Abstract
We examined the population dynamics of human immunodeficiency virus type 1 pro variants during the evolution of resistance to the protease inhibitor ritonavir (RTV) in vivo. pro variants were followed in subjects who had added RTV to their previously failed reverse transcriptase inhibitor therapy using a heteroduplex tracking assay designed to detect common resistance-associated mutations. In most cases the initial variant appeared rapidly within 2 to 3 months followed by one or more subsequent population turnovers. Some of the subsequent transitions between variants were rapid, and some were prolonged with the coexistence of multiple variants. In several cases variants without resistance mutations persisted despite the emergence of new variants with an increasing number of resistance-associated mutations. Based on the rate of turnover of pro variants in the RTV-treated subjects we estimated that the mean fitness of newly emerging variants was increased 1.2-fold (range, 1.02 to 1.8) relative to their predecessors. A subset of pro genes was introduced into infectious molecular clones. The corresponding viruses displayed impaired replication capacity and reduced susceptibility to RTV. A subset of these clones also showed increased susceptibility to two nonnucleoside reverse transcriptase inhibitors and the protease inhibitor saquinavir. Finally, a significant correlation between the reduced replication capacity and reduced processing at the gag NC-p1 processing site was noted. Our results reveal a complexity of patterns in the evolution of resistance to a protease inhibitor. In addition, these results suggest that selection for resistance to one protease inhibitor can have pleiotropic effects that can affect fitness and susceptibility to other drugs.
Protease inhibitors (PIs) potently inhibit human immunodeficiency virus type 1 (HIV-1) replication, and their initial introduction into combination antiretroviral therapy was associated with a significant decrease of HIV-1-associated mortality and morbidity (41, 45). However, in clinical practice, virologic failure of protease inhibitor-containing therapy occurs in a significant fraction of the treated population (19, 22, 42, 46, 52, 70). Virologic failure of protease inhibitor treatment is often due to the selection of HIV-1 strains with mutations in the pro gene, which encodes the viral protease (PR), and in specific protease cleavage sites encoded in the gag gene that collectively confer reduced drug susceptibility to the harboring strain (11, 12, 28, 36, 43, 47, 72).
The resistance mutations are hypothesized to be largely preexisting, generated by the highly error-prone and rapid replication of HIV-1 in large virus populations (5, 10, 57). However, in PI-naïve subjects, some critical resistance mutations cannot be detected or can only be detected at very low levels, suggesting that these mutations may confer a selective disadvantage relative to wild-type HIV-1 strains (3, 18, 25, 32, 34, 63). Indeed, it has been shown that the catalytic efficiency of protease is reduced by PI resistance-associated mutations, that polyprotein processing in strains bearing resistance mutations is incomplete, and that replication capacity, fitness, and infectivity are reduced compared to those in drug-susceptible strains (4, 9, 13, 18, 27, 29, 36, 39, 40, 44, 51, 58, 62, 64, 71). Some mutations in pro and protease cleavage sites in gag have been found to partially compensate for the loss of fitness and enzymatic activity (12, 36, 40, 62, 72).
In vivo, PI resistance mutations accumulate in an ordered fashion (18, 43, 72). However, the population genetics that underlies this macroscopically observed accumulation of mutations in pro remains poorly understood. In the present study, we characterized the population genetics of pro variants during the evolution of ritonavir (RTV) resistance in subjects who participated in an RTV efficacy trial (7) by using a multiple site-specific heteroduplex tracking assay (MSS-HTA) capable of detecting point mutations at positions 46, 48, 54, 82, 84, and 90 (55). This method allows pro genotypes with different numbers of mutations, mostly resistance related, to be quantitated. Moreover we analyzed the drug susceptibility and replication capacity of molecular clones with pro sequences derived from treated subjects at different times. These studies reveal complexity in the patterns of evolution of RTV resistance and demonstrate pleiotropic effects of these resistance mutations on viral fitness and susceptibility to other inhibitors. Finally, we correlated the loss in fitness with underprocessing of the NC-p1 protease cleavage site.
MATERIALS AND METHODS
Blood plasma samples.
Blood plasma samples were obtained with institutional review board approval unlinked to subject identifiers and as excess tissue samples. The blood plasma samples are from subjects who participated in the virology substudy of the ritonavir efficacy trial reported by Cameron et al. (7) and were provided by Dale Kempf (Abbott Laboratories). In this trial, ritonavir therapy was initiated in protease inhibitor-naïve subjects while maintaining their ongoing failed nucleoside reverse transcriptase inhibitor regimen. Under these circumstances the added RTV approximated monotherapy and most subjects experienced only a transient reduction in virus load followed by rebound. All subjects had CD4+ T-cell counts below 100/μl at entry.
RNA isolation, RT-PCR, and heteroduplex tracking assay.
Procedures for RNA isolation, reverse transcriptase PCR (RT-PCR), and the HTA were carried out as described previously (55). Briefly, viral RNA was isolated from 140 μl of blood plasma or from a pellet containing virus particles obtained by centrifugation of 1 ml of blood plasma (25,000 × g, 1.5 h). The RNA was purified using the QIAmp Viral RNA kit (QIAGEN) according to the manufacturer's instructions. All nucleotide numbering is based on the HIV-1 Hxb molecularly cloned genome. Primers PRAMPUP (5′-[Hxb2305]-AACTAAAGGAAGCTCTATTAGATACAGGAG-[Hxb2334]-3′) and PRAMPDW (5′-[Hxb2551]-GGAAAATTTAAAGTGCAACCAATCTGA-[Hxb2525]-3′) were used for reverse transcription (avian myeloblastosis virus reverse transcriptase, 42°C, 45 min) and PCR (1 cycle of 95°C for 3 min, 56°C for 60 s, and 68°C for 60 s; 40 cycles of 95°C for 40 s, 56°C for 60 s, and 68°C for 60 s to 4 min) to obtain an amplified product spanning codons 18 to 99 of the pro gene. For the heteroduplex tracking assay, 5-μl aliquots of the unpurified PCR-amplified DNA fragments were mixed with 0.1 μg 33P-labeled, multiple site-specific probe 6.1 (for sequence, see reference 55) in annealing buffer (100 mM NaCl, 10 mM Tris-HCl, pH 7.5, and 2 mM EDTA) in a total volume of 10 μl. This mixture was heat denatured, and DNA strands were allowed to anneal at room temperature to form probe-target heteroduplexes. The heteroduplexes were subjected to native polyacrylamide gel electrophoresis (PAGE) (12%; acrylamide-bisacrylamide ratio = 61.5:1) in 1× Tris-borate-EDTA gel buffer (60). After separation, the heteroduplexes were visualized by autoradiography. Quantitative analysis was performed by storage phosphor autoradiography with a Storm 840 PhosphorImager using the ImageQuant software (Molecular Dynamics).
Estimation of relative fitness of variants during ritonavir treatment.
Relative fitness of two variants or one variant compared to all other variants present concurrently was estimated as described previously (20). In brief, the logarithm of the ratio of the abundance of two variants (ln[variant 1/variant 2]) is plotted against time in generations. The slope of this line is an estimate of the selective advantage of variant 1 (s). The fold change of the ratio per generation (eslope) is referred to as relative fitness.
Sequence determination.
The sequence of virus variants corresponding to HTA bands was determined by sequencing a PCR product directly if, by HTA, there was only a single variant present in the amplified product. If multiple variants were present, the amplified DNA fragment was cloned into the PCR cloning vector pT7blue (Novagen), individual clones were screened by HTA and assigned to groups corresponding to HTA bands, and several clones from each group were sequenced. RT-PCR products for sequence determination were either the same as the RT-PCR products used for HTA analysis or were generated using primers USPR (5′-[Hxb2147]-CAGAGCCAACAGCCCCAC-[Hxb2164]-3′) and DSPR (5′-[Hxb2605]-GGGCCATCCATTCCTGGC-[Hxb2588]-3′), which amplify a fragment from the p6 coding region in gag to the 5′ end of the RT coding domain in pol.
Construction of recombinant viral genomes.
DNA fragments spanning nucleotides 2147 to 2605 (Hxb numbering, from the 3′ end of the p6 coding domain to the 5′ end of the RT coding domain) were generated using primers USPR and DSPR from viral RNA prepared from three subjects (three time points each) as described above. In a second round of amplification, this DNA fragment was mixed with a DNA fragment derived from the HIV-1 Hxb cloned genome (54) spanning nucleotides 1994 to 2174, the primers PRBSP (5′-[Hxb1994]-CCAGAAATTGCAGGGCCCC-[Hxb2012]-3′; underlined nucleotides indicate a Bsp120I site), and PRRSR (5′-[Hxb2607]-TCGGACCGTCCAATTCCTGGC-[Hxb2588]-3′; underlined nucleotides indicate a RsrII site) at 2 μM each, in a solution containing 0.2 mM each deoxynucleoside triphosphate (dNTP), 10 mM KCl, 10 mM (NH4)2SO4, 20 mM Tris-HCl (pH 8.8), 2 mM MgSO4, 0.1% Triton X-100, and 2 U of Vent DNA polymerase (New England BioLabs). PCR was performed in a Stratagene 40 Robocycler (1 cycle of 95°C for 3 min, 54°C for 60 s, and 72°C for 90 s; 25 cycles of 95°C for 45 s, 54°C for 60 s, and 72°C for 90 s; 1 cycle of 95°C for 45 s, 54°C for 60 s, and 72°C for 4 min), and the resulting 612-nucleotide fragment, flanked by Bsp120I and RsrII sites, was cloned blunt into pT7blue. Clones were screened by HTA as described above, and the sequence of several clones representing each HTA band was determined. The Bsp120I/RsrII fragment of a clone with a sequence close to the consensus sequence of the corresponding HTA band was ligated into the Bsp120I and RsrII sites of the pCadNΔBspBcl plasmid (S. Pettit and R. Swanstrom, unpublished). This molecular clone contains the XbaI (Hxb1089; p17 coding domain)-to-EcoRV (Hxb2980; RT coding domain) fragment of Hxb within the NL4-3 backbone (1). The Bsp120I (Hxb2007)-to-BclI (Hxb2430) fragment has been deleted from this clone. From the three subjects, a total of 10 infectious clones were constructed.
Drug susceptibility determination and single-cycle replication capacity assay.
Single-cycle replication capacity and drug susceptibility were measured using a replication-incompetent HIV-1 vector in which a luciferase gene has been inserted into a deleted portion of the env gene as described previously (8, 50). The patient-derived sequences in these vectors span nucleotides 2147 to 2605 (Hxb numbering) as described in the previous section.
Virus stocks.
For transfection, 293 cells were seeded at 4 × 105 cells/well in six-well plates. One day after seeding, the cultures were transfected with 0.8 μg of plasmid DNA using Effectene reagents (QIAGEN) according to the manufacturer's instructions. Virus-containing supernatants were collected 48 h after transfection, clarified by centrifugation for 1 min at 3,000 × g, and stored in aliquots at −80°C.
RT activity assay.
The activity of reverse transcriptase in culture supernatants was measured as described previously (6) with the following modifications. The radionucleotide incorporated into the nascent DNA strand was 33P, a vacuum manifold (Bio-Rad) was used to capture the newly synthesized DNA on anion-exchange paper, and storage phosphor autoradiography (Storm 840 PhosphorImager; Molecular Devices) was used to quantitate incorporated radioactivity.
Quantitative determination of capsid protein p24.
The amount of the p24 capsid protein (CA) in transfection supernatants was measured with an enzyme-linked immunosorbent assay using a commercially available set of reagents (New England Nuclear) according to the manufacturer's instructions.
Specific infectivity assay.
Virus stocks were generated by transfection of plasmid DNA into 293 cells in triplicate. For each transfection, RT activity and the amount of CA were measured as described above. Virus titers in transfection supernatants were determined with the multinuclear activation of a galactosidase indicator (MAGI) assay (31). Specific infectivity was calculated as the number of infectious units per ng of CA or per unit of RT activity and normalized to the parental strain.
Analysis of Gag processing.
A 500-μl aliquot of transfection supernatant containing HIV-1 virions was centrifuged at 25,000 × g for 1.5 h in a microcentrifuge. Pellets containing virus particles were resuspended and lysed in 50 μl sodium dodecyl sulfate (SDS) sample buffer (45 mM Tris-HCl, pH 8.45, 12% [vol/vol] glycerol, 4% [wt/vol] sodium dodecyl sulfate, 50 mM dithiothreitol, 0.0075% Coomassie blue G, 0.0025% phenol red). A 5-μl aliquot of the lysed virus particles was subjected to denaturing SDS-PAGE as described previously (61). Proteins were transferred to polyvinylidene difluoride membrane (HybondP; Amersham) and viral proteins were detected immunologically with either a polyclonal rabbit anti-p15 antibody (provided by S. Erickson-Viitanen) or an anti-HIV-1 human antiserum, using enhanced chemiluminescence (ECLplus; Amersham). ECLplus reagents allow the detection of luminescence as well as blue-excited fluorescence (Storm 840 PhosphorImager; Molecular Devices) for quantitation.
Statistical analysis.
Statistical analysis was carried out with the R language (http://www.r-project.org). Associations between different parameters were estimated with Spearman's correlation coefficient ρ or linear regression. The significance of differences in mean values was estimated with Student's t test.
Nucleotide sequence accession number.
All sequences have been submitted to GenBank and have been assigned the consecutive accession numbers AY678610 to AY679067.
RESULTS
Dynamics of protease variants during the evolution of ritonavir resistance.
We employed an MSS-HTA capable of detecting mutations at HIV-1 protease codon positions 46, 48, 54, 82, 84, and 90 (55) to investigate the population dynamics of pro variants underlying the evolution of RTV resistance in a set of subjects failing RTV therapy. The MSS-HTA separates heteroduplexes formed between RT-PCR products of pro variants and a radioactively labeled DNA probe in a native polyacrylamide gel largely by changes at these target positions and allows reliable quantitation of the frequency of protease alleles exceeding an abundance of 3% (55). All amplifications and MSS-HTAs were carried out in duplicate, and any samples with discordant HTA banding patterns were excluded from this study. This assured that the amplifications contained enough template strands to form a representative sample of the total population.
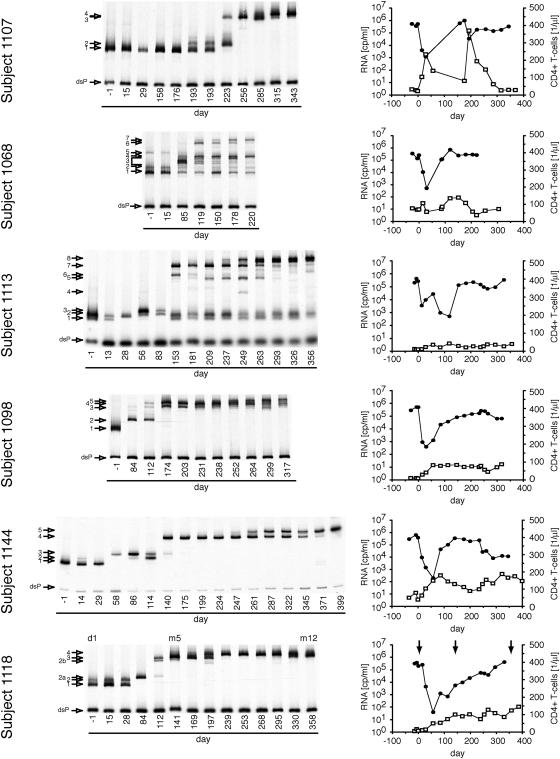
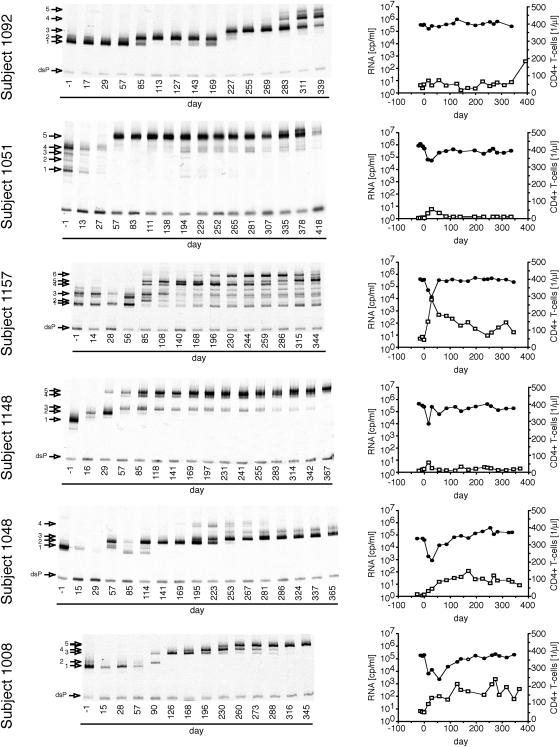
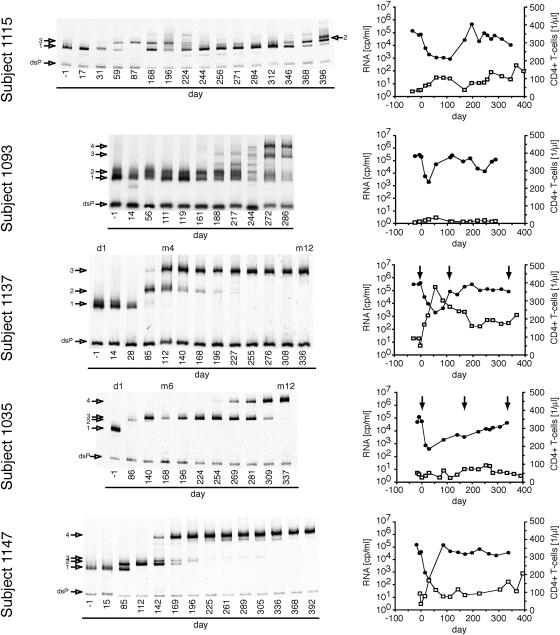
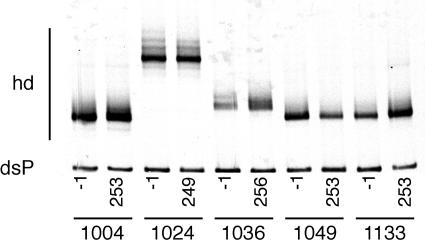
Figures 1 through 3 summarize the patterns of evolution observed in 17 subjects on RTV, as well as the changes in HIV-1 RNA load and CD4+ T-cell levels over the course of the treatment period. Figure 1 shows subjects with a 300-fold or greater initial reduction of virus load in response to RTV therapy, Fig. 2 shows subjects with a 100- to 300-fold initial reduction of virus load, and Fig. 3 shows subjects with a 2- to 100-fold initial reduction of virus load. All subjects, irrespective of the magnitude of virus load reduction, displayed one to several steps of adaptive evolution following the introduction of RTV. In most subjects, the initial change in the virus population was concurrent with virus load rebound or nadir and occurred a mean of 90 days (range, 29 to 193 days) after treatment initiation. The timing and rates of population turnovers as well as the number of detectable variants present concurrently (range, 1 to 7; for example, compare subjects 1157 and 1148 in Fig. 1) varied considerably between subjects. No sequence changes were detected by the MSS-HTA (Fig. 4) and sequence analysis (data not shown) in matched subjects on the placebo arm of the same study over the same period of time.
FIG. 1.
Dynamics of protease variants during ritonavir resistance evolution in subjects with a transient virus load reduction of greater than 300-fold. Dynamics were analyzed with a multiple site-specific HTA which displays mobility shifts in response to mutations at positions 46, 48, 54, 82, 84, and 90 in protease. Each band represents a protease variant and is assigned a number on the left. The position of the labeled probe strand annealed to a fully complementary strand is indicated (double-stranded probe; dsP). In addition, the viral load (filled circle) and the concentration of CD4+ T cells (open squares) are shown for each subject in the right column. Arrows in the viral load graphs indicate protease alleles that were introduced into molecular clones.
FIG. 3.
Dynamics of protease variants during ritonavir resistance evolution in subjects with a transient virus load reduction of more than 2- to 100-fold. Otherwise as described in the legend to Fig. 2.
FIG. 2.
Dynamics of protease variants during ritonavir resistance evolution in subjects with a transient 100- to 300-fold virus load reduction. Otherwise as described in the legend to Fig. 1.
FIG. 4.
Dynamics of protease variants in the absence of ritonavir analyzed by a multiple site-specific HTA. For five subjects, the protease populations at the beginning of the trial (day −1) and the end of the blinded phase (around day 250) were compared. No significant changes were detected over the observation period. hd, heteroduplex; dsP, double-stranded probe.
Maintenance of populations without primary resistance mutations.
In 6 of the 17 studied subjects (1051, 1157, 1115, 1093, 1068, and 1113) detectable amounts of pro variants with MSS-HTA mobility identical to pretreatment variants were maintained late in the trial period in the face of the emergence of variants with considerable mobility shifts. Sequence analysis of clones representing each HTA band showed that the high-mobility bands represented variants that were closely related or identical to pretreatment variants and had not acquired any known RTV resistance mutations over the course of treatment. However, the low-mobility bands represented variants that had acquired RTV resistance mutations. This indicated that variants with low resistance could be maintained at a level of 4 to 20% in the face of the evolution of more resistant variants under selective pressure from RTV.
Sequence evolution of pro variants.
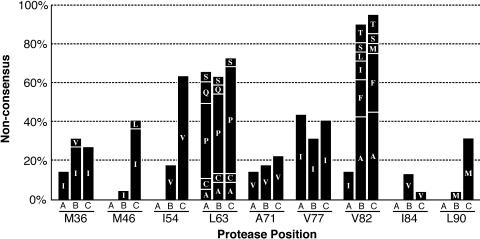
The sequence corresponding to pro codons 30 to 95 of variants corresponding to a number of MSS-HTA bands was determined. For homogenous populations, the RT-PCR product was sequenced directly. If more than one pro variant was present, RT-PCR products were cloned and several clones corresponding to each MSS-HTA band were subjected to sequencing. As has been described previously for subjects failing RTV therapy (18, 43), this first step of evolution included a mutation at position 82 in all 17 virus variants that emerged between days 29 and 120 after treatment start (Fig. 5). The I54V mutation was present in approximately 20% of early rebound variants as well. Subsequent turnovers were associated with the emergence of strains with additional mutations at positions 46, 54, 84, and 90 (Fig. 5).
FIG. 5.
Changes in the coding sequence of protease over the course of treatment. The sequences of 53 protease variants were determined as the consensus sequence of multiple clones per variant. Indicated in this stacked bar graph are the percentage of variants that showed differences with the clade B consensus sequence at the indicated resistance-associated positions. The first bar (A) in each group reflects amino acid changes present at trial entry, the second bar (B) reflects the amino acid composition of variants that arose between entry and day 120, and the third bar (C) reflects the composition of variants that arose after day 120. Letters indicate mutant amino acids. Note the increase of mutations at positions 82, 46, 54, 84, and 90.
Each new MSS-HTA band represented a variant with a mean of 1.8 new coding mutations compared to variants present at the preceding time point. A mean of 1.3 of these mutations was found at positions associated with reduced RTV susceptibility (positions 36, 46, 54, 71, 77, 82, 84, 90). The cumulative number of RTV resistance-associated mutations increased significantly with time (ρ = 0.58; P < 0.0001), but the number of mutations outside these positions did not. The rate of accumulation was significantly higher during the first 120 days of treatment (0.035 mutation per day) than during the remainder of the trial period (0.016 mutation per day; P = 0.047).
Evolution of viral fitness in vivo.
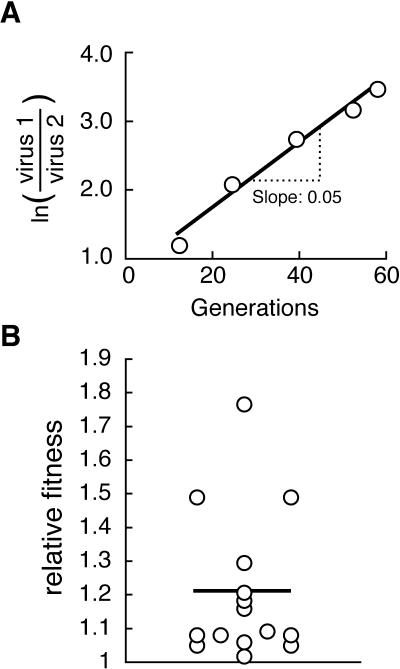
The relative fitness of newly emerging pro variants was determined using a model proposed by Holland et al. (27a). In this model, the slope of the line obtained by plotting the logarithm of the ratio of two variants or one variant and all other variants against time in generations is interpreted as the selective advantage of the variant (20). The relative fitness, or fold change of the ratio per generation, is equal to eslope. A representative example for this analysis is shown in Fig. 6A. When the analysis was carried out for all emerging variants that coexisted with the preceding variants for more than two consecutive time points, the mean relative fitness of new variants compared to the preceding variants was 1.2 (range, 1.02 to 1.76; Fig. 6B). Note that this analysis excluded the first step of resistance evolution due to its speed, which did not allow for coexisting variants. Note also that the estimate used for the HIV-1 generation time was 2.3 days (26, 38, 48, 49, 53, 68).
FIG. 6.
(A) Determination of relative fitness of pro variant 5 in subject 1008. The logarithm of the ratio of variant 5 (virus 1) and the other variants present at the same time (virus 2) was plotted against time in generations. The rate of change was determined by linear regression, and eslope was computed as an estimate for the relative fitness of variant 5 compared to the other variants present. (B) Fold fitness increases associated with the steps of adaptive evolution. Relative fitness of all newly emerging pro variants was determined in vivo as described above and plotted as a scatter graph. The mean of all observations is shown as a horizontal line.
Evolution of drug susceptibility.
To examine the evolution of phenotypes, a fragment spanning protease from the 3′ end of p6 to the 5′ end of reverse transcriptase was amplified from three time points from each of three subjects and a single representative sequence for each HTA band present in the samples was selected and subjected to drug susceptibility testing as described previously (50). In Table 1 the fold susceptibility reduction is shown for all tested drugs. In all cases but one, the drug susceptibility of the entry sample was within twofold of the NL4-3 control for all tested inhibitors.
TABLE 1.
Resistance profile of infectious clones
| Clone | Genotype | Fold reduction of susceptibilitya
|
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RTV | SQV | APV | NFV | IDV | LPV | NVP | EFV | DLV | ABC | ADV | ddI | 3TC | d4T | TFV | ddC | ZDV | ||
| 1035d1-3 | T12S K43R L63P | 0.5 | 0.4 | 0.5 | 0.6 | 0.5 | 0.5 | 1.1 | 0.5 | 1.5 | 0.6 | 0.7 | 0.8 | 0.7 | 0.8 | 0.8 | 0.9 | 0.7 |
| 1035m6-5 | T12S K43R M46I L63P V82F | 14.8 | 0.2 | 2.2 | 7.4 | 2.3 | 4.5 | 0.2 | 0.3 | 0.6 | 0.5 | 0.7 | 0.8 | 1.0 | 0.8 | 0.7 | 0.9 | 0.5 |
| 1035m12-10 | T12S K43R M46I I54V Q61H L63P V82F | 30.8 | 0.1 | 1.7 | 19.2 | nd | 5.8 | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd |
| 1118d1-15 | N37S L63P V77I I93L | 1.2 | 0.9 | 1.0 | 1.9 | 1.4 | 0.9 | 0.6 | 0.6 | 1.6 | 0.8 | 0.8 | 0.9 | 0.9 | 1.0 | 0.9 | 1.0 | 0.7 |
| 1118m5-1 | N37S I54V L63P V77I V82T I93L | 6.5 | 0.6 | nd | 2.3 | 2.3 | 3.3 | 0.6 | 0.4 | 0.6 | 0.7 | 0.8 | 1.0 | 0.6 | 1.1 | 0.6 | 0.8 | 0.4 |
| 1118m12-6 | L33F E34Q N37S I54V Q58E L63P A71V V771 V82A L90M I93L | 59.5 | 1.8 | 1.6 | 9.0 | 4.1 | 2.9 | 0.4 | 0.4 | 1.0 | 0.7 | 0.8 | 0.8 | 0.9 | 0.9 | 0.8 | 1.0 | 0.6 |
| 1137d1-12 | N37E D60E I64V | 1.2 | 1.0 | 1.1 | 1.6 | 1.1 | 1.0 | 0.7 | 0.7 | 1.8 | 0.8 | 0.8 | 0.9 | 0.9 | 0.9 | 0.8 | 0.9 | 0.7 |
| 1137M4-9 | N37E I54V D60E 164V V82A | 3.4 | 0.4 | 0.3 | 0.5 | 0.6 | 1.2 | 0.4 | 0.4 | 0.4 | 0.6 | 1.1 | 0.7 | 1.8 | 1.5 | 0.9 | 0.8 | 0.6 |
| 1137m4-13 | N37E F53I D60E I64V V82A | 7.7 | 0.7 | 1.1 | 1.8 | 1.6 | 1.7 | 0.4 | 0.4 | 1.0 | 0.6 | 0.6 | 0.8 | 0.8 | 0.8 | 0.7 | 0.9 | 0.5 |
| 1137m12-6 | L24I N37E M46I I54V D60E L63P I64V V82A | 33.9 | 1.3 | 1.6 | 6.7 | 6.9 | 13.6 | 0.2 | 0.4 | 0.7 | 0.6 | 0.7 | 0.8 | 0.7 | 0.8 | 0.8 | 0.8 | 0.5 |
| wtb | N37S | 0.6 | 0.6 | 0.6 | 0.5 | 0.5 | 0.6 | 0.5 | 0.6 | 1.4 | 0.8 | 0.8 | 0.8 | 0.9 | 0.9 | 0.8 | 0.9 | 0.6 |
Relative to NL4-3. RTV, ritonavir; SQV, saquinavir; APV, amprenavir; NFV, nelfinavir; IDV, indinavir; LPV, lopinavir; NPV, nevirapine; EFV, efavirenz; DLV, delavirdine; ABC, abacavir; ADV, adefovir; ddI, didanosine; 3TC, lamivudine; d4T, stavudine; TFV, tenofovir; ddC, zalcitabine; ZDV, zidovudine. Underlined numbering indicates greater than 2.5-fold deviation from control before rounding; italics indicates low luciferase activity resulting in less reliable measurements. nd, not determined (low luciferase activity).
Parental strain for generation of infectious clones.
RTV resistance increased in successive time points and was associated with significant cross-resistance to lopinavir (P < 0.0001), nelfinavir (P = 0.003), and indinavir (P = 0.0001). Hypersusceptibility (50% inhibitory concentration of <0.4) to two protease inhibitors (saquinavir and amprenavir) was observed with sequences derived from two subjects. Note, however, that only one sequence displayed amprenavir hypersusceptibility and that this sequence also displayed low overall luciferase activity in the target cells, making the measured drug susceptibility less reliable. Hypersusceptibility to either of the two nonnucleoside reverse transcriptase inhibitors nevirapine (P = 0.01) and efavirenz (P = 0.02) was associated with RTV resistance. Note that only the first 18 amino acids of reverse transcriptase in the resistance assays were encoded by patient-derived sequences, suggesting that the observed RT inhibitor hypersusceptibility is attributable to the changes in protease.
Evolution of replication capacity and specific infectivity in the absence of RTV.
The protease sequences tested for drug susceptibility were introduced into infectious molecular clones and used to test specific infectivity (infectious units [IU]/ng CA or IU/RT activity) and RT/CA ratio in supernatant virus particles. In addition, the vector system used for drug susceptibility testing was also employed to determine replication capacity during a single replication cycle (luciferase activity in infected cells/luciferase activity in transfected cells). We have shown previously that these single-cycle assays can reflect relative fitness accurately (56). The results of these analyses are summarized in Fig. 7.
FIG. 7.
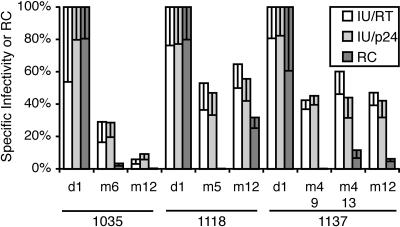
Specific infectivity and replication capacity of clones bearing protease sequences derived from study subjects. A protease sequence representative of each variant present at the beginning (day 1 [d1]), a time point close to virus load rebound (month 4 [m4], m5, or m6), and the last available time point (m12) in subjects 1035, 1118, and 1137 was introduced into a molecular infectious clone. To determine specific infectivity, molecular clones were transfected into 293 cells in triplicate, virus supernatants were collected, and the titer, the reverse transcriptase activity, and the concentration of the capsid protein were determined. Specific infectivity is given relative to the parental strain as the ratio of titer to RT activity (open bars) or the ratio of titer to capsid protein (light gray bars). Replication capacity (RC [dark gray bars]) was determined in a single-cycle replication assay with a luciferase-encoding pseudotyped vector system as described previously (8, 50).
There were two patterns of evolution of specific infectivity. In one subject (1035), specific infectivity decreased with the initial rebound of virus load and with subsequent selection of higher resistance in the face of continued ritonavir selection. In the other two subjects, specific infectivity was reduced at the time of virus load rebound but remained stable during subsequent decreases in ritonavir susceptibility. Wild-type-like specific infectivity was not restored in any subject by mutations in the pro-spanning fragment used in this experiment. No significant differences between the RT/CA ratios were found between the infectious clones with patient-derived protease sequences. Single-cycle replication capacity, as measured with the resistance-testing vectors, showed similar patterns, but in several cases the absolute values were lower for pro sequences carrying resistance mutations when compared to the specific infectivity assay, as has been noted previously (56).
We observed a negative correlation between reduced RTV susceptibility and specific infectivity for 9 of the 11 clones in this study (ρ = −0.83; P = 0.008). The two clones that displayed the greatest reduction of ritonavir resistance did not fit this pattern. This may suggest that the protease sequences in these clones had undergone some compensation that attenuated the negative fitness effect of further resistance mutations. Note that all fitness measurements were carried out in the absence of protease inhibitors and that by definition resistant variants have a selective advantage in the presence of drug.
Evolution of sequence changes in Gag.
The evolution of high-level resistance to protease inhibitors can be accompanied by compensatory mutations in the processing sites flanking the Gag p1 domain (17, 72). We determined near-full-length sequence for the gag gene at entry and day of termination for five subjects who displayed multistep evolution of the viral pro gene (1137, 1035, 1098, 1144, 1147). In three cases the p2 (the position 2 amino acid upstream of the cleavage site) Ala of the NC-p1 processing site was mutated to Val in comparing the entry to the termination sequences; in one case the p1′ (the position just downstream of the cleavage site) Leu of the p1/p6 site was mutated to Phe (1144), and in the remaining case there were no processing site mutations (1147). These observations are consistent with the evolution of high-level resistance while the subjects were maintained on RTV therapy after virus rebound.
It has also been reported that nonprocessing site mutations are selected in the gag gene during selection for protease inhibitor resistance in cell culture using four different inhibitors (21). We have confirmed this observation in part by examining viruses that were selected in cell culture for high levels of resistance to protease inhibitors (67). However, in examining the gag genes of the five subjects cited above, we found no evidence for a similar pattern of mutations after selection of protease inhibitor resistance in vivo.
The extent of processing at the NC-p1 junction correlates with specific infectivity.
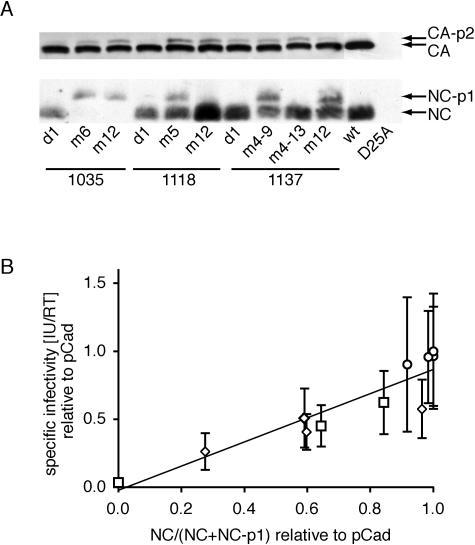
The processing of the polyproteins Gag and Gag-Pro-Pol by protease is an obligatory step in the formation of infectious particles. We analyzed the processing of the NC-p1 and CA-p2 sites which have been implicated in contributing to the loss of fitness associated with protease inhibitor resistance. Incomplete processing of the CA-p2 and NC-p1 cleavage sites was apparent (Fig. 8A). The impact of the resistance mutations on CA-p2 processing was subtler than the impact on NC-p1 processing. We hypothesized that the aberrant processing of cleavage sites flanking NC was closely linked to the observed loss of fitness, consistent with these sites being the most frequently mutated cleavage sites in Gag-Pro-Pol (12, 36, 72). In agreement with this hypothesis, there was a significant correlation (ρ = 0.94; P < 0.0001) between the specific infectivity and the fraction of NC-p1 sites processed (Fig. 8B).
FIG. 8.
Correlation of the fraction of processed NC-p1 sites and specific infectivity. (A) Virions were concentrated by purification, and pelleted virus proteins were separated by SDS-PAGE, blotted to polyvinylidene difluoride membranes, and detected immunologically with human serum from an HIV-1-infected person (top blot) or a polyclonal anti-NC-p1-p6 antibody. Mutant proteases are described by subject of origin and time point of sampling. wt, parental infectious clone; D25A, parental infectious clone with the protease-inactivating D25A mutation as a negative control for processing. (B) The fraction of processed NC-p1 sites was normalized to the parental strain and correlated with the specific infectivity (see Fig. 7) for each variant. Also shown is the best-fit linear regression line.
DISCUSSION
We investigated the population genetics of ritonavir resistance evolution in subjects who participated in a ritonavir efficacy trial (7) using an MSS-HTA (55). The MSS-HTA revealed multiple steps of sequence evolution among all of the examined subjects on the RTV treatment arm (Fig. 1 to 3) and did not detect any sequence changes among five subjects on the placebo arm (Fig. 4). Thus the MSS-HTA proved to be a sensitive tool for resolving pro sequence populations during the evolution of RTV resistance. One advantage of this pro MSS-HTA was that distinct sequence variants were separated based on linked sequence differences, allowing the quantitation of discrete genotypic variants. However, consistent with previous observations (data not shown), the initial mutation in a pro variant generally resulted in a larger heteroduplex mobility shift than subsequent mutations occurring in the background of an already mutated protease.
Sequence analysis showed that each new protease variant that emerged carried a mean of 1.8 (range 1 to 4) new coding mutations compared to previously existing variants, 1.4 (range 1 to 3) of which were at positions previously implicated in ritonavir resistance. We were not able to determine if more than one mutation emerged in a single step or if mutations accumulated one at a time. The mutations observed here match those previously described both in sequence identity and in the order of accumulation (18, 43). The accumulation rate of resistance-associated mutations was highest following the onset of RTV therapy and with time asymptotically approached an estimated 0.016 mutation/day. Each of the evolutionary steps was associated with a mean 1.2-fold increase in relative fitness in the presence of RTV. This is an underestimate of the actual gains because the analysis excluded for technical reasons turnovers that occurred rapidly without displaying any coexistence of the emerging variant with the preceding variant. This analysis assumes an excess of target cells in vivo; however, the validity of this assumption is not known.
We demonstrated the maintenance of detectable levels of drug-susceptible HIV-1 variants concurrently with the evolution of resistant strains in 6 of 17 subjects. The high degree of RTV susceptibility of these variants was inferred from the unchanged MSS-HTA mobility as well as by the absence of known resistance mutations from protease codons 30 to 95. Mutations outside this region are generally considered compensatory for the mutations in this fragment of protease. Moreover, in some cases complete protease sequences were available and did not show any resistance-associated mutations in the strains with high MSS-HTA mobility. Thus we believe that we are observing the persistence of nonresistant variants in a mixture with resistant variants in up to one-third of the subjects. There are several possible explanations for the persistence of wild-type-like pro alleles concurrently with alleles with reduced drug susceptibility. We do not believe that these sequences represent archived viral genomes present in contaminating DNA given their steady levels in the longitudinal samples from specific subjects and complete absence in others. Therefore, there must be one or more mechanisms for the maintenance of actively replicating parental sequences in the face of drug selective pressure. First, poor compliance or pharmacokinetic issues may lead to intermittently suboptimal RTV concentrations allowing residual replication of more highly susceptible variants. In two cases (subjects 1115 and 1113) the reappearance of a variant with wild-type mobility and the viral load changes suggest that poor compliance might indeed account for the persistence of the wild-type pro sequences. Second, variants with high RTV susceptibility could be replicating in RTV-inaccessible compartments, such as the central nervous system (23, 33, 66). And third, the fitness loss associated with the evolution of resistance in the sequence background of these pro genes may have resulted in fitness levels that were similar to those of the wild type in the presence of RTV. The patterns of evolution in the four remaining subjects in which pro variants with resistance-associated mutations evolved with rates comparable to those of the rest of the 17 subjects despite the maintenance of wild-type variants are consistent with the latter two possibilities.
Susceptibility to ritonavir decreased during the treatment period, as expected. Other protease inhibitors with overlapping resistance mutation profiles had reduced activity against the ritonavir-resistant strains as well. However, susceptibility to two protease inhibitors, saquinavir and amprenavir, increased in three of the analyzed variants that had significant reduction of RTV susceptibility. A more consistent negative correlation between RTV resistance and susceptibility to the reverse transcriptase inhibitors nevirapine and efavirenz was observed as well. One possible explanation is that the hypersusceptibilities were due to a reduction of virion-associated protease activity and hence reduced fitness. In this model, the reduced protease activity in the virus particles would lead to incomplete processing of the Gag-Pro-Pol polyprotein leading to hypersusceptibility to a tightly binding reverse transcriptase inhibitor, in a way that is analogous to the observation that resistance to a protease inhibitor increases susceptibility to zidovudine (16, 35). The same model could also be used to explain the hypersusceptibility to two of the protease inhibitors where the selected mutations did not provide cross-resistance. However, we cannot exclude the possibility that some RTV resistance-associated mutations simply enhance binding of some inhibitors, as has been suggested for some reverse transcriptase inhibitors (69).
Specific infectivity (a surrogate marker for fitness) of recombinant virus strains bearing pro sequences derived from three subjects after treatment was reduced to between 4 to 65% of the pretreatment infectivity (Fig. 7). Even after up to 12 months of RTV treatment and in the presence of several pro mutations that could be considered compensatory in nature, none of the resistant variants recovered pretreatment-specific infectivity levels, consistent with observations made by others (2). The reversion of resistant virus populations to drug-susceptible populations even after prolonged treatment failure (≥12 months) also suggests that compensation is indeed incomplete (15, 18, 24, 30). The infectivity measurements presented here are consistent with the infectivity of single-, double-, and triple-mutant strains described by Mammano et al. (37) but lower than the fitness of clones harboring multiple resistance mutations measured by coculture experiments (40, 44). This difference may be partially accounted for by compensatory alanine-to-valine mutations at the P2 position of the NC-p1 cleavage site seen in two of the three subjects tested here at trial termination (subjects 1035 and 1137) but not included in the sequences used to test replication capacity. In contrast to previously reported findings (16), we did not detect significant differences in the amount of particle-associated RT activity in the infectious clones presented here.
We found the processing of Gag to be incomplete in infectious clones harboring protease sequences derived from the RTV-treated subjects. This has been noted previously (13, 14, 59, 71). However, here we showed that the reduction in processing of the NC-p1 cleavage site correlated significantly with the reduction in specific infectivity (Fig. 8). This is consistent with the finding that mutations in the protease cleavage sites at NC-p1 and p1-p6 are selected in vivo by a protease inhibitor-containing regimen (12, 17, 37, 72) and that these mutations can partially compensate for the loss of fitness associated with protease resistance mutations (37, 72).
It will be important to examine a larger set of clinical isolates with resistance to protease inhibitors to confirm the correlation between incomplete NC-p1 processing and decreases in fitness of these viruses. Loss of fitness through this mechanism may contribute to the disconnect phenotype in which CD4+ cell gains are maintained in the face of an actively replicating drug-resistant virus variant (2, 15, 65). The mechanism by which underprocessed NC-p1 reduces infectivity remains to be explored.
Acknowledgments
This work was supported by NIH grants R01-AI25321 and R01-AI50485 to R.S. and by the UNC Center for AIDS Research (NIH P30-AI50410).
S. Erickson-Viitanen graciously provided the anti-p15 antiserum.
REFERENCES
- 1.Adachi, A., H. E. Gendelman, S. Koenig, T. Folks, R. Willey, A. Rabson, and M. A. Martin. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59:284-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barbour, J. D., T. Wrin, R. M. Grant, J. N. Martin, M. R. Segal, C. J. Petropoulos, and S. G. Deeks. 2002. Evolution of phenotypic drug susceptibility and viral replication capacity during long-term virologic failure of protease inhibitor therapy in human immunodeficiency virus-infected adults. J. Virol. 76:11104-11112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barrie, K. A., E. E. Perez, S. L. Lamers, W. G. Farmerie, B. M. Dunn, J. W. Sleasman, and M. M. Goodenow. 1996. Natural variation in HIV-1 protease, Gag p7 and p6, and protease cleavage sites within gag/pol polyproteins: amino acid substitutions in the absence of protease inhibitors in mothers and children infected by human immunodeficiency virus type 1. Virology 219:407-416. [DOI] [PubMed] [Google Scholar]
- 4.Bleiber, G., M. Munoz, A. Ciuffi, P. Meylan, and A. Telenti. 2001. Individual contributions of mutant protease and reverse transcriptase to viral infectivity, replication, and protein maturation of antiretroviral drug-resistant human immunodeficiency virus type 1. J. Virol. 75:3291-3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonhoeffer, S., and M. A. Nowak. 1997. Pre-existence and emergence of drug resistance in HIV-1 infection. Proc. R. Soc. Lond. B Biol. Sci. 264:631-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buckheit, R. W., Jr., and R. Swanstrom. 1991. Characterization of an HIV-1 isolate displaying an apparent absence of virion-associated reverse transcriptase activity. AIDS Res. Hum. Retrovir. 7:295-302. [DOI] [PubMed] [Google Scholar]
- 7.Cameron, D. W., M. Heath-Chiozzi, S. Danner, C. Cohen, S. Kravcik, C. Maurath, E. Sun, D. Henry, R. Rode, A. Potthoff, J. Leonard, et al. 1998. Randomised placebo-controlled trial of ritonavir in advanced HIV-1 disease. Lancet 351:543-549. [DOI] [PubMed] [Google Scholar]
- 8.Campbell, T. B., K. Schneider, T. Wrin, C. J. Petropoulos, and E. Connick. 2003. Relationship between in vitro human immunodeficiency virus type 1 replication rate and virus load in plasma. J. Virol. 77:12105-12112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clavel, F., E. Race, and F. Mammano. 2000. HIV drug resistance and viral fitness. Adv. Pharmacol. 49:41-66. [DOI] [PubMed] [Google Scholar]
- 10.Coffin, J. M. 1995. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science 267:483-489. [DOI] [PubMed] [Google Scholar]
- 11.Condra, J. H., D. J. Holder, W. A. Schleif, O. M. Blahy, R. M. Danovich, L. J. Gabryelski, D. J. Graham, D. Laird, J. C. Quintero, A. Rhodes, H. L. Robbins, E. Roth, M. Shivaprakash, T. Yang, J. A. Chodakewitz, P. J. Deutsch, R. Y. Leavitt, F. E. Massari, J. W. Mellors, K. E. Squires, R. T. Steigbigel, H. Teppler, and E. A. Emini. 1996. Genetic correlates of in vivo viral resistance to indinavir, a human immunodeficiency virus type 1 protease inhibitor. J. Virol. 70:8270-8276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cote, H., Z. Brumme, K. Hertogs, B. Larder, and P. R. Harrigan. 2000. Cleavage site mutations in patients with HIV highly resistant to several protease inhibitors. 7th Conf. Retroviruses Opportunistic Infect., abstr. 722.
- 13.Croteau, G., L. Doyon, D. Thibeault, G. McKercher, L. Pilote, and D. Lamarre. 1997. Impaired fitness of human immunodeficiency virus type 1 variants with high-level resistance to protease inhibitors. J. Virol. 71:1089-1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dauber, D. S., R. Ziermann, N. Parkin, D. J. Maly, S. Mahrus, J. L. Harris, J. A. Ellman, C. Petropoulos, and C. S. Craik. 2002. Altered substrate specificity of drug-resistant human immunodeficiency virus type 1 protease. J. Virol. 76:1359-1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deeks, S. G., T. Wrin, T. Liegler, R. Hoh, M. Hayden, J. D. Barbour, N. S. Hellmann, C. J. Petropoulos, J. M. McCune, M. K. Hellerstein, and R. M. Grant. 2001. Virologic and immunologic consequences of discontinuing combination antiretroviral-drug therapy in HIV-infected patients with detectable viremia. N. Engl. J. Med. 344:472-480. [DOI] [PubMed] [Google Scholar]
- 16.de la Carriere, L. C., S. Paulous, F. Clavel, and F. Mammano. 1999. Effects of human immunodeficiency virus type 1 resistance to protease inhibitors on reverse transcriptase processing, activity, and drug sensitivity. J. Virol. 73:3455-3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doyon, L., G. Croteau, D. Thibeault, F. Poulin, L. Pilote, and D. Lamarre. 1996. Second locus involved in human immunodeficiency virus type 1 resistance to protease inhibitors. J. Virol. 70:3763-3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eastman, P. S., J. Mittler, R. Kelso, C. Gee, E. Boyer, J. Kolberg, M. Urdea, J. M. Leonard, D. W. Norbeck, H. Mo, and M. Markowitz. 1998. Genotypic changes in human immunodeficiency virus type 1 associated with loss of suppression of plasma viral RNA levels in subjects treated with ritonavir (Norvir) monotherapy. J. Virol. 72:5154-5164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fatkenheuer, G., A. Theisen, J. Rockstroh, T. Grabow, C. Wicke, K. Becker, U. Wieland, H. Pfister, M. Reiser, P. Hegener, C. Franzen, A. Schwenk, and B. Salzberger. 1997. Virological treatment failure of protease inhibitor therapy in an unselected cohort of HIV-infected patients. AIDS 11:F113-F116. [DOI] [PubMed] [Google Scholar]
- 20.Frost, S. D. W., M. Nijhuis, R. Schuurman, C. A. Boucher, and A. J. L. Brown. 2000. Evolution of lamivudine resistance in human immunodeficiency virus type 1-infected individuals: the relative roles of drift and selection. J. Virol. 74:6262-6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gatanaga, H., Y. Suzuki, H. Tsang, K. Yoshimura, M. F. Kavlick, K. Nagashima, R. J. Gorelick, S. Mardy, C. Tang, M. F. Summers, and H. Mitsuya. 2002. Amino acid substitutions in Gag protein at non-cleavage sites are indispensable for the development of a high multitude of HIV-1 resistance against protease inhibitors. J. Biol. Chem. 277:5952-5961. [DOI] [PubMed] [Google Scholar]
- 22.Grabar, S., C. Pradier, E. Le Corfec, R. Lancar, C. Allavena, M. Bentata, P. Berlureau, C. Dupont, P. Fabbro-Peray, I. Poizot-Martin, and D. Costagliola. 2000. Factors associated with clinical and virological failure in patients receiving a triple therapy including a protease inhibitor. AIDS 14:141-149. [DOI] [PubMed] [Google Scholar]
- 23.Granda, B. W., G. M. Giancarlo, L. L. von Moltke, and D. J. Greenblatt. 1998. Analysis of ritonavir in plasma/serum and tissues by high-performance liquid chromatography. J. Pharmacol. Toxicol. Methods 40:235-239. [DOI] [PubMed] [Google Scholar]
- 24.Hance, A. J., V. Lemiale, J. Izopet, D. Lecossier, V. Joly, P. Massip, F. Mammano, D. Descamps, F. Brun-Vezinet, and F. Clavel. 2001. Changes in human immunodeficiency virus type 1 populations after treatment interruption in patients failing antiretroviral therapy. J. Virol. 75:6410-6417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Havlir, D. V., S. Eastman, A. Gamst, and D. D. Richman. 1996. Nevirapine-resistant human immunodeficiency virus: kinetics of replication and estimated prevalence in untreated patients. J. Virol. 70:7894-7899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ho, D. D., A. U. Neumann, A. S. Perelson, W. Chen, J. M. Leonard, and M. Markowitz. 1995. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature 373:123-126. [DOI] [PubMed] [Google Scholar]
- 27.Ho, D. D., T. Toyoshima, H. Mo, D. J. Kempf, D. Norbeck, C. M. Chen, N. E. Wideburg, S. K. Burt, J. W. Erickson, and M. K. Singh. 1994. Characterization of human immunodeficiency virus type 1 variants with increased resistance to a C2-symmetric protease inhibitor. J. Virol. 68:2016-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27a.Holland, J. J., J. C. de la Torre, D. K. Clarke, and E. Duarte. 1991. Quantitation of relative fitness and great adaptability of clonal populations of RNA viruses. J. Virol. 65:2960-2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jacobsen, H., M. Hanggi, M. Ott, I. B. Duncan, S. Owen, M. Andreoni, S. Vella, and J. Mous. 1996. In vivo resistance to a human immunodeficiency virus type 1 proteinase inhibitor: mutations, kinetics, and frequencies. J. Infect. Dis. 173:1379-1387. [DOI] [PubMed] [Google Scholar]
- 29.Kaufmann, D., M. Munoz, G. Bleiber, S. Fleury, B. Lotti, R. Martinez, W. Pichler, P. Meylan, and A. Telenti. 2000. Virological and immunological characteristics of HIV treatment failure. AIDS 14:1767-1774. [DOI] [PubMed] [Google Scholar]
- 30.Kijak, G. H., V. Simon, P. Balfe, J. Vanderhoeven, S. E. Pampuro, C. Zala, C. Ochoa, P. Cahn, M. Markowitz, and H. Salomon. 2002. Origin of human immunodeficiency virus type 1 quasispecies emerging after antiretroviral treatment interruption in patients with therapeutic failure. J. Virol. 76:7000-7009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kimpton, J., and M. Emerman. 1992. Detection of replication-competent and pseudotyped human immunodeficiency virus with a sensitive cell line on the basis of activation of an integrated β-galactosidase gene. J. Virol. 66:2232-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kozal, M. J., N. Shah, N. Shen, R. Yang, R. Fucini, T. C. Merigan, D. D. Richman, D. Morris, E. Hubbell, M. Chee, and T. R. Gingeras. 1996. Extensive polymorphisms observed in HIV-1 clade B protease gene using high-density oligonucleotide arrays. Nat. Med. 2:753-759. [DOI] [PubMed] [Google Scholar]
- 33.Lafeuillade, A., C. Solas, P. Halfon, S. Chadapaud, G. Hittinger, and B. Lacarelle. 2002. Differences in the detection of three HIV-1 protease inhibitors in non-blood compartments: clinical correlations. HIV Clin. Trials 3:27-35. [DOI] [PubMed] [Google Scholar]
- 34.Lech, W. J., G. Wang, Y. L. Yang, Y. Chee, K. Dorman, D. McCrae, L. C. Lazzeroni, J. W. Erickson, J. S. Sinsheimer, and A. H. Kaplan. 1996. In vivo sequence diversity of the protease of human immunodeficiency virus type 1: presence of protease inhibitor-resistant variants in untreated subjects. J. Virol. 70:2038-2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leigh Brown, A. J., S. D. W. Frost, B. Good, E. S. Daar, V. Simon, M. Markowitz, A. C. Collier, E. Connick, B. Conway, J. B. Margolick, J.-P. Routy, J. Corbeil, N. S. Hellmann, D. D. Richman, and S. J. Little. 2004. Genetic basis of hypersusceptibility to protease inhibitors and low replicative capacity of human immunodeficiency virus type 1 strains in primary infection. J. Virol. 78:2242-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mammano, F., C. Petit, and F. Clavel. 1998. Resistance-associated loss of viral fitness in human immunodeficiency virus type 1: phenotypic analysis of protease and gag coevolution in protease inhibitor-treated patients. J. Virol. 72:7632-7637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mammano, F., V. Trouplin, V. Zennou, and F. Clavel. 2000. Retracing the evolutionary pathways of human immunodeficiency virus type 1 resistance to protease inhibitors: virus fitness in the absence and in the presence of drug. J. Virol. 74:8524-8531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Markowitz, M., M. Louie, A. Hurley, E. Sun, M. Di Mascio, A. S. Perelson, and D. D. Ho. 2003. A novel antiviral intervention results in more accurate assessment of human immunodeficiency virus type 1 replication dynamics and T-cell decay in vivo. J. Virol. 77:5037-5038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martinez-Picado, J., A. V. Savara, L. Shi, L. Sutton, and R. T. D'Aquila. 2000. Fitness of human immunodeficiency virus type 1 protease inhibitor-selected single mutants. Virology 275:318-322. [DOI] [PubMed] [Google Scholar]
- 40.Martinez-Picado, J., A. V. Savara, L. Sutton, and R. T. D'Aquila. 1999. Replicative fitness of protease inhibitor-resistant mutants of human immunodeficiency virus type 1. J. Virol. 73:3744-3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mocroft, A., S. Barry, C. A. Sabin, A. C. Lepri, S. Kinloch, A. Drinkwater, M. Lipman, M. Youle, M. A. Johnson, A. N. Phillips et al. 1999. The changing pattern of admissions to a London hospital of patients with HIV: 1988-1997. AIDS 13:1255-1261. [DOI] [PubMed] [Google Scholar]
- 42.Mocroft, A., M. J. Gill, W. Davidson, and A. N. Phillips. 1998. Predictors of a viral response and subsequent virological treatment failure in patients with HIV starting a protease inhibitor. AIDS 12:2161-2167. [DOI] [PubMed] [Google Scholar]
- 43.Molla, A., M. Korneyeva, Q. Gao, S. Vasavanonda, P. J. Schipper, H. M. Mo, M. Markowitz, T. Chernyavskiy, P. Niu, N. Lyons, A. Hsu, G. R. Granneman, D. D. Ho, C. A. Boucher, J. M. Leonard, D. W. Norbeck, and D. J. Kempf. 1996. Ordered accumulation of mutations in HIV protease confers resistance to ritonavir. Nat. Med. 2:760-766. [DOI] [PubMed] [Google Scholar]
- 44.Nijhuis, M., R. Schuurman, D. de Jong, J. Erickson, E. Gustchina, J. Albert, P. Schipper, S. Gulnik, and C. A. Boucher. 1999. Increased fitness of drug resistant HIV-1 protease as a result of acquisition of compensatory mutations during suboptimal therapy. AIDS 13:2349-2359. [DOI] [PubMed] [Google Scholar]
- 45.Palella, F. J., Jr., K. M. Delaney, A. C. Moorman, M. O. Loveless, J. Fuhrer, G. A. Satten, D. J. Aschman, S. D. Holmberg et al. 1998. Declining morbidity and mortality among patients with advanced human immunodeficiency virus infection. N. Engl. J. Med. 338:853-860. [DOI] [PubMed] [Google Scholar]
- 46.Paris, D., B. Ledergerber, R. Weber, J. Jost, M. Flepp, M. Opravil, C. Ruef, and S. Zimmerli. 1999. Incidence and predictors of virologic failure of antiretroviral triple-drug therapy in a community-based cohort. AIDS Res. Hum. Retrovir. 15:1631-1638. [DOI] [PubMed] [Google Scholar]
- 47.Patick, A. K., M. Duran, Y. Cao, D. Shugarts, M. R. Keller, E. Mazabel, M. Knowles, S. Chapman, D. R. Kuritzkes, and M. Markowitz. 1998. Genotypic and phenotypic characterization of human immunodeficiency virus type 1 variants isolated from patients treated with the protease inhibitor nelfinavir. Antimicrob. Agents Chemother. 42:2637-2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Perelson, A. S., P. Essunger, Y. Cao, M. Vesanen, A. Hurley, K. Saksela, M. Markowitz, and D. D. Ho. 1997. Decay characteristics of HIV-1-infected compartments during combination therapy. Nature 387:188-191. [DOI] [PubMed] [Google Scholar]
- 49.Perelson, A. S., A. U. Neumann, M. Markowitz, J. M. Leonard, and D. D. Ho. 1996. HIV-1 dynamics in vivo: virion clearance rate, infected cell life-span, and viral generation time. Science 271:1582-1586. [DOI] [PubMed] [Google Scholar]
- 50.Petropoulos, C. J., N. T. Parkin, K. L. Limoli, Y. S. Lie, T. Wrin, W. Huang, H. Tian, D. Smith, G. A. Winslow, D. J. Capon, and J. M. Whitcomb. 2000. A novel phenotypic drug susceptibility assay for human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 44:920-928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Picchio, G. R., H. Valdez, R. Sabbe, A. L. Landay, D. R. Kuritzkes, M. M. Lederman, and D. E. Mosier. 2000. Altered viral fitness of HIV-1 following failure of protease inhibitor-based therapy. J. Acquir. Immune Defic. Syndr. 25:289-295. [DOI] [PubMed] [Google Scholar]
- 52.Puig, T., M. Perez-Olmeda, A. Rubio, L. Ruiz, C. Briones, J. M. Franco, M. Gomez-Cano, L. Stuyver, L. Zamora, C. Alvarez, M. Leal, B. Clotet, V. Soriano et al. 2000. Prevalence of genotypic resistance to nucleoside analogues and protease inhibitors in Spain. AIDS 14:727-732. [DOI] [PubMed] [Google Scholar]
- 53.Ramratnam, B., S. Bonhoeffer, J. Binley, A. Hurley, L. Zhang, J. E. Mittler, M. Markowitz, J. P. Moore, A. S. Perelson, and D. D. Ho. 1999. Rapid production and clearance of HIV-1 and hepatitis C virus assessed by large volume plasma apheresis. Lancet 354:1782-1785. [DOI] [PubMed] [Google Scholar]
- 54.Ratner, L., A. Fisher, L. L. Jagodzinski, H. Mitsuya, R. S. Liou, R. C. Gallo, and F. Wong-Staal. 1987. Complete nucleotide sequences of functional clones of the AIDS virus. AIDS Res. Hum. Retrovir. 3:57-69. [DOI] [PubMed] [Google Scholar]
- 55.Resch, W., N. Parkin, E. L. Stuelke, T. Watkins, and R. Swanstrom. 2001. A multiple-site-specific heteroduplex tracking assay as a tool for the study of viral population dynamics. Proc. Natl. Acad. Sci. USA 98:176-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Resch, W., R. Ziermann, N. Parkin, A. Gamarnik, and R. Swanstrom. 2002. Nelfinavir-resistant, amprenavir-hypersusceptible strains of human immunodeficiency virus type 1 carrying an N88S mutation in protease have reduced infectivity, reduced replication capacity, and reduced fitness and process the Gag polyprotein precursor aberrantly. J. Virol. 76:8659-8666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ribeiro, R. M., and S. Bonhoeffer. 2000. Production of resistant HIV mutants during antiretroviral therapy. Proc. Natl. Acad. Sci. USA 97:7681-7686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Robinson, L. H., R. E. Myers, B. W. Snowden, M. Tisdale, and E. D. Blair. 2000. HIV type 1 protease cleavage site mutations and viral fitness: implications for drug susceptibility phenotyping assays. AIDS Res. Hum. Retrovir. 16:1149-1156. [DOI] [PubMed] [Google Scholar]
- 59.Rose, R. E., Y. F. Gong, J. A. Greytok, C. M. Bechtold, B. J. Terry, B. S. Robinson, M. Alam, R. J. Colonno, and P. F. Lin. 1996. Human immunodeficiency virus type 1 viral background plays a major role in development of resistance to protease inhibitors. Proc. Natl. Acad. Sci. USA 93:1648-1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 61.Schagger, H., and G. von Jagow. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166:368-379. [DOI] [PubMed] [Google Scholar]
- 62.Schock, H. B., V. M. Garsky, and L. C. Kuo. 1996. Mutational anatomy of an HIV-1 protease variant conferring cross-resistance to protease inhibitors in clinical trials. Compensatory modulations of binding and activity. J. Biol. Chem. 271:31957-31963. [DOI] [PubMed] [Google Scholar]
- 63.Shafer, R. W., D. Stevenson, and B. Chan. 1999. Human Immunodeficiency Virus Reverse Transcriptase and Protease Sequence Database. Nucleic Acids Res. 27:348-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stoddart, C. A., T. J. Liegler, F. Mammano, V. D. Linquist-Stepps, M. S. Hayden, S. G. Deeks, R. M. Grant, F. Clavel, and J. M. McCune. 2001. Impaired replication of protease inhibitor-resistant HIV-1 in human thymus. Nat. Med. 7:712-718. [DOI] [PubMed] [Google Scholar]
- 65.Sufka, S. A., G. Ferrari, V. E. Gryszowka, T. Wrin, S. A. Fiscus, G. D. Tomaras, H. F. Staats, D. D. Patel, G. D. Sempowski, N. S. Hellmann, K. J. Weinhold, and C. B. Hicks. 2003. Prolonged CD4+ cell/virus load discordance during treatment with protease inhibitor-based highly active antiretroviral therapy: immune response and viral control. J. Infect. Dis. 187:1027-1037. [DOI] [PubMed] [Google Scholar]
- 66.van Der Sandt, I. C., C. M. Vos, L. Nabulsi, M. C. Blom-Roosemalen, H. H. Voorwinden, A. G. de Boer, and D. D. Breimer. 2001. Assessment of active transport of HIV protease inhibitors in various cell lines and the in vitro blood-brain barrier. AIDS 15:483-491. [DOI] [PubMed] [Google Scholar]
- 67.Watkins, T., W. Resch, D. Irlbeck, and R. Swanstrom. 2003. Selection of high-level resistance to human immunodeficiency virus type 1 protease inhibitors. Antimicrob. Agents Chemother. 47:759-769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wei, X., S. K. Ghosh, M. E. Taylor, V. A. Johnson, E. A. Emini, P. Deutsch, J. D. Lifson, S. Bonhoeffer, M. A. Nowak, B. H. Hahn, M. S. Saag, and G. M. Shaw. 1995. Viral dynamics in human immunodeficiency virus type 1 infection. Nature 373:117-122. [DOI] [PubMed] [Google Scholar]
- 69.Whitcomb, J. M., W. Huang, K. Limoli, E. Paxinos, T. Wrin, G. Skowron, S. G. Deeks, M. Bates, N. S. Hellmann, and C. J. Petropoulos. 2002. Hypersusceptibility to non-nucleoside reverse transcriptase inhibitors in HIV-1: clinical, phenotypic and genotypic correlates. AIDS 16:F41-F47. [DOI] [PubMed] [Google Scholar]
- 70.Wit, F. W., R. van Leeuwen, G. J. Weverling, S. Jurriaans, K. Nauta, R. Steingrover, J. Schuijtemaker, X. Eyssen, D. Fortuin, M. Weeda, F. de Wolf, P. Reiss, S. A. Danner, and J. M. Lange. 1999. Outcome and predictors of failure of highly active antiretroviral therapy: one-year follow-up of a cohort of human immunodeficiency virus type 1-infected persons. J. Infect. Dis. 179:790-798. [DOI] [PubMed] [Google Scholar]
- 71.Zennou, V., F. Mammano, S. Paulous, D. Mathez, and F. Clavel. 1998. Loss of viral fitness associated with multiple Gag and Gag-Pol processing defects in human immunodeficiency virus type 1 variants selected for resistance to protease inhibitors in vivo. J. Virol. 72:3300-3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang, Y. M., H. Imamichi, T. Imamichi, H. C. Lane, J. Falloon, M. B. Vasudevachari, and N. P. Salzman. 1997. Drug resistance during indinavir therapy is caused by mutations in the protease gene and in its Gag substrate cleavage sites. J. Virol. 71:6662-6670. [DOI] [PMC free article] [PubMed] [Google Scholar]