Abstract
Infection by some rotavirus strains requires the presence of sialic acid on the cell surface, its infectivity being reduced in cells treated with neuraminidase. A neuraminidase treatment-resistant mutant was isolated from the porcine rotavirus strain OSU. In reassortant strains, the neuraminidase-resistant phenotype segregated with the gene coding for VP4. The mutant retained its capacity to bind to sialic acid. The VP4 sequence of the mutant differed from that of the parental OSU strain in an Asp-to-Asn substitution at position 100. Neutralization escape mutants selected from an OSU neuraminidase-sensitive clone by monoclonal antibodies that failed to recognize the neuraminidase-resistant mutant strain carried the same mutation at position 100 and were also neuraminidase resistant. Neuraminidase sensitivity was restored when the mutation at position 100 was compensated for by a second mutation (Gln to Arg) at position 125. Molecular mechanics simulations suggest that the neuraminidase-resistant phenotype associated with mutation of OSU residue 100 from Asp to Asn reflects the conformational changes of the sialic acid cleft that accompany sialic acid binding.
Rotaviruses are the leading cause of severe infantile gastroenteritis in the world, infecting practically every child by the fifth year of age (22). These viruses can cause severe dehydrating diarrhea, leading to significant morbidity and mortality. Furthermore, rotaviruses are also a major pathogen in animals of veterinary importance (13).
The rotavirus virion is composed of three concentric layers of proteins and 11 segments of double-stranded RNA (13). The outer layer is composed of two proteins, VP4 and VP7. VP4 forms multimeric spikes that project from the surface of the virus (7, 32, 34). VP7 is a structural glycoprotein, the major constituent of the outer protein layer. VP4 is the viral attachment protein (6, 28). In the presence of trypsin, VP4 is cleaved into two polypeptides, VP8* and VP5*; this cleavage is not necessary for cell binding (23) but is associated with an increase in infectivity and entry (5, 11, 12). VP8*, the N-terminal fragment, is responsible for the hemagglutination activity of the rotaviruses (14, 16), while the C-terminal portion, VP5*, has been associated with membrane permeabilization (8, 19, 33) and recognition of cellular receptors (36). Both VP4 and VP7 induce neutralizing and protective antibodies (22).
Rotaviruses bind to the surface of cells in culture by at least two distinct mechanisms: some rotaviruses of animal origin require the presence of sialic acid residues for efficient binding and infectivity, whereas infection by most animal and almost all human rotavirus strains is independent of sialic acid (2, 11, 28). The infectivity of the former type of strains may be reduced up to 95% by treatment of cells with neuraminidase, while the infectivity of the latter strains is unaffected. Dependence on sialic acid has not been linked to cell or tissue tropism, suggesting that other receptors are involved: binding to sialic acid probably increases the efficiency of the entry process by facilitating virus interaction with another cellular ligand(s) (27).
Sialic acid-independent and neuraminidase-resistant mutants have been derived from the sialic acid-dependent and neuraminidase-sensitive simian strains RRV and SA11, respectively, indicating that binding to sialic acid is not an essential step for rotavirus infection (2, 31, 29). While the infectivity of the variants is no longer dependent on the presence of sialic acid on the cell surface, generally they retain the capacity to bind to sialic acid and to be recognized by monoclonal antibodies directed to the parental strain (2, 29, 30). The mutations responsible for the altered phenotype have been located in VP8* (2, 28, 30). The rotavirus VP8* is a compactly folded globular domain featuring two β-sheets. The sialic acid binding domain lies on a shallow groove located above the cleft between the two β-sheets (10).
The isolation of neuraminidase-resistant mutants has helped in the understanding of the early events of rotavirus infection and in the identification of VP4 residues that play a role in these events. In this study, we report the isolation of neuraminidase-resistant mutants derived from the porcine rotavirus strain OSU. We have identified a new VP8* amino acid position responsible for the neuraminidase-resistant phenotype. The importance of this position was confirmed by isolating escape mutant strains that carry the same mutation and are also neuraminidase resistant. Molecular mechanics modeling suggests that small conformational changes in VP8* may be responsible for the altered phenotype.
MATERIALS AND METHODS
Cells and viruses.
MA-104 cells were grown in minimal essential medium (MEM) supplemented with 10% fetal calf serum. Virus stocks of the OSU strain of porcine rotavirus, the RF strain of bovine rotavirus, and the H-2 strain of equine rotavirus were all propagated in MA-104 cells in the presence of trypsin (1 μg/ml) as previously described (20). Before infection, viruses were treated with trypsin (10 μg/ml) for 30 min at 37°C.
Neuraminidase-resistant mutants.
To select neuraminidase-resistant mutants, confluent monolayers of MA-104 cells grown in 96-well plates were treated with phosphate-buffered saline (PBS) solution containing 50 mU/ml of neuraminidase from Vibrio cholerae for 90 min at 37°C. Previous experiments had shown that neuraminidase treatment of MA-104 cells under these conditions reduced the infectivity of the RF and OSU strains of rotavirus by more than 90%. After neuraminidase treatment, cells were washed once with PBS and incubated with OSU virus at a multiplicity of infection of approximately 10. After 90 min absorption at 37°C, the inoculum was removed; cells were washed once with PBS-1 mM EGTA and replenished with 100 μl/well of maintenance MEM. Infected cells were incubated at 37°C under a CO2 atmosphere until full cytopathic effect was evident. Progeny viruses were recovered, treated with 10 μg/ml trypsin for 30 min at 37°C, and used to inoculate neuraminidase-treated MA-104 cells again. After three rounds of selection, progeny viruses were plaque purified twice and tested for neuraminidase resistance in infectivity assays. The OSU origin of the mutants was confirmed by electropherotype analysis. Mutants were designed OSUm followed by the number of the clone.
Infectivity assays.
Confluent monolayers of MA-104 cells grown on 96-well plates were treated with 50 μl of a solution containing 50 mU/ml of neuraminidase from Vibrio cholerae for 90 min at 37°C or, as a control, PBS. After neuraminidase treatment, cells were washed with PBS and inoculated with 10-fold serial dilutions of virus. The RF bovine and H-2 equine rotaviruses, known to be neuraminidase sensitive and resistant, respectively (4), were included in the assays as controls. The next day, the cells were fixed with methanol and immunostained for focus-forming units (FFU) according to a standard protocol (1). Infectivity in neuraminidase-treated cells was expressed as a percentage of the infectivity obtained in control cells treated with PBS.
Reassortants.
Reassortants of the parental strain RF × OSU-derived neuraminidase-resistant mutants were generated by coinfection of MA-104 cells. To select against the parental strains, the progeny reassortants were incubated for 4 h at 37°C with neutralizing monoclonal antibodies (MAbs) directed to RF VP4 and to OSU VP7. The neutralization mix was inoculated onto MA-104 cells grown on 96-well plates, and infection was allowed to proceed until cytopathic effect was observed. The selection with the MAbs was repeated three times. Finally, the progeny reassortants were plaque purified after an additional round of incubation with the neutralizing MAbs. The parental origin of each gene was determined by electrophoresis in polyacrylamide gels and by neutralization assays using selected MAbs (25). The neuraminidase-sensitive or -resistant phenotype of the reassortant strains was tested in infectivity assays as described above.
Nucleotide sequencing.
The entire gene 4 of the neuraminidase-resistant mutants and the VP8* coding region of the neutralization escape mutants (see below) were sequenced. RNA was extracted using Trizol-LS (Invitrogen, Carlsbad, California) following the manufacturer's instructions. The VP8* coding region was amplified by reverse transcription-PCR from in vitro-produced transcripts (20) using the negative-sense primer 794 (5′ TATATTGCATTTCTTTCC 3′, nucleotide positions 794 to 811 from porcine OSU strain) and positive-sense primer D10 (5′ GGCTATAAAATGGCTTCGG 3′, complementary to the conserved first 20 nucleotides of gene 4). The gene 4 portion encoding VP5* was amplified by reverse transcription-PCR directly from genomic RNA using the negative-sense primers 2366 (5′ TCACAACCTCTAGACACTACTTACA 3′, nucleotide positions 2366 to 2390), 2044 (5′ CCAGCT/CTCAAATACTTCATC 3′, nucleotide positions 2044 to 2063), and 1611 (5′ TCATTACA/GTTTGTTGCCATAGATTT 3′, nucleotide positions 1611 to 1635) and the positive-sense primers 1575 (5′ TTGCCATTAGATATGTTTTCAATGGTT 3′, nucleotide positions 1575 to 1601) and 1081 (5′ TGGGATGATTCACAGGCATT 3′, nucleotide positions 1081 to 1100). Primers 2366 and 1611 were used in combination with primer 1081. Primer 2044 was used in combination with primer 1575.
For automated sequencing, purified DNA fragments were analyzed directly by the cycle-sequencing dye terminator method (Big Dye terminator cycle sequencing ready reaction kit, Perkin-Elmer Applied Biosystems, Foster City, California) using an ABI-377 automatic DNA sequencer (Perkin-Elmer Applied Biosystems). Amplicons were sequenced in both directions. Nucleic acid sequences were analyzed and edited using the software DNAman, version 3.2 (Lynnon Biosoft, Quebec, Canada). In addition, sequences were compared with strains deposited in GenBank using the BLAST program (http:\\www.ncbi.nlm.nih.gov).
Characterization of the mutants.
The capacity of the mutant viruses to agglutinate human type O red blood cells was tested as described by Lizano et al. (26). In addition, the capacity of the mutants to bind to sialic acid was measured in infectivity assays in the presence of glycophorin A. Single dilutions of viruses containing 100 to 200 FFU were incubated for 1 h at 4°C with increasing concentrations (from 6.25 μg/ml up to 200 μg/ml) of glycophorin A (Sigma Chemical Co., St. Louis, MI) prepared in MEM. MA-104 cells grown in 96-well plates were inoculated with 100 μl/well of the virus-glycophorin mixture and incubated for 1 h at 4°C. After incubation, cells were washed once with PBS-1 mM EGTA and replenished with 100 μl/well of maintenance MEM. The next day, cells were fixed and stained by FFU as described above. Infectivity was expressed as a percentage of the infectivity obtained with viruses not treated with glycophorin A.
The reactivity of the neuraminidase-resistant mutants against a panel of selected MAbs directed to OSU VP8* was tested by immunohistochemistry and by a modified neutralization assay. For the immunohistochemistry assays, MA-104 cells grown on 96-well plates were infected with 100 to 200 FFU/well, and the next day cells were fixed and stained for fluorescent focus-forming units using selected anti VP8* MAbs as the primary antibody or an MAb directed to VP6 as a control. Reactivity was defined as the presence of fluorescent foci in the wells. For the neutralization assay, 50 μl of ascites fluid diluted 1:100 in MEM was mixed with 50 μl of undiluted virus stock. After incubation for 3 h at 37°C, the mixtures were serially diluted and inoculated onto MA-104 cells grown in 96-well plates. The next day the cells were fixed and stained for FFU. Neutralization was defined as a reduction of the initial infectivity by at least 66%.
Neutralization escape mutants.
Neutralization-resistant mutants were selected by neutralizing MAbs directed to OSU VP8* (25). Mutants were selected by incubating the parental virus for 4 h at 37°C in the presence of a 1:100 final dilution of the ascites fluid of the respective MAb before inoculation onto MA-104 cells. The grown virus was reexposed to the antibody under the same conditions nine times prior to the inoculation into six-well plates for plaque production. Selected clones were plaque purified twice before further analysis. Reactivity between the neutralizing MAbs and the mutants was tested by immunofluorescence in infected MA-104 cells using fluorescence focus assays. Each mutant is designed by the name of the parental strain followed by the name of the neutralizing MAb. The neutralization escape mutants were tested for neuraminidase resistance in infectivity assays and for their capacity to bind sialic acid using glycophorin A. Their OSU origin was confirmed by electropherotype analysis.
Molecular mechanics calculations.
All calculations were performed with the Insight II and Discover set of programs (Accelrys, San Diego, California) using the AMBER interatomic force field (Accelrys). The summation cutoff distance employed was 9.5 Å (24). At the start of a run, a minimization was first done in order to relax any initial strain left from the construction of the molecules (300 steps, each of 10 picoseconds). To better explore the molecular conformational space, two types of calculations were used. First, dynamical runs were performed in vacuum at 298 K for 10,000 steps and then a second one was applied for 100,000 steps at 310 K. In order to explore other possible configurations of lower energy, a simulated annealing procedure (24) was applied to the molecules in vacuum. The starting point was the conformation obtained at 300 K. A production run of 100,000 steps was set at 1,200 K and 15 snapshots equally spaced in time were taken. These molecular conformations were cooled down to 310 K in steps of 300 K and subjected to a minimization process until the derivative was <0.001 kcal/(mol Å). Again, the lowest-energy conformation was used.
RESULTS
Isolation of OSU-derived neuraminidase-resistant mutants.
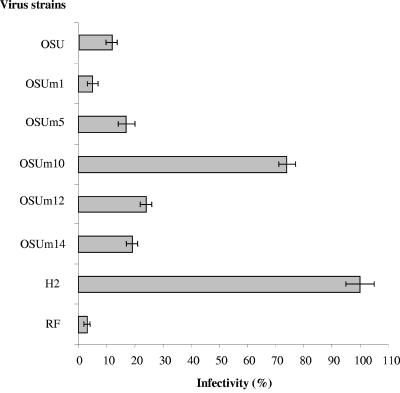
After serial passages of the porcine OSU strain in neuraminidase treated MA-104 cells, five clones were finally recovered, amplified, and tested for sialic acid dependence. Figure 1 shows that the infectivity titers of four of these strains, named OSUm1, OSUm5, OSUm12, and OSUm14, were reduced between 95% and 75% by treatment with neuraminidase. However, the infectivity of strain OSUm10 was reduced only 25% in relation to that obtained in control untreated cells, indicating that strain OSUm10 was no longer neuraminidase sensitive. Electropherotype analysis of the five derived strains and the OSU parental strain showed identical patterns (data not shown). Thus, strain OSUm10 was considered an OSU-derived neuraminidase-resistant mutant. Strain OSUm1, which showed the highest sensitivity to neuraminidase treatment, was used in subsequent experiments as a control.
FIG. 1.
Infectivity of OSU-derived mutants (OSUm) and control strains (RF and H-2) in MA-104 cells treated with Vibrio cholerae neuraminidase. Virus infectivity is expressed as a percentage of the FFU per milliliter in cells treated only with PBS. Results represent the mean of at least three experiments with standard deviations indicated.
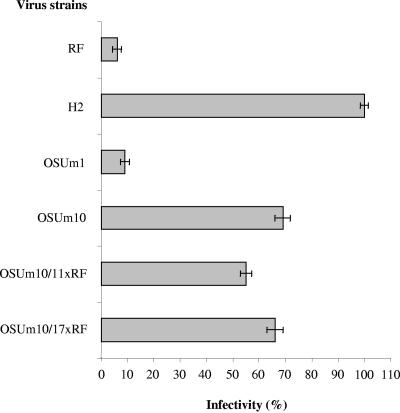
In order to map the gene segment encoding resistance to neuraminidase treatment in OSUm10, a series of reassortants between the neuraminidase-sensitive strain RF and strain OSUm10 were derived. The parental origin of each segment in the reassortants was determined by electrophoresis and neutralization assays (data not shown). Two reassortants, named OSUm10/11xRF and OSUm10/17xRF, which derived genes 2, 3, 4, and 10 from strain OSUm10 and genes 5, 7, 8, and 9 from strain RF were obtained. The origins of genes 1, 6, and 11 could not be determined. As shown in Fig. 2, the infectivity of both reassortant strains was neuraminidase resistant, confirming that differences in neuraminidase sensitivity between strains cosegregate with gene segment 4, not gene 9 (4, 29). Genes segments 2 and 3 encode inner core structural proteins (VP2 and VP3), and gene 10 encodes a nonstructural protein (NSP4), and they are unlikely to be responsible for the mutant phenotype.
FIG. 2.
Infectivity of parental strains (OSUm10 and RF), derived reassortants (OSUm10/11xRF and OSUm10/17xRF), and control strains (OSUm1 and H-2) in MA-104 cells treated with neuraminidase. Virus infectivity is expressed as a percentage of the FFU per milliliter in cells treated only with PBS. Results represent the mean of three experiments with standard deviations indicated.
Effect of glycophorin A on the infectivity of strains OSUm1 and OSUm10.
In order to test if strain OSUm10 was still able to bind sialic acid, infectivity assays were performed in the presence of increasing concentrations of glycophorin A. Figure 3 shows that the infectivity of strain OSUm10 is as susceptible to glycophorin A as the infectivity of the neuraminidase-sensitive strains OSUm1 and RF. Incubation of strains OSUm10, OSUm1, and RF with glycophorin A at a concentration of 50 μg/ml reduced their infectivity by more than 60%, while the infectivity of the neuraminidase-resistant strain H-2 was unaffected, even at the highest concentration used (200 μg/ml). In addition, strain OSUm10 was still capable of hemagglutinating human type O red blood cells (data not shown).
FIG. 3.
Effect of treatment with increasing concentrations of glycophorin A on the infectivity of neuraminidase-sensitive and -resistant OSU-derived mutants (OSUm1 and OSUm10) and control strains (RF and H-2) in MA-104 cells. Virus infectivity is expressed as a percentage of the FFU per milliliter of viruses treated only with MEM. The result of one experiment is shown. Experiments were repeated two times with identical results.
Reactivity of strains OSUm1 and OSUm10 with a panel of neutralizing monoclonal antibodies directed to OSU VP8* and generation of neutralization escape mutants.
To evaluate possible changes in antigenicity associated with the mutated phenotype, the reactivity of strains OSUm10, OSUm1, OSUm5, OSUm12, and OSUm14 plus the parental OSU strain was evaluated against a panel of 14 neutralizing monoclonal antibodies directed to OSU VP8* (25), using a fluorescence focus assay. The reactivity of strains OSUm1, OSUm5, OSUm12, and OSUm14 against the panel of MAbs used was identical to the reactivity shown by the parental OSU strain, that is to say, they were recognized by all the MAbs tested. In contrast, strain OSUm10 showed total loss of reactivity with three MAbs (4B2, 3E7, and 1C11), and decreased reactivity with three other MAbs (2D5, 2B6, and 2C9) (data not shown).
To gain further information on the nature of the mutation that changed the phenotype of strain OSUm10, neutralization escape mutants of strain OSUm1 were generated using three MAbs that showed loss or partial loss of reactivity with strain OSUm10 (4B2, 3E7, and 2D5) plus two additional MAbs (5G7 and 3G5) also directed to OSU VP8* and still capable of recognizing OSUm10. Electrophoretic analysis showed that all escape mutants had an electropherotype identical to that of the parental strain OSUm1. The results of the reactivity between the MAbs and one clone representative of each group of neutralization escape mutants are shown in Table 1. Mutants were clearly divided into three nonoverlapping groups of reactivity. One group included the mutants that were resistant to MAbs 4B2 and 3E7, the second group included mutants resistant to MAbs 5G7 and 3G5, and the third group included mutants resistant to MAb 2D5.
TABLE 1.
Reactivity of neutralizing monoclonal antibodies directed against OSU VP4 region VP8* with neutralization escape mutants derived from strain OSUm1
| Virus strain | Reactivityawith monoclonal antibody:
|
Neuraminidase sensitiveb | ||||
|---|---|---|---|---|---|---|
| 5G7 | 3G5 | 2D5 | 3E7 | 4B2 | ||
| OSUm1 | + | + | + | + | + | Yes |
| OSUm1/3E7 | + | + | +/− | − | − | No |
| OSUm1/4B2 | + | + | +/− | − | − | No |
| OSUm1/3G5 | − | − | + | + | + | Yes |
| OSUm1/5G7 | − | − | + | + | + | Yes |
| OSUm1/2D5 | − | − | − | − | − | Yes |
| OSUm10 | + | + | +/− | − | − | No |
Reactivity determined by immunofluorescence focus assays, +, reactive; −, nonreactive; +/−, reduced reactivity.
Neuraminidase sensitivity was determined in MA-104 cells treated with neuraminidase from Vibrio cholerae.
The infectivity in neuraminidase-treated cells of neutralization escape mutants representative of each group in shown in Fig. 4. Mutants from the first group, selected with MAbs 4B2 and 3E7, showed a reduction in infectivity of approximately 30% in neuraminidase-treated cells, indicating that these mutants were not neuraminidase sensitive, while the rest of the mutants were still neuraminidase sensitive. The infectivity of both neuraminidase-resistant mutants was reduced by more than 80% by incubating the viruses with 50 μg/ml of glycophorin A but not with asialoglycophorin, indicating that the mutants were still capable of binding to sialic acid (data not shown).
FIG. 4.
Infectivity of parental strain (OSUm1), neutralization escape mutants OSUm1/4B2, and OSUm1/3E7, and control strains (RF and H-2) in MA-104 cells treated with neuraminidase. Virus infectivity is expressed as a percentage of the FFU per milliliter in cells treated only with PBS. Results represent the mean of three experiments with standard deviations indicated.
Nucleotide sequence of gene 4 of strains OSUm1 and OSUm10.
To identify the mutations responsible for the change in phenotype observed for strain OSUm10, the entire coding region of gene 4 of strains OSUm1 and OSUm10 was sequenced. A comparison of the nucleotide sequence of strain OSUm10 with parental strain OSU showed a single nucleotide change (Gly to Ala) at position 300, which resulted in change of an Asp to Asn at position 100 of the deduced amino acid sequence of strain OSUm10. Strain OSUm1 showed a single amino acid change (Gln to His) at position 137 in relation to parental strain OSU (Table 2). Thus, strains OSUm1 and OSU m10 differed at positions 100 and 137.
TABLE 2.
Mutations found in the neuraminidase-resistant mutants and neutralization escape mutantsc
| Virus strain | Residue at position:
|
Neuraminidase sensitive | |||
|---|---|---|---|---|---|
| 100 | 125 | 137 | 191 | ||
| OSUGBa | Asp | Gln | Gln | Ala | Yes |
| OSU 2245b | Asp | Gln | Gln | Thr | Yes |
| OSUm1 | Asp | Gln | His | Thr | Yes |
| OSUm10 | Asn | Gln | Gln | Thr | No |
| OSUm1/3E7 | Asn | Gln | His | Thr | No |
| OSUm1/4B2 | Asn | Gln | His | Thr | No |
| OSUm1/3G5 | Asp | Arg | His | Thr | Yes |
| OSUm1/5G7 | Asp | Arg | His | Thr | Yes |
| OSUm1/2D5 | Asn | Arg | His | Thr | Yes |
OSU VP4 sequence deposited in GenBank (X13190).
VP4 sequence of OSU grown in our laboratory.
Boldface type indicates changes in amino acids from those of OSUGB.
Amino acid sequences of VP8* of the neutralization escape mutants.
To establish the molecular basis of the epitope recognized by the neutralizating MAbs used, the VP8* sequences of one antigenic mutant selected with each MAb were determined. A single amino acid substitution compared to the parental strain OSUm1 was identified in each case, with the exception of the mutant selected with MAb 2D5, which possessed two amino acid changes (Table 2). Mutants obtained with MAbs 3E7 and 4B2, which became neuraminidase resistant, showed an amino acid substitution (Asp to Asn) at position 100, while mutants obtained with MAbs 3G5 and 5G7, which remained neuraminidase sensitive, showed a change (Gln to Arg) at position 125. Interestingly, the mutant obtained with MAbs 2D5, which is neuraminidase sensitive, showed mutations at both positions 100 and 125. All escape mutants showed the change at position 137 (Gln to His) present in parental strain OSUm1.
Molecular mechanics calculations.
To gain information on the mechanisms associated with the change in phenotype of the neuraminidase-resistant mutants, molecular mechanic calculations of a series of VP8* molecules were performed. A crystal structure of the RRV VP8* core in complex with sialic acid shows that a hydrogen bond between OD1 of the Asp 100 side chain and the backbone amide of Thr 192 spans the sialic acid binding cleft (10). Comparison of the nuclear magnetic resonance structures of the RRV VP8* core in the absence of sialic acid with the crystal structure of the complex suggests a slight narrowing of the cleft upon binding to sialic acid (10). A new crystal structure of the RRV VP8* core without sialic acid confirms that the cleft narrows upon sialic acid binding and indicates that the hydrogen bond between the Asp 100 side chain and the Thr 192 main chain is absent in the unbound state (Monnier and Dormitzer, personal communication).
Because an Asp-to-Asn mutation at position 100 of VP8* produces a neuraminidase-resistant phenotype in strain OSU, we hypothesize that Asn at position 100 of OSUm10 might form a cleft-narrowing hydrogen bond to the Thr 192 backbone amide even in the absence of sialic acid, whereas Asp100 only forms this bond upon sialic acid binding. To test this hypothesis, all the amino acid changes (46 total) between strains rhesus and OSU were changed on the atomic structure of the rhesus rotavirus VP8* core domain (Protein Data Bank number 1KQR). The amino acid residues directly involved in sialic acid binding are well conserved between the rhesus and OSU strains. Mutations were replaced in the OSU model and the molecules were subjected to dynamic minimization and simulated annealing to obtain the most stable conformations.
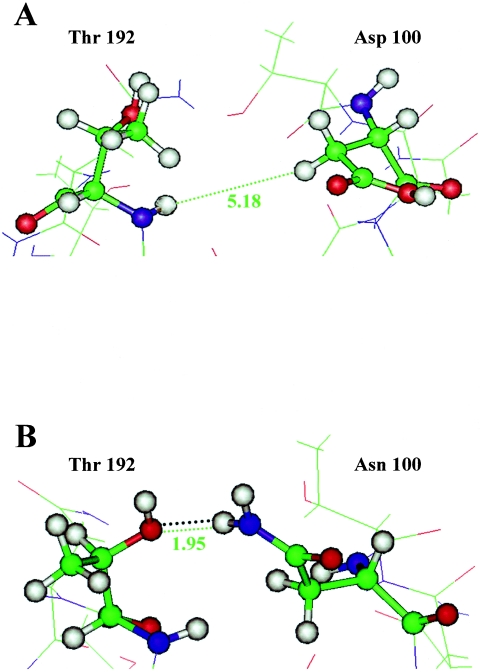
The minimal measured distances between side chain hydrogen atoms of residue 100 and the backbone amide of residue Thr 192 for the different molecules analyzed are shown in Table 3. Calculations for the OSU and OSUm10 strains showed distances of 5.2 and 3.5 Å, respectively (Table 3). The distance obtained for OSUm10 suggests a narrowing of the cleft in the mutant strain and is compatible with the formation of one hydrogen bond between the nitrogen of the side chain amide of the residue 100 and the oxygen of the side chain hydroxyl of the residue 192 (Fig. 5). The distances observed with the remaining strains were in each case compatible with the observed phenotype. The distance obtained with strain OSUm1 (7.1Å) is wider than the distance obtained for the parental strain OSU. Similarly, the distance obtained for the neutralization escape mutant OSUm1/4B2 was 3.3 Å, compatible with the formation of one hydrogen bond. On the other hand, the neuraminidase-sensitive double mutant OSUm1/2D5 showed a distance in the minimal energy conformation of 4.1 Å, incompatible with an hydrogen bond, suggesting that the second mutation at position 125 results in a reopening of the end of the cleft. The measured distance for mutant OSUm1/3G5, with a single Gln-to-Arg mutation at position 125, was 10.9 Å.
TABLE 3.
Minimal calculated distances between side chain hydrogen atoms of residue 100 and the backbone amide of residue T192 of rotavirus VP8*
| Virus strain | Neuraminidase sensitive | Distance (Å)a
|
|
|---|---|---|---|
| Dynamic minimization | Simulated annealing | ||
| OSU | Yes | 4.0 | 5.2 |
| OSUm1 | Yes | 3.7 | 7.1 |
| OSUm10 | No | 4.0 | 3.5 |
| OSUm1/4B2 | No | 3.9 | 3.3 |
| OSUm1/3G5 | Yes | 3.9 | 10.9 |
| OSUm1/2D5 | Yes | 3.9 | 4.1 |
Values given are for vacuum conditions. Values obtained in solution (water) conditions did not differ significantly from values obtained in vacuum (data not shown).
FIG. 5.
Ball-and-stick diagram of the open end of the sialic acid binding cleft of VP8*, showing the minimal measured distances (green dashed line, in Å) between side chain hydrogen atoms at position 100 and residue Thr 192 for the OSU parental strain (A) and the OSUm10 neuraminidase-resistant mutant strain (B). The distance in B is compatible with the formation of a hydrogen bond (black dotted lines). Models were constructed using the atomic structure of rhesus rotavirus VP8* (Protein Data Bank identification code 1KQR) as the frame. Gray, hydrogen; blue, nitrogen; red, oxygen; green, carbon.
In addition to the distance, the orientation of the H-N bond with regard to the acceptor oxygen atom is also a key factor for the formation of hydrogen bonds. These orientations were determined for strains OSUm10 and OSUm1/4B2 and found to be close to the ideal values for hydrogen bond formation (data not shown).
DISCUSSION
Several animal rotavirus strains require the presence of sialic acid on the cell surface to infect cells in vitro and in vivo (2, 17, 28), and their infectivity is reduced when cells are treated with neuraminidase. This requirement segregates with VP4 and correlates with the VP4 genotype, not the species of origin (4, 28). To investigate further the interactions between VP4 and sialic acid residues on the cell surface, we generated neuraminidase-resistant mutants of porcine strain OSU. Our results suggest that, despite retaining the capacity to bind to sialic acid, the mutant viruses do not require this binding for infectivity, possibly indicating that they bind directly to a second receptor that mediates infectivity. Presumably, conformational changes in the VP8* region of the viral attachment protein VP4 allow the mutant viruses to bypass the very early, neuraminidase-dependent steps of infection. The characterization of the OSU mutants confirms and extends the results obtained previously with neuraminidase-resistant mutants obtained from simian strains RRV and SA11 and adds support to the notion that sequential multiple binding steps take place during rotavirus entry (27).
In this work, we identified position 100 in the VP8* region of the VP4 as responsible for the change in phenotype of the porcine OSU strain from neuraminidase sensitive to resistant. An Asp-to-Asn mutation at this position was the only difference detected between the neuraminidase-resistant strain OSUm10 and the parental strain OSU. The Gln-to -His mutation at position 137 in the OSUm1 strain did not alter neuraminidase sensitivity. These findings were corroborated independently by the observation that neutralization escape mutants carrying the same change at position 100 became neuraminidase resistant. However, additional changes in VP8* may be required to fully mimic the changes induced by actual sialic acid binding, since, at variance with the neuraminidase-resistant strain derived from simian rotaviruses (2, 29, 30), none of the OSU-derived neuraminidase-resistant strains selected showed 100% infectivity in cells treated with neuraminidase.
Residue Asp100 is conserved in most neuraminidase-sensitive strains with the exception of strain RF (4), which, despite carrying an Asn at this position, is still neuraminidase sensitive. Furthermore, the same mutation reported in this work, Asp100Asn, in the RRV strain did not result in a neuraminidase-resistant phenotype (29). Taken together, these results suggest that the role played by position 100 in resistance to neuraminidase treatment depends on the virus strain and is probably influenced by other regions of the molecule, as directly evidenced by the compensatory effect on this phenotype of mutation at position 125.
The exact amino acids involved in sialic acid binding by OSU have not been determined. However, the amino acid residues of RRV that bind sialic acid are highly conserved in other neuraminidase-sensitive animal strains (10, 18, 21). Positions Arg101, Tyr188, Tyr189, and Ser190 are conserved in the OSU strain, and position 155 shows a conservative change (Tyr to His). Thus, it is reasonable to assume that these positions are also involved in binding to the sialic acid molecule in OSU. Interestingly, none of the positions reported so far to be associated with changes in phenotype, that is, mutations Gly150Glu and Lys187Arg in RRV; Gln180Arg, Asn183Asp, and Tyr194Cys in SA11; and Asp100Asn in OSU (2, 29, 31; this work), are located in the precise positions reported to be involved directly in sialic acid binding, but rather are adjacent to them. This is in line with the notion that the changes in phenotype are due to small conformational changes in VP8* that do not directly affect the sialic acid binding site. In agreement, most of the neuraminidase-resistant mutants isolated so far retain their capacity to bind sialic acid. The observation that preincubation of the virus with glycophorin A blocks infectivity suggest that the sialic acid of the glycophorin A is bound by the virion with high affinity, resulting in an esteric inhibition of the viral interaction with other molecules of the cell surface.
Molecular mechanics data suggest that the Asp-to-Asn change in the OSUm10 mutant results in small conformational changes that will allow the formation of a hydrogen bond between position Asn100 and position Thr192, even in the absence of sialic acid, presumably locking the mutant in a “bound” conformation. Thus, the mutation may allow the virus to obviate the conformational change induced by the binding to sialic acid required to recognize the second receptor. How such a slight and localized change in an exposed area of VP8* could translate into the capacity of the virus to recognize its second receptor is not known.
Zhou et al. (37) noted that MAbs selecting mutations at VP8* RRV residues E180 and N183 are capable of destabilizing the outer capsid of the virion, indicating that local alterations in VP8* can induce alterations in the whole virus structure. An escape mutant generated by MAb 2D5 carrying mutations at positions 100 and 125 retained the original neuraminidase-sensitive phenotype, showing that the Asp-to-Asn mutation at position 100 can be compensated for by a Gln-to-Arg mutation at position 125. The Gln-to-Arg mutation is a nonconservative change that will reintroduce a hydrophilic residue lost by the Asp-to-Asn change. However, the location of residue 125 in an extended β-ribbon (strand βF) on top of the VP8* core and away from position 100, argues against this explanation. A simple restoration of the original conformation of the protein seems unlikely, since the double mutant did not regain reactivity with MAb 4B2 or 3E7.
Early events in rotavirus infections are complex (27). Presumably, the initial step of neuraminidase-sensitive rotaviruses infection is attachment to sialic acid moieties sensitive to hydrolysis by neuraminidases present on the cell surface. Next, the virus will attach to the secondary receptor(s) that will finally lead to penetration. The broad specificity and low affinity of the interaction between sialic acid and rotavirus VP8* are consistent with this notion (9). While human and most animal rotavirus strains do not show sensitivity to neuraminidase treatment of the cell surface to infect cells in vitro (4), this observation does not rule out that these strains may bind to a different type of sugar, to modified sugars, or to sugars presented in a different context in vivo.
The enterocyte, the main viral cell target for rotavirus, is covered by the glycocalyx and a thick mucus layer which covers the intestinal mucosa (15). Both of these layers are rich in sialic acid and other carbohydrates. Thus, binding to sialic acid or to any other sugar may allow the virus to remain longer on the cell surface, increasing the chances for the virus to encounter its secondary receptor. In addition, binding to sialic acid may prevent the virus from being washed off by the peristaltic movements of the intestine (35). The significance of the dichotomy between neuraminidase-sensitive and -resistant strains for in vivo infection and enteropathogenicity of rotaviruses remains an open question.
Acknowledgments
We thank Philip Dormitzer and Nilah Monnier for extensive sharing of unpublished results, helpful discussions, and critical reading of the manuscript.
This work was partially financed by Proyecto Iniciativa Cientifica del Milenio, FONACIT (Proyecto No. 2001001312).
REFERENCES
- 1.Bass, D. M., M. R. Baylor, C. Chen, E. M. Mackow, M. Bremont, and H. B. Greenberg. 1992. Liposome-mediated transfection of intact viral particles reveals that plasma membrane penetration determines permissivity of tissue culture cells to rotavirus. J. Clin. Investig. 90:2313-2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ciarlet, M., and M. K. Estes. 1999. Hum. and most animal rotavirus strains do not require the presence of sialic acid on the cell surface for efficient infectivity. J. Gen. Virol. 80:943-948. [DOI] [PubMed] [Google Scholar]
- 3.Ciarlet, M., M. Hidalgo, M. Gorziglia, and F. Liprandi. 1994. Characterization of neutralization epitopes on the VP7 surface protein of serotype G11 porcine rotaviruses. J. Gen. Virol 75:1867-1873. [DOI] [PubMed] [Google Scholar]
- 4.Ciarlet, M., J. E. Ludert, M. Iturriza-Gomara, F. Liprandi, J. J. Gray, U. Desselberger, and M. K. Estes. 2002. Initial interaction of rotavirus strains with N-acetylneuraminic (sialic) acid residues on the cell surface correlates with VP4 genotype, not species of origin. J. Virol. 76:4087-4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clark, S. M., J. R. Roth, M. L. Clark, B. B. Barnett, and R. S. Spendlove. 1981. Trypsin enhancement of rotavirus infectivity: mechanism of enhancement. J. Virol. 39:816-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crawford, S. E., M. Labbe, J. Cohen, M. H. Burroughs, Y. J. Zhou, and M. K. Estes. 1994. Characterization of virus-like particles produced by the expression of rotavirus capsid proteins in insect cells. J. Virol. 68:5945-5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crawford, S. E., S. K. Mukherjee, M. K. Estes, J. A. Lawton, A. L. Shaw, R. F. Ramig, and B. V. Prasad. 2001. Trypsin cleavage stabilizes the rotavirus VP4 spike. J. Virol. 75:6052-6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dormitzer, P. R., E. B. Nason, B. V. Prasad, and S. C. Harrison. 2004. Structural rearrangements in the membrane penetration protein of a non-enveloped virus. Nature 430:1053-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dormitzer, P. R., Z. Y. Sun, O. Blixt, J. C. Paulson, G. Wagner, and S. C. Harrison. 2002. Specificity and affinity of sialic acid binding by the rhesus rotavirus VP8* core. J. Virol. 76:10512-10517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dormitzer, P. R., Z. Y. Sun, G. Wagner, and S. C. Harrison. 2002. The rhesus rotavirus VP4 sialic acid binding domain has a galectin fold with a novel carbohydrate binding site. EMBO J. 21:885-897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Espejo, R. T., S. Lopez, and C. Arias. 1981. Structural polypeptides of simian rotavirus SA11 and the effect of trypsin. J. Virol. 37:156-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Estes, M. K., D. Y. Graham, and B. B. Mason. 1981. Proteolytic enhancement of rotavirus infectivity: molecular mechanisms. J. Virol. 39:879-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Estes. M.K. 2001. Rotaviruses and their replication, p. 1747-1785. In D. M. Knipe and P.M. Howley (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 14.Fiore, L., H. B. Greenberg, and E. R. Mackow. 1991. The VP8 fragment of VP4 is the rhesus rotavirus hemagglutinin. Virology 181:553-563. [DOI] [PubMed] [Google Scholar]
- 15.Forstner, J. F., G. G. Forstner. 1994. Gastrointestinal mucus, p. 1255-12843. In L. R. Johnson et al. (ed.), Physiology of the gastrointestinal trac, 3rd ed. Raven Press, New York, NY.
- 16.Fuentes-Panana, E. M., S. Lopez, M. Gorziglia, and C. F. Arias. 1995. Mapping of the hemagglutination domain of rotaviruses. J. Virol. 69:2629-2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fukudome, K., O. Yoshie, and T. Konno. 1989. Comparison of human, simian, and bovine rotaviruses for requirement of sialic acid in hemagglutination and cell adsorption. Virology 172:196-205. [DOI] [PubMed] [Google Scholar]
- 18.Giammarioli, A. M., E. R. Mackow, L. Fiore, H. B. Greenberg, and F. M. Ruggeri. 1996. Production and characterization of murine IgA monoclonal antibodies to the surface antigens of rhesus rotavirus. Virology 225:97-110. [DOI] [PubMed] [Google Scholar]
- 19.Golantsova, N. E., E. E. Gorbunova, and E. R. Mackow. 2004. Discrete domains within the rotavirus VP5* direct peripheral membrane association and membrane permeability. J. Virol. 78:2037-2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gorziglia, M., C. Larrea, F. Liprandi, and J. Esparza. 1985. Biochemical evidence for the oligomeric (possibly trimeric) structure of the major inner capsid polypeptide (45K) of rotaviruses. J. Gen. Virol. 66:1889-1900. [DOI] [PubMed] [Google Scholar]
- 21.Isa, P., S. Lopez, L. Segovia, and C. F. Arias. 1997. Functional and structural analysis of the sialic acid-binding domain of rotaviruses. J. Virol. 71:6749-6756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kapikian, A. Z., Y. Hoshino, and R. M. Chanock. 2001. Rotaviruses, p. 1787-1833. In D. M. Knipe and P.M. Howley (ed.). Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 23.Keljo, D. J., and A. K. Smith. 1988. Characterization of binding of simian rotavirus SA-11 to cultured epithelial cells. J. Pediatr. Gastroenterol. Nutr. 7:249-256. [DOI] [PubMed] [Google Scholar]
- 24.Leach, A. R. 2001. Molecular modelling: principles and applications, 2nd ed. Pearson Education Limited, Essex, England.
- 25.Liprandi, F., I. Rodriguez, C. Pina, G. Larralde, and M. Gorziglia. 1991. VP4 monotype specificities among porcine rotavirus strains of the same VP4 serotype. J. Virol. 65:1658-1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lizano, M., S. Lopez, and C. F. Arias. 1991. The amino-terminal half of rotavirus SA114fM VP4 protein contains a hemagglutination domain and primes for neutralizing antibodies to the virus. J. Virol. 65:1383-1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.López, S., and C. F. Arias. 2003. Attachment and post-attachment receptors for rotavirus, p. 143-163. In U. Desselberger and J. Grey (ed.), Viral gastroenteritis, Elsevier, Amsterdam, The Netherlands.
- 28.Ludert, J. E., N. Feng, J. H. Yu, R. L. Broome, Y. Hoshino, and H. B. Greenberg. 1996. Genetic mapping indicates that VP4 is the rotavirus cell attachment protein in vitro and in vivo. J. Virol. 70:487-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ludert, J. E., B. B. Mason, J. Angel, B. Tang, Y. Hoshino, N. Feng, P. T. Vo, E. M. Mackow, F. M. Ruggeri, and H. B. Greenberg. 1998. Identification of mutations in the rotavirus protein VP4 that alter sialic-acid-dependent infection. J. Gen. Virol. 79:725-729. [DOI] [PubMed] [Google Scholar]
- 30.Mendez, E., C. F. Arias, and S. Lopez. 1993. Binding to sialic acids is not an essential step for the entry of animal rotaviruses to epithelial cells in culture. J. Virol. 67:5253-5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mendez, E., C. F. Arias, and S. Lopez. 1996. Interactions between the two surface proteins of rotavirus may alter the receptor-binding specificity of the virus. J. Virol. 70:1218-1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prasad, B. V., J. W. Burns, E. Marietta, M. K. Estes, and W. Chiu. 1990. Localization of VP4 neutralization sites in rotavirus by three-dimensional cryo-electron microscopy. Nature 343:476-479. [DOI] [PubMed] [Google Scholar]
- 33.Ruiz, M. C., M. J. Abad, A. Charpilienne, J. Cohen, and F. Michelangeli. 1997. Cell lines susceptible to infection are permeabilized by cleaved and solubilized outer layer proteins of rotavirus. J. Gen. Virol. 78:2883-2893. [DOI] [PubMed] [Google Scholar]
- 34.Yeager, M., J. A. Berriman, T. S. Baker, and A. R. Bellamy. 1994. Three-dimensional structure of the rotavirus haemagglutinin VP4 by cryo-electron microscopy and difference map analysis. EMBO J. 13:1011-1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yuan, Q., and W. A. Walker. 2004. Innate immunity of the gut: mucosal defense in health and disease. J. Pediatr. Gastroenterol. Nutr. 38:463-473. [DOI] [PubMed] [Google Scholar]
- 36.Zarate, S., M. A. Cuadras, R. Espinosa, P. Romero, K. O. Juarez, M. Camacho-Nuez, C. F. Arias, and S. Lopez. 2003. Interaction of rotaviruses with Hsc70 during cell entry is mediated by VP5. J. Virol. 77:7254-7260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou, Y. J., J. W. Burns, Y. Morita, T. Tanaka, and M. K. Estes. 1994. Localization of rotavirus VP4 neutralization epitopes involved in antibody-induced conformational changes of virus structure. J. Virol. 68:3955-3964. [DOI] [PMC free article] [PubMed] [Google Scholar]