Abstract
The presumed broad tropism of poxviruses has stymied attempts to identify both the cellular receptor(s) and the viral determinant(s) for binding. Detailed studies of poxvirus binding to and infection of primary human cells have not been conducted. In particular, the determinants of target cell infection and the consequences of infection for cells involved in the generation of antiviral immune responses are incompletely understood. In this report, we show that vaccinia virus (VV) exhibits a more restricted tropism for primary hematolymphoid human cells than has been previously recognized. We demonstrate that vaccinia virus preferentially infects antigen-presenting cells (dendritic cells, monocytes/macrophages, and B cells) and activated T cells, but not resting T cells. The infection of activated T cells is permissive, with active viral replication and production of infectious progeny. Susceptibility to infection is determined by restricted expression of a cellular receptor that is induced de novo upon T-cell activation and can be removed from the cell surface by either trypsin or pronase treatment. The VV receptor expressed on activated T cells displays unique characteristics that distinguish it from the receptor used to infect cell lines in culture. The observed restricted tropism of VV may have significant consequences for the understanding of natural poxvirus infection and immunity and for poxvirus-based vaccine development.
Poxviruses have recently generated significant interest as vectors for vaccines against infectious diseases such as AIDS, malaria, and tuberculosis and as possible agents of bioterrorism (1, 7, 17, 35, 38). However, many features of the interaction of poxviruses with the immune system that are relevant to the illumination of pathogenic mechanisms of poxvirus infection, as well as the development of improved poxvirus vaccine vectors and immunomodulatory strategies, are poorly understood. In particular, the cellular targets for infection within the human immune system have not been clearly delineated. The ability of vaccinia virus (VV), the virus used for immunization against smallpox and the best-studied laboratory model for poxvirus biology and immunity, to infect almost any cell line in culture has resulted in descriptions of its broad cellular tropism and presumed widespread expression of a virus binding receptor (40). However, no cell surface molecule(s) on primary cells or cell lines has been convincingly demonstrated to be essential for poxvirus infection and, in fact, the viral determinants for binding and entry of any form of VV remain unknown.
VV belongs to the family Poxviridae, the subfamily Chordopoxvirinae, and the genus Orthopoxvirus. The Orthopoxvirus genome is composed of 167 to 224 kbp of linear, double-stranded DNA and includes the viruses variola (the causative agent of smallpox), monkeypox, cowpox, ectromelia, and vaccinia (40). There are different infectious forms of Orthopoxviruses; importantly, the virus form responsible for transmission in the setting of natural poxvirus infection has not been determined. The durability of the outer envelope of the intracellular mature virion (IMV) has led some to suggest that this viral form serves an important role in interhost transmission (40). Cell-associated enveloped virions (CEV) and, upon release from the cell membrane, extracellular enveloped virions (EEV) contain a second trans-Golgi-derived membrane surrounding the IMV particle and are proposed to mediate cell-to-cell spread within a host and in vitro (2, 6, 49). In contrast to IMV, the outer membrane of CEV/EEV is extremely fragile, and pure populations of this virus form are difficult to prepare (26, 44). Common preparations of the smallpox vaccine and all vaccine vectors being developed using the vaccinia system, in addition to virus preparations used for studies of vaccinia immunology, are derived from infected cells and are thus composed of a mixture of virion forms, including intracellular virions and CEV with a damaged outer membrane. When the outer CEV/EEV membrane is ruptured, the particle resembles an IMV with portions of the CEV/EEV membrane surrounding it like a shroud (G. C. Carter, M. Hollinshead, M. Law, and G. Smith, XVth Intl. Poxvirus and Iridovirus Conf., abstr. W1.1, 2004). This “IMV with retained EEV membrane proteins” remains fully infective (37, 56).
It has been suggested that IMV and EEV enter cells by different mechanisms, perhaps reflecting their use of different cellular receptors (53). The IMV surface proteins encoded by L1R and A28L have been implicated in cell penetration but not attachment (27, 48, 57), and it is thought that cell surface proteoglycans serve as virus attachment sites but are dispensable for virus infection (12, 24, 31). Earlier studies proposing that the epidermal growth factor receptor and several chemokine receptors serve as receptors for VV and myxoma virus, respectively, have not been validated (19, 25, 30, 32, 33). Overall, based on studies almost exclusively conducted in tissue culture where a broad host tropism has been demonstrated, it is assumed that the receptor for VV infection is widely, if not ubiquitously, expressed and that the events that make a cell nonpermissive for VV infection occur after virus binding and entry (36, 40).
In this report, we demonstrate that the cellular tropism of VV for cells involved in the generation of immune responses is more restricted than previously imagined and show that the key determinant for infection is virus binding to the cell surface. We hypothesized that studying primary human cells of hematolymphoid origin would provide a clearer and more biologically relevant picture of the tropism of poxviruses used for vaccines, would increase our understanding of the processing and presentation of poxvirus (and poxvirus-vectored) antigens, and might, in fact, lead to the identification of the poxvirus cellular receptor(s). Using recombinant VV expressing enhanced green fluorescent protein (rVV-EGFP), we have determined that these viruses infect antigen-presenting cells (dendritic cells [DCs], monocytes/macrophages, and B cells to a more variable extent) and activated, but not resting, T cells. Using recombinant VV with EGFP fused to a virion membrane component (rVV-B5R-EGFP), we further show that VV is not able to bind to unstimulated T cells, suggesting that T-cell activation results in the upregulation of a cell surface molecule (or molecules) that acts as a VV receptor (or receptors). We provide evidence that expression of the VV receptor on activated T cells requires de novo gene expression, that the VV receptor can be removed by cell surface protease treatment, and that the described VV receptor on activated T cells is a unique molecule that differs from the receptor used to enter cell lines in culture. In all primary, hematolymphoid human cells examined, we find that restricted expression of a virus binding receptor is precisely correlated with and likely determines cell tropism.
MATERIALS AND METHODS
Mice.
6-week-old female BALB/c mice were purchased from the Jackson Laboratories (Bar Harbor, ME) and maintained in the Yerkes National Primate Research Center vivarium following Institutional Animal Care and Use Committee procedures.
Human subjects.
Whole-blood or leukapheresis samples were obtained from healthy human volunteers with informed consent in accordance with Emory University's Institutional Review Board policies.
Mononuclear cell subsets. (i) PBMCs.
Peripheral blood mononuclear cells (PBMCs) were isolated by standard density gradient centrifugation on lymphocyte separation medium (ICN Biomedicals, Aurora, OH).
(ii) T cells.
T cells were sorted using (i) anti-CD3 microbeads (Miltenyi Biotec, Auburn, CA), (ii) anti-CD3 biotin (BD Pharmingen, San Diego, CA) in combination with anti-biotin microbeads (Miltenyi Biotec), or (iii) the Pan T Cell Isolation Kit II (Miltenyi Biotec). T-cell purity was determined by monoclonal antibody (MAb) staining for CD3 and was always >95%.
T-cell activation.
T cells were activated with plate-bound anti-CD3 (BD Discovery Labware, Bedford, MA) with or without the addition of 2 μg/ml each of anti-CD28 and anti-CD49d MAbs (BD Pharmingen). Alternatively, total PBMCs were stimulated with 10 μg/ml phytohemagglutinin (PHA) (Sigma, St. Louis, MO). In some experiments, T cells were treated with 0.5 μg/ml actinomycin D (Calbiochem, San Diego, CA), 8 μg/ml cycloheximide (Calbiochem), or 10 μg/ml brefeldin A (BD Pharmingen) at the time of activation.
Viruses. (i) Virus stocks.
rVV-EGFP (NYCBH strain) expressing EGFP under the control of the p7.5 early/late promoter was a gift of L. Corey (University of Washington). Recombinant modified vaccinia virus Ankara was constructed to contain a recombinant GFP-ZEO fusion protein (Invitrogen, Carlsbad, CA) that expresses GFP under the control of the early H5 promoter and confers resistance to Zeocin (rMVA-GFP). rVV-B5R-EGFP (WR strain), nonfluorescent rVV control (WR strain), and rVV (IHD-J strain) were a gift of B. Moss (National Institutes of Health). rVV stocks were expanded in HeLa cells (American Type Culture Collection [ATCC], Manassas, VA), and the titers were determined on BSC40 cells (ATCC). rMVA stocks were expanded in and titers were determined on the chicken embryo fibroblast cell line DF-1 (ATCC). Viruses were purified from both infected cells and medium and pelleted through a 36% (wt/vol) sucrose cushion. Titers were determined by serial dilutions and plaque assays. For all viruses, both intracellular and extracellular particles were collected; however, stocks were subjected to at least one freeze-thaw cycle to disrupt the outer membrane of CEV/EEV and transform it into an IMV with retained EEV membrane proteins (26).
(ii) Virus infection.
Cells were infected with rVV-EGFP or rMVA-GFP at a multiplicity of infection (MOI) of 10 for 1 h at 37°C, washed with either RPMI-10% fetal bovine serum (FBS; HyClone, Logan, UT) or Dulbecco's modified Eagle medium (DMEM)-10% FBS, depending on the cell type, and then incubated in RPMI-10% FBS or DMEM-10% FBS in six-well plates at 37°C.
(iii) Virus binding.
Cells were incubated with rVV-B5R-EGFP or a nonfluorescently tagged rVV control (WR strain) at an MOI of 50 for 1 h on ice, washed extensively with ice-cold phosphate-buffered saline (PBS)-10% FBS, and fixed with 1% paraformaldehyde (PFA). In some cases, soluble heparin (Sigma) at 2, 10, or 50 μg/ml was added concurrently with virus, and cells were incubated at 4°C to remain consistent with a previous report (12).
Cell surface enzyme treatment.
Cells were treated with either 1.25 mg/ml trypsin (from bovine pancreas; Sigma) or 1 mg/ml pronase (from Streptomyces griseus, Roche, Mannheim, Germany) in PBS for 30 min and then washed extensively with PBS-10% FBS.
Activated T-cell and monocyte immune sera.
Monocytes were sorted from human PBMCs with anti-CD14 microbeads (Miltenyi Biotec). The monocyte-depleted fraction was then stimulated for 18 h with PHA, and activated T cells were isolated by selection of the negative population after anti-CD56 microbead (Miltenyi Biotec) incubation, followed by selection of the positive population after anti-CD2 microbead (Miltenyi Biotec) incubation. The purity of each T-cell preparation was assessed by MAb staining and flow cytometry and was always >89%, with >70% activation as determined by CD69 or CD25 positivity. Monocyte preparations were always >95% pure as determined by CD14 MAb staining. BALB/c mice were injected intraperitoneally with 1.45× 107 to 2 × 107 activated T cells or monocytes in 500 μl of PBS on days 0, 10, and 20 and were bled on day 33. Unimmunized mice were bled for control serum (preimmune). Serum was frozen at −80°C and was heat inactivated for 30 min at 56°C before use.
Antibody staining and flow cytometry.
The following MAbs were used: phycoerythrin (PE)-, CyChrome-, or allophycocyanin (APC)-conjugated mouse anti-human CD2, CD3, CD14, CD16, CD19, CD56, CD123, HLA-DR, and isotype controls (all from BD Pharmingen); PE-, peridinin chlorophyll protein-, or APC-conjugated mouse anti-human CD4, CD8, CD11c, CD19, CD20, CD25, CD45RA, CD62L, CD69, HLA-DR, and isotype controls (all from BD Immunocytometry Systems, San Jose, CA); and APC-conjugated mouse anti-human CD16 (Caltag Laboratories, Burlingame, CA). PE-conjugated goat anti-mouse immunoglobulin (Ig)-specific polyclonal Ab (pAb) was purchased from Biosource International (Camarillo, CA). Fluorescein isothiocyanate-conjugated cholera toxin B subunit was purchased from Sigma. The MAbs VVI-6B6, VV4-2F6, and VVI-4G9 that recognize a 29-kDa early protein of VV, VV D8L, and VV hemagglutinin (HA) A56R, respectively, were a gift of A. Schmaljohn (National Institutes of Health). MAb 2D5 directed against VV L1R was a gift of I. Damon (Centers for Disease Control and Prevention).
Staining was performed as previously described (50). Samples were acquired on a FACSCalibur flow cytometer (BD Immunocytometry Systems) and analyzed using FlowJo software (TreeStar, San Carlos, CA). PHA-activated T cells for confocal microscopy were sorted using a MoFlo sorter (Cytomation, Fort Collins, CO), following staining with PE-conjugated CD14, CD16, and CD20 MAbs to deplete monocytes, NK cells, and B cells, respectively.
Confocal microscopy.
A total of 5 × 105 cells were adhered to Alcian blue-coated glass coverslips, fixed in 4% PFA, washed, and mounted in Vectashield (Vector Laboratories, Burlingame, CA) on glass slides. All analysis was performed on a LSM510 confocal microscope (Zeiss Microimaging, Thornwood, NY).
Vaccinia uracil DNA glycosylase (UDG) real-time PCR.
Cells were frozen at −80°C at 0, 1.5, 4, and 24 h after infection. DNA was extracted using the MagNA Pure LC DNA Isolation Kit I (Roche) and quantified by UV spectrometry.
The probe and primers for real-time PCR were designed by the use of Primer Express software (Applied Biosystems, Foster City, CA) within the conserved UDG region. The TaqMan probe (5′-CGAGACGAGACGTCGCCTATTCCTG-3′) (Applied Biosystems) was labeled at the 5′ end with the reporter dye FAM (6-carboxyfluorescein) and at the 3′ end with the quencher dye TAMRA (6-carboxytetramethylrhodamine) with a melting temperature (Tm) of 68°C. Primer sequences were as follows: 5′GGTAGAGTTTTATAACGAAGTAGCCAGTT-3′ (sense; length, 29 bases, Tm = 58°C) and 5′-CTCGTTTATTTCTAAGCGGTTGTTT-3′ (antisense; length, 25 bases, Tm = 58°C).
Real-time PCR was performed using the ABI Prism 7700HT sequence detection system (Applied Biosystems) with the TaqMan Gold kit under the following conditions: the 50-μl reaction mixtures contained 5 μl of 10× TaqMan buffer A, 4 mM MgCl2, 200 μM deoxynucleoside triphosphates, 200 nM each primer, 125 nM fluorogenic probe, and 1.25 U of AmpliTaq Gold DNA polymerase. Universal thermal cycling conditions consisted of 10 min at 95°C, followed by 45 cycles, each consisting of 15 s at 95°C and 1 min at 60°C. A total of 200 ng of DNA was analyzed as a template for amplification, and the results obtained were expressed as the number of UDG copies per 200 ng of DNA. Each reaction was carried out in duplicate.
Progeny virus titration.
Cells were frozen at −80°C at 0, 1.5, 8, and 24 h postinfection. Repetitive freeze-thaw cycles were used to lyse the cells, followed by sonication for 1 min. Titers were determined for the supernatant dilutions on BSC40 monolayers for 48 h, and plaques were stained with 0.04% neutral red (Sigma).
RESULTS
VV targets specific cell subsets for infection.
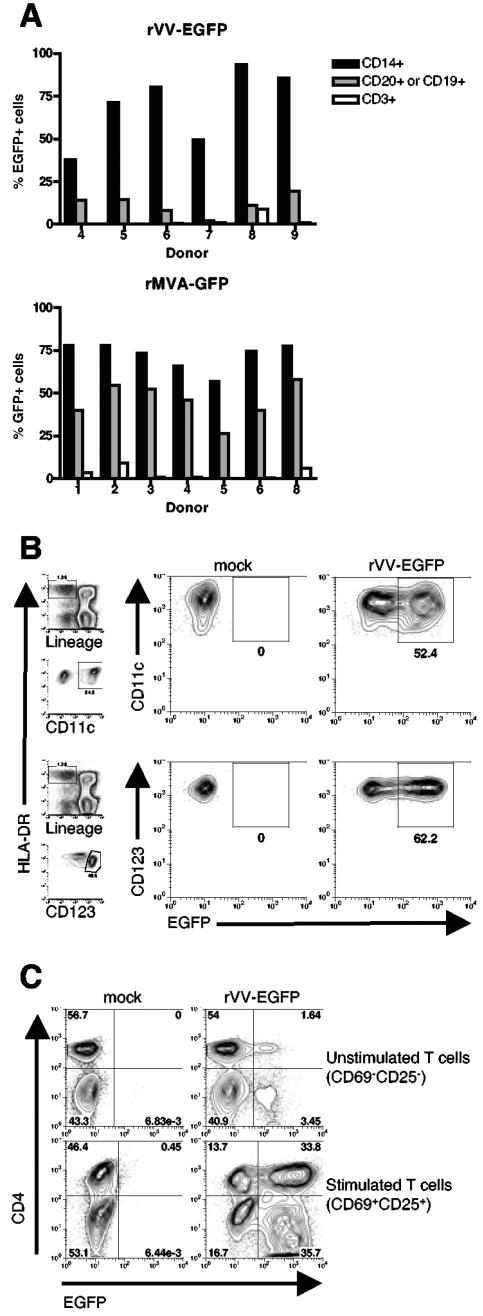
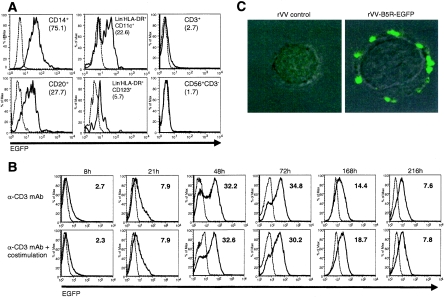
To investigate the tropism of VV for primary cells of the human immune system, we first determined the range of cell types present in the peripheral blood that could be infected by VV. For this purpose, we utilized an rVV encoding EGFP under the control of an early viral promoter (rVV-EGFP) to identify cells that internalized virus and expressed a viral early protein by flow cytometry. PBMCs from multiple donors were isolated and infected with rVV-EGFP at an MOI of 10; after 6 h, cells were stained with MAbs specific for monocytes (CD14), B cells (CD19 or CD20), and T cells (CD3). We observed that virus infection was not uniform in all cell types, and EGFP was detected in 70% of monocytes, 12% of B cells, and only 2% of T cells (mean of six samples), following exposure to rVV-EGFP (Fig. 1A, top). VV infection of specific cell subsets within PBMCs was also documented using MAb VV1-6B6, which recognizes a 29-kDa early protein of VV, with similar results (data not shown). To confirm this cellular tropism using a strain of VV most relevant to current poxvirus-vectored vaccines (1, 17, 35, 38), the same experiment was repeated with a recombinant modified vaccinia virus Ankara encoding GFP under the control of a virus early promoter (rMVA-GFP). rMVA-GFP infection resulted in GFP expression in 72% of monocytes, 46% of B cells, and 3% of T cells (mean of seven samples) (Fig. 1A, bottom). As the ability of VV to infect peripheral blood DC subsets has not been examined, we next infected PBMCs with rVV-EGFP for 6 h and gated on EGFP+ myeloid (Lin− HLA-DR+ CD11c+) and plasmacytoid (Lin− HLA-DR+ CD123+) DCs (Fig. 1B; gating strategy shown in panels on left). We found that both subsets of blood DCs were susceptible to infection, with 52% of myeloid and 62% of plasmacytoid DCs expressing EGFP (Fig. 1B). CD56+ NK cells were not infected by rVV-EGFP (data not shown).
FIG. 1.
VV infects APCs and activated T cells. (A) PBMCs were incubated with rVV-EGFP (top) or rMVA-GFP (bottom) at an MOI of 10; after 6 h, EGFP expression was measured by flow cytometry in conjunction with staining for the cell surface markers CD14, CD20, or CD19 and CD3 to identify monocytes, B cells, and T cells, respectively. (B) PBMCs were incubated with rVV-EGFP at an MOI of 10 and cultured for 6 h in the presence of recombinant interleukin 3 (20 ng/ml), a survival factor for plasmacytoid dendritic cells. EGFP expression was then measured by flow cytometry in myeloid (identified as Lin− HLA-DR+ CD11c+) (top) and plasmacytoid (identified as Lin− HLA-DR+ CD123+) (bottom) dendritic cell subsets. Panels on the left show the gating strategy used (the lineage cocktail consisted of CD3, CD14, CD16, CD19, CD20, and CD56 MAbs). (C) T cells were sorted from PBMCs using anti-CD3 microbeads (to >98% purity) and were stimulated with plate-bound anti-CD3 plus anti-CD28 and anti-CD49d for 68 h. These stimulated T cells and unstimulated T cells isolated from the same donor were incubated with rVV-EGFP at an MOI of 10 for 12 h. Within the stimulated T cells, a gate was set on cells expressing the activation markers CD69 and CD25; within the unstimulated cells, a gate was set on cells lacking CD69 and CD25. EGFP expression is shown for CD4+ and CD4− (i.e., CD8+) T cells. These data are representative of at least six independent experiments.
VV infects activated, but not resting, T cells.
We observed that only a small fraction of T cells found in peripheral blood were directly susceptible to infection with VV. As some viruses preferentially infect cells in an activated state, we next tested the ability of rVV-EGFP to infect T cells following their activation. Purified T cells were activated with plate-bound anti-CD3 plus anti-CD28 and anti-CD49d MAbs for 3 days, and these cells were then infected with rVV-EGFP at an MOI of 10. EGFP expression was measured after 12 h in conjunction with staining for T-cell activation markers. Figure 1C shows that 69.5% of activated, CD69+ CD25+ T cells were infected by VV, compared to 5.1% of purified peripheral blood T cells prior to activation. Infection was seen in both CD4+ and CD8+ T cells, as indicated. In addition, both naïve (CD45RA+ CD62L+) and memory (i.e., nonnaïve) T cells were susceptible to VV infection upon activation (data not shown). The observed cellular tropism of VV presented in Fig. 1A to C is extremely reproducible, having been demonstrated in a large number of donors, with a representative group shown here. This restricted cellular tropism has also been confirmed by studies conducted with mice (L. Liu et al., unpublished data).
Virus binding predicts susceptibility to infection.
The cellular tropism of VV may be influenced by a restriction in certain cell types of any one of a number of early stages in the virus life cycle. Specifically, it is possible that all cells bind to VV and that a postbinding event, such as a conformational change in a receptor or coreceptor, a fusion event, or a postentry mechanism such as nucleotide pool availability (20) or expression of an essential cellular cofactor, allows the infection to proceed in certain cell subsets. Alternatively, it may be that only cells that are able to bind the virus via expression of a specific receptor are susceptible to infection. To investigate what determines the observed differential susceptibility to infection of different primary cell lineages and of specific cell lineages (e.g., T cells) in response to activation, we studied the initial binding reaction between virus and cell surface.
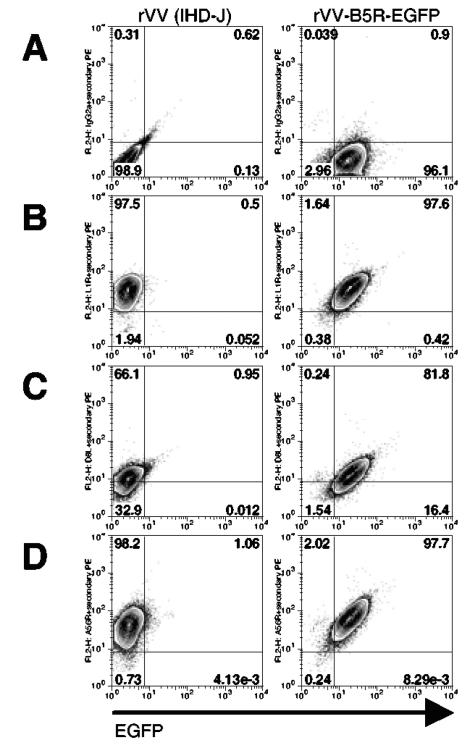
To determine if susceptibility to infection is conferred by the ability to bind virus, we used an rVV containing EGFP fused to the open reading frame of an EEV membrane protein (encoded by the B5R gene), making the input virus particle fluorescent (55). As with all of our virus preparations, we subjected the rVV-B5R-EGFP to at least one freeze-thaw cycle to disrupt the EEV outer envelope and expose IMV surface proteins (26). Therefore, in this system, the EGFP signal detects binding of a particle perhaps best described as an IMV with retained EEV membrane proteins (Carter et al., XVth Intl. Poxvirus and Iridovirus Conf.), with all cells that appear EGFP+ by flow cytometry also exhibiting positive staining for the IMV membrane proteins encoded by L1R and D8L and the EEV membrane protein encoded by A56R (Fig. 2). This binding assay thus allows for either IMV or EEV surface proteins to bind to the cell. PBMCs were incubated with rVV-B5R-EGFP at an MOI of 50:1, in keeping with other virus binding assays (16, 53), for 1 h on ice, a condition that permits virus binding but not entry (15); virus binding to the cell surface was detected by flow cytometry. Monocytes, B cells, and DCs bound rVV-B5R-EGFP, but resting T cells and NK cells did not (Fig. 3A). No green fluorescence was observed when a nonfluorescently tagged WR strain control virus was used.
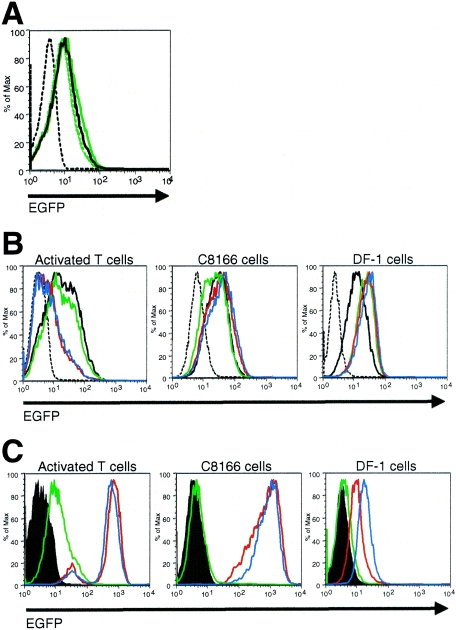
FIG. 2.
Characterization of an IMV with retained EEV membrane proteins. rVV-B5R-EGFP and a nonfluorescently tagged rVV (strain IHD-J, producing predominately EEV) (42) were prepared as described in Materials and Methods and incubated with BSC40 cells at an MOI of 50 for 1 h on ice. After washing, cells were stained with an isotype control (A) or with anti-VV MAbs recognizing L1R (B) or D8L (C), specific for the IMV surface, or A56R (D), specific for the EEV surface; the cells were then analyzed by flow cytometry. BSC40 cells alone did not exhibit above-background fluorescence when stained with anti-VV MAbs (data not shown).
FIG. 3.
Virus binding to the cell membrane predicts susceptibility to infection. (A) Flow cytometric detection of rVV-B5R-EGFP (solid line) and a nonfluorescently tagged rVV control (dashed line) binding to different subsets of PBMCs. PBMCs were incubated with virus at an MOI of 50 for 1 h on ice, stained with phenotyping MAbs for flow cytometry, and identified as follows: monocytes, CD14+; B cells, CD20+; myeloid DCs, Lin− HLA-DR+ CD11c+; plasmacytoid DCs, Lin− HLA-DR+ CD123+; T cells, CD3+; and NK cells, CD56+ CD3−. The mean fluorescence intensities of rVV-B5R-EGFP minus rVV control are shown in parentheses. (B) T cells were sorted from PBMCs using the Pan T Cell Isolation Kit II (to >95% purity) and were then activated with plate-bound anti-CD3 with (bottom) or without (top) anti-CD28 and anti-CD49d for 9 days. At each time point, cells were incubated with rVV-B5R-EGFP or rVV control at an MOI of 50 for 1 h on ice. Flow cytometric detection of rVV-B5R-EGFP (solid line) and rVV control (dashed line) binding to T cells undergoing activation (gated as CD2+ CD56− cells) is shown. The mean fluorescence intensities of rVV-B5R-EGFP minus rVV control are shown. (C) PBMCs were stimulated with PHA for 72 h, and activated T cells were purified by FACSorting for the CD14− CD16− CD20− population. Activated T cells were then incubated with rVV-B5R-EGFP or rVV control at an MOI of 50 for 1 h on ice, washed, adhered to glass coverslips, and fixed in 4% PFA. Visualization of rVV-B5R-GFP and rVV control binding to the activated T-cell membrane was performed by confocal microscopy. These data are representative of at least six independent experiments.
We then repeated the virus binding assay with purified T cells at several time points over the course of 9 days of activation. T cells were activated with plate-bound anti-CD3 MAb alone or anti-CD3 plus anti-CD28 and anti-CD49d MAbs. T-cell activation in the presence or absence of costimulation (i.e., anti-CD28 and anti-CD49d) results in the induction of different patterns of mRNA transcript expression (14). Therefore, we analyzed the effects of these two activation regimens on the induction of VV binding to T cells as a way of guiding future efforts to molecularly clone the requisite binding receptor. We observed maximal and similar levels of binding 48 to 72 h after activation with either regimen that decreased by 1 week (when the cells have returned to a resting state) (Fig. 3B; 0 h shown in Fig. 3A). CD69, CD25, and HLA-DR expression on T cells was used as confirmation of their activation status (data not shown). Stimulation with anti-CD28 and anti-CD49d without anti-CD3 did not result in an increase of either CD25 expression or rVV-B5R-EGFP binding above that seen with unstimulated T cells (data not shown).
Direct visualization of VV binding to activated T cells was performed by confocal microscopy. PBMCs were stimulated with PHA for 72 h; monocytes, B cells, and NK cells were depleted by FACSorting for the CD14− CD20− CD56− population. Figure 3C shows a representative image of an activated T cell with green fluorescent virus particles decorating its surface, following incubation with rVV-B5R-EGFP (and no green fluorescence after exposure to the rVV control). These data indicate that VV does not ubiquitously bind to all cell types and that there is a precise correlation between those cells that are able to bind VV and those cells that become infected by VV (and express early viral genes). Therefore, VV binding is a specific event that predicts its cellular tropism.
Upregulation of receptor expression requires de novo gene expression.
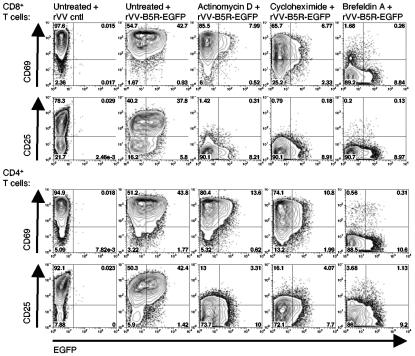
To determine the level at which VV receptor expression on T cells is controlled, we observed binding of rVV-B5R-EGFP to T cells following the simultaneous administration of an activating stimulus with general inhibitors of transcription, protein synthesis, or intracellular protein transport. Specifically, purified T cells were treated with plate-bound anti-CD3 plus anti-CD28 and anti-CD49d MAbs in the presence of actinomycin D, cycloheximide, or brefeldin A to block transcription, translation, and protein transport to the cell surface, respectively; virus binding was assessed after 21 h. Although we found higher levels of VV binding to activated T cells after 48 and 72 h of stimulation (Fig. 3B), 21 h was chosen for this experiment due to drug toxicity observed at later time points. Virus binding to activated T cells in the absence of any of the above treatments was found to be 44% for CD8+ T cells and 45% for CD4+ T cells (Fig. 4). All three treatments significantly reduced rVV-B5R-EGFP binding (by an average of 80% for CD8+ T cells and 73% for CD4+ T cells) (Fig. 4). Examination of control gene production yielded expected results, with cell surface expression of the T cell activation marker CD25 requiring de novo RNA and protein synthesis and protein transport (and thus was blocked by all three treatments) (13, 29), whereas CD69 expression required only transport of the preformed molecule from intracellular vesicles (and thus was blocked by brefeldin A only) (41) (Fig. 4) .
FIG. 4.
Upregulation of the VV receptor on activated T cells requires de novo gene expression. T cells were sorted from PBMCs using anti-CD3 biotin MAb and biotin microbeads (to >97% purity) and were then activated for 21 h with plate-bound anti-CD3 plus anti-CD28 and anti-CD49d MAbs in the presence or absence of actinomycin D, cycloheximide, or brefeldin A. Cells were then incubated with rVV-B5R-EGFP or rVV control at an MOI of 50 for 1 h on ice, and binding was assessed in conjunction with staining for the T-cell activation markers CD69 and CD25. The percentages of EGFP+ cells were determined for both CD4+ and CD8+ T-cell subsets. These data are representative of three independent experiments.
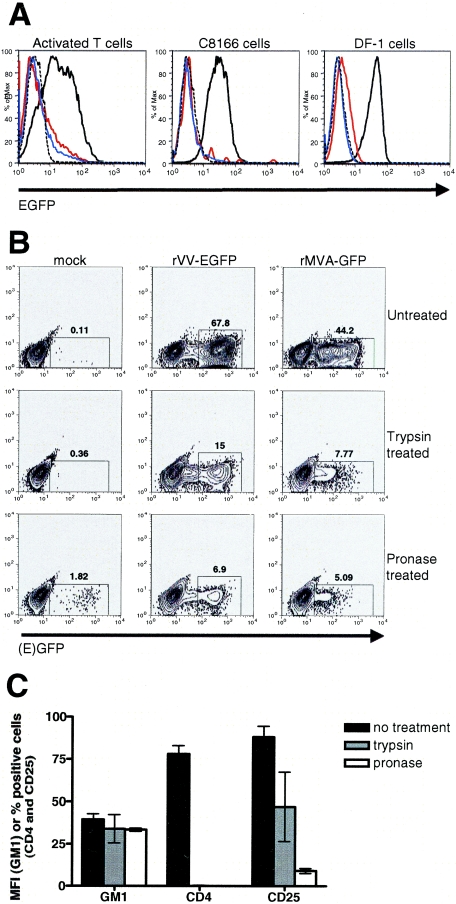
Cell surface protease treatment inhibits VV binding and infection.
Additional experiments were conducted to determine the physical nature of the VV receptor upregulated on activated T cells. First, rVV-B5R-EGFP binding was assessed following pretreatment with the enzymes trypsin (1.25 mg/ml) or pronase (1 mg/ml). Activated T cells, C8166 cells (a human T-cell leukemia virus type 1 (HTLV-1)-transformed activated CD4+ T-cell line) (45), and DF-1 cells (a chicken embryo fibroblast cell line) (23, 46) were analyzed. Both treatments resulted in a loss of binding in all three cell types (Fig. 5A). Second, activated T cells were pretreated with trypsin or pronase and then infected with rVV-EGFP or rMVA-GFP at an MOI of 10. After 6 h of infection, the protease-treated cells displayed decreased EGFP expression compared to untreated cells (a reduction of 78 to 90%) as determined by flow cytometry (Fig. 5B). The observed reduction in VV binding following enzyme treatment of activated T cells was thus also reflected in a reduction of both VV and MVA infection after the same treatment. As a positive control for protease treatment, activated T cells were stained with MAbs to CD4 and CD25 following exposure to trypsin or pronase; as shown in Fig. 5C, expression of these surface proteins was greatly reduced compared to untreated cells. By contrast, expression of the ganglioside GM1 was monitored using the cholera toxin B subunit (47); importantly, this binding event that does not utilize a protein receptor remained intact after both trypsin and pronase treatment of activated T cells (Fig. 5C). In all, these data indicate that the cellular receptor for VV found on activated T cells is likely a protein whose surface expression depends on its de novo transcription, translation, and intracellular transport.
FIG. 5.
The VV receptor is removed from the cell surface by trypsin or pronase digestion. (A) T cells were sorted from PBMCs using anti-CD3 microbeads (to >98% purity) and were then activated for 42 h with plate-bound anti-CD3 plus anti-CD28 and anti-CD49d MAbs. Activated T cells, C8166 cells, and DF-1 cells were treated with trypsin or pronase for 30 min on ice and then incubated with rVV-B5R-EGFP or rVV control at an MOI of 50 for 1 h on ice. Virus binding was assessed by flow cytometry. Dashed black histograms, untreated cells, rVV control; solid black histograms, untreated cells, rVV-B5R-EGFP; red histograms, trypsin-treated cells, rVV-B5R-EGFP; blue histograms, pronase treated cells, rVV-B5R-EGFP. (B) Activated T cells were prepared as described in panel A; after 65 h, cells were treated with trypsin or pronase for 30 min at 25°C and then incubated with rVV-EGFP or rMVA-GFP at an MOI of 10 for 6 h at 37°C. EGFP expression was measured by flow cytometry. (C) Activated T cells were prepared as described in panel A; after 60 h, cells were treated with trypsin or pronase for 30 min on ice and then stained with MAbs to CD4 and CD25 and a fluorescein isothiocyanate-conjugated cholera toxin B subunit that binds ganglioside GM1. Expression of these molecules before and after enzyme treatment was measured by flow cytometry. These data are representative of three independent experiments.
VV binds to a unique receptor on primary cells.
As previously discussed, the presumed wide tropism of VV has led some to suggest that its cellular receptor is ubiquitously expressed (40). However, our initial studies indicated that VV was not able to bind to and infect every cell it encounters; specifically, resting T cells and NK cells did not bind VV and are resistant to infection. The restricted cellular tropism we present here may be a reflection of VV's use of different receptors to infect primary human cells compared to cell lines in culture. Alternatively, the same receptor may be expressed on susceptible primary cells and cell lines, yet may be absent from certain primary cells that are resistant to infection. The glycosaminoglycan heparan sulfate has been proposed as an attachment site for the VV proteins A27L and H3L, and soluble heparin (at 50 μg/ml) has been shown to inhibit VV binding to BSC40 cells (12, 31). We found, conversely, that soluble heparin (at 2, 10, or 50 μg/ml) does not inhibit the binding of rVV-B5R-EGFP to activated T cells (Fig. 6A).
FIG. 6.
VV uses a unique receptor to bind to activated T cells. (A) T cells were sorted from PBMCs using anti-CD3 microbeads (to >98% purity) and were then activated for 52 h with plate-bound anti-CD3 plus anti-CD28 and anti-CD49d MAbs. Activated T-cells were incubated with rVV-B5R-EGFP or rVV control at an MOI of 50 in the presence or absence of heparin for 1 h at 4°C. Virus binding was assessed by flow cytometry. Dashed black histogram, rVV control; solid black histogram, rVV-B5R-EGFP; solid green histogram, 2 μg/ml heparin, rVV-B5R-EGFP; dashed green histogram, 10 μg/ml heparin, rVV-B5R-EGFP; dotted green histogram, 50 μg/ml heparin, rVV-B5R-EGFP. (B) T cells were sorted from PBMCs using the Pan T Cell Isolation Kit II (to >95% purity) and were then activated with plate-bound anti-CD3 plus anti-CD28 and anti-CD49d for 48 h. Activated T cells, C8166 cells, and DF-1 cells were incubated with preimmune serum or immune sera from mice immunized with activated T cells or monocytes at a 1:10 dilution for 30 min at 25°C and then incubated with rVV-B5R-EGFP or rVV control at an MOI of 50 for 1 h on ice. Virus binding was assessed by flow cytometry. Dashed black histograms, no serum, rVV control; solid black histograms, no serum, rVV-B5R-EGFP; green histograms, preimmune serum, rVV-B5R-EGFP; red histograms, activated T-cell immune serum, rVV-B5R-EGFP; blue histograms, monocyte immune serum, rVV-B5R-EGFP. (C) Cells were incubated with mouse sera as described in the legend to panel B and then stained with a pAb directed against mouse Ig. The ability of the preimmune and immune sera to recognize cellular epitopes was assessed by flow cytometry. Filled histograms, no serum; green histograms, preimmune serum; red histograms, activated T-cell immune serum; blue histograms, monocyte immune serum. These data are representative of three independent experiments.
To further investigate the hypothesis that VV may utilize a unique receptor to bind to primary human cells, we immunized BALB/c mice with either purified activated T cells or monocytes (two cell types that bind VV with high efficiency) (Fig. 3) to generate immune sera capable of recognizing the virus binding receptor(s) present on these hematolymphoid cells. We then used various dilutions of the immune sera to pretreat cells prior to performing the rVV-B5R-EGFP binding assay. Two cell lines were studied: the human, HTLV-1-transformed, activated, CD4+ T-cell line C8166 and the chicken embryo fibroblast cell line DF-1, which were compared to primary activated T cells. We discovered that rVV-B5R-EGFP binding to activated T cells was significantly reduced by pretreatment with a 1:10 dilution of the activated T-cell immune serum, but levels of binding were unchanged or even slightly enhanced when C8166 or DF-1 cells were studied (Fig. 6B). Interestingly, a 1:10 dilution of the monocyte immune serum also blocked VV binding to activated T cells, but again no change in binding was seen when C8166 or DF-1 cells were used (Fig. 6B). Preimmune serum had no effect on rVV-B5R-EGFP binding at this dilution (Fig. 6B).
To confirm that the immune sera were able to recognize epitopes on the cell surface, all three cell types were incubated with preimmune, activated T-cell, or monocyte immune sera and stained with a pAb directed against mouse Ig. As expected, the chicken-derived DF-1 cells did not display significant staining above background after incubation with any type of serum (Fig. 6C); as such, VV binding was not affected by serum pretreatment (Fig. 6B). In contrast, activated T cells and C8166 cells of human origin both displayed high levels of pAb staining after incubation with activated T-cell and monocyte immune sera and background levels after incubation with preimmune serum (Fig. 6B). This result indicates that activated T cells and monocytes share a number of cell surface molecules, as would be predicted, and that some of these molecules are also present on the C8166 cell surface. As pretreatment with both types of immune sera reduced VV binding to primary activated T cells but had no effect on VV binding to the activated T-cell line C8166, it may be postulated that VV utilizes different receptors to bind to primary cells compared to cell lines.
Activated T cells support permissive viral infection.
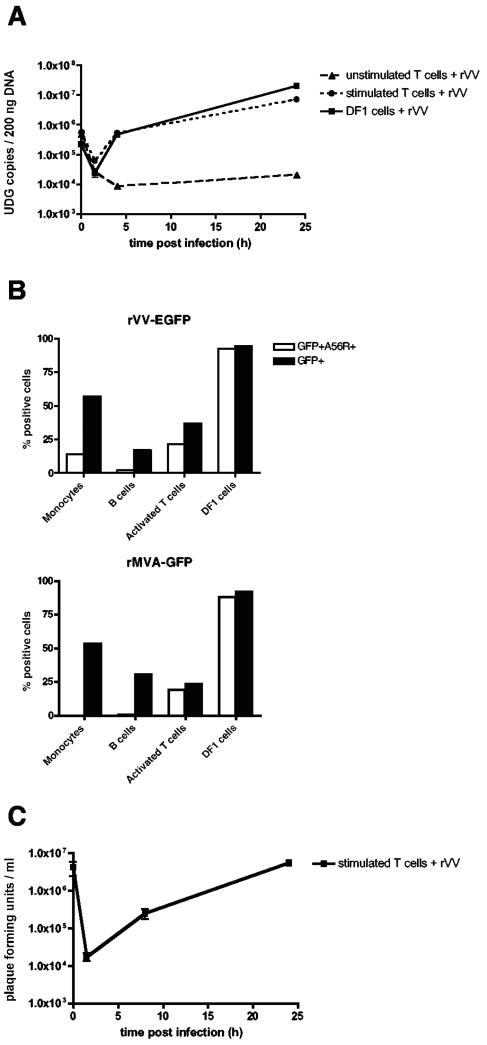
Our initial assessments of VV infection of activated T cells were predicated on evaluation of VV early gene expression alone. We further sought to determine if the virus completes its life cycle in these cells, in contrast to what has been observed for APCs (3, 8, 16, 18, 28). Importantly, permissive infection of activated T cells could represent a mechanism for virus dissemination throughout the lymphatic system. Towards this end, purified activated T cells were infected with rVV-EGFP, and viral DNA replication was monitored over a 24-h period by a real-time PCR assay to detect the conserved UDG gene, encoded by D4R (52). Input virus copy number was measured at time zero and, after an eclipse phase, both activated T cells and a VV-permissive cell line (DF-1) successfully replicated the virus, showing a 1.11- and 1.95-log increase, respectively, in UDG copy number at 24 h (Fig. 7A). Unstimulated T cells, which did not display evidence of VV binding or early gene expression, were also not able to replicate VV (showing a 1.38-log decrease in UDG copy number at 24 h compared to 0 h) (Fig. 7A), further demonstrating their resistance to infection.
FIG. 7.
VV infection of activated T cells is permissive. (A) DF-1 cells (solid line, squares), unstimulated T cells (dashed line, triangles), and T cells stimulated with plate-bound anti-CD3 plus anti-CD28 and anti-CD49d for 68 h (dotted line, circles) were infected in duplicate with rVV-EGFP at an MOI of 1 for 0, 1.5, 4, and 24 h. At each time point, cells were lysed, total DNA was extracted, and viral replication was assessed by real-time PCR with primers specific for the VV UDG gene. The mean and standard deviation are plotted. (B) Cells were infected with rVV-EGFP or rMVA-GFP and expression of a virus late gene product was measured by cell surface staining for the VV/MVA hemagglutinin (HA, encoded by A56R) in conjunction with detection of the early EGFP signal at 16 h postinfection. Cells were identified as follows: monocytes, CD14+ PBMCs; B cells, CD19+ PBMCs; PHA-activated T cells, CD3+CD25+. DF-1 cells were included as a positive control. (C) T cells were activated as described in the legend to panel A were infected with rVV-EGFP at an MOI of 1; at 0, 1.5, 8, and 24 h postinfection, cells were lysed and titers were determined for supernatants on BSC40 monolayers in duplicate. After 2 days, plates were stained with neutral red, and plaques were counted the following day. Titers are presented as PFU per milliliter, and the mean and standard deviation are plotted. These data are representative of three independent experiments.
We next looked for the expression of a viral late gene in infected cells. PBMCs, activated T cells, and DF-1 cells were infected with rVV-EGFP or rMVA-GFP at an MOI of 10 for 16 h, followed by MAb staining for viral HA, encoded by A56R. The A56R gene, which is intact in MVA, is under the control of early and late promoters, but the majority of HA protein accumulates late (9). Although MVA nonproductively infects nonavian cells, the block to virus replication is believed to occur after late gene expression, at the stage of virion assembly (10, 34, 51). By flow cytometry, we found that both DF-1 cells and activated T cells express levels of HA that are comparable to the levels of EGFP (controlled by an early promoter), but that, in CD14+ monocytes and CD20+ B cells, A56R staining was very low or absent compared to EGFP, following either VV or MVA infection (Fig. 7B). This loss of late gene expression in monocytes and B cells confirms previous reports of abortive infection in APCs (3, 8, 16, 18, 28). The low level of A56R staining seen in VV-infected monocytes presumably resulted from the minority of HA that is expressed early.
To confirm their ability to support a permissive VV infection, rVV-EGFP-infected activated T-cell lysates were titered on BSC40 cell monolayers. Progeny virus production from resting T cells and APCs was not tested, due to their lack of active DNA replication or late gene expression, respectively (Fig. 7A and B). Figure 7C shows the input virus titer at 0 h, followed by an eclipse phase as virus enters activated T cells and uncoats (1.5 h) and subsequent production of infectious viral progeny at 8 and 24 h postinfection. Thus, VV undergoes successful transit through all stages of a productive poxvirus life cycle in activated T cells.
DISCUSSION
In recent years, the interest in poxviruses has increased enormously, largely due to their potential as vectors for vaccines and their possible use as agents of bioterrorism (1, 7, 17, 35, 38, 43). Optimization of poxvirus vectors for the derivation of effective and safer smallpox vaccines and as vehicles for the induction of immune responses against other pathogens will depend on a better understanding of the processes by which VV leads to the generation of antiviral immunity. Knowledge of the cellular tropism of VV may help us to further understand the mechanisms of poxvirus infection and immunity. In this report, we show that VV does not indiscriminately infect all cell types it encounters but instead demonstrates a more limited tropism when primary human cells of hematolymphoid origin, rather than cell lines, are considered. Among hematolymphoid cell lineages, VV preferentially targets APCs (peripheral blood DCs, monocytes/macrophages, and B cells to a lesser extent) and activated T cells for infection but does not infect resting T cells or NK cells. This restricted tropism has been seen with a number of diverse VV isolates. The reproducibility of these findings across multiple donors and the concordance of data generated from studies of both VV infection and VV binding lead us to conclude that, for primary human cells, susceptibility to infection is determined by the restricted expression of a poxvirus cellular receptor.
The observation that most activated, but not resting, T cells bind and are able to be permissively infected by VV is suggestive of a poxvirus receptor whose expression is induced upon T-cell activation. Although it is possible that cells of different lineages possess different VV receptors, the same receptor found on activated T cells may also be constitutively expressed on the surface of monocytes, DCs, and B cells, as these APCs bound rVV-B5R-EGFP with high efficiency (Fig. 3A), as well as epithelial cells and fibroblasts, two cell subsets that are major sites of poxvirus infection in vivo (58). In addition, VV may use this cellular receptor to infect immortalized cell lines in culture, but it is also possible that during tissue culture propagation VV preferentially utilizes one or many receptors that are not representative of the predominant one used for infection of primary cells in culture (or in vivo, as exemplified by the exclusive use of CXCR4 by tissue culture cell line-adapted human immunodeficiency virus type 1 isolates) (21). Support for the latter hypothesis comes from our studies of VV binding to primary T cells activated ex vivo compared to the HTLV-1-transformed, activated T-cell line C8166. Mouse serum raised against human activated T cells was able to coat the surface of both cell types, due to shared cell surface molecules, but only inhibited VV binding to the primary cells (Fig. 6B and C). Significantly, mouse serum raised against human monocytes also inhibited VV binding to activated T cells, suggesting that VV uses a common receptor to bind to primary cells of different lineages in vivo (Fig. 6B). The finding that the receptor expressed on activated T cells and monocytes appears to be distinct from the one VV employs to infect cell lines in culture emphasizes the absolute importance of using primary cells for investigations aimed at determining the identity of the VV receptor relevant to in vivo infection.
As mentioned previously, to detect VV binding we used a form of the virus best described as an IMV with retained EEV membrane proteins (Carter et al., XVth Intl. Poxvirus and Iridovirus Conf.). The B5R-EGFP fusion protein is localized to the CEV/EEV membrane, but one freeze-thaw cycle results in disruption of this membrane and consequent exposure of IMV surface proteins, resulting in a particle that is fully infectious as an IMV (5, 37, 56) and that, when bound to cells, is recognized by MAbs to both IMV and EEV membrane proteins (Fig. 2). Thus, rVV-B5R-EGFP binding to the cell surface could be mediated via EEV or IMV membrane proteins, although the EGFP fluorescence signal derives only from the B5R-EGFP fusion protein resident in the EEV membrane. Our rVV-B5R-EGFP preparation likely also contains pure IMV (with no EGFP on its membrane), which may bind to the cell surface as well, although no IMV without EGFP was observed on the surface of BSC40 cells (Fig. 2). Pure IMV binding may be more apparent at an MOI of <50 (which was used here to see levels of binding similar to that for infection with an MOI of 10). Even with this caveat in mind, the correlation between cells that bind rVV-B5R-EGFP and cells that are infected by rVV-EGFP (or rMVA-GFP) emphasizes the previously unappreciated restricted cellular tropism of VV. Still, to identify the cellular receptor(s) specific for either the IMV or EEV form of VV, more pure preparations of virus should be utilized.
In additional studies comparing the binding of VV to primary cells and cell lines, we observed that cell surface enzymatic digestion with both pronase and trypsin abrogated binding to activated T cells, C8166 cells, and DF-1 cells (Fig. 5A). A MAb (B2) that prevents VV binding to the surface of BSC40 cells is believed to target a trypsin- and pronase-sensitive molecule (11), in keeping with the loss of VV binding we observed following cell surface treatment with both enzymes. Others have shown that the IMV and EEV forms of VV behave differently in their binding to trypsin- or pronase-treated RK13 cells and HeLa cells. Specifically, pronase treatment reduced IMV binding to the cell surface and increased EEV binding, whereas trypsin treatment also increased EEV binding but had no effect on IMV (54). It is difficult to reconcile the reduction in binding seen following trypsin digestion of all three cell types we studied (and reduction in infection of activated T cells) with these findings, regardless of IMV or EEV membrane proteins being responsible for rVV-B5R-EGFP binding in our system.
Further research will be needed to identify the poxvirus receptor (or receptors) on primary, human hematolymphoid cells. These studies should be greatly facilitated by the ability to induce receptor expression on activated T cells by defined stimuli within a defined time period (and to block its expression at the levels of transcription, translation, and transport). Towards this end, we have shown that a CD3 agonist and the mitogen PHA can induce VV receptor expression on T cells but that costimulation is not necessary (Fig. 3B and C). The genomic expression program of T-cell activation induced by different stimuli has been described previously (14); elucidation of the role of specific transcription factors in upregulating the VV receptor may help define the subset of candidate genes likely to be useful for its identification.
We observed a discrepancy in the percentage of B cells (54%) able to bind rVV-B5R-EGFP and the percentage (12%) that expressed EGFP after 6 h of infection with rVV-EGFP (Fig. 1A and 2A), although the ability to bind to VV still predicted susceptibility to infection. Interestingly, 42% of B cells expressed GFP after rMVA-GFP infection (Fig. 1A). Similar levels of binding and infection have been observed in murine splenic B cells exposed to VV or MVA (L. Liu et al., unpublished data). These results may reflect a biologic mechanism that distinguishes the postbinding infectious process or the timing of early gene expression of the two virus strains in certain cell types.
Most of the available data that describe poxvirus pathogenesis come from studies of experimental infections in animals. The sites of primary infection and virus replication in cynomologus monkeys exposed to lethal doses of aerosolized monkeypox, a model of naturally acquired Orthopoxvirus infection, were shown to be the lower airway epithelium and neighboring lymphoid tissues (58). Consistent with what has been postulated for variola virus using a variety of poxvirus infection models, Zaucha et al. speculate that the systemic spread of virus occurred via macrophages and DCs carried in the lymph (7, 22, 58). However, our results raise the alternative possibility that activated T cells may be an important and perhaps primary agent of virus replication and dissemination of virus infection via the lymphatics. VV infection of monocytes/macrophages and DCs (and, in fact, B cells) is abortive and results in apoptosis (Fig. 7B) (3, 8, 16, 18, 28, 50). Thus, unless they serve as passive carriers of virus, macrophages and DCs are unlikely to represent primary niduses of infection in vivo. Although VV replication in PHA-stimulated (and not unstimulated) leukocytes was first observed over 30 years ago (4, 39), immunologic and virologic tools necessary to characterize the infectious process in detail were not available at that time, and no subsequent publications have addressed this interesting issue.
We have shown that vaccinia virus tropism for cells encountered in vivo is far more limited than has been presumed based on tissue culture studies alone. Determining the specific sequence of events involved in VV and other poxvirus binding to and entry into primary host target cells may reveal important aspects of virus dissemination and pathogenesis in vivo. Further, such studies may also aid in the design of antiviral therapies that could protect susceptible individuals from infection with monkeypox, a potential emerging pathogen, or smallpox, in the case of accidental or bioterrorist release. If receptor binding is the main determinant of virus entry, then anti-receptor antibodies or small molecule inhibitors of the binding reaction may prove useful in preventing infection. In addition, a better appreciation of the issue of viral tropism in vivo may facilitate the development of vaccines that are specifically engineered to target pathways of antigen presentation and T-cell priming that result in maximal immunogenicity.
Acknowledgments
We thank Bernard Moss for rVV-B5R-EGFP, rVV (WR strain), and rVV (IHD-J strain); Lawrence Corey for rVV-EGFP; Inger Damon for MAb 2D5; and Alan Schmaljohn for VVI-6B6, VV4-2F6, and VVI-4G9. We thank David Lee for technical assistance, Dirk Hunt for help with confocal microscopy, and David Garber and Richard Moyer for critical review of the manuscript.
This work was supported by grants from the National Institutes of Health (P01-AI46007 and U19-AI061728) and the Elizabeth Glaser Scientist Award from the Pediatric AIDS Foundation (6-98 EGSA), all awarded to M.B.F.
REFERENCES
- 1.Amara, R. R., F. Villinger, J. D. Altman, S. L. Lydy, S. P. O'Neil, S. I. Staprans, D. C. Montefiori, Y. Xu, J. G. Herndon, L. S. Wyatt, M. A. Candido, N. L. Kozyr, P. L. Earl, J. M. Smith, H. L. Ma, B. D. Grimm, M. L. Hulsey, J. Miller, H. M. McClure, J. M. McNicholl, B. Moss, and H. L. Robinson. 2001. Control of a mucosal challenge and prevention of AIDS by a multiprotein DNA/MVA vaccine. Science 292:69-74. [DOI] [PubMed] [Google Scholar]
- 2.Appleyard, G., A. J. Hapel, and E. A. Boulter. 1971. An antigenic difference between intracellular and extracellular rabbitpox virus. J. Gen. Virol. 13:9-17. [DOI] [PubMed] [Google Scholar]
- 3.Baixeras, E., A. Cebrian, J. P. Albar, J. Salas, A. C. Martinez, E. Vinuela, and Y. Revilla. 1998. Vaccinia virus-induced apoptosis in immature B lymphocytes: role of cellular Bcl-2. Virus Res. 58:107-113. [DOI] [PubMed] [Google Scholar]
- 4.Benda, R., J. Cinatl, and V. Plaisner. 1975. Reproduction of vaccinia virus (strain neurolapina) in rabbit blood leucocytes in vitro. J. Hyg. Epidemiol. Microbiol. Immunol. 19:93-104. [PubMed] [Google Scholar]
- 5.Bjorck, P. 2001. Isolation and characterization of plasmacytoid dendritic cells from Flt3 ligand and granulocyte-macrophage colony-stimulating factor-treated mice. Blood 98:3520-3526. [DOI] [PubMed] [Google Scholar]
- 6.Blasco, R., and B. Moss. 1992. Role of cell-associated enveloped vaccinia virus in cell-to-cell spread. J. Virol. 66:4170-4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Breman, J. G., and D. A. Henderson. 2002. Diagnosis and management of smallpox. N. Engl. J. Med. 346:1300-1308. [DOI] [PubMed] [Google Scholar]
- 8.Broder, C. C., P. E. Kennedy, F. Michaels, and E. A. Berger. 1994. Expression of foreign genes in cultured human primary macrophages using recombinant vaccinia virus vectors. Gene 142:167-174. [DOI] [PubMed] [Google Scholar]
- 9.Brown, C. K., P. C. Turner, and R. W. Moyer. 1991. Molecular characterization of the vaccinia virus hemagglutinin gene. J. Virol. 65:3598-3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carroll, M. W., and B. Moss. 1997. Host range and cytopathogenicity of the highly attenuated MVA strain of vaccinia virus: propagation and generation of recombinant viruses in a nonhuman mammalian cell line. Virology 238:198-211. [DOI] [PubMed] [Google Scholar]
- 11.Chang, W., J. C. Hsiao, C. S. Chung, and C. H. Bair. 1995. Isolation of a monoclonal antibody which blocks vaccinia virus infection. J. Virol. 69:517-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chung, C. S., J. C. Hsiao, Y. S. Chang, and W. Chang. 1998. A27L protein mediates vaccinia virus interaction with cell surface heparan sulfate. J. Virol. 72:1577-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Depper, J. M., W. J. Leonard, M. Kronke, P. D. Noguchi, R. E. Cunningham, T. A. Waldmann, and W. C. Greene. 1984. Regulation of interleukin 2 receptor expression: effects of phorbol diester, phospholipase C, and reexposure to lectin or antigen. J. Immunol. 133:3054-3061. [PubMed] [Google Scholar]
- 14.Diehn, M., A. A. Alizadeh, O. J. Rando, C. L. Liu, K. Stankunas, D. Botstein, G. R. Crabtree, and P. O. Brown. 2002. Genomic expression programs and the integration of the CD28 costimulatory signal in T cell activation. Proc. Natl. Acad. Sci. USA 99:11796-11801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doms, R. W., R. Blumenthal, and B. Moss. 1990. Fusion of intra- and extracellular forms of vaccinia virus with the cell membrane. J. Virol. 64:4884-4892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Drillien, R., D. Spehner, A. Bohbot, and D. Hanau. 2000. Vaccinia virus-related events and phenotypic changes after infection of dendritic cells derived from human monocytes. Virology 268:471-481. [DOI] [PubMed] [Google Scholar]
- 17.Earl, P. L., J. L. Americo, L. S. Wyatt, L. A. Eller, J. C. Whitbeck, G. H. Cohen, R. J. Eisenberg, C. J. Hartmann, D. L. Jackson, D. A. Kulesh, M. J. Martinez, D. M. Miller, E. M. Mucker, J. D. Shamblin, S. H. Zwiers, J. W. Huggins, P. B. Jahrling, and B. Moss. 2004. Immunogenicity of a highly attenuated MVA smallpox vaccine and protection against monkeypox. Nature 428:182-185. [DOI] [PubMed] [Google Scholar]
- 18.Engelmayer, J., M. Larsson, M. Subklewe, A. Chahroudi, W. I. Cox, R. M. Steinman, and N. Bhardwaj. 1999. Vaccinia virus inhibits the maturation of human dendritic cells: a novel mechanism of immune evasion. J. Immunol. 163:6762-6768. [PubMed] [Google Scholar]
- 19.Eppstein, D. A., Y. V. Marsh, A. B. Schreiber, S. R. Newman, G. J. Todaro, and J. J. Nestor, Jr. 1985. Epidermal growth factor receptor occupancy inhibits vaccinia virus infection. Nature 318:663-665. [DOI] [PubMed] [Google Scholar]
- 20.Fairbanks, L. D., M. Bofill, K. Ruckemann, and H. A. Simmonds. 1995. Importance of ribonucleotide availability to proliferating T-lymphocytes from healthy humans. Disproportionate expansion of pyrimidine pools and contrasting effects of de novo synthesis inhibitors. J. Biol. Chem. 270:29682-29689. [PubMed] [Google Scholar]
- 21.Feng, Y., C. C. Broder, P. E. Kennedy, and E. A. Berger. 1996. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science 272:872-877. [DOI] [PubMed] [Google Scholar]
- 22.Fenner, F., D. A. Henderson, I. Arita, Z. Jezek, and I. D. Ladnyi. 1988. Smallpox and its eradication. World Health Organization, Geneva, Switzerland.
- 23.Himly, M., D. N. Foster, I. Bottoli, J. S. Iacovoni, and P. K. Vogt. 1998. The DF-1 chicken fibroblast cell line: transformation induced by diverse oncogenes and cell death resulting from infection by avian leukosis viruses. Virology 248:295-304. [DOI] [PubMed] [Google Scholar]
- 24.Hsiao, J. C., C. S. Chung, and W. Chang. 1999. Vaccinia virus envelope D8L protein binds to cell surface chondroitin sulfate and mediates the adsorption of intracellular mature virions to cells. J. Virol. 73:8750-8761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hugin, A. W., and C. Hauser. 1994. The epidermal growth factor receptor is not a receptor for vaccinia virus. J. Virol. 68:8409-8412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ichihashi, Y. 1996. Extracellular enveloped vaccinia virus escapes neutralization. Virology 217:478-485. [DOI] [PubMed] [Google Scholar]
- 27.Ichihashi, Y., T. Takahashi, and M. Oie. 1994. Identification of a vaccinia virus penetration protein. Virology 202:834-843. [DOI] [PubMed] [Google Scholar]
- 28.Jenne, L., C. Hauser, J. F. Arrighi, J. H. Saurat, and A. W. Hugin. 2000. Poxvirus as a vector to transduce human dendritic cells for immunotherapy: abortive infection but reduced APC function. Gene Ther. 7:1575-1583. [DOI] [PubMed] [Google Scholar]
- 29.Karlsson, H., and L. Nassberger. 1995. Influence of compounds affecting synthesis, modification and transport of proteins on the expression and release of interleukin-2 receptor. Immunol. Cell Biol. 73:81-88. [DOI] [PubMed] [Google Scholar]
- 30.Lalani, A. S., J. Masters, W. Zeng, J. Barrett, R. Pannu, H. Everett, C. W. Arendt, and G. McFadden. 1999. Use of chemokine receptors by poxviruses. Science 286:1968-1971. [DOI] [PubMed] [Google Scholar]
- 31.Lin, C. L., C. S. Chung, H. G. Heine, and W. Chang. 2000. Vaccinia virus envelope H3L protein binds to cell surface heparan sulfate and is important for intracellular mature virion morphogenesis and virus infection in vitro and in vivo. J. Virol. 74:3353-3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marsh, Y. V., and D. A. Eppstein. 1987. Vaccinia virus and the EGF receptor: a portal for infectivity? J. Cell Biochem. 34:239-245. [DOI] [PubMed] [Google Scholar]
- 33.Masters, J., A. A. Hinek, S. Uddin, L. C. Platanias, W. Zeng, G. McFadden, and E. N. Fish. 2001. Poxvirus infection rapidly activates tyrosine kinase signal transduction. J. Biol. Chem. 276:48371-48375. [DOI] [PubMed] [Google Scholar]
- 34.Mayr, A., V. Hochstein-Mintzel, and H. Stickl. 1975. Abstammung, Eigenschaften und Verwendung des attenuierten Vaccinia-Stammes MVA. Infection 3:6-16. [Google Scholar]
- 35.McConkey, S. J., W. H. Reece, V. S. Moorthy, D. Webster, S. Dunachie, G. Butcher, J. M. Vuola, T. J. Blanchard, P. Gothard, K. Watkins, C. M. Hannan, S. Everaere, K. Brown, K. E. Kester, J. Cummings, J. Williams, D. G. Heppner, A. Pathan, K. Flanagan, N. Arulanantham, M. T. Roberts, M. Roy, G. L. Smith, J. Schneider, T. Peto, R. E. Sinden, S. C. Gilbert, and A. V. Hill. 2003. Enhanced T-cell immunogenicity of plasmid DNA vaccines boosted by recombinant modified vaccinia virus Ankara in humans. Nat. Med. 9:729-735. [DOI] [PubMed] [Google Scholar]
- 36.McFadden, G. 2005. Poxvirus tropism. Nat. Rev. Microbiol. 3:201-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McIntosh, A. A., and G. L. Smith. 1996. Vaccinia virus glycoprotein A34R is required for infectivity of extracellular enveloped virus. J. Virol. 70:272-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McShane, H., A. A. Pathan, C. R. Sander, S. M. Keating, S. C. Gilbert, K. Huygen, H. A. Fletcher, and A. V. Hill. 2004. Recombinant modified vaccinia virus Ankara expressing antigen 85A boosts BCG-primed and naturally acquired antimycobacterial immunity in humans. Nat. Med. 10:1240-1244. [DOI] [PubMed] [Google Scholar]
- 39.Miller, G., and J. F. Enders. 1968. Vaccinia virus replication and cytopathic effect in cultures in phytohemagglutinin-treated human peripheral blood leukocytes. J. Virol. 2:787-792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moss, B. 2001. Poxviridae: the viruses and their replication, p. 2849-2883. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus, Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 41.Nylander, S., and I. Kalies. 1999. Brefeldin A, but not monensin, completely blocks CD69 expression on mouse lymphocytes: efficacy of inhibitors of protein secretion in protocols for intracellular cytokine staining by flow cytometry. J. Immunol. Methods 224:69-76. [DOI] [PubMed] [Google Scholar]
- 42.Payne, L. G. 1979. Identification of the vaccinia hemagglutinin polypeptide from a cell system yielding large amounts of extracellular enveloped virus. J. Virol. 31:147-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Robinson, H. L. 2003. Prime boost vaccines power up in people. Nat. Med. 9:642-643. [DOI] [PubMed] [Google Scholar]
- 44.Roos, N., M. Cyrklaff, S. Cudmore, R. Blasco, J. Krijnse-Locker, and G. Griffiths. 1996. A novel immunogold cryoelectron microscopic approach to investigate the structure of the intracellular and extracellular forms of vaccinia virus. Embo. J. 15:2343-2355. [PMC free article] [PubMed] [Google Scholar]
- 45.Salahuddin, S. Z., P. D. Markham, F. Wong-Staal, G. Franchini, V. S. Kalyanaraman, and R. C. Gallo. 1983. Restricted expression of human T-cell leukemia-lymphoma virus (HTLV) in transformed human umbilical cord blood lymphocytes. Virology 129:51-64. [DOI] [PubMed] [Google Scholar]
- 46.Schaefer-Klein, J., I. Givol, E. V. Barsov, J. M. Whitcomb, M. VanBrocklin, D. N. Foster, M. J. Federspiel, and S. H. Hughes. 1998. The EV-O-derived cell line DF-1 supports the efficient replication of avian leukosis-sarcoma viruses and vectors. Virology 248:305-311. [DOI] [PubMed] [Google Scholar]
- 47.Schon, A., and E. Freire. 1989. Thermodynamics of intersubunit interactions in cholera toxin upon binding to the oligosaccharide portion of its cell surface receptor, ganglioside GM1. Biochemistry 28:5019-5024. [DOI] [PubMed] [Google Scholar]
- 48.Senkevich, T. G., B. M. Ward, and B. Moss. 2004. Vaccinia virus entry into cells is dependent on a virion surface protein encoded by the A28L gene. J. Virol. 78:2357-2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smith, G. L., A. Vanderplasschen, and M. Law. 2002. The formation and function of extracellular enveloped vaccinia virus. J. Gen. Virol. 83:2915-2931. [DOI] [PubMed] [Google Scholar]
- 50.Subklewe, M., A. Chahroudi, A. Schmaljohn, M. G. Kurilla, N. Bhardwaj, and R. M. Steinman. 1999. Induction of Epstein-Barr virus-specific cytotoxic T-lymphocyte responses using dendritic cells pulsed with EBNA-3A peptides or UV-inactivated, recombinant EBNA-3A vaccinia virus. Blood 94:1372-1381. [PubMed] [Google Scholar]
- 51.Sutter, G., and B. Moss. 1992. Nonreplicating vaccinia vector efficiently expresses recombinant genes. Proc. Natl. Acad. Sci. USA 89:10847-10851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Upton, C., D. T. Stuart, and G. McFadden. 1993. Identification of a poxvirus gene encoding a uracil DNA glycosylase. Proc. Natl. Acad. Sci. USA 90:4518-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vanderplasschen, A., M. Hollinshead, and G. L. Smith. 1998. Intracellular and extracellular vaccinia virions enter cells by different mechanisms. J. Gen. Virol. 79:877-887. [DOI] [PubMed] [Google Scholar]
- 54.Vanderplasschen, A., and G. L. Smith. 1997. A novel virus binding assay using confocal microscopy: demonstration that the intracellular and extracellular vaccinia virions bind to different cellular receptors. J. Virol. 71:4032-4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ward, B. M., and B. Moss. 2001. Visualization of intracellular movement of vaccinia virus virions containing a green fluorescent protein-B5R membrane protein chimera. J. Virol. 75:4802-4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wolffe, E. J., E. Katz, A. Weisberg, and B. Moss. 1997. The A34R glycoprotein gene is required for induction of specialized actin-containing microvilli and efficient cell-to-cell transmission of vaccinia virus. J. Virol. 71:3904-3915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wolffe, E. J., S. Vijaya, and B. Moss. 1995. A myristylated membrane protein encoded by the vaccinia virus L1R open reading frame is the target of potent neutralizing monoclonal antibodies. Virology 211:53-63. [DOI] [PubMed] [Google Scholar]
- 58.Zaucha, G. M., P. B. Jahrling, T. W. Geisbert, J. R. Swearengen, and L. Hensley. 2001. The pathology of experimental aerosolized monkeypox virus infection in cynomolgus monkeys (Macaca fascicularis). Lab. Investig. 81:1581-1600. [DOI] [PMC free article] [PubMed] [Google Scholar]