 1999. Interference, p. 850-854. In A. Granoff and R. G. Webster (ed.), Encyclopedia of virology, vol. 2. Academic Press, San Diego, Calif. [Google Scholar]
- 92.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]