Abstract
Hepatitis C virus (HCV) infection is a major global health problem. Hepatic expression of immune costimulatory signaling molecules (e.g., B7) is known to be associated with ongoing liver injury in hepatitis C patients. However, due to the general lack of viral culture systems and adequate animal models, the function of these molecules in disease pathogenesis is poorly understood. To investigate the role of CD86 in HCV-related liver injury, we developed two transgenic mouse lineages with inducible expression of HCV structural proteins and constitutive expression of the costimulatory molecule CD86/B7.2 in the liver. Using a hydrodynamic-based, nonviral delivery protocol, we induced HCV transgene expression in the livers of HCV and CD86 single- and double-transgenic mice. We found that hepatic CD86 expression resulted in increased activation of and cytokine production (e.g., interleukin-2 and gamma interferon) by CD4+ T cells and that the retention of these cells was associated with more pronounced necroinflammatory lesions in the liver. Taken together, these data suggest that augmented, parenchymal antigen presentation conferred by hepatocyte CD86 expression alters homeostasis and effector functions of CD4+ T cells and contributes to liver injury. This study provides an additional rationale for exploring immunomodulation-based therapies that could reduce disease progression in individuals with chronic HCV infection.
Liver disease due to hepatitis C virus (HCV) infection is an emerging public health problem, as persistent viral infection leads to liver cirrhosis and cancer in some patients (63a). No vaccine for prevention of HCV infection exists, and current interferon-based therapies result in a sustained antiviral response in only about 50% of patients (39). The mechanisms of pathogenesis are not fully understood. Since immune-based therapies that could improve viral clearance and reduce disease progression in chronically infected individuals would be of great benefit (21, 41, 65), a better understanding of the mechanisms contributing to liver injury and T-cell clearance of HCV infection are important goals for HCV research.
The body maintains tolerance to many gut-derived antigens (54). Although the mechanisms underlying this process remain unclear, it is thought that immune responses within the liver are associated with tolerance. In the liver, hepatocytes express low levels of major histocompatibility complex (MHC) and virtually no immune costimulatory molecules (such as CD80/B7.1, CD86/B7.2, and CD40). These conditions ensure that T cells “ignore” antigens expressed by the parenchymal cells. To mount an efficient immune response, costimulatory molecules on antigen-presenting cells need to engage their ligands on T cells, and this interaction provides a crucial signal permitting the activation and differentiation of T cells into effector cells. In human HCV infection, high levels of MHC class II and costimulatory molecules are expressed on the activated Kupffer cells and hepatocytes, and their levels are also correlated with the extent of intrahepatic inflammation and elevation of serum alanine aminotransferase (ALT) a biochemical marker of liver injury (10, 37). Despite this association between costimulatory molecules and these clinical parameters of disease, the functional relationship between expression of these molecules and HCV-related liver injury has not been directly examined, partly due to a lack of suitable animal models.
To examine the pathogenesis of HCV in vivo, several groups have established transgenic mice that constitutively express HCV proteins in the liver and have found increased micro- and macrovesicular hepatic steatosis and an increased incidence of hepatocellular carcinoma in these animals (32, 38, 55). However, since the constitutive expression of HCV proteins begins in utero in these mice, they do not develop HCV-specific cellular responses or hepatic inflammation due to immune tolerance to the HCV antigens. To overcome this obstacle, we developed transgenic mouse lineages with inducible expression of HCV structural proteins and/or constitutive expression of the costimulatory molecule CD86/B7.2 in the liver, and we investigated the relationship between costimulatory signaling molecule CD86 expression and HCV-related hepatic inflammation in double-transgenic animals. Using a hydrodynamic-based, nonviral delivery protocol (33), we induced expression of the HCV transgene in mouse liver with Cre DNA recombinase (30) and found that animals coexpressing CD86 developed more pronounced inflammatory responses within the liver. Our results suggest that increased parenchymal antigen presentation conferred by hepatocyte CD86 expression prolongs the presence and augments the functions of effector T cells in the liver, thus contributing to HCV-related liver injury.
MATERIALS AND METHODS
Transgenic mice.
The transgene construct carrying loxP sequences was derived from pAlbSVPA-HCV-S, containing the core, E1, and E2-p7 sequences of genotype 1b HCV (32). A 1.4-kb spacer, containing stop codons in all three reading frames and flanked by loxP sequences at both ends, was copied by PCR from pMA19 (2) and inserted between the promoter and the HCV structural genes (Fig. 1A). Thus, the HCV sequences in the construct are transcribed, but not translated, unless the spacer is removed by Cre-mediated DNA recombination. Transgenic mice were derived from C57BL/6 eggs by using a standard procedure at the University of Texas Medical Branch (UTMB) Transgenic Mouse Core Facility, which has been described in detail elsewhere (32). Genotyping was routinely done by PCR analysis of tail DNA, which was validated by Southern blot analysis. Out of 14 founders, 4 tested positive for the expression of HCV mRNA in the liver by reverse transcription-PCR. A transgenic founder (SL-24), which expressed the highest level, was mated with C57BL/6 mice (H-2b; Jackson Laboratories, Bar Harbor, ME) to produce transgenic offspring. All mice were housed in a specific-pathogen-free facility, and their serology profiles were monitored at regular intervals. The Institutional Animal Care and Use Committee of the UTMB approved the study.
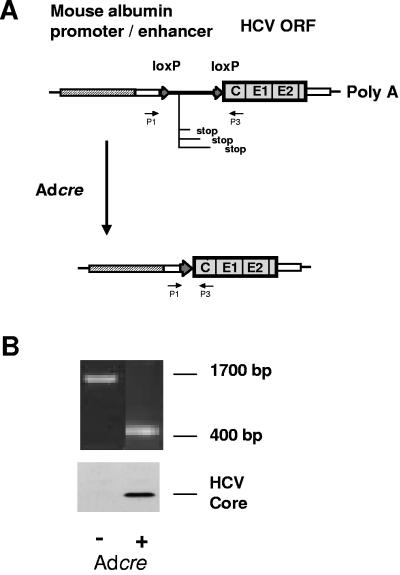
FIG. 1.
Construction of conditional HCV transgenic mouse lines. (A) The transgene construct was prepared by inserting a 1.4-kb spacer that is flanked by two loxP sequences between the promoter and structural genes. A 1.7-kb PCR fragment is detectable with the primer pair P1-P3 in the transgenic mouse. (B) When a recombinant adenovirus carrying the Cre DNA recombinase was injected, the spacer was removed by recombination between the two loxP sequences, as evidenced by the presence of a 400-bp band amplified by PCR using the same primers. Expression of the HCV core protein at 21 kDa in the liver was detected by Western blot analysis.
AT-B7.2 transgenic mice expressing CD86 (B7.2) under the control of the human anti-thrombin III promoter were generated at the Centenary Institute. The construct was obtained by inserting the mouse CD86 cDNA (kindly provided by Sylvie Guerder, Marseille Luminy, France) into the pAT3-Lack plasmid (a kind gift of Nicolas Glaichenhaus, Nice, France) containing the human anti-thrombin III promoter and regulatory intronic sequences of the MHC class II gene. Constructs were linearized, injected into C57BL/6 embryonic stem cells, and transferred into pseudopregnant female mice. Offspring were then screened for incorporation of the construct into the cellular DNA by PCR. Of the 12 lines incorporating the CD86 transgene, only one (AT-B7.2, C line) expressed the CD86 protein on hepatocytes.
Cre DNA recombinase expression vector.
Rearrangement of chromosomally integrated HCV structural genes leading to protein expression was mediated by Cre DNA recombinase from bacteriophage P1 (25). In initial experiments, conditional HCV transgenic mice were induced by infusion via the tail vein of 3 × 109 PFU AdCre, an E1-deleted adenovirus vector carrying the Cre recombinase gene under the control of a cytomegalovirus (CMV) immediate-early promoter. Two Cre plasmids (pCMVCre and pNLSCre) used for the remaining studies were generated by Brian Sauer (Stowers Institute, Kansas City, MO) and are described in detail elsewhere (30, 47). pCMVCre (pBS185) was constructed by fusing the CMV promoter to the N terminus of Cre (47). For more efficient Cre entry into the nuclei of hepatocytes, we used pNLSCre (pBS391), which expresses a modified Cre molecule with a nuclear localization signal (NLS) (30).
Animal experiments.
Hydrodynamic delivery of DNA into mouse liver was carried out as described by Liu et al. (33, 66). Fifty micrograms Cre DNA was diluted in normal saline. A bolus of the solution equivalent to 8 to 10% of the body weight was injected through the tail vein within approximately 5 to 10 s. At days 1, 4 to 7, 14, and 45 postinduction, two to four mice in each group were anesthetized via intraperitoneal (i.p.) injection of 0.25 mg/g pentobarbital. The liver was perfused with 10 to 20 ml of Hanks balanced salts via the portal vein at a flow rate of 10 ml/min with a 21-gauge needle and luer-lock syringe (11). The perfused liver was mechanically disrupted on a 50-mesh, 280-μm stainless steel sieve (Sigma, St. Louis, MO) in Dulbecco's modified Eagle's medium. The cell suspension was filtered through 85-μm nylon mesh and centrifuged at 1,000 rpm at 4°C for 5 min. The pellet was resuspended in Dulbecco's modified Eagle's medium, centrifuged again, and overlaid on Lympholyte-M density medium (Cedarlane Laboratories, Ontario, Canada). After centrifugation, interface mononuclear cells were harvested, washed, and counted. A portion of the isolated lymphocyte fraction was immediately stained with fluorescein isothiocyanate-, phycoerythrin (PE)-, or TriColor-labeled anti-CD3, -CD4, and -CD8 monoclonal antibodies (MAb) (BD PharMingen, San Diego, CA) and analyzed with a FACScan flow cytometer (Becton Dickinson, San Jose, CA). Serum samples were collected and stored at −70°C for ALT measurements. All data were combined from two to four independent experiments.
Determination of Cre-mediated DNA recombination.
Total genomic DNA from the mouse tail and liver was extracted using a QIAamp DNA minikit (QIAGEN Inc., Valencia, CA). Conventional PCR was used for routine analysis of Cre-mediated recombination of the transgene. For this purpose, a pair of primers, P1 (ACC TTT CCG GCA TGC AAG) and P3 (CTT TGA GGT TTA GGA TTC GTG CTC), were used to identify the transgenic and recombination status (Fig. 1A). Copy numbers of recombined transgene in the liver were assessed by real-time quantitative PCR with the same pair of primers. The RS18 gene, which encodes the small ribosomal protein 18, was used as an internal control for all samples. Standard curves were generated for the unrecombined and recombined transgene with pALB.HCV.S-N.lox and pALB.HCV.S-N, respectively, and copy numbers of both amplicons were normalized to that of RS18. Analysis was carried out on an ABI PRISM 7700 sequence detector with a core reagent kit (QuantiTect SYBR Green PCR). The extracted DNA samples were diluted to 1:10 with Tris-EDTA buffer, and amplification reactions were performed with 5-μl sample aliquots in a 25-μl final volume. Reaction cycles for both the target and RS18 included denaturation at 95°C for 15 s, annealing at 60°C for 20 s, and extension at 72°C for 30 s. The standard curve of the threshold cycles versus the template abundance was linear over 6 orders of magnitude, with a regression correlation typically greater than 0.98 for both curves. All data were combined from two independent experiments.
Western blotting and histopathology.
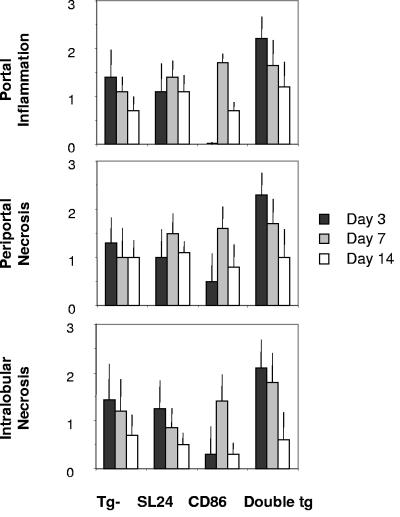
For immunoblotting, liver tissues were snap frozen in liquid nitrogen, and 20 μg of protein extracted from the livers was electrophoresed in a precast 4 to 12% NuPage polyacrylamide gel (Invitrogen, San Diego, CA) in a buffer containing 1% sodium dodecyl sulfate. The proteins were electrotransferred to a polyvinylidene difluoride membrane (Bio-Rad, Hercules, CA) and probed with a MAb for HCV core (Anogen, Mississauga, Canada) or polyclonal Ab for HCV E2 (Biodesign, Saco, ME), followed by a horseradish peroxidase (HRP)-conjugated anti-mouse or anti-goat immunoglobulin G (IgG) (Southern Biotechnology Associates, Birmingham, AL), respectively, and ECL detection reagents (Amersham Pharmacia, Buckinghamshire, England). For evidence of histopathological changes, liver tissues were fixed in 10% buffered formalin, embedded in paraffin, and stained with hematoxylin-eosin. Samples were evaluated by three individuals in a randomized, double-blinded fashion and characterized with respect to (i) periportal degeneration and focal necrosis, (ii) intralobular degeneration and focal necrosis, and (iii) portal inflammation (28). Each parameter was scored from 0 (no pathology) to 4 (severe pathology) (64).
Function of hepatic CD86 in vitro.
Hepatocytes from CD86 and BALB/c mice were isolated as described by Isom (22). Briefly, mouse liver was perfused with liver perfusion medium (Invitrogen/GIBCO) and digested with liver digest medium (Invitrogen/GIBCO) at 10 ml/min for 2 to 3 min each. After being filtered through 45-μm (Tetko, Kansas City, MO; no. 3-45/29) and 80-μm (no. 3-80/42) nylon mesh, the cells were washed with hepatocyte wash medium twice, purified by Percoll (Sigma) density gradient separation, and resuspended in HepatoZYME serum-free medium. Hepatocytes (1 × 104/well) were then seeded into 96-well V-bottom plates in duplicate as potential antigen-presenting cells and pulsed with 10 μM of antigenic peptide (hen egg ovalbumin323-339) for 30 min. T cells used in this study were prepared from the spleens of DO11.10 mice (40). In these mice, a majority of CD4+ T cells was specific for the ovalbumin323-339 peptide on the I-Ad molecules. Splenocytes (1 × 106, 3 × 105, 1 × 105, and 3 × 104) were added to the wells containing loaded hepatocytes. After incubation at 37°C with 5% CO2 for 72 h, cells and supernatant were collected and subjected to flow cytometric, gamma interferon (IFN-γ), and interleukin-2 (IL-2) analyses (54), respectively.
Statistical analysis.
The three-way analysis of variance procedure was used to evaluate the histopathological effects of HCV transgene (factor 1), CD86 transgene (factor 2), and days postinduction (factor 3) independently and interactively. Kruskal-Wallis tests were used for post hoc comparisons as warranted. All statistics were computed using NCSS (Number Cruncher Statistical System) software.
RESULTS
Conditional HCV transgenic mice.
The HCV transgenic mouse was designed to express the structural proteins of HCV only within the liver and in a Cre recombinase-regulated fashion. The transgenic construct was derived from pALB.HCV.S-N, which contains the HCV core-, E1-, E2-, and p7-coding sequences of genotype 1b HCV (32). A 1.4-kb spacer, containing stop codons in all three reading frames and flanked by loxP sequences at both ends, was inserted between the albumin promoter and the HCV structural genes (Fig. 1A). Thus, the HCV sequence in the construct is transcribed, but not translated, unless the spacer is removed by Cre-mediated DNA recombination. Out of 14 founders, 4 tested positive for the expression of HCV mRNA by reverse transcription-PCR. We chose to further characterize founder SL24 and to focus on it in this study, since this lineage expressed the highest level of the transgene. In preliminary studies to confirm Cre regulation of the transgene, these mice were inoculated via the tail vein with an E1-deleted adenovirus vector expressing Cre under the control of a cytomegalovirus immediate-early promoter (AdCre). Genomic DNA was extracted from the liver and analyzed with a PCR that amplified a 1.7-kb segment of the transgenic construct in the uninduced animals (Fig. 1). The same pair of primers generated a 400-bp amplicon at 3 days post-AdCre injection, which suggested that Cre expressed from AdCre recognized loxP sites, thereby removing the spacer containing the translational stop codons and inducing expression of the HCV structural proteins. These transgenic mice expressed high levels of HCV core protein for 2 weeks, with expression decreasing to an undetectable level by day 21 (Fig. 1B and data not shown). Basal-level synthesis of the HCV proteins in mouse liver was not detectable by Western blot analysis (Fig. 1B), indicating tight control of transgene expression in this system.
Nonviral induction of transgene expression.
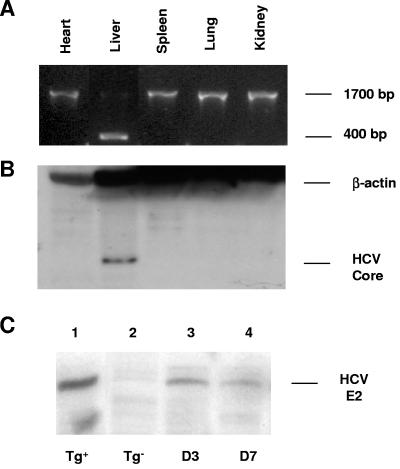
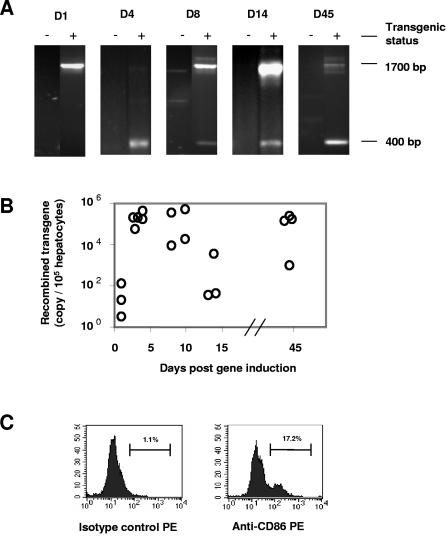
While the adenovirus-based vector is relatively efficient in inducing Cre-mediated DNA recombination, several studies have shown that the viral vector itself can induce strong inflammatory responses in mouse and human livers (24, 48, 52, 62). To avoid this problem, we explored the possibility of delivering Cre using a novel hydrodynamic-based protocol, thereby avoiding the expression of exogenous viral DNA and proteins. This procedure requires a 5- to 10-s intravenous injection of plasmid DNA resuspended in saline equivalent to 8 to 10% of the body weight. This approach has been shown to be efficient in the delivery of DNA into the liver (33, 66). We first injected male and female transgenic mice and their nontransgenic littermates with 50 μg pNLSCre plasmid DNA (30). Following induction, two to four animals were sacrificed at various times postinjection. Tissue samples were collected and analyzed for DNA recombination and protein expression. Since the hydrodynamic injection favors gene delivery to the liver (33, 66), DNA recombination of the HCV transgene took place only in that organ and not in the heart, lungs, spleen, and kidneys, as indicated by the presence or absence of a 400-bp PCR fragment (Fig. 2A). As a result, HCV core as well as E2 protein became detectable in the liver (Fig. 2B and C). The recombined transgenic fragment was clearly detectable by PCR 4 days following pNLSCre injection and persisted through day 45, at which time all of the mice were sacrificed (Fig. 3A). The recombination events mediated by pNLSCre were further quantified by highly sensitive, real-time quantitative PCR. The level of the recombined transgene was no higher than background (32 copies/105 hepatocytes) in HCV transgenic animals 1 day post-pNLSCre injection (Fig. 3B). However, consistent with the results obtained from the qualitative PCR assays (Fig. 3A), there was a 1,000-fold increase in the recombined transgene copy number by day 3, and this remained elevated at very high levels in most animals through day 45. These levels of recombination efficiency were comparable to those mediated by viral vector AdCre (1.1 × 106 copies/105 hepatocytes at day 3) and far superior to those obtained with pCMVCre, a first-generation Cre DNA recombinase that lacks the nuclear localization signal (1.1 × 103 copies/105 hepatocytes at day 3) (45, 46). The reason for low copy numbers in two animals at 14 days postinjection was not clear (Fig. 3B), but it could reflect the possible immune clearance of hepatocytes expressing viral proteins or variations in hydrodynamic injections. These results demonstrate that the hydrodynamic delivery of naked pNLSCre plasmid DNA is effective in inducing chromosomal recombination of HCV transgenic DNA in vivo.
FIG. 2.
Hydrodynamic induction of HCV structural transgenes with a nonviral vector, pNLSCre. (A) Recombination of the HCV transgene took place in the liver following hydrodynamic injection of 50 μg pNLSCre. The 400-bp fragment was detected by PCR with primers P1 and P3 (Fig. 1). (B) High levels of HCV core expression in the liver were detected by Western blotting with an anti-core MAb, followed by HRP-conjugated anti-mouse antibody. Shown are results for a representative HCV transgenic mouse 10 days following pNLSCre injection. (C) HCV E2 expression in the liver at day 3 (lane 3) and day 7 (lane 4) postinduction was detected by Western blotting with an anti-E2 polyclonal Ab, followed by an HRP-conjugated anti-goat IgG. A constitutive HCV transgenic (Tg) mouse (lineage 139) (lane 1) and a nontransgenic mouse (lane 2) were used as positive and negative controls, respectively.
FIG. 3.
Recombined HCV transgene in HCV transgenic mice and CD86 expression in AT-7.2 transgenic mice. Mice were injected with 50 μg pNLSCre. Genomic DNA was extracted from the liver at the indicated days postinduction. (A) PCR specific for Cre and the HCV transgene was performed as for Fig. 1 and 2. Recombined chromosomal DNA remained detectable through day 45 post-pNLSCre injection. (B) Recombination efficiency of the transgene was confirmed by highly sensitive real-time quantitative PCR (see Materials and Methods). Copy numbers of the recombined transgene were measured after induction. DNA recombination took place at day 3 and persisted through day 45. The values are representative of two real-time PCR assays with similar results. (C) Primary hepatocytes were isolated from the liver of a CD86 transgenic mouse and stained with a PE-conjugated MAb against murine CD86 (clone GL-3) along with a rat IgG2a isotype control MAb. The values are representative of two independent experiments with similar results.
Exacerbation of hepatic inflammation due to CD86 expression in HCV transgenic mice.
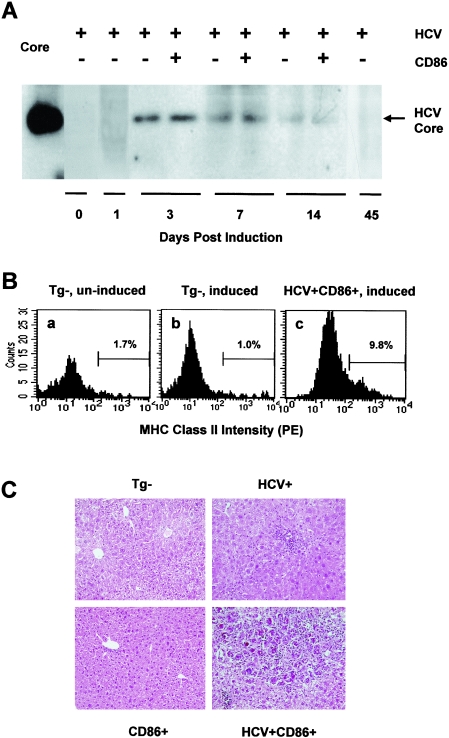
To directly address the role of CD86 in HCV-related hepatic inflammation, we generated a transgenic mouse lineage that expresses CD86 under control of the hepatocyte-specific human anti-thrombin III promoter (AT-B7.2) and crossed these mice with the conditional HCV transgenic (SL-24) mice. The CD86 mice were healthy, with normal liver functions and hepatic histology (D. G. Bowen and P. Bertolino, unpublished results), and approximately 16% of their hepatocytes expressed CD86 molecules, as assessed by flow cytometry (Fig. 3C). Mice from both sexes were divided into nontransgenic, HCV and CD86 single-transgenic, and HCV/CD86 double-transgenic groups, and all received a hydrodynamic injection of 50 μg pNLSCre. Two to four animals from each group were sacrificed at 3, 7, and 14 days. Cre-mediated DNA recombination in the liver was confirmed with PCR as described for Fig. 3A (data not shown). Western blot analyses of liver extracts demonstrated high levels of HCV core and E2 protein expression at days 3 to 14 after transgene induction (Fig. 4A and data not shown). Using an HCV core-specific enzyme-linked immunosorbent assay (trak-C; Ortho, Raritan, NJ), we determined that the expression level of HCV core was approximately 2.1 pg per μg of liver protein extract on day 3, which was sustained for at least 11 days before becoming undetectable at day 45. Although CD86 did not affect HCV transgene induction and expression in the liver (Fig. 4A), coexpression of CD86 and HCV core and envelope proteins resulted in enhanced expression of MHC class II molecules in hepatocytes, as demonstrated by fluorescence-activated cell sorter (FACS) analysis (Fig. 4B).
FIG. 4.
Expression of HCV core and MHC class II as well as hepatic inflammation in double-transgenic mice after gene induction. All mice were injected with 50 μg pNLSCre. (A) Liver protein was extracted at the indicated days. Protein samples (100 μg) were loaded in each lane, and HCV core on a polyvinylidene difluoride membrane was detected with a core-specific MAb. On the left, 6 ng recombinant HCV core (positions 1 to 179) was used as a control. (B) Hepatic MHC class II accumulation following HCV/CD86 coexpression in the liver. Nontransgenic (Tg-) and double-transgenic mice were induced to express HCV structural proteins (or were uninduced). Five days later, the livers were perfused, and hepatocytes were purified and double stained intracellularly with PE-conjugated anti-class II (clone M5/114.15.2) and fluorescein isothiocyanate-conjugated anti-CD11c (clone HL3) MAbs. In hepatocytes free from dendritic cell contamination, the mean fluorescence intensities in uninduced nontransgenic (a), induced nontransgenic (b), and induced double-transgenic (c) mice were 23.5, 28.7, and 68.6, respectively. These results are representative of those from two experiments with similar results. (C) Hepatic inflammation following expression of HCV structural proteins. Liver samples were collected and fixed in 10% buffered formalin. Paraffin-embedded sections were stained with hematoxylin and eosin. The photomicrographs were taken on day 7 after gene induction and are representative of four experiments with similar results (objectives, ×20).
As previously reported (33, 66), hydrodynamic injection of pNLSCre resulted in a mild, transient cellular infiltration in nearly all mice. Histopathological examination of the liver indicated that HCV and CD86 double-transgenic mice developed more severe inflammation in the liver than did those expressing a single or no transgene (Fig. 4C). Hepatic lesions were infiltrated by CD4+, CD8+, B220+, Mac-1+ cells and granulocytes, as determined by immunohistochemical staining (data not shown), and were accompanied by various degrees of apoptotic bodies, ballooning degeneration, and hepatocellular necrosis. In double-transgenic mice, aggregations of multinuclear giant cells were present in foci characteristic of granulomatous inflammation at day 7. As HCV antigen levels declined gradually (Fig. 4A), the severity of pathological changes lessened at day 14 (Fig. 5), and resolved completely at day 45 (data not shown). As in some hepatitis C patients, ALT levels remained within the normal range, except for a mild, injection-induced elevation (up to 120 units/ml) at day 3 in all groups of animals (data not shown). Using three-way variance analysis, we examined the effect of HCV transgene, CD86 transgene, and days postinduction on liver pathological changes, as measured by histological activity indices (HAI) (28, 64). We found that expression of HCV structural proteins, but not CD86 (P > 0.05 for all three anatomical areas), was associated with higher HAIs than those observed in nontransgenic mice. These differences were significant in the portal (P < 0.01) and periportal (P < 0.04) areas and statistically insignificant in the intralobular area (P < 0.11). There was a synergistic effect of HCV structural proteins and CD86 molecules on liver HAIs. This effect was particularly significant in the intralobular area (P < 0.01) and less obvious in the portal (P < 0.06) and periportal (P < 0.07) areas.
FIG. 5.
Histopathological changes of mouse livers in response to HCV core induction. Liver tissues were harvested following injection of 50 μg pNLSCre and evaluated for evidence of pathological changes by light microscopic examination of hematoxylin-and-eosin-stained sections. Samples were characterized with respect to (i) periportal degeneration and focal necrosis, (ii) intralobular degeneration and focal necrosis, and (iii) portal inflammation. The extent of pathology was scored from 0 (no pathology) to 4 (severe pathology). The results from two to four experiments are summarized and depict the mean scores and standard deviations for two to four animals at each data point. Tg, transgenic.
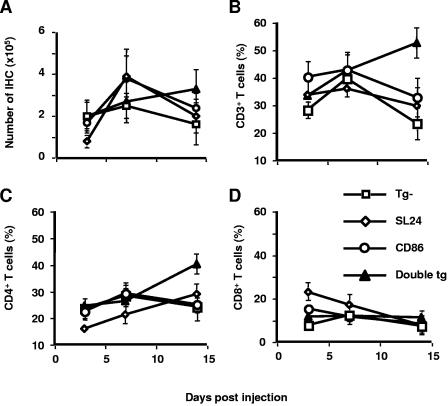
Along with the histopathological changes in the liver, total numbers of inflammatory cells in the HCV and CD86 single-transgenic mice rose sharply at day 7 and then fell at day 14 (Fig. 6A), while those in the double-transgenic mice continued to climb beyond day 7. A considerable increase in the percentage of CD3+ CD4+ T cells in the double-transgenic mice, but not in other groups (Fig. 6B to D), suggested a major expansion of the total number of CD4+ T cells in the liver. This net increase in CD4+ T cells could result from increased recruitment or prolonged survival of these cells, or both, in the liver. These data indicate that hepatocyte CD86 expression results in the increased retention of CD4+ T cells in the liver, which is associated with HCV-related pathological changes (Fig. 4 and 5).
FIG. 6.
Intrahepatic cellular response to HCV transgene induction in mice. After injection of pNLSCre, mice were sacrificed and liver tissues were collected. (A) Intrahepatic lymphocytes were isolated from the liver perfused with Hanks balanced salts, purified with Lympholyte-M separation medium, and counted. (B to D) The infiltrating lymphocytes were stained with MAb specific for murine CD3, CD4, and CD8 and analyzed with three-color FACS analysis. The results from two to four experiments are summarized, and the data points depict the mean percentages ± standard errors of the means of T-cell subsets among two to four animals.
Effect of hepatic CD86 expression on T cells.
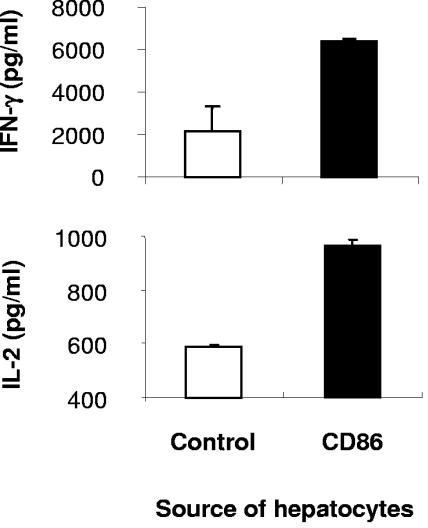
To test whether, upon CD86 expression, hepatocytes could present antigen in an immunogenic form to T cells, we measured T-cell responses to a well-defined antigenic peptide, ovalbumin323-339. In this coculture system, the CD4+ transgenic T cells derived from DO11.10 T-cell receptor transgenic mice are specific for the ovalbumin peptide (40, 44), and hepatocytes with or without CD86 expression were pulsed with the peptide (AT-7.2, H-2d, and BALB/c mice). In the presence of CD86 molecules, the mean forward scatter of DO11 T cells increased from 101.4 to 124.4, consistent with blast formation. During coculture, their percentage expanded from 36.6% to 69.7% among total splenic lymphocytes, and these cells secreted more than 6 ng/ml IFN-γ and nearly 1 ng/ml IL-2 (Fig. 7). In contrast, the same T cells failed to form blasts (mean forward scatter, 106.8) or to expand (33.4%) following coculture with normal nontransgenic hepatocytes, and they produced far less IFN-γ and IL-2 (2.2 and 0.59 ng/ml, respectively). This effect cannot be attributed to dendritic cells, since CD86+ hepatocyte preparations contained equal or slightly fewer CD11c+ cells (clone HL3) detected by FACS (data not shown). A MAb specific for MHC class II (clone M5-114) nearly completely blocked IL-2 secretion in both CD86+ and CD86− control groups (96.7% and 95.4%, respectively), while a MAb against CD86 (clone GL-1) resulted in a partial block (45.7% and 24.5%, respectively). Overall, these results suggest that increased, parenchymal antigen presentation conferred by hepatocyte CD86 expression augments T-cell effector functions and retention, contributing to HCV pathogenesis in the liver.
FIG. 7.
Hepatic expression of CD86 results in increased antigen presentation and T-cell activation. Primary hepatocytes were isolated from CD86 transgenic (H-2d) and BALB/c mice. The hepatocytes were pulsed with 10 μM ovalbumin323-339 peptide and cocultured with the splenic lymphocytes from DO11.10 T-cell receptor transgenic mice for 72 h. The values are representative of two experiments with similar results and show average values and standard deviations from duplicate wells.
DISCUSSION
Hepatitis C research has been hindered by the absence of suitable animal models that mimic the liver injury associated with human HCV infection (1, 9). Chimpanzees have proved invaluable for the characterization of the natural history of infection, immunity, and pathogenesis (15, 29, 50). However, these animals are not widely available and are extraordinarily expensive and difficult to handle. In previous studies, Wakita and coworkers generated a transgenic mouse that can conditionally express HCV proteins in the liver (62) and showed that HCV-specific cytotoxic-T-lymphocyte (CTL) responses in these animals contribute to liver injury (63). Since the adenoviral vector used to induce transgene expression in these mice can, by itself, elicit strong inflammatory responses in the liver, however, this mode of gene induction is not suitable for studies concerning the involvement of immune molecules in the HCV-specific inflammatory process (24, 52).
Over the last several years, a number of nonviral Cre delivery systems have been reported (23, 33). In preliminary studies, we tested a cell-permeative Cre fusion protein (23, 51) and, like others (43), failed to demonstrate any recombinase activity in the livers of HCV transgenic mice following i.p. injections (data not shown). We also found that a first-generation Cre construct (pCMVCre) (47) did not mediate efficient DNA recombination in mouse liver, due primarily to its failure to gain critical access to the chromosomally integrated transgene within the nucleus (data not shown). In this study, injection of an improved pNLSCre containing nuclear localization signals resulted in efficient Cre translocation to the nucleus and, thus, transgene recombination in the liver (Fig. 2, 3, and 4A). The ensuing immune responses, free from those against adenoviral antigens, afford a cleaner system and a viable alternative to virus-based Cre expression strategies in the mouse (62, 63).
Although HCV-specific CD8+ CTLs are capable of migrating to the liver and producing IFN-γ, thereby contributing to viral clearance, they can also cause liver injury (16, 56, 58, 59). Among many factors, the hepatic expression of costimulatory signaling molecules is known to be associated with enduring liver injury. High levels of CD80, CD86, and CD40 are expressed in hepatocytes and activated Kupffer cells in hepatitis C patients, and their levels are closely correlated with those of intrahepatic inflammation and serum ALT (10, 37). Using T-cell receptor transgenic mice, we previously showed that hepatocytes are capable of activating naive T cells but are unable to promote their survival beyond the first 3 days, causing premature T-cell death and tolerance development (3, 4, 8). In this study, we further demonstrated that hepatic CD86 expression resulted in a sustained expansion of CD4+ T cells in the liver and that the increased retention of these cells in the liver was associated with an exacerbation of the necroinflammatory injury in these animals (Fig. 4 to 6). These results are consistent with recent studies in which the liver has been shown to selectively retain large numbers of CD4+ or CD8+ T cells under various experimental conditions (27, 35, 36). Activation and retention of CD8+ T cells in the liver can inflict damage to neighboring hepatocytes through IFN-γ and TNF-α (7, 13). In HCV patients, CTLs specific for the virus can kill uninfected hepatocytes through the Fas-mediated lysis of hepatocytes (16). It has been demonstrated that 0.8 to 1.5% of cells presenting HCV antigens suffice to induce lysis of 10 to 29% of bystander cells, suggesting that this mechanism may be operative when only small numbers of hepatocytes are infected in vivo. This mode of killing has also been implicated in CD4+ Th1-mediated liver injury in a murine hepatitis B model (42).
In this study, we asked whether or not costimulatory molecule CD86 contributes to HCV-related liver damage. Our data suggest that augmented, parenchymal antigen presentation conferred by hepatocyte CD86 expression promotes proliferation and differentiation of antigen-specific CD4+ T cells (Fig. 7) and that the altered effector functions and homeostasis of these cells contribute to liver injury (Fig. 4C, 5, and 6). Additional studies suggest that transient expression of the CD80 costimulatory molecule in the livers of these mice is also associated with hepatic inflammatory lesions (data not shown). These results indicate that HCV-related liver injury may depend not only on T-cell-priming events but also on the ability of the local environment to promote T-cell survival. Indeed, the requirement for a local factor in T-cell-mediated tissue injury is not peculiar to the liver. It has been demonstrated that a viral protein expressed in mouse pancreatic β cells, by itself, cannot induce autoimmune diabetes. However, in transgenic mice coexpressing a costimulatory or local inflammatory molecule, CTLs were activated, and spontaneous insulitis and diabetes occurred (18, 19, 61).
In HCV infection, strong CD4+ T-helper and CD8+ CTL responses are critical in viral clearance or disease progression (17, 26, 49, 50, 60). For reasons that are not clear, the coexpression of HCV and CD86 proteins in the livers of double-transgenic mice elicited neither a major CD8+ T-cell expansion nor persistent ALT elevation (Fig. 6 and data not shown). This lack of a CD8+ expansion could either result from a genetic restriction on recognition of MHC class I epitopes of HCV in this strain of mouse (H-2b/k) or be attributed to insufficient cross-priming due to the absence of viral replication. It has been reported that cross-priming in dendritic cells requires an activation or “licensing” step and that IFN-α, which is induced effectively by viral double-stranded RNA or DNA (6, 14, 34, 53), is an important “licensor.” This action is independent of CD40 and MHC class II molecules (hence CD4+ T helper cells) (12, 31). The lack of prolonged transgene expression following a single Cre injection is another notable limitation of this animal model. Recently, transgenic mice carrying a Cre fused to an estrogen receptor have been shown to bypass the need for hydrodynamic Cre injection (20, 57). In these animals, single or multiple i.p. injections of tamoxifen induced efficient and prolonged transgene recombination, without obvious adverse effects, making this induction mode suitable for small-animal models of hepatitis C, such as ours.
In summary, our results demonstrate that hepatocyte CD86 expression can alter effector functions and homeostasis of CD4+ T cells, resulting in liver injury. These data underscore the potential benefits of future immune modulatory therapeutics aimed at minimizing pathogenesis in chronically infected individuals (21, 41, 65).
Acknowledgments
We gratefully acknowledge John Taylor of Fox Chase Cancer Center for sharing his early insights concerning hydrodynamic delivery; Brian Sauer and H. E. Ruley (Vanderbilt University) and Gregg Milligan for providing Cre plasmids pBS185 and pBS391, Cre fusion protein, and ovalbumin323-399 peptide, respectively; Michelle Amesbury for microinjection that generated the AT-B7.2 mice; Elbert Whorton for expertise in statistical analysis; Steve Weinman and Lynn Soong for critical reading of the manuscript; Yixiao Sun for technical assistance; and Mardelle Susman for assistance with manuscript preparation.
This work was supported by grants from the National Institutes of Health (AI60560, AI53638, and AI40035), the UTMB GRIP program, the Texas Gulf Coast DDC (DK56338), and the National Health and Medical Research Council of Australia.
REFERENCES
- 1.Abe, K., T. Kurata, Y. Teramoto, J. Shiga, and T. Shikata. 1993. Lack of susceptibility of various primates and woodchucks to hepatitis C virus. J. Med. Primatol. 22:433-434. [PubMed] [Google Scholar]
- 2.Anton, M., and F. L. Graham. 1995. Site-specific recombination mediated by an adenovirus vector expressing the Cre recombinase protein: a molecular switch for control of gene expression. J. Virol. 69:4600-4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bertolino, P., M. C. Trescol-Biemont, and C. Rabourdin-Combe. 1998. Hepatocytes induce functional activation of naive CD8+ T lymphocytes but fail to promote survival. Eur. J. Immunol. 28:221-236. [DOI] [PubMed] [Google Scholar]
- 4.Bertolino, P., D. G. Bowen, G. W. McCaughan, and B. Fazekas de St Groth. 2001. Antigen-specific primary activation of CD8+ T cells within the liver. J. Immunol. 166:5430-5438. [DOI] [PubMed] [Google Scholar]
- 5.Reference deleted.
- 6.Bigger, C. B., K. M. Brasky, and R. E. Lanford. 2001. DNA microarray analysis of chimpanzee liver during acute resolving hepatitis C virus infection. J. Virol. 75:7059-7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bowen, D. G., A. Warren, T. Davis, M. W. Hoffmann, G. W. McCaughan, B. F. De St Groth, and P. Bertolino. 2002. Cytokine-dependent bystander hepatitis due to intrahepatic murine CD8 T-cell activation by bone marrow-derived cells. Gastroenterology 123:1252-1264. [DOI] [PubMed] [Google Scholar]
- 8.Bowen, D. G., M. Zen, L. Holz, T. Davis, G. W. McCaughan, and P. Bertolino. 2004. The site of primary T cell activation is a determinant of the balance between intrahepatic tolerance and immunity J. Clin. Investig. 114:701-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bukh, J. 2001. Failure to infect rhesus monkeys with hepatitis C virus strains of genotypes 1a, 2a or 3a. J. Viral Hepat. 8:228-231. [DOI] [PubMed] [Google Scholar]
- 10.Burgio, V. L., G. Ballardini, M. Artini, M. Caratozzolo, F. B. Bianchi, and M. Levrero. 1998. Expression of co-stimulatory molecules by Kupffer cells in chronic hepatitis of hepatitis C virus etiology. Hepatology 27:1600-1606. [DOI] [PubMed] [Google Scholar]
- 11.Carvalho, L. H., J. C. Hafalla, and F. Zavala. 2001. Elispot Assay to Measure Antigen-Specific Murine CD8+ T Cell Responses. J. Immunol. Methods 252:207-218. [DOI] [PubMed] [Google Scholar]
- 12.Cho, H. J., T. Hayashi, S. K. Datta, K. Takabayashi, J. H. Van Uden, A. Horner, M. Corr, and E. Raz. 2002. IFN-alpha beta promote priming of antigen-specific CD8+ and CD4+ T lymphocytes by immunostimulatory DNA-based vaccines. J. Immunol. 168:4907-4913. [DOI] [PubMed] [Google Scholar]
- 13.Crispe, I. N. 2003. Hepatic T cells and liver tolerance. Nat. Rev. Immunol. 3:51-62. [DOI] [PubMed] [Google Scholar]
- 14.Diebold, S. S., M. Montoya, H. Unger, L. Alexopoulou, P. Roy, L. E. Haswell, A. Al-Shamkhani, R. Flavell, P. Borrow, and C. Reis e Sousa. 2003. Viral infection switches non-plasmacytoid dendritic cells into high interferon producers. Nature 424:324-328. [DOI] [PubMed] [Google Scholar]
- 15.Grakoui, A., N. H. Shoukry, D. J. Woollard, J. H. Han, H. L. Hanson, J. Ghrayeb, K. K. Murthy, C. M. Rice, and C. M. Walker. 2003. HCV persistence and immune evasion in the absence of memory T Cell help. Science 302:659-662. [DOI] [PubMed] [Google Scholar]
- 16.Gremion, C., B. Grabscheid, B. Wolk, D. Moradpour, J. Reichen, W. Pichler, and A. Cerny. 2004. Cytotoxic T lymphocytes derived from patients with chronic hepatitis C virus infection kill bystander cells via Fas-Fasl interaction. J. Virol. 78:2152-2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gruener, N. H., F. Lechner, M. C. Jung, H. Diepolder, T. Gerlach, G. Lauer, B. Walker, J. Sullivan, R. Phillips, G. R. Pape, and P. Klenerman. 2001. Sustained dysfunction of antiviral CD8+ T lymphocytes after infection with hepatitis C virus. J. Virol. 75:5550-5558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guerder, S., D. E. Picarella, P. S. Linsley, and R. A. Flavell. 1994. Costimulator B7-1 confers antigen-presenting-cell function to parenchymal tissue and in conjunction with tumor necrosis factor alpha leads to autoimmunity in transgenic mice. Proc. Natl. Acad. Sci. USA 91:5138-5142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guerder, S., E. E. Eynon, and R. A. Flavell. 1998. Autoimmunity without diabetes in transgenic mice expressing beta cell-specific CD86, but not CD80: parameters that trigger progression to diabetes. J. Immunol. 161:2128-2140. [PubMed] [Google Scholar]
- 20.Hayashi, S., and A. P. McMahon. 2002. Efficient recombination in diverse tissues by a tamoxifen-inducible form of Cre: a tool for temporally regulated gene activation/inactivation in the mouse. Dev. Biol. 244:305-318. [DOI] [PubMed] [Google Scholar]
- 21.Inoue, K., K. Sekiyama, M. Yamada, T. Watanabe, H. Yasuda, and M. Yoshiba. 2003. Combined interferon alpha2b and cyclosporin a in the treatment of chronic hepatitis C: controlled trial. J. Gastroenterol. 38:567-572. [DOI] [PubMed] [Google Scholar]
- 22.Isom, H. C. 1980. DNA synthesis in isolated hepatocytes infected with herpesviruses. Virology 103:199-216. [DOI] [PubMed] [Google Scholar]
- 23.Jo, D., A. Nashabi, C. Doxsee, Q. Lin, D. Unutmaz, J. Chen, and H. E. Ruley. 2001. Epigenetic regulation of gene structure and function with a cell-permeable Cre recombinase. Nat. Biotechnol. 19:929-933. [DOI] [PubMed] [Google Scholar]
- 24.Kafri, T., D. Morgan, T. Krahl, N. Sarvetnick, L. Sherman, and I. Verma. 1998. Cellular immune response to adenoviral vector infected cells does not require de novo viral gene expression: implications for gene therapy. Proc. Natl. Acad. Sci. USA 95:11377-11382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kanegae, Y., G. Lee, Y. Sato, M. Tanaka, M. Nakai, T. Sakaki, S. Sugano, and I. Saito. 1995. Efficient gene activation in mammalian cells by using recombinant adenovirus expressing site-specific Cre recombinase. Nucleic Acids Res. 23:3816-3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kantzanou, M., M. Lucas, E. Barnes, H. Komatsu, G. Dusheiko, S. Ward, G. Harcourt, and P. Klenerman. 2003. Viral escape and T cell exhaustion in hepatitis C virus infection analysed using class I peptide tetramers. Immunol. Lett. 85:165-171. [DOI] [PubMed] [Google Scholar]
- 27.Klugewitz, K., F. Blumenthal-Barby, A. Schrage, P. A. Knolle, A. Hamann, and I. N. Crispe. 2002. Immunomodulatory effects of the liver: deletion of activated CD4+ effector cells and suppression of IFN-gamma-producing cells after intravenous protein immunization. J. Immunol. 169:2407-2413. [DOI] [PubMed] [Google Scholar]
- 28.Knodell, R. G., K. G. Ishak, W. C. Black, T. S. Chen, R. Craig, N. Kaplowitz, T. W. Kiernan, and J. Wollman. 1981. Formulation and application of a numerical scoring system for assessing histological activity in asymptomatic chronic active hepatitis. Hepatology 1:431-435. [DOI] [PubMed] [Google Scholar]
- 29.Lanford, R. E., C. Bigger, S. Bassett, and G. Klimpel. 2001. The chimpanzee model of hepatitis C virus infections. ILAR J. 42:117-126. [DOI] [PubMed] [Google Scholar]
- 30.Le, Y., S. Gagneten, D. Tombaccini, B. Bethke, and B. Sauer. 1999. Nuclear targeting determinants of the phage P1 Cre DNA recombinase. Nucleic Acids Res. 27:4703-4709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Le Bon, A., N. Etchart, C. Rossmann, M. Ashton, S. Hou, D. Gewert, P. Borrow, and D. F. Tough. 2003. Cross-priming of CD8+ T cells stimulated by virus-induced type I interferon. Nat. Immunol. 4:1009-1015. [DOI] [PubMed] [Google Scholar]
- 32.Lerat, H., M. Honda, M. R. Beard, K. Loesch, J. Sun, Y. Yang, M. Okuda, R. Gosert, S. Y. Xiao, S. A. Weinman, and S. M. Lemon. 2002. Steatosis and liver cancerin transgenic mice expressing the structural and nonstructural proteins of hepatitis C virus. Gastroenterology 122:352-365. [DOI] [PubMed] [Google Scholar]
- 33.Liu, F., Y. Song, and D. Liu. 1999. Hydrodynamics-based transfection in animals by systemic administration of plasmid DNA. Gene Ther. 6:1258-1266. [DOI] [PubMed] [Google Scholar]
- 34.Lund, J., A. Sato, S. Akira, R. Medzhitov, and A. Iwasaki. 2003. Toll-like receptor 9-mediated recognition of herpes simplex virus-2 by plasmacytoid dendritic cells. J. Exp. Med. 198:513-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mehal, W. Z., A. E. Juedes, and I. N. Crispe. 1999. Selective retention of activated Cd8+ T cells by the normal liver. J. Immunol. 163:3202-3210. [PubMed] [Google Scholar]
- 36.Mehal, W. Z., F. Azzaroli, and I. N. Crispe. 2001. Antigen presentation by liver cells controls intrahepatic T cell trapping, whereas bone marrow-derived cells preferentially promote intrahepatic T cell apoptosis. J. Immunol. 167:667-673. [DOI] [PubMed] [Google Scholar]
- 37.Mochizuki, K., N. Hayashi, K. Katayama, N. Hiramatsu, T. Kanto, E. Mita, T. Tatsumi, N. Kuzushita, A. Kasahara, H. Fusamoto, T. Yokochi, and T. Kamada. 1997. B7/Bb-1 expression and hepatitis activity in liver tissues of patients with chronic hepatitis C. Hepatology 25:713-718. [DOI] [PubMed] [Google Scholar]
- 38.Moriya, K., H. Fujie, Y. Shintani, H. Yotsuyanagi, T. Tsutsumi, K. Ishibashi, Y. Matsuura, S. Kimura, T. Miyamura, and K. Koike. 1998. The core protein of hepatitis C virus induces hepatocellular carcinoma in transgenic mice. Nat. Med. 4:1065-1067. [DOI] [PubMed] [Google Scholar]
- 39.Muir, A. J., J. D. Bornstein, and P. G. Killenberg. 2004. Peginterferon alfa-2b and ribavirin for the treatment of chronic hepatitis C in blacks and non-hispanic whites. N. Engl. J. Med. 350:2265-2271. [DOI] [PubMed] [Google Scholar]
- 40.Murphy, K. M., A. B. Heimberger, and D. Y. Loh. 1990. Induction by antigen of intrathymic apoptosis of CD4+CD8+TCRlo thymocytes in vivo. Science 250:1720-1723. [DOI] [PubMed] [Google Scholar]
- 41.Nelson, D. R., G. Y. Lauwers, J. Y. Lau, and G. L. Davis. 2000. Interleukin 10 treatment reduces fibrosis in patients with chronic hepatitis C: a pilot trial of interferon nonresponders. Gastroenterology 118:655-660. [DOI] [PubMed] [Google Scholar]
- 42.Ohta, A., M. Sekimoto, M. Sato, T. Koda, S.-i. Nishimura, Y. Iwakura, K. Sekikawa, and T. Nishimura. 2000. Indispensable role for TNF-alpha and IFN-gamma at the effector phase of liver injury mediated by Th1 cells specific to hepatitis B virus surface antigen. J. Immunol. 165:956-961. [DOI] [PubMed] [Google Scholar]
- 43.Peitz, M., K. Pfannkuche, K. Rajewsky, and F. Edenhofer. 2002. Ability of the hydrophobic Fgf and basic tat peptides to promote cellular uptake of recombinant Cre recombinase: a tool for efficient genetic engineering of mammalian genomes. Proc. Natl. Acad. Sci. USA 99:4489-4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Robertson, J. M., P. E. Jensen, and B. D. Evavold. 2000. DO11.10 and OT-II T cells recognize a C-terminal ovalbumin 323-339 epitope. J. Immunol. 164:4706-4712. [DOI] [PubMed] [Google Scholar]
- 45.Sauer, B., and N. Henderson. 1988. Site-specific DNA recombination in mammalian cells by the Cre recombinase of bacteriophage P1. Proc. Natl. Acad. Sci. USA 85:5166-5170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sauer, B., and N. Henderson. 1989. Cre-stimulated recombination at LoxP-containing DNA sequences placed into the mammalian genome. Nucleic Acids Res. 17:147-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sauer, B., and N. Henderson. 1990. Targeted insertion of exogenous DNA into the eukaryotic genome by the Cre recombinase. New Biol. 2:441-449. [PubMed] [Google Scholar]
- 48.Savulescu, J. 2001. Harm, ethics committees and the gene therapy death. J. Med. Ethics 27:148-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shata, M. T., D. D. Anthony, N. L. Carlson, L. Andrus, B. Brotman, N. Tricoche, P. McCormack, and A. Prince. 2002. Characterization of the immune response against hepatitis C infection in recovered, and chronically infected chimpanzees. J. Viral Hepat. 9:400-410. [DOI] [PubMed] [Google Scholar]
- 50.Shoukry, N. H., A. Grakoui, M. Houghton, D. Y. Chien, J. Ghrayeb, K. A. Reimann, and C. M. Walker. 2003. Memory CD8+ T cells are required for protection from persistent hepatitis C virus infection. J. Exp. Med. 197:1645-1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Soriano, P. 1999. Generalized LacZ expression with the Rosa26 Cre reporter strain. Nat. Genet. 21:70-71. [DOI] [PubMed] [Google Scholar]
- 52.Sparer, T. E., S. G. Wynn, D. J. Clark, J. M. Kaplan, L. M. Cardoza, S. C. Wadsworth, A. E. Smith, and L. R. Gooding. 1997. Generation of cytotoxic T lymphocytes against immunorecessive epitopes after multiple immunizations with adenovirus vectors is dependent on haplotype. J. Virol. 71:2277-2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Su, A. I., J. P. Pezacki, L. Wodicka, A. D. Brideau, L. Supekova, R. Thimme, S. Wieland, J. Bukh, R. H. Purcell, P. G. Schultz, and F. V. Chisari. 2002. Genomic analysis of the host response to hepatitis C virus infection. Proc. Natl. Acad. Sci. USA 99:15669-15674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sun, J., B. Dirden-Kramer, K. Ito, P. B. Ernst, and N. Van Houten. 1999. Antigen-specific T cell activation and proliferation during oral tolerance induction. J. Immunol. 162:5868-5875. [PubMed] [Google Scholar]
- 55.Sun, J. R., F. Bodola, X. G. Fan, H. Irshad, L. Soong, S. M. Lemon, and T. S. Chan. 2001. Hepatitis C virus core and envelope proteins do not suppress the host's ability to clear a hepatic viral infection. J. Virol. 75:11992-11998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sun, J. R., K. Li, M. T. Shata, and T. Chan. 2004. The immunologic basis for hepatitis C infection. Curr. Opin. Gastroenterol. 20:598-602. [DOI] [PubMed] [Google Scholar]
- 57.Tannour-Louet, M., A. Porteu, S. Vaulont, A. Kahn, and M. Vasseur-Cognet. 2002. A tamoxifen-inducible chimeric Cre recombinase specifically effective in the fetal and adult mouse liver. Hepatology 35:1072-1081. [DOI] [PubMed] [Google Scholar]
- 58.Thimme, R., D. Oldach, K. M. Chang, C. Steiger, S. C. Ray, and F. V. Chisari. 2001. Determinants of viral clearance and persistence during acute hepatitis C virus infection. J. Exp. Med. 194:1395-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thimme, R., J. Bukh, H. C. Spangenberg, S. Wieland, J. Pemberton, C. Steiger, S. Govindarajan, R. H. Purcell, and F. V. Chisari. 2002. Viral and immunological determinants of hepatitis C virus clearance, persistence, and disease. Proc. Natl. Acad. Sci. USA 99:15661-15668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tsai, S. L., Y. F. Liaw, M. H. Chen, C. Y. Huang, and G. C. Kuo. 1997. Detection of type 2-like T-helper cells in hepatitis C virus infection: implications for hepatitis C virus chronicity. Hepatology 25:449-458. [DOI] [PubMed] [Google Scholar]
- 61.von Herrath, M. G., S. Guerder, H. Lewicki, R. A. Flavell, and M. B. Oldstone. 1995. Coexpression of B7-1 and viral (“self”) transgenes in pancreatic beta cells can break peripheral ignorance and lead to spontaneous autoimmune diabetes. Immunity 3:727-738. [DOI] [PubMed] [Google Scholar]
- 62.Wakita, T., C. Taya, A. Katsume, J. Kato, H. Yonekawa, Y. Kanegae, I. Saito, Y. Hayashi, M. Koike, and M. Kohara. 1998. Efficient conditional transgene expression in hepatitis C virus cDNA transgenic mice mediated by the Cre/LoxP system. J. Biol. Chem. 273:9001-9006. [DOI] [PubMed] [Google Scholar]
- 63.Wakita, T., A. Katsume, J. Kato, C. Taya, H. Yonekawa, Y. Kanegae, I. Saito, Y. Hayashi, M. Koike, M. Miyamoto, Y. Hiasa, and M. Kohara. 2000. Possible role of cytotoxic T cells in acute liver injury in hepatitis C virus cDNA transgenic mice mediated by Cre/Loxp system. J. Med. Virol. 62:308-317. [DOI] [PubMed] [Google Scholar]
- 63a.World Health Organization. 2000. Fact sheet 164. World Health Organization, Geneva, Switzerland.
- 64.Yang, Y., H. C. Ertl, and J. M. Wilson. 1994. Mhc class I-restricted cytotoxic T lymphocytes to viral antigens destroy hepatocytes in mice infected with E1-deleted recombinant adenoviruses. Immunity 1:433-442. [DOI] [PubMed] [Google Scholar]
- 65.Zeuzem, S., and V. Carreno. 2001. Interleukin-12 in the treatment of chronic hepatitis B and C. Antiviral Res. 52:181-188. [DOI] [PubMed] [Google Scholar]
- 66.Zhang, G., Y. K. Song, and D. Liu. 2000. Long-term expression of human alpha1-antitrypsin gene in mouse liver achieved by intravenous administration of plasmid DNA using a hydrodynamics-based procedure. Gene Ther. 7:1344-1349. [DOI] [PubMed] [Google Scholar]