Abstract
Transcription regulators STAT1 and STAT2 are key components of the interferon signaling system leading to innate antiviral immunity. The related STAT3 protein is a regulator of interleukin-6-type cytokine signals and can contribute to both cell growth and death important for cancer gene regulation and tumor survival. These three STAT proteins are targeted for proteasome-mediated degradation by RNA viruses in the Rubulavirus genus of the Paramyxoviridae. A single viral protein, the V protein, assembles STAT-specific ubiquitin ligase complexes from cellular components. Simian virus 5 (SV5) targets STAT1, human parainfluenza virus 2 targets STAT2, and mumps virus targets both STAT1 and STAT3. Analysis of the V-dependent degradation complex (VDC) composition and assembly revealed several features contributing to targeting specificity. SV5 and mumps V proteins require STAT2 to recruit the STAT1 target, yet mumps V protein binds STAT3 independent of STAT1 and STAT2. All Rubulavirus V proteins tested require cellular DDB1 to target STATs for degradation but differ in the use of Roc1, which is essential for mumps V STAT3 targeting. Protein interaction analysis reveals that paramyxovirus V proteins can homo- and heterooligomerize and that the conserved cysteine-rich zinc-binding C-terminal domain is necessary and sufficient for oligomerization. Purified SV5 V protein spontaneously assembles into spherical macromolecular particles, and similar particles constitute SV5 and mumps VDC preparations.
Signal transducer and activator of transcription (STAT) proteins are well known as regulators of cytokine and growth factor-activated gene expression. STAT-dependent pathways have been implicated in numerous biological responses including innate and adaptive immunity and regulation of cell growth and apoptosis. In addition to their normal functions, STAT pathways are also known to be inappropriately activated in human diseases such as inflammation, autoimmunity, and cancer (11, 15).
While some of the seven mammalian STAT proteins are expressed and activated in a tissue-specific fashion, three of the family members, STAT1, STAT2, and STAT3, are ubiquitously expressed. STAT1 and STAT2 mediate responses to interferon (IFN). STAT1 is activated by IFN-γ to form a homodimeric transcription factor complex called the gamma IFN-activated factor, which binds directly to the promoters of IFN-γ target genes. IFN-α/β, the principal innate antiviral cytokines induced in response to virus infection, induce tyrosine phosphorylation of latent STAT1 and STAT2. The activated STAT proteins combine with IFN regulatory factor 9 to form a transcription factor complex known as IFN-stimulated gene factor 3. This factor binds to the promoters of antiviral target genes and activates their expression to produce a broadly effective cellular state resistant to virus infection (1, 54).
The role of STAT3 in the innate antiviral response is not well understood, but it can be activated in response to IFNs and has been identified as a target for virus host evasion (42, 60). STAT3 is more widely known for being activated in cells by cytokines such as interleukin-6 (IL-6) (22), growth factors like epidermal growth factor and platelet-derived growth factor (53, 61), or nonreceptor tyrosine kinases like Src (10, 67). STAT3 also has the properties of an oncogene and is found constitutively activated in many types of human cancers, tumor-derived tissues, and cancer cell lines (15). In a fashion similar to IFN-γ-activated STAT1, STAT3 activation leads to dimerization to produce a DNA-binding factor that can induce target gene transcription.
In all cases, the STAT-containing transcription factors are inactivated by the action of nuclear protein tyrosine phosphatase enzymes, and the dephosphorylated STAT proteins are recycled to the cytoplasm where they can participate in further activation cycles (56). Consistent with their regulation by posttranslational modifications rather than degradation, these STATs have long half-lives (21, 31, 44). However, in conspicuous contrast to their normal stability, STAT1, STAT2, and STAT3 can be rapidly degraded during infection of cells with certain species of paramyxoviruses in the Rubulavirus genus or by expression in cells of a single Rubulavirus protein called V.
The Rubulavirus V proteins catalyze the degradation of STAT proteins with remarkable targeting specificity, a countermeasure to evade host IFN and immune responses. The simian virus 5 (SV5) V protein destroys STAT1, the type 2 human parainfluenza virus (HPIV2) V protein targets STAT2, and the mumps virus V protein can eliminate both STAT1 and STAT3 (16, 17, 29, 44, 60, 66). The Rubulavirus V proteins achieve STAT degradation by coordinating the assembly of an ubiquitin ligase (E3) complex with cellular components including the UV-damaged DNA binding protein 1 (DDB1), cullin 4A (Cul4A), and both target and nontarget STATs (3, 58, 60). These complexes, with the components V, DDB1, and Cul4A, are collectively termed V-dependent degradation complexes (VDC). Cellular partners DDB1, STAT2, and Cul4A are essential for SV5-mediated STAT1 degradation (3, 45, 59), but while these proteins copurify with all the Rubulavirus V proteins, the generality of their importance to other VDC activities has not been reported. The V proteins also copurify with other cellular proteins that are unrelated to STAT targeting or that have yet to be characterized (59, 60).
DDB1 and Cul4A are components of cellular E3 ubiquitin ligase complexes in the subset known as SCF (Skp/cullin/F-box) (5, 20, 23, 35, 36, 63, 65). Crystallographic analysis and functional studies of SCF components suggest that the cullin component serves as a scaffold, with its N terminus bound to DDB1 and its C terminus bound to regulator of cullins 1 (Roc1, also known as Rbx1) (reviewed in reference 47). Roc1 is a RING domain-containing protein that is thought to recruit the E2 ubiquitin-conjugating enzyme to facilitate ubiquitin transfer onto substrates (13, 24, 37, 58, 63, 64). The role of Roc1, if any, in STAT targeting by VDC has not been reported.
The STAT proteins targeted by rubulaviruses are highly homologous to one another, and the Rubulavirus V proteins themselves are also very similar, inviting inquiry into the determinants of targeting specificity and selectivity of each V protein. Some insight into the complexity of this problem was provided by the discovery that, in some cases, accessory STAT proteins are required for the V protein to destroy its target STAT protein. For example, STAT1 is needed by HPIV2 to target STAT2, while STAT2 is absolutely required for both SV5 and mumps V proteins to target STAT1. However, neither STAT1 nor STAT2 is required for mumps V to target STAT3 (45, 60). These differences imply that distinct structural features and assembly mechanisms might be used by the V proteins to target a particular STAT.
An unappreciated intricacy in VDC action involves the problem of how the approximately 220-amino-acid V protein can interact with and coordinate the assembly of several large cellular proteins to affect ubiquitylation of STAT targets. The most highly conserved region of the V protein is its cysteine-rich C-terminal domain (CTD). Although the V protein CTD is virus designed and is not homologous in sequence to known cellular proteins, there are some seemingly superficial similarities to RING domains that might indicate functional convergence. Both RING domains and V CTDs bind to two atoms of zinc per polypeptide (7, 8, 46) and act in multimolecular complexes to facilitate Ub transfer to degradation substrates (47). Physical and microscopic analysis of RING domains has revealed their propensity for self-assembly to produce macromolecular particles that are visible in the electron microscope as 50- to 100-nm spheres. These particles are thought to serve as molecular scaffolds to support enzymatic reactions (6, 26, 27), similar to the reactions carried out by V proteins in STAT targeting (59, 60).
In this report, Rubulavirus VDC STAT targeting activities were compared to elucidate the accessory roles of cellular proteins STAT2, DDB1, and Roc1. Results clarify the role of STAT2 in STAT1 targeting by the SV5 and mumps VDCSTAT1, and RNAinterference experiments demonstrate that DDB1 is essential to the targeting of all STATs by all V proteins tested. Roc1 is found to copurify with both SV5 and mumps V proteins, bound both directly and indirectly. However, Roc1 is required only for STAT3 targeting by mumps V. Furthermore, results demonstrate that paramyxovirus V proteins can oligomerize via the CTD and are able to self-assemble into macromolecular spherical particles, providing a plausible model to support the coordination of multiple V protein-mediated activities.
MATERIALS AND METHODS
Cell culture.
293T, 2fTGH, and 2fTGH-derivative cell line U6A (STAT2 deficient) were grown in Dulbecco's modified Eagle's medium (Gibco-BRL) supplemented with 10% cosmic calf serum (HyClone) and 1% penicillin-streptomycin (Gibco-BRL). To produce V-expressing stable cell lines, 2fTGH cells were infected with replication-deficient lentiviruses engineered to express individual V proteins and selected for resistance to blasticidin. Positive clones were tested by immunoblotting for FLAG-V expression and STAT protein targeting. IFN-dependent luciferase reporter gene assays were performed to confirm loss of STAT protein function. V proteins expressing 2fTGH stable cell lines were maintained in medium supplemented with 5 μg/ml blasticidin (Calbiochem).
Plasmids and transfections.
Expression plasmids pEF-FLAG, pEF-FLAG-SV5-V, pEF-FLAG-SVN, pEF-FLAG-SVC, and pEF-FLAG-mumps-V have been described previously (44, 59, 60). Similar constructs were created with the open reading frame for each V protein downstream of and in frame with the hemagglutinin (HA) tag in the pEF vector (pEF-HA-SV5-V and pEF-HA-mumps-V). Plasmid pEF-FLAG-thioredoxin (FLAG-Trx) was a gift of Patricia Cortes (Mount Sinai School of Medicine). FLAG-Roc1, myc-Cul4A, and T7-DDB1 expression plasmids were a gift of Zhen-Qiang Pan (Mount Sinai School of Medicine). The pCDNA-STAT2 expression plasmid has been described previously (45). The glutathione transferase (GST)-SV fusion protein expression plasmid (pGEX-5X-SV5-V) has been described previously (45, 59).
293T cells were transfected by a standard calcium phosphate method. For affinity purification experiments, 2 μg of pEF-FLAG-SV5 V or pEF-FLAG-mumps-V per 100-mm plate was transfected, and 20 plates were harvested. For combinatorial expression experiments, 60-mm plates were transfected using 0.5 μg of pEF-FLAG-Trx, 5 μg of FLAG-Roc1, 5 μg of myc-Cul4A, 5 μg of T7-DDB1, 0.5 μg of pEF-HA-SV5-V, and/or 0.5 μg of pEF-HA-mumps-V per plate in the combinations indicated (see Fig. 3). In experiments for V self-association, 9 μg (each) of pEF-FLAG-SV5-V, pEF-FLAG-mumps-V, pEF-HA, pEF-HA-SV5-V, and pEF-HA-mumps-V was transfected per 60-mm plate, while 5 μg of pEF-HA-SV5-V and 14 μg (each) of pEF-FLAG-SV5-V, pEF-FLAG-SVN, pEF-FLAG-SVC were transfected per plate for domain analysis. For all 293T transfections, cells were harvested at 24 h posttransfection for immunoprecipitation and analysis.
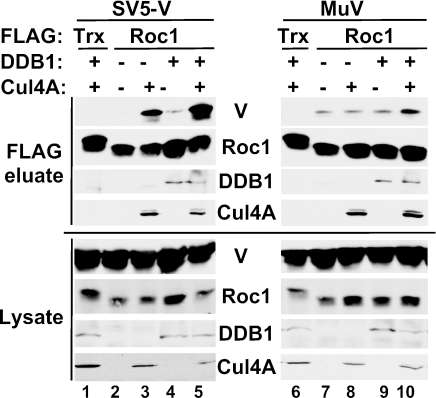
FIG. 3.
Interaction of Roc1 with SV5 V, but not mumps V, depends on Cul4A. Roc1 coprecipitates SV5 V only when Cul4A is also expressed. HA-SV5 V (left panel) or HA-Mu V (right panel) were expressed with FLAG-thioredoxin control (Trx) or FLAG-Roc1 in combination with T7-DDB1 and myc-Cul4A as indicated. FLAG-associated proteins were immunoprecipitated, washed extensively, and eluted with FLAG peptide.
2fTGH and derivative cell lines were transfected by Superfect (QIAGEN) according to the manufacturer's protocol. Plates (100 mm) of 2fTGH or U6A cells were transfected with 2.5 μg of pEF-FLAG, 2.5 μg of pEF-FLAG-SV5-V, or 2.5 μg pEF-FLAG-mumps-V with or without 2.5 μg of pCDNA-STAT2 per plate.
Affinity purification, immunoprecipitations, and immunoblotting.
Affinity purification of SV5 and mumps VDC was exactly as described (59, 60). For sequential immunoprecipitations (IP), eluate of affinity purification of mumps VDC was precleared with protein A agarose beads (Roche) for 1 h at 4°C; the supernatant was divided into four samples for IP with 2 μg of rabbit anti-STAT1 (Santa Cruz sc-345), 2 μg of rabbit anti-STAT2 (Santa Cruz sc-476), 2 μg of rabbit anti-STAT3 (Santa Cruz sc-482), or 2 μg of goat anti-actin (Santa Cruz sc-1616) overnight at 4°C. Immune complexes were captured on protein A agarose for 1 h at 4°C and washed three times in whole-cell extract buffer, and bound proteins were eluted by boiling in protein gel loading buffer and separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) for immunoblotting.
For IP of FLAG-tagged proteins in 2fTGH and U6A cells and from 293T transfections in 60-mm plates, 40 μl of anti-FLAG-M2 affinity agarose (Sigma A2220) was used for 4 h (2fTGH and U6A cells) or overnight (293T cells). Agarose beads with immune complexes were washed five times with whole-cell extract buffer, and bound proteins were eluted in protein gel loading buffer (2fTGH and U6A), or the complexes were eluted with 150 μg/ml of 3× FLAG tripeptide (Sigma) and then boiled in protein gel loading buffer and separated by SDS-PAGE for immunoblot analysis.
Samples separated by SDS-PAGE were transferred to nitrocellulose for immunoblotting. Antibodies and dilutions for blotting are as follows: rabbit anti-STAT1 (1:4,000; Santa Cruz sc-345), rabbit anti-STAT2 (1:5,000; Santa Cruz sc-476), rabbit anti-STAT3 (1:3,000; Santa Cruz sc-482), rabbit anti-FLAG tag (1:5,000; Sigma F7425), mouse anti-DDB1 (1:500; BD Transduction 612488), rabbit anti-Cul4A (1:200; Santa Cruz sc-10782), rabbit anti-Roc1 (1:100; Zymed 34-2500), mouse anti-T7 tag (1:10,000; Novagen 69522-3), mouse anti-myc tag (1:1,000, clone 9E10; Mount Sinai Hybridoma Facility), and mouse anti-HA tag conjugated to peroxidase (1:500, clone 3F10; Roche 1815016). Primary antibody incubations were overnight at 4°C. Secondary antibodies used were goat anti-mouse conjugated to horseradish peroxidase and goat anti-rabbit-horseradish peroxidase (VWR). Detection was with enhanced chemiluminescence reagent (Perkin-Elmer).
RNA interference.
2fTGH V protein-expressing stable cell lines were transfected in 24-well dishes with 60 pmol of small interfering RNA (siRNA) using 0.75 μl of siLentFect reagent (Bio-Rad 170-3361) per well according to the manufacturer's protocol. Cells were harvested and lysates were processed at 72 h posttransfection, and 20 μg of total protein was separated by SDS-PAGE and processed for immunoblotting. All siRNAs were from Dharmacon. DDB1 siRNA was custom designed and synthesized as previously described (59). Roc1 siRNA was purchased as a SMARTpool. Control siRNA was siCONTROL nontargeting control number 2.
Luciferase assay.
2fTGH/mumps V protein-expressing cells were transfected in a 24-well dish in triplicate for each condition. Per well, cells were transfected with 0.07 μg of 4× M67-luciferase reporter gene plasmid, 0.005 μg of Renilla luciferase plasmid (as internal transcriptional control), and either with or without 60 pmol of the indicated siRNA using 0.75 μl of siLentFect. Cells were treated at 48 h posttransfection with IL-6 and the soluble IL-6 receptor (400 ng/ml and 500 ng/ml, respectively; Calbiochem) overnight and then harvested in luciferase assay lysis buffer (Promega dual-luciferase kit). Dual-luciferase assays were carried out: luminescence of both firefly and Renilla luciferase was measured, and the ratio of the two values was calculated (normalized luciferase activity). Data represent average values with standard deviations of triplicate samples for each condition.
Bacterial expression and purification of SV5-V, gel filtration, and electron microscopy analysis.
GST-SV5-V was induced in 6 liters of Escherichia coli culture by the addition of isopropyl-β-d-thiogalactopyranoside (IPTG) and purified with glutathione Sepharose (Pharmacia) by standard procedures (4). SV5-V was cleaved from GST tag by Factor Xa digest, and cleaved material was further purified by application to a Sepharose Q anion exchange column (Amersham Pharmacia) and eluted with a linear gradient of NaCl in 20 mM Na2PO4 (pH 7.5) at 4°C. Purified SV5-V protein was fractionated by gel filtration through a Superose 6 HR column on the ΔKTAexplorer chromatograph (Amersham Pharmacia). Purified SV5 V was injected directly onto the column and separated at 6°C at a rate of 0.2 to 0.5 ml/min in under 1 h while UV absorbance was recorded at 214, 254, and 280 nm. Formvar-coated nickel grids (Electron Microscopy Sciences, Fort Washington, PA) were glow discharged, floated on sample drops (fraction 17 or 19 of purified SV5-V or affinity-purified green fluorescent protein, SV5-VDC, or mumps-VDC), blotted, and air dried. They were then negatively stained with 1% (wt/vol) aqueous uranyl acetate. Grids were photographed by using a JEOL JEM 100-CX electron microscope under low-dose conditions at 80 kV.
RESULTS
STAT2 is required to recruit STAT1.
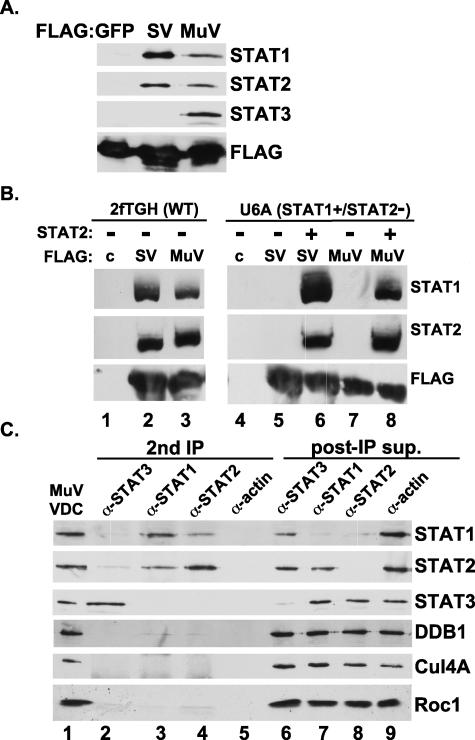
The SV5 and mumps V proteins cannot degrade STAT1 in cells that are lacking STAT2 (45, 60), and VDCs that copurify with SV5 and mumps V proteins contain both STAT1 and STAT2. In contrast, only the mumps V protein copurifies STAT3, consistent with its additional targeting capacity (60) (Fig. 1A). To investigate the reason STAT2 is required for STAT1 targeting, VDC purification was executed in wild-type (2fTGH) and STAT2-deficient (U6A) cells. SV5 and mumps V proteins could copurify both STAT1 and STAT2 from wild-type cells (Fig. 1B, lanes 2 and 3). In contrast, neither SV5 nor mumps V proteins could bind STAT1 in the absence of STAT2 (Fig. 1B, lanes 5 and 7). Expression of STAT2 complemented the STAT1-binding defect, enabling both SV5 and mumps V to copurify their STAT1 target as well as the expressed STAT2 protein (Fig. 1B, lanes 6 and 8). This pattern of copurification indicates that an important role for STAT2 is to facilitate efficient recruitment of STAT1 and suggests that STAT2 is stabilizing STAT1 interaction with the VDC.
FIG. 1.
Distinct copurification of STATs in VDC subcomplexes. (A) Immunoblot illustrating STAT components of SV5 and mumps VDC after FLAG affinity purification. (B) STAT2 is needed for efficient binding to STAT1. Wild-type 2fTGH or STAT2-deficient U6A cells were used for expression and purification of SV5 and mumps VDC. STAT1 and STAT2 are only precipitated by V protein from cells reconstituted with STAT2 by cDNA transfection. (C) Mumps VDC contains separate STAT1- and STAT3-containing complexes. FLAG affinity-purified mumps VDC (MuVDC) was subjected to a second IP with antiserum for STAT1, STAT2, STAT3, or α-actin control. After capture of immune complexes with protein A agarose, supernatant was retained for analysis (post-IP sup). All fractions were probed by immunoblotting for STATs, DDB1, Cul4A, and Roc1.
Mumps V protein assembles separate STAT1 and STAT3 targeting complexes.
In contrast to the requirement for STAT2 in STAT1 targeting, mumps V protein requires neither STAT1 nor STAT2 to target STAT3 (60). This observation implies that the mumps V protein engages separate machinery for targeting the two proteins. To test the concept of separate targeting complexes, affinity-purified VDC (Fig. 1C, lane 1) were fractionated further by a second IP for STAT1, STAT2, or STAT3 (Fig. 1C, lanes 2 to 5). Carrying out the second IP for STAT1 consistently yielded STAT2 but not STAT3 (Fig. 1C, lane 3). Complementary results were obtained for the second IP for STAT2, which yielded STAT1 but not STAT3 (Fig. 1C, lane 4). In these cases, the STAT3 protein was recovered in the post-IP supernatants. In agreement with these results, the second IP for STAT3 yielded neither STAT1 nor STAT2 (Fig. 1C, lane 2). These results verify the role of the STAT1-STAT2 interaction within VDC and indicate that the mumps V-interacting material contains separate VDC subcomplexes for targeting STAT1 and STAT3. One complex, VDCSTAT1, contains STAT1 and STAT2, while a separate complex, VDCSTAT3, contains STAT3.
General and specific roles for cellular DDB1 and Roc1 in VDC activity.
Cellular SCF ubiquitin ligase complexes uniformly contain Roc1, which associates with cullin family proteins including Cul4A (13, 18, 19, 24, 40, 57). As Rubulavirus V proteins bind DDB1 and Cul4A, it seemed likely that Roc1 might also be in complex with the viral proteins. Immunoblotting of affinity-purified SV5 and mumps V protein complexes demonstrated the copurification of endogenous Roc1 protein (Fig. 2A).
FIG. 2.
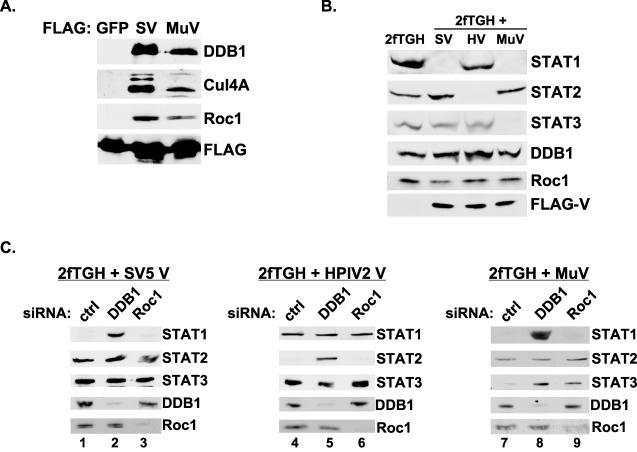
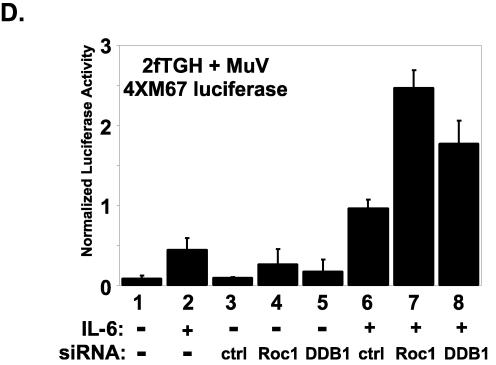
General and specific requirements for cellular VDC components in STAT targeting. (A) Immunoblot illustrating non-STAT components of SV5 and mumps VDC after FLAG affinity purification. DDB1, Cul4A, and Roc1 are present in each VDC preparation. (B) Extracts from parental 2fTGH and V protein expression cell lines were subjected to immunoblotting for VDC components. SV, SV5 V; HV, HPIV2 V; MuV, mumps V. (C) RNA interference to test the function of cellular proteins in VDC activities. Cells were transfected with 60 pmol of the indicated siRNA and harvested 72 h posttransfection for immunoblotting. Ctrl, control nontargeting siRNA. (D) Recovery of biologically active STAT3 by interference with Roc1 or DDB1. IL-6-responsive 4XM67 firefly luciferase reporter gene assay activation is expressed as the ratio of firefly luciferase to control Renilla luciferase activity. For each condition the average normalized luciferase units are plotted with the standard deviations indicated.
RNA interference was used previously to definitively test the roles of VDC components in STAT1 degradation by SV5 (3, 59). To extend this method to the other Rubulavirus V proteins, stable cell lines were created expressing the V proteins from SV5, HPIV2, and mumps virus. The V-expressing cell lines exhibit specific STAT protein degradation patterns. SV5 V-expressing cells lack STAT1, HPIV2 V-expressing cells lack STAT2, and mumps V-expressing cells lack both STAT1 and STAT3 (Fig. 2B). Stable expression of the V proteins did not alter the steady-state expression levels of the nontarget STATs, DDB1, or Roc1.
To test the requirement for DDB1 and Roc1 in STAT degradation by V proteins, the stable cell lines were transfected with siRNAs, and after 72 h lysates were subjected to immunoblotting. Specific immunoblotting verified the loss of steady-state DDB1 or Roc1 in the cells transfected with the corresponding siRNAs. Control siRNAs had no effect on STAT abundance. In agreement with prior results, interference with DDB1 in SV5 V protein-expressing cells resulted in recovery of STAT1 (Fig. 2C, lane 2). Interference with DDB1 in HPIV2 V-expressing cells recovered STAT2 (Fig. 2C, lane 5), and interference with DDB1 in mumps V-expressing cells recovered both STAT1 and STAT3 (Fig. 2, lane 8). These data clearly demonstrate that DDB1 is a central component of VDC activities, as it is generally required for STAT degradation by all V proteins.
Similar experiments were carried out in parallel to test the requirement for Roc1 in STAT degradation. In contrast to the general effects of DDB1, interference with Roc1 in both SV5 and HPIV2 V protein-expressing cells did not facilitate recovery of either STAT1 or STAT2, respectively (Fig. 2C, lanes 3 and 6). These findings indicate that Roc1 is dispensable for SV5 and HPIV2 VDC actions. A different pattern was observed in the mumps V-expressing cells. Interference with Roc1 did not result in the recovery of STAT1 protein, similar to the lack of effects on SV5-STAT1 targeting (Fig. 2C, lane 9). Strikingly, interference with Roc1 in mumps V cells resulted in the recovery of STAT3 (Fig. 2C, lane 9). These results indicate that Roc1 is required for STAT3 targeting by the mumps V protein.
To confirm that interference with Roc1 expression leads to the recovery of functional STAT3, RNA interference was carried out in the context of a STAT3-dependent IL-6 reporter gene assay. Mumps V protein-expressing 2fTGH cells were treated with or without siRNA for 48 h, which was followed by overnight IL-6 treatment. The mumps V stable cell lines have little residual response to IL-6 in the absence of siRNA as they lack STAT3 expression (Fig. 2D, lanes 1 and 2). Slightly enhanced reporter gene activity was observed in cells transfected with the control nonspecific siRNA in combination with IL-6 treatment (Fig. 2D, lane 6). However, the siRNA for Roc1 resulted in the activation of IL-6 responses 2.5-fold greater than the control (Fig. 2D, lane 7), and DDB1 RNA interference activated the reporter gene twofold above the control (Fig. 2D, lane 8). These data demonstrate that loss of Roc1 or DDB1 leads to recovery of STAT3 signaling in mumps V protein-expressing stable cell lines, confirming their essential roles in VDCSTAT3 activity. Furthermore, these results provide evidence that cytokine signaling activity can be recovered by using RNA interference to counteract viral evasion mechanisms.
SV5 V binds Roc1 through Cul4A, while mumps V binds Roc1 independent of Cul4A.
Both SV5 and mumps VDC eluates contain Roc1, but it is required only for mumps-induced STAT3 degradation. To explain this apparent conundrum, the nature of the V protein interaction with Roc1 was analyzed by combinatorial expression of epitope-tagged proteins. In these experiments, FLAG-Roc1 and HA-V proteins were expressed either alone or in combination with T7-DDB1 or myc-Cul4A. FLAG-Roc1 and associated proteins were immunoprecipitated, separated by SDS-PAGE, and subjected to immunoblotting for the other tagged proteins. A control protein, FLAG-thioredoxin, did not copurify any VDC components (Fig. 3, lanes 1 and 6). FLAG-Roc1 did not coprecipitate SV5 V unless Cul4A was also present (Fig. 3, compare lanes 2 and 3). This result suggests that the presence of Roc1 in the SV5 VDC is the result of indirect association via Cul4A. In contrast, the mumps V protein was found in association with Roc1 in the absence of expressed of Cul4A (Fig. 3, lane 7). This finding indicates that mumps V interacts with Roc1 independent of Cul4A, consistent with the differential requirements for Roc1 in VDC activities of SV5 and mumps.
Homo- and heterooligomerization of V proteins.
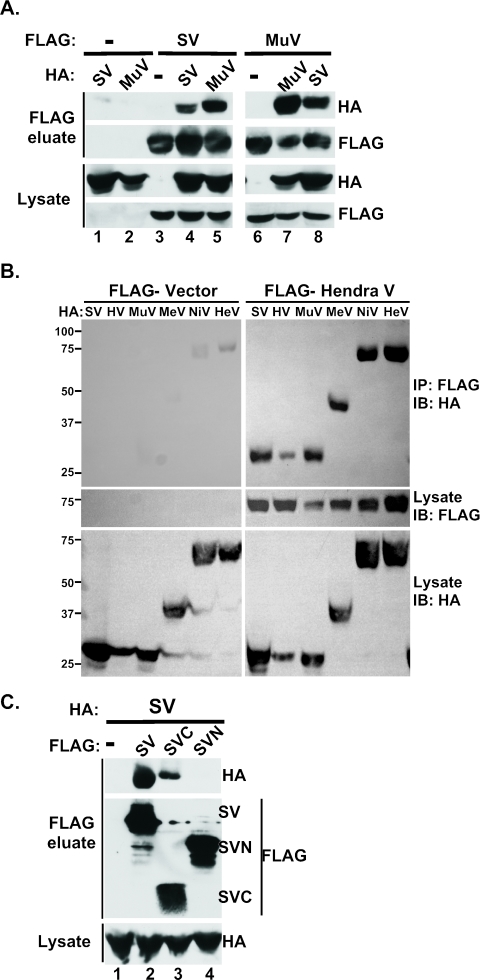
The data indicate that V proteins are engaging cellular factors through distinct targeting subcomplexes. A question arises as to how the ∼220-amino-acid V proteins can assemble these relatively large subunits into a STAT-targeting ubiquitin ligase entity. To test the possibility of V protein oligomerization, differentially epitope-tagged SV5 or mumps V proteins were coexpressed in cells, and lysates were immunoprecipitated with FLAG affinity resin and eluted with FLAG tripeptide. Eluates were then subjected to immunoblotting to detect the presence of HA-V protein. No nonspecific associations were detected with the HA-V proteins (Fig. 4A, lanes 1 and 2). However, the purified FLAG V proteins were able to coprecipitate both the homotypic and heterotypic HA-V protein (Fig. 4A, lanes 4 and 5 and 7 and 8). These results demonstrate that SV5 and mumps V proteins can oligomerize with both themselves and each other.
FIG. 4.
Paramyxovirus V proteins homo- and heterooligomerize via the conserved C terminus. (A) SV5 and mumps V proteins form oligomers. 293T cells were transfected with FLAG vector (−), FLAG-SV5 V (SV), or FLAG-Mumps V (MuV) with or without HA vector (−), HA-SV, or HA-MuV. FLAG-associated proteins were immunoprecipitated, washed extensively, and eluted with FLAG peptide. FLAG eluates and fractions of total lysates prior to IP (lysate) were separated by SDS-PAGE and subjected to immunoblotting to detect coprecipitated HA- and FLAG-tagged proteins. (B) Example of generality of paramyxovirus V protein homo- and heterooligomerization. Methodology is the same as described in panel A but with FLAG-Hendra virus V protein used in conjunction with HA-tagged SV5 (SV), HPIV2 (HV), mumps (MuV), measles (MeV), Nipah virus (NiV), and Hendra virus (HeV) V proteins. (C) SV5 V protein CTD mediates oligomerization. FLAG vector (−), FLAG-SV, FLAG-SVC (amino acids 162 to 222 of SV5 V protein), or FLAG-SVN (amino acids 1 to 162 of SV5 V protein) were expressed in 293T cells with HA-SV and coprecipitated as described above.
The discovery of heterotypic V-V interactions between SV5 and mumps was unexpected and implies a previously overlooked level of V protein organization that might be preserved across the paramyxovirus family. To test the generality of V protein oligomerization within the Rubulavirus genus and beyond, similar coprecipitation assays were conducted with V proteins from Rubulavirus (SV5, HPIV2, and mumps), Henipavirus (Nipah virus and Hendra virus), and Morbillivirus (measles virus) genera. Astonishingly, all of the V proteins interacted not only with themselves homotypically but also with one another heterotypically (Fig. 4B and Table 1). These results establish oligomerization as a general property of paramyxovirus V proteins and also suggest that oligomerization is mediated by the only conserved domain in these proteins, the CTD.
TABLE 1.
Oligomerization of paramyxovirus V proteinsa
| SV5 | HPIV2 | Mumps | Measles | Nipah | Hendra | |
|---|---|---|---|---|---|---|
| SV5 | + | + | + | + | + | + |
| HPIV2 | + | + | + | + | + | |
| Mumps | + | + | + | + | ||
| Measles | + | + | + | |||
| Nipah | + | + | ||||
| Hendra | + |
Interactions tested by expressing two differentially epitope-tagged V proteins in 293T cells followed by immunoprecipitation from whole-cell lysates with antiserum to first tag and immunoblotting for second tag. +, interaction detected.
Cysteine-rich CTD is necessary and sufficient for homooligomerization.
To better define the V protein self-association, FLAG-tagged expression constructs were created containing the C-terminal cysteine-rich domain (amino acids 162 to 222) or the N-terminal region (amino acids 1 to 162). These vectors or the full-length FLAG-SV5 V were coexpressed individually with full-length HA-SV5 V protein and subjected to coprecipitation assays. As in Fig. 4A, full-length SV5 V can coprecipitate itself (Fig. 4C, lane 2), but the SV5 N terminus did not (Fig. 4C, lane 4), indicating that the C terminus is needed for self-association. Conducting the experiment with the SV5 C terminus revealed that this region by itself was sufficient to coprecipitate full-length SV5 V protein (Fig. 4C, lane 3). These results demonstrate that the V protein self-associations are mediated through the conserved cysteine-rich CTD, which we now define as an oligomerization domain.
Macromolecular spherical particles are formed by V proteins alone and in VDC.
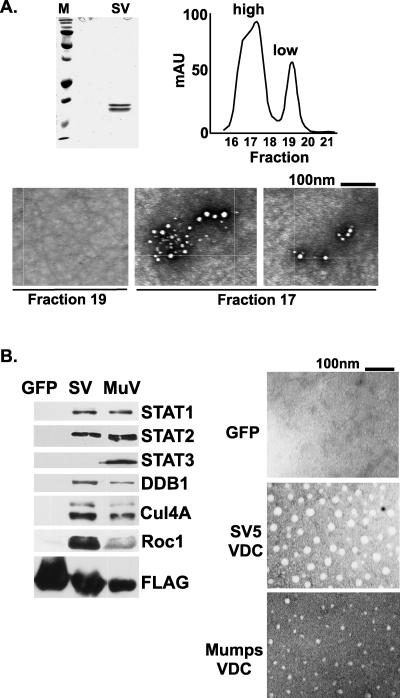
Although the V protein C termini are not homologous to any known cellular proteins, they share several features with cellular zinc-binding proteins. Prominently, they share a 2:1 zinc-protein stoichiometry that is found in the RING domain. Like V proteins, many RING domain proteins are components of Ub ligase complexes. RING domains have been demonstrated to self-assemble into macromolecular structures thought to serve as molecular scaffolds for catalyzing polyubiquitylation (26-28). To test if the isolated V protein could undergo a similar self-assembly, SV5 V protein was expressed in E. coli, purified, and subjected to gel filtration chromatography. The purified SV5 V protein preparation migrated as a doublet on SDS-PAGE gels, likely the result of proteolytic cleavage (Fig. 5A, left). The gel filtration revealed the V protein eluted in two distinct peaks indicative of low- and high-molecular-weight forms (Fig. 5A, right). The peak fractions (17 and 19) were analyzed by negative staining and electron microscopy. No distinct structures were observed for the material in the low-molecular-weight fraction 19, but an abundance of spherical particles was detected in the high-molecular-weight fraction 17 (Fig. 5A, bottom). These particles strikingly resemble those formed by purified RING domain proteins and indicate that the isolated V protein can similarly self-assemble to form large spherical particles.
FIG. 5.
V proteins and VDC assemble into macromolecular structures. (A) Purified SV5 V protein forms macromolecular structures. SV5 V (SV) expressed in E. coli was purified, separated by SDS-PAGE, and detected by Coomassie blue staining (top left). Size fractionation by gel filtration chromatography (top right) produced two peaks subjected to electron microscopy (bottom). (B) Affinity-purified SV5 and mumps VDC preparations contain spherical particles. FLAG eluates (left) were subjected to negative staining and electron microscopy (right).
To determine if these particles are present in the more complicated arrangement of purified VDC, affinity preparations of SV5 VDC and mumps VDC were isolated from transfected cells and analyzed by electron microscopy (Fig. 5B). Both VDC preparations contained spherical structures similar to those observed with pure high-molecular-weight SV5 V protein, and no complexes were observed in a control FLAG-green fluorescent protein preparation. By analogy to the RING domain proteins, these data suggest that the V proteins form a molecular scaffold for the execution of polyubiquitylation reactions.
DISCUSSION
Rubulavirus V proteins assemble STAT-targeting VDC ubiquitin ligase complexes from cellular components. The results presented here demonstrate that the VDCs assembled by SV5, HPIV2, and mumps virus V proteins have several features in common yet differ in their engagement of cellular machinery in order to catalyze degradation of their specific target STATs.
DDB1 was the first cellular protein reported to interact with V, through a direct interaction that requires the CTD cysteine residues (33). All of the VDC activities—SV5 and mumps VDCSTAT1, HPIV2 VDCSTAT2, and mumps VDCSTAT3—require DDB1 in order to degrade their STAT targets, indicating its central role in the VDC E3 ubiquitylation machinery that the V proteins utilize to target STATs. As DDB1 is known to interact with Cul4A (14, 35, 55), we hypothesize that it is the key recruiter of cellular E3 subunits.
Both SV5 and mumps V proteins can copurify Roc1, but for SV5 this association is mediated by Cul4A, and Roc1 is nonessential. In contrast, mumps V interacts with Roc1 independent of Cul4A and is needed for STAT3 destruction. Perhaps relevant to the Roc1 interaction with mumps V is the observation that several cullin family proteins can copurify with mumps V (C. M. Ulane, unpublished observations), any of which might bind to Roc1 and function in STAT3 targeting. Apparently, Roc1 is dispensable for VDCSTAT1 and VDCSTAT2. The possibility that Roc1 participates in targeting unknown Rubulavirus VDC substrates cannot be excluded based on available evidence. The present evidence contrasts with the importance of Roc1 in a cellular E3 complex that uses DDB1, Cul4A, and Roc1 to target the transcription factor c-Jun for degradation (63). For this complex, results indicate that Roc1 is required for c-Jun targeting activity (63). Unlike the VDCSTAT1 and VDCSTAT2, Roc1 is an essential component of the VDCSTAT3 machinery used by mumps virus to catalyze STAT3 destruction. Accordingly, differential use of Roc1 enables the mumps V protein to expand its targeting range.
Both SV5 and mumps V proteins require STAT2 in order to target STAT1, and this requirement restricts the host range of SV5, as murine STAT2 fails to support STAT1 targeting (43). Results indicate that the role of STAT2 in these complexes is to provide a vehicle for high-affinity recruitment of STAT1 to the VDCSTAT1. A recent report using an in vitro system to analyze VDCSTAT1 demonstrated that purified V protein could not efficiently bind to STAT1 in the absence of STAT2 (48). Several interpretations can be applied to this STAT1-STAT2 codependence. For example, a unique conformational arrangement between STAT1 and STAT2 could be mediated by the V protein. Alternatively, sequential binding of STAT2 followed by STAT1 may occur, as is the case for the V protein from Nipah virus, a member of the Henipavirus genus (52). Furthermore, V proteins may have evolved to recognize a STAT1-STAT2 dimer. Although STAT activation and dimerization by tyrosine phosphorylation are not required for the V protein to target and degrade STATs (45), it is possible that a previously unrecognized interaction between latent nonphosphorylated STAT1 and STAT2 is the target of V proteins. Indeed, several groups of investigators have reported detection of nonphosphorylated STAT dimers in either cell lysates or preparations of purified STATs (9, 30, 38, 39, 41). A very recent crystallographic structure of nonphosphorylated STAT1 demonstrates an antiparallel dimer with a unique dimerization interface (34). The ability to form this antiparallel nonphosphorylated STAT1 homodimer is required for proper regulation of the STAT1 activation-inactivation cycle (68). It seems reasonable to speculate that a latent unphosphorylated STAT1-STAT2 heterodimer is the entity recruited to VDCSTAT1 and that following STAT1 ubiquitylation, the remnant STAT2 can either dissociate or recruit an additional latent STAT1. Consistent with the use of distinct STAT-targeting mechanisms, mumps V protein forms a separate subcomplex for the recruitment of STAT3 to VDCSTAT3, and STAT3 targeting does not require STAT2 or STAT1 as a cofactor. Notably, DDB1, Cul4A, and Roc1 do not copurify in any of the STAT subcomplexes formed by mumps V protein but are instead found in the post-IP supernatant (Fig. 1C). Thus, V protein can interact with cellular E3 machinery separate from its STAT-binding capability.
Results from coimmunoprecipitation studies indicate that V proteins from all paramyxovirus genera tested can homo- and heterooligomerize. As these viruses and their V proteins did not coevolve in doubly infected cells, it is most logical that oligomerization is mediated by a common conserved domain. In fact, results indicate that the SV5 V protein self-association is mediated through the most conserved domain, the cysteine-rich CTD, which is both necessary and sufficient for self-association. This study defines the paramyxovirus V protein cysteine-rich, zinc-binding CTD as an oligomerization domain, which may explain the diverse roles for this highly conserved domain in virus replication and host evasion.
The ability of V proteins to participate in several seemingly unrelated and independent protein complexes that need to be integrated with one another for substrate targeting can be largely explained by self-assembly. Purified SV5 V protein oligomerizes spontaneously to form high-molecular-weight species that form spherical macromolecular particles visible with an electron microscope. Similar spherical structures are observed in preparations of SV5 and mumps VDC purified from cells. This self-assembly property is in common with cellular zinc-binding domains in the RING domain family, many of which are known to function in ubiquitin ligase reactions. Mutations that disrupt self-assembly also eliminate RING domain E3 enzymatic activity (27). Like RING proteins, Rubulavirus V proteins exhibit ubiquitin ligase activity in vitro and catalyze polyubiquitylation and degradation of targets in vivo (17, 29, 59, 60). Nonetheless, other than a superficial resemblance with respect to cysteine-mediated zinc binding, the V protein CTD shares little sequence conservation that would suggest a RING domain-like fold. The self-assembly of Rubulavirus V proteins into RING-like spherical particles reveals structural and functional similarities that were not apparent from amino acid sequence comparisons, indicating an evolutionary convergence between V protein CTDs and cellular RING domains. The use of self-assembling macromolecular structures for catalysis of ubiquitylation and other enzymatic reactions may be a universal theme important for both cell biology and virus replication (12, 47, 50).
Model for VDC assembly and activity.
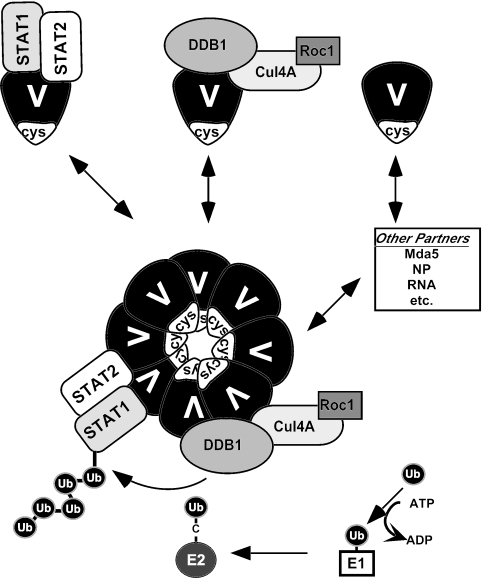
The Rubulavirus V proteins are capable of many protein interactions both with viral partners, including the nucleocapsid protein (NP) and viral RNA (32), and with cellular machinery, like Mda5 or the VDC components. Based on the ability of V proteins to self-assemble, a working model emerges from these studies where the V protein mediates associations between subsets of macromolecular complexes to manifest polyubiquitylation of STAT (Fig. 6). It is sensible to imagine that V proteins are in equilibrium between low- and high-molecular-weight states, and evidence presented here indicates that they can form discrete interactions with several cellular collaborators. A subset of interactions is indirect, as in the case of Roc1 recruitment by SV5 V, and others require stabilizing interactions between binding partners, as is the case for STAT2-dependent STAT1 binding.
FIG. 6.
Working model for VDC structure and function. For simplicity, only VDCSTAT1 is depicted. V proteins can interact with STAT1+STAT2 or with DDB1+Cul4A+Roc1. These subcomplexes can associate with each other through V protein oligomerization via the C-terminal domain (cys) to form spherical particles that act as a scaffold for E3 ubiquitin ligase activity. Additional interactions (Mda5, NP, RNA, etc.) mediated by V can either participate in the VDC superstructure or not, and the interdependence of these interactions must be tested independently.
The results demonstrate that the highly conserved V protein cysteine-rich CTD functions as an oligomerization domain, allowing independently assembled V protein subcomplexes to interact. The V proteins can oligomerize to form large spherical structures, which we hypothesize to be the enzymatically active component for STAT polyubiquitylation. The surfaces of the macromolecular particles allow V proteins that are bound to STATs to interact more efficiently with V proteins bound to cellular ubiquitylation machinery (e.g., DDB1, Cul4A, and Roc1). Juxtaposition of the enzymatic subcomplex and the substrate subcomplex allows more efficient ubiquitin transfer from E2 ubiquitin-conjugating enzyme to the STAT substrate and might enhance sequential cycles of ubiquitin transfer to favor polyubiquitylation. The equilibrium inherent in this model also allows the V protein to freely interact with additional cellular or virus-encoded cofactors needed for its VDC-independent functions, such as inhibition of cellular Mda-5 signaling or roles in viral RNA replication (2, 25, 49, 51, 62). Furthermore, the V protein-containing particles may function as enzymatic reaction centers providing a supporting role for multiple outcomes other than STAT polyubiquitylation. While further work will be needed to resolve all the details of this model, it is certain that the V protein is a multifunctional anticellular factor with an intricate and dynamic action against STAT signaling based on coordinated protein interactions.
In addition to revealing mechanisms underlying pathogen replication, analysis of animal viruses has long served the study of molecular and cell biology by revealing basic principles of life processes. Understanding the cellular requirements for V protein-induced STAT targeting and the mechanistic basis of VDC assembly for STAT targeting demonstrates how a viral protein can manipulate cellular partners to block the host antiviral response. The protein interaction sites identified are valid targets for creation of specific antiviral compounds. Moreover, as inappropriately activated STAT protein pathways are known effectors of numerous human diseases including cancer and inflammation, understanding how V proteins specifically remove STATs from cells will identify opportunities for therapeutic intervention and may aid the development of novel STAT-directed therapeutics (11, 15).
Acknowledgments
This work is supported by NIH grant AI50707 to C.M.H. C.M.U. was supported by Mount Sinai Immunobiology Training Grant 5T32AI07605-04.
REFERENCES
- 1.Aaronson, D. S., and C. M. Horvath. 2002. A road map for those who don't know JAK-STAT. Science 296:1653-1655. [DOI] [PubMed] [Google Scholar]
- 2.Andrejeva, J., K. S. Childs, D. F. Young, T. S. Carlos, N. Stock, S. Goodbourn, and R. E. Randall. 2004. The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, Mda-5, and inhibit its activation of the IFN-beta promoter. Proc. Natl. Acad. Sci. USA 101:17264-17269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andrejeva, J., E. Poole, D. F. Young, S. Goodbourn, and R. E. Randall. 2002. The p127 subunit (DDB1) of the UV-DNA damage repair binding protein is essential for the targeted degradation of STAT1 by the V protein of the paramyxovirus simian virus 5. J. Virol. 76:11379-11386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1994. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 5.Bondar, T., A. Ponomarev, and P. Raychaudhuri. 2004. Ddb1 is required for the proteolysis of the Schizosaccharomyces pombe replication inhibitor Spd1 during S phase and after DNA damage. J. Biol. Chem. 279:9937-9943. [DOI] [PubMed] [Google Scholar]
- 6.Borden, K. L. 2000. RING domains: master builders of molecular scaffolds? J. Mol. Biol. 295:1103-1112. [DOI] [PubMed] [Google Scholar]
- 7.Borden, K. L. 1998. RING fingers and B-boxes: zinc-binding protein-protein interaction domains. Biochem. Cell Biol. 76:351-358. [DOI] [PubMed] [Google Scholar]
- 8.Borden, K. L., M. N. Boddy, J. Lally, N. J. O'Reilly, S. Martin, K. Howe, E. Solomon, and P. S. Freemont. 1995. The solution structure of the RING finger domain from the acute promyelocytic leukaemia proto-oncoprotein PML. EMBO J. 14:1532-1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Braunstein, J., S. Brutsaert, R. Olson, and C. Schindler. 2003. STATs dimerize in the absence of phosphorylation. J. Biol. Chem. 278:34133-34140. [DOI] [PubMed] [Google Scholar]
- 10.Bromberg, J., C. M. Horvath, D. Besser, W. W. Lathem, and J. J. E. Darnell. 1998. Stat3 activation is required for cellular transformation by v-SRC. Mol. Cell. Biol. 18:2553-2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buettner, R., L. B. Mora, and R. Jove. 2002. Activated STAT signaling in human tumors provides novel molecular targets for therapeutic intervention. Clin. Cancer Res. 8:945-954. [PubMed] [Google Scholar]
- 12.Cardozo, T., and M. Pagano. 2004. The SCF ubiquitin ligase: insights into a molecular machine. Nat. Rev. Mol. Cell Biol. 5:739-751. [DOI] [PubMed] [Google Scholar]
- 13.Chen, A., K. Wu, S. Y. Fuchs, P. Tan, C. Gomez, and Z. Q. Pan. 2000. The conserved RING-H2 finger of ROC1 is required for ubiquitin ligation. J. Biol. Chem. 275:15432-15439. [DOI] [PubMed] [Google Scholar]
- 14.Chen, X., Y. Zhang, L. Douglas, and P. Zhou. 2001. UV-damaged DNA-binding proteins are targets of CUL-4A-mediated ubiquitination and degradation. J. Biol. Chem. 276:48175-48182. [DOI] [PubMed] [Google Scholar]
- 15.Darnell, J. E. 2002. Transcription factors as targets for cancer therapy. Nat. Rev. Cancer 2:740-749. [DOI] [PubMed] [Google Scholar]
- 16.Didcock, L., D. F. Young, S. Goodbourn, and R. E. Randall. 1999. Sendai virus and simian virus 5 block activation of interferon-responsive genes: importance for virus pathogenesis. J. Virol. 73:3125-3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Didcock, L., D. F. Young, S. Goodbourn, and R. E. Randall. 1999. The V protein of simian virus 5 inhibits interferon signaling by targeting STAT1 for proteasome-mediated degradation. J. Virol. 73:9928-9933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Furukawa, M., T. Ohta, and Y. Xiong. 2002. Activation of UBC5 ubiquitin-conjugating enzyme by the RING finger of ROC1 and assembly of active ubiquitin ligases by all cullins. J. Biol. Chem. 277:15758-15765. [DOI] [PubMed] [Google Scholar]
- 19.Furukawa, M., Y. Zhang, J. McCarville, T. Ohta, and Y. Xiong. 2000. The CUL1 C-terminal sequence and ROC1 are required for efficient nuclear accumulation, NEDD8 modification, and ubiquitin ligase activity of CUL1. Mol. Cell. Biol. 20:8185-8197.11027288 [Google Scholar]
- 20.Groisman, R., J. Polanowska, I. Kuraoka, J. Sawada, M. Saijo, R. Drapkin, A. F. Kisselev, K. Tanaka, and Y. Nakatani. 2003. The ubiquitin ligase activity in the DDB2 and CSA complexes is differentially regulated by the COP9 signalosome in response to DNA damage. Cell 113:357-367. [DOI] [PubMed] [Google Scholar]
- 21.Haspel, R. L., M. Salditt-Georgieff, and J. E. Darnell, Jr. 1996. The rapid inactivation of nuclear tyrosine phosphorylated Stat1 depends on a protein tyrosine phosphatase. EMBO J. 15:6262-6268. [PMC free article] [PubMed] [Google Scholar]
- 22.Hirano, T., K. Ishihara, and M. Hibi. 2000. Roles of STAT3 in mediating the cell growth, differentiation and survival signals relayed through the IL-6 family of cytokine receptors. Oncogene 19:2548-2556. [DOI] [PubMed] [Google Scholar]
- 23.Hu, J., C. M. McCall, T. Ohta, and Y. Xiong. 2004. Targeted ubiquitination of CDT1 by the DDB1-CUL4A-ROC1 ligase in response to DNA damage. Nat. Cell Biol. 6:1003-1009. [DOI] [PubMed] [Google Scholar]
- 24.Jackson, P. K., and A. G. Eldridge. 2002. The SCF ubiquitin ligase: an extended look. Mol. Cell 9:923-925. [DOI] [PubMed] [Google Scholar]
- 25.Kawano, M., M. Kaito, Y. Kozuka, H. Komada, N. Noda, K. Nanba, M. Tsurudome, M. Ito, M. Nishio, and Y. Ito. 2001. Recovery of infectious human parainfluenza type 2 virus from cDNA clones and properties of the defective virus without V-specific cysteine-rich domain. Virology 284:99-112. [DOI] [PubMed] [Google Scholar]
- 26.Kentsis, A., and K. L. Borden. 2000. Construction of macromolecular assemblages in eukaryotic processes and their role in human disease: linking RINGs together. Curr. Protein Pept. Sci. 1:49-73. [DOI] [PubMed] [Google Scholar]
- 27.Kentsis, A., R. E. Gordon, and K. L. Borden. 2002. Control of biochemical reactions through supramolecular RING domain self-assembly. Proc. Natl. Acad. Sci. USA 99:15404-15409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kentsis, A., R. E. Gordon, and K. L. Borden. 2002. Self-assembly properties of a model RING domain. Proc. Natl. Acad. Sci. USA 99:667-672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kubota, T., N. Yokosawa, S. Yokota, and N. Fujii. 2001. C terminal CYS-RICH region of mumps virus structural V protein correlates with block of interferon alpha and gamma signal transduction pathway through decrease of STAT 1-alpha. Biochem. Biophys. Res. Commun. 283:255-259. [DOI] [PubMed] [Google Scholar]
- 30.Lackmann, M., A. G. Harpur, A. C. Oates, R. J. Mann, A. Gabriel, W. Meutermans, P. F. Alewood, I. M. Kerr, G. R. Stark, and A. F. Wilks. 1998. Biomolecular interaction analysis of IFN gamma-induced signaling events in whole-cell lysates: prevalence of latent STAT1 in high-molecular-weight complexes. Growth Factors 16:39-51. [DOI] [PubMed] [Google Scholar]
- 31.Lee, C. K., H. A. Bluyssen, and D. E. Levy. 1997. Regulation of interferon-alpha responsiveness by the duration of Janus kinase activity. J. Biol. Chem. 272:21872-21877. [DOI] [PubMed] [Google Scholar]
- 32.Lin, G. Y., R. G. Paterson, and R. A. Lamb. 1997. The RNA binding region of the paramyxovirus SV5 V and P proteins. Virology 238:460-469. [DOI] [PubMed] [Google Scholar]
- 33.Lin, G. Y., R. G. Paterson, C. D. Richardson, and R. A. Lamb. 1998. The V protein of the paramyxovirus SV5 interacts with damage-specific DNA binding protein. Virology 249:189-200. [DOI] [PubMed] [Google Scholar]
- 34.Mao, X., Z. Ren, G. N. Parker, H. Sondermann, M. A. Pastorello, W. Wang, J. S. McMurray, B. Demeler, J. E. Darnell, Jr., and X. Chen. 2005. Structural bases of unphosphorylated STAT1 association and receptor binding. Mol. Cell 17:761-771. [DOI] [PubMed] [Google Scholar]
- 35.McCall, C. M., J. Hu, and Y. Xiong. 2005. Recruiting substrates to cullin 4-dependent ubiquitin ligases by DDB1. Cell Cycle 4:27-29. [DOI] [PubMed] [Google Scholar]
- 36.Nag, A., S. Bagchi, and P. Raychaudhuri. 2004. Cul4A physically associates with MDM2 and participates in the proteolysis of p53. Cancer Res. 64:8152-8155. [DOI] [PubMed] [Google Scholar]
- 37.Nag, A., T. Bondar, S. Shiv, and P. Raychaudhuri. 2001. The xeroderma pigmentosum group E gene product DDB2 is a specific target of cullin 4A in mammalian cells. Mol. Cell. Biol. 21:6738-6747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ndubuisi, M. I., G. G. Guo, V. A. Fried, J. D. Etlinger, and P. B. Sehgal. 1999. Cellular physiology of STAT3: where's the cytoplasmic monomer? J. Biol. Chem. 274:25499-25509. [DOI] [PubMed] [Google Scholar]
- 39.Novak, U., H. Ji, V. Kanagasundaram, R. Simpson, and L. Paradiso. 1998. STAT3 forms stable homodimers in the presence of divalent cations prior to activation. Biochem. Biophys. Res. Commun. 247:558-563. [DOI] [PubMed] [Google Scholar]
- 40.Ohta, T., J. J. Michel, A. J. Schottelius, and Y. Xiong. 1999. ROC1, a homolog of APC11, represents a family of cullin partners with an associated ubiquitin ligase activity. Mol. Cell 3:535-541. [DOI] [PubMed] [Google Scholar]
- 41.Ota, N., T. J. Brett, T. L. Murphy, D. H. Fremont, and K. M. Murphy. 2004. N-domain-dependent nonphosphorylated STAT4 dimers required for cytokine-driven activation. Nat. Immunol. 5:208-215. [DOI] [PubMed] [Google Scholar]
- 42.Palosaari, H., J. P. Parisien, J. J. Rodriguez, C. M. Ulane, and C. M. Horvath. 2003. STAT protein interference and suppression of cytokine signal transduction by measles virus V protein. J. Virol. 77:7635-7644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Parisien, J.-P., J. F. Lau, and C. M. Horvath. 2002. STAT2 acts as a host range determinant for species-specific paramyxovirus interferon antagonism and simian virus 5 replication. J. Virology 76:6435-6441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parisien, J.-P., J. F. Lau, J. J. Rodriguez, B. M. Sullivan, A. Moscona, G. D. Parks, R. A. Lamb, and C. M. Horvath. 2001. The V protein of human parainfluenza virus 2 antagonizes type I interferon responses by destabilizing signal transducer and activator of transcription 2. Virology 283:230-239. [DOI] [PubMed] [Google Scholar]
- 45.Parisien, J.-P., J. F. Lau, J. J. Rodriguez, C. M. Ulane, and C. M. Horvath. 2002. Selective STAT protein degradation induced by paramyxoviruses requires both STAT1 and STAT2 but is independent of alpha/beta interferon signal transduction. J. Virol. 76:4190-4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Paterson, R. G., G. P. Leser, M. A. Shaughnessy, and R. A. Lamb. 1995. The paramyxovirus SV5 V protein binds two atoms of zinc and is a structural component of virions. Virology 208:121-131. [DOI] [PubMed] [Google Scholar]
- 47.Petroski, M. D., and R. J. Deshaies. 2005. Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 6:9-20. [DOI] [PubMed] [Google Scholar]
- 48.Precious, B., D. F. Young, L. Andrejeva, S. Goodbourn, and R. E. Randall. 2005. In vitro and in vivo specificity of ubiquitination and degradation of STAT1 and STAT2 by the V proteins of the paramyxoviruses simian virus 5 and human parainfluenza virus type 2. J. Gen. Virol. 86:151-158. [DOI] [PubMed] [Google Scholar]
- 49.Precious, B., D. F. Young, A. Bermingham, R. Fearns, M. Ryan, and R. E. Randall. 1995. Inducible expression of the P, V, and NP genes of the paramyxovirus simian virus 5 in cell lines and an examination of NP-P and NP-V interactions. J. Virol. 69:8001-8010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ptak, C., J. A. Prendergast, R. Hodgins, C. M. Kay, V. Chau, and M. J. Ellison. 1994. Functional and physical characterization of the cell cycle ubiquitin-conjugating enzyme CDC34 (UBC3). Identification of a functional determinant within the tail that facilitates CDC34 self-association. J. Biol. Chem. 269:26539-26545. [PubMed] [Google Scholar]
- 51.Randall, R. E., and A. Bermingham. 1996. NP:P and NP:V interactions of the paramyxovirus simian virus 5 examined using a novel protein:protein capture assay. Virology 224:121-129. [DOI] [PubMed] [Google Scholar]
- 52.Rodriguez, J. J., C. D. Cruz, and C. M. Horvath. 2004. Identification of the nuclear export signal and STAT-binding domains of the Nipah virus V protein reveals mechanisms underlying interferon evasion. J. Virol. 78:5358-5367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sadowski, H. B., K. Shuai, J. E. Darnell, Jr., and M. Z. Gilman. 1993. A common nuclear signal transduction pathway activated by growth factor and cytokine receptors. Science 261:1739-1744. [DOI] [PubMed] [Google Scholar]
- 54.Samuel, C. E. 2001. Antiviral actions of interferons. Clin. Microbiol. Rev. 14:778-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shiyanov, P., A. Nag, and P. Raychaudhuri. 1999. Cullin 4A associates with the UV-damaged DNA-binding protein DDB. J. Biol. Chem. 274:35309-35312. [DOI] [PubMed] [Google Scholar]
- 56.Shuai, K., and B. Liu. 2003. Regulation of JAK-STAT signalling in the immune system. Nat. Rev. Immunol. 3:900-911. [DOI] [PubMed] [Google Scholar]
- 57.Swaroop, M., M. Gosink, and Y. Sun. 2001. SAG/ROC2/Rbx2/Hrt2, a component of SCF E3 ubiquitin ligase: genomic structure, a splicing variant, and two family pseudogenes. DNA Cell Biol. 20:425-434. [DOI] [PubMed] [Google Scholar]
- 58.Tan, P., S. Y. Fuchs, A. Chen, K. Wu, C. Gomez, Z. Ronai, and Z. Q. Pan. 1999. Recruitment of a ROC1-CUL1 ubiquitin ligase by Skp1 and HOS to catalyze the ubiquitination of I kappa B alpha. Mol. Cell 3:527-533. [DOI] [PubMed] [Google Scholar]
- 59.Ulane, C. M., and C. M. Horvath. 2002. Paramyxoviruses SV5 and HPIV2 assemble STAT protein ubiquitin ligase complexes from cellular components. Virology 304:160-166. [DOI] [PubMed] [Google Scholar]
- 60.Ulane, C. M., J. J. Rodriguez, J. P. Parisien, and C. M. Horvath. 2003. STAT3 ubiquitylation and degradation by mumps virus suppress cytokine and oncogene signaling. J. Virol. 77:6385-6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vignais, M. L., H. B. Sadowski, D. Watling, N. C. Rogers, and M. Gilman. 1996. Platelet-derived growth factor induces phosphorylation of multiple JAK family kinases and STAT proteins. Mol. Cell. Biol. 16:1759-1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Watanabe, N., M. Kawano, M. Tsurudome, M. Nishio, M. Ito, S. Ohgimoto, S. Suga, H. Komada, and Y. Ito. 1996. Binding of the V proteins to the nucleocapsid proteins of human parainfluenza type 2 virus. Med. Microbiol. Immunol. 185:89-94. [DOI] [PubMed] [Google Scholar]
- 63.Wertz, I. E., K. M. O'Rourke, Z. Zhang, D. Dornan, D. Arnott, R. J. Deshaies, and V. M. Dixit. 2004. Human De-etiolated-1 regulates c-Jun by assembling a CUL4A ubiquitin ligase. Science 303:1371-1374. [DOI] [PubMed] [Google Scholar]
- 64.Wu, K., S. Y. Fuchs, A. Chen, P. Tan, C. Gomez, Z. Ronai, and Z. Q. Pan. 2000. The SCF(HOS/beta-TRCP)-ROC1 E3 ubiquitin ligase utilizes two distinct domains within CUL1 for substrate targeting and ubiquitin ligation. Mol. Cell. Biol. 20:1382-1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yanagawa, Y., J. A. Sullivan, S. Komatsu, G. Gusmaroli, G. Suzuki, J. Yin, T. Ishibashi, Y. Saijo, V. Rubio, S. Kimura, J. Wang, and X. W. Deng. 2004. Arabidopsis COP10 forms a complex with DDB1 and DET1 in vivo and enhances the activity of ubiquitin conjugating enzymes. Genes Dev. 18:2172-2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yokosawa, N., T. Kubota, and N. Fujii. 1998. Poor induction of interferon-induced 2′,5′-oligoadenylate synthetase (2-5 AS) in cells persistently infected with mumps virus is caused by decrease of STAT-1 alpha. Arch. Virol. 143:1985-1992. [DOI] [PubMed] [Google Scholar]
- 67.Yu, C. L., D. J. Meyer, G. S. Campbell, A. C. Larner, C. Carter-Su, J. Schwartz, and R. Jove. 1995. Enhanced DNA-binding activity of a Stat3-related protein in cells transformed by the Src oncoprotein. Science 269:81-83. [DOI] [PubMed] [Google Scholar]
- 68.Zhong, M., M. A. Henriksen, K. Takeuchi, O. Schaefer, B. Liu, J. T. Hoeve, Z. Ren, X. Mao, X. Chen, K. Shuai, and J. E. Darnell, Jr. 2005. Implications of an antiparallel dimeric structure of nonphosphorylated STAT1 for the activation-inactivation cycle. Proc. Natl. Acad. Sci. USA 102:3966-3971. [DOI] [PMC free article] [PubMed] [Google Scholar]