Abstract
De novo infection of cultured cells with Kaposi's sarcoma-associated herpesvirus (KSHV) typically results in a latent infection. Recently, however, it has been reported that a subset of lytic mRNAs can be detected in cells shortly after KSHV infection; this expression is transient and eventually subsides, leading to latent infection (H. H. Krishnan et al., J. Virol 78:3601-3620, 2004). Since it has been shown that viral RNAs can be packaged into other herpesvirus virions, we sought to determine if KSHV virions contained RNAs and, if so, whether these RNAs contributed to the pool of lytic transcripts detected immediately after infection. Using DNA microarray, reverse transcription (RT)-PCR, and Northern blotting analyses, we identified 11 virally encoded RNAs in KSHV virions. These corresponded in size to the full-length mRNAs found in cytoplasmic RNA, and at least one was directly demonstrated to be translated upon infection in the presence of actinomycin D. Ten of these RNAs correspond to transcripts reported by Krishnan et al. at early times of infection, representing ca. 30% of such RNAs. Thus, import of RNAs in virions can account for some but not all of the early-appearing lytic transcripts. Quantitative RT-PCR analysis of infected-cell RNA demonstrated that most of the virion RNAs were very abundant at late times of infection, consistent with nonspecific incorporation during budding. However, the intracellular levels of one virion mRNA, encoding the viral protease, were much lower than those of transcripts not packaged in the virus particle, strongly suggesting that it may be incorporated by a specific mechanism.
Herpesviruses are a family of large DNA viruses capable of establishing persistent infections. Members of this family share several structural characteristics: an icosahedrally symmetric capsid containing the viral genome, a lipid envelope studded with virally encoded glycoproteins, and a structured layer of proteins termed the tegument that resides between the capsid and the envelope. In addition to these conserved features, two herpesviruses have been shown to encapsidate RNAs into the virus particle (7, 18, 30). These transcripts are released into the newly infected cell during virus entry and have the potential to influence the cellular milieu prior to transcription from the viral genome. Human cytomegalovirus (HCMV) packages several RNAs (7, 18, 27). These represent only a subset of viral transcripts found in infected cells, but recent work reports that the levels of RNA encapsidated into particles are proportional to transcript levels at late times of infection, suggesting that HCMV may simply incorporate RNAs nonspecifically during the assembly and budding steps (7, 18, 33). Herpes simplex virus type 1 also incorporates several viral RNAs into the virus particle (30). Some of these RNAs are not abundantly expressed during late times of infection, when the virus particles are maturing, suggesting specificity in packaging (30). Further work established that three herpes simplex virus tegument proteins are capable of binding RNA, offering one potential explanation for how the RNAs might be packaged into the virus particle (31).
Kaposi's sarcoma-associated herpesvirus (KSHV, or human herpesvirus type 8) is a gammaherpesvirus associated with the endothelium-based neoplasm Kaposi's sarcoma, as well as two B-cell-proliferative diseases, primary effusion lymphoma and a subset of multicentric Castleman's disease (9, 10, 13). Like other herpesviruses, KSHV is able to establish both latent and lytic infections. Latent infection is characterized by the expression of a small subset of the viral genes and by genome maintenance as a nuclear episome (3, 29). During lytic replication, the full repertoire of viral genes is expressed in a temporally regulated cascade leading to virus production. Latently infected cells can be stimulated to enter the lytic cycle by the addition of butyrate or phorbol esters or by the overexpression of the KSHV switch protein, RTA (4, 8, 9, 15, 17, 20, 24, 25, 32, 36).
KSHV establishes a latent infection after de novo infection of cultured cells (1, 2, 4, 14, 15, 22, 26, 34, 37), with latency usually being achieved by 24 h postinoculation. However, a recent report revealed that when newly infected cells are examined at very early times (2 to 8 h) postinfection, the patterns of viral-gene expression are more complex (21). Using reverse transcription (RT)-PCR and microarray analysis, Krishnan and colleagues (21) have shown that in addition to latent gene expression, there is also a transient accumulation of selected mRNAs that are normally considered lytic cycle specific. The full lytic program, however, is not engaged, and this initial burst of lytic transcript accumulation eventually subsides, with supervention of the classical latent gene expression program (21). The lytic genes detected include immunomodulatory molecules, as well as antiapoptotic molecules that could play important roles during the establishment of KSHV infection. These observations raise important questions about how these transcripts arise after introduction of the genome and how this unusual genetic program, which includes transient expression of the lytic switch protein RTA, is terminated in favor of latency.
One possible explanation for the early appearance of selected lytic transcripts in newly infected cells is that some or all of them were delivered there by incoming virions that had incorporated them on the prior round of replication. To explore this possibility, we examined KSHV virions for the presence of viral mRNAs, and we report here that 11 such transcripts are packaged into the virus particles. A number of these RNAs indeed correspond to those observed by Krishnan et al. (21) in newly infected cells, but their findings cannot be entirely explained in this fashion.
MATERIALS AND METHODS
Isolation and purification of KSHV.
KSHV virions were gradient purified from the supernatant of sodium butyrate-induced BCBL-1 cells 6 to 7 days postinduction as previously described (4, 5). In brief, cultures of BCBL-1 cells induced with 0.3 mM sodium butyrate for 6 to 7 days were clarified of cells by centrifugation (10 min; 2,000 rpm) and then concentrated in a GSA rotor for 2 h at 24,000 × g. The pellets were resuspended in phosphate-buffered saline (PBS) (GIBCO), pooled, and centrifuged in a SW28 rotor for 2 h at 29,000 × g. The pellets were resuspended in PBS and then layered onto 9 ml 20 to 35% histodenz (Sigma) gradients and centrifuged for 2 h at 71,000 × g. Fractions were collected from the gradients by puncturing the bottoms of the tubes and then assayed for protein and DNA content as described previously (5). The desired fractions were then diluted in PBS and centrifuged for 2 h at 29,000 × g in an SW28 rotor. The virus was then diluted with PBS, resuspended in PBS, and stored at −80°C.
Isolation of virion RNA.
Gradient-purified virions were supplemented with 5 mM MgCl2 and 1 mM CaCl2 and then treated with micrococcal nuclease (15 U/μl) for 15 min at 37°C. The reaction was quenched by the addition of EGTA (Calbiochem), and RNA was isolated by Trizol LS (Invitrogen) extraction (according to the manufacturer's instructions). The RNA was then treated with DNase I (Ambion) for 30 min at 37°C. The DNase was inactivated according to the manufacturer's instructions.
RNA isolation and poly(A) purification.
BJAB, human foreskin fibroblast (HFF), and uninduced and induced BCBL-1 cell RNAs were isolated using RNA-BEE (Tel-Test, Inc.) according to the manufacturer's instructions; then, poly(A) was purified using the Oligotex mRNA kit (QIAGEN).
Constructing dot blots.
Nylon membranes (Hybond N+; Amersham) were spotted with plasmids containing the KSHV open reading frames (ORFs) (16) or PCR products using a Schleicher and Schuell dot blot apparatus. The DNA was fixed to the membranes by washing them with 0.4 N NaOH. The membranes were then rinsed in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and then dried and stored at room temperature.
DNA arrays.
DNA microarrays were produced by robotic spotting on poly-l-lysine-coated slides. The oligonucleotide array contained ∼22,700 oligonucleotides derived from ∼20,700 human genes (Illumina, San Diego, CA), as well as specific 70-mers designed to detect all open reading frames of cytomegalovirus and KSHV. Virus-specific oligonucleotides were designed using ArrayOligoSelector (6). For the generation of the KSHV tile array, 306 primer pairs were selected that amplify the entire long unique region of the KSHV genome in small, nonoverlapping fragments with an average length of 500 bp. KSHV sequences were amplified from a genomic KSHV cosmid library generated from BCBL-1 cells (A. Polson and D. Ganem, unpublished data). Genomic regions refractory to PCR amplification (e.g., DR1 and DR2 repeats), as well as the terminal repeats, were excised from the cosmids by restriction digestion. The PCR products and restriction fragments were subsequently purified and spotted on poly-l-lysine-coated slides.
Northern blots.
RNA was separated on a 1% agarose formaldehyde gel and then transferred to nylon membranes using Turbo Blot kits (Schleicher and Schuell) according to the manufacturer's instructions. The transferred membranes were rinsed in 2× SSC, autocrosslinked twice (UV Stratalinker 2400; Stratagene), and then stained with methylene blue. The blots were prehybridized in ultrahyb (Ambion) and then hybridized with 32P-labeled probes. Riboprobes were generated using Maxiscript kits (Ambion), and DNA probes were made using RediPrimeII (Amersham Biosciences) according to the manufacturers' instructions.
RT-PCR.
Fifty nanograms of poly(A)-purified RNA from BJAB and uninduced and induced BCBL-1 cells and 100 ng of RNA isolated from KSHV virions were reverse transcribed with Superscript II reverse transcriptase according to the manufacturer's instructions using an oligo(dT) primer (Invitrogen). The RNA was then degraded by the addition of 1 μl RNase H (Invitrogen) and incubation at 37°C for 20 min. One microliter of the cDNA was then subjected to PCR using KSHV gene-specific primers. The PCR conditions were 94°C for 1 min, 60°C for 1 min, and 72°C for 1 min for 25 to 28 cycles. The PCR products were then separated on 1.5 to 2% agarose gels.
RT-PCR primers.
Primers for K7 and PAN (38) and K8.1 (28) have been described elsewhere. Other primers were ORF17, 5′ TCAGCGGCTCGGTCTCACAC; ORF17, 3′ AGCTACTTACGTGCTGGAGG; K12, 5′ CGGGATCCATGGATAGAGGCTTAACGGTGTTTGTG; K12, 3′ GGAATTCTCAGTGCGCGCCCGTTGAACTCGTGTC; ORF58, 5′ GGCAGCCAGAAAACGCCGGA; ORF58, 3′ TGCCAGTCACGCACTTGGCC; ORF59, 5′ GACAGCGTCTCGCTGACAGA; ORF59, 3′ CACACGCGTGAGCTATTCGG; ORF54, 5′ AACCCCACGTGGCTCTAGCA; ORF54, 3′ CCCTTGAGGATGTGTCTGCG; K2, 5′ TGGACGTCAGGAGTCACGTC; K2, 3′ CTGGTTCAAGTTGTGGTCTC K4, 5′ GATCCGTCGCGTAAATGCGC; K4, 3′ ACCGCGCCTGACCTAACATC; K6, 5′ AGTTGGGCCGCAGTGATATC; K6, 3′ TGCGAACTTGGCACCTCCAG; K5IR, 5′ CGTGTTAGTGTCACCCACTG; K5IR, 3′ GACGGAGAATAGACCAGCCT; ORF73, 5′ GAAGTGGATTACCCTGTTGTTAGC; ORF73, 3′ TTGGATCTCGTCTTCCATCC; ORF50, 5′ GCCCTCTGCCTTTTGGTT; and ORF50, 3′ GATGATGCTGACGGTGTG.
Probing and analyzing DNA arrays.
RNA isolated from KSHV virions (1 μg) was reverse transcribed in the presence of [32P]dUTP and then hybridized to nylon membrane arrays as described for dot blotting. The probes were hybridized overnight at 42°C in ultrahyb (Ambion). The blots were washed one time for 15 min with each of the following: 2× SSC, room temperature; 2× SSC plus 1% sodium dodecyl sulfate (SDS), 65°C; 0.4× SSC plus 1% SDS, 65°C; and 0.2× SSC, room temperature. The blot was then analyzed on a phosphorimager. RNA isolated from KSHV virions and BJAB cells was reverse transcribed in the presence of amino-allyl dUTP and then coupled to Cy3 or Cy5 dye (Amersham). In brief, 2 μg poly(A)-purified BJAB RNA or 1 μg KSHV virion RNA was incubated with 1 μg of oligo(dT) and random hexamers at 65°C for 5 min and then cooled on ice. Reverse transcription was carried out at 42°C for 2 hours after the addition of 100 U Stratascript superscript II, reaction buffer, dithiothreitol, and aa-dNTP nucleotide mix (50× aa-dNTP mix is composed of 25 mM dATP, 25 mM dCTP, 25 mM dTGP, 10 mM TTP, and 15 mM amino-allyl dUTP). The RNA was degraded by adding NaOH and EDTA to final concentrations of 100 mM and 10 mM, respectively; the reaction was neutralized by the addition of HEPES, pH 7, to 500 mM. cDNA was concentrated using a Zymo spin column (Zymo Research). The cDNA was buffered to 100 mM sodium bicarbonate, pH 9, and then used to resuspend Cy3 or Cy5 (Amersham) dehydrated dye and incubated for 1 h at room temperature. Uncoupled dye was removed using Zymo spin columns (Zymo Research). For the KSHV tile array, equal amounts of BJAB and virion RNA cDNAs were mixed together; adjusted to 3× SSC, 25 mM HEPES, pH 7, and 0.25% SDS; and then boiled for 2 minutes. Cooled samples were applied to an array, and the array was incubated overnight at 65°C. For the large human array, equal amounts of BJAB and virion cDNAs were mixed; adjusted to 20% formamide, 5.5× SSC, 0.25% SDS, and 25 mM HEPES, pH 7; and then boiled for 2 min. Cooled samples were applied to the microarray and incubated overnight at 55°C. After incubation, the coverslips were removed and the samples were washed by plunging the arrays up and down first in a solution of 0.6× SSC and 0.03% SDS and then in a 0.06× SSC solution. The arrays were dried by brief centrifugation in a tabletop centrifuge and then analyzed on an Axon 4000B scanner using GenePix Pro 3.0 software.
Primer pairs for riboprobes.
Primer pairs for riboprobes were as follows: APORF17, 3′TTCAGGAGCTCCTCGCAGAA; K2, 3′ TGGACGTCAGGAGTAACGTC; K2, 5′ CTGGTTCAAGTTGTGGTCTC; K4, 3′ GATCCGTCGCGTAAATGCGC; K4, 5′ACCGCGCCTGACCTAACATC; K6, 3′ AGTTGGGCCGCAGTGATATC; K6, 5′ TGCGAACTTGGCACCTCCAG; PRORF17, 3′ AGCTACTTACGTGCTGGAGG; PRORF17, 5′TCAGCGGCTCGGTCTCACAC; K5INT, 3′ TCAAGAGCCTATGCTGGGAC; K5INT, 5′ GGCAACAACCTGTTGCCATG; ORF54, 3′ CCCTTGAGGATGTGTCTGCG; and ORF54, 5′AGGCTCCAAGCCGTCTATCC.
Generating riboprobes.
Sequences for KSHV ORFs were amplified using the above-mentioned primer pairs or those listed for RT-PCR and then TA cloned into pCR4-TOPO (Invitrogen). The clones were sequenced and then linearized using either PmeI or NotI (New England Biolabs) and gel purified. DNA was recovered using a Qiaquick gel extraction kit (QIAGEN). Riboprobes were generated using a MAXIscript in vitro transcription kit (Ambion).
Translation experiment.
Dishes (60 mm2) of 293 cells were starved for 30 minutes by incubating them in Dulbecco's modified Eagle's medium lacking methionine and cysteine. During this period, the cells were also treated with dimethyl sulfoxide (DMSO), actinomycin D (2 μg/ml), or cycloheximide (100 μg/ml). After 30 minutes, the cells were mock infected or KSHV infected in the presence of 200 mCi of [35S]methionine and cysteine. Each plate infected with KSHV received the equivalent of 4 liters of unconcentrated BCBL-1 supernatants. After 4 or 8 h, the cells were washed, resuspended in PBS containing 1% Ipegal, and then incubated on ice for 30 min. The lysates were cleared of debris by centrifugation (16,000 × g; 10 min; 4°C). The lysates were then precleared with Protein A/G agarose by rotation for 1 h at 4°C; 5 μl of monoclonal ORF59 antibody (Advanced Biotechnologies Inc.) was added to each lysate, and the lysates were rotated for 1 h at 4°C. Protein A/G was then added, and the lysates were further rotated at 4°C overnight. The lysates were cleared of beads by quick centrifugation and transferred to new tubes, and then 5 μl of rabbit polyclonal LANA antibody was added to each. The lysates were rotated for 1 h at 4°C, at which time protein A/G agarose was added, and rotation continued overnight at 4°C. The beads were washed five times with lysis buffer, and protein was removed from the beads by the addition of 2× Laemmli sample buffer, followed by boiling. The boiled samples were separated on 7.5% or 10% SDS-polyacrylamide gel electrophoresis (PAGE) gels. Gels for autoradiography were fixed overnight in 30% methanol-10% acetic acid. The gels were then treated with Enhance (Perkin-Elmer) and dried for 2 h. The dried gels were exposed to film. The gels for immunoblotting were transferred to polyvinylidene difluoride membranes at 80 V for 2 h at 4°C. The membranes were blocked in 5% milk in Tris-buffered saline-Tween (TBST) for 1 hour and then incubated with primary antibody (rat monoclonal ORF73 antibody diluted 1:1,000 in 1% milk in TBST) overnight at 4°C. The membranes were washed three times in TBST and then incubated with goat anti-rat-horseradish peroxidase (Jackson Laboratories; diluted 1:10,000 in 1% milk in TBST) for 30 minutes. The membranes were then washed with TBST three times and incubated with equal volumes of ECL reagent (Amersham) and exposed to film.
TaqMan.
RNA isolated from induced BCBL-1 cells (3 days postinoculation with sodium butyrate) was subjected to reverse transcription as described above and then analyzed by real-time PCR (for the primers and probes, see Table 2). The GAPDH (glyceraldehyde-3-phosphate dehydrogenase) primer/probe set was purchased from Applied Biosystems. cDNA was mixed with 2× TaqMan universal PCR master mix (Applied Biosystems), primers (300 nM), and 6-carboxyfluorescein-labeled probes (200 nM). The probes had 5′ 6-carboxyfluorescein and 3′ 6-carboxytetramethylrhodamine modifications (Integrated DNA Technologies). Standard curves were prepared for each primer/probe set using either 10-fold serial dilutions of a plasmid bearing a GAPDH cDNA fragment or the KSHV bacterial artificial chromosome (BAC) (40). Negative controls for each primer and probe set included water and no-reverse-transcriptase samples. The relative amounts of transcripts were determined from the standard curves and then normalized to the level of GAPDH. The results are shown as increases (n-fold) over GAPDH.
TABLE 2.
Real-time PCR primers and probes
| Gene | 5′ Primer | 3′ Primer | Probe |
|---|---|---|---|
| K2 | TGGATGCTATGGGTGATCGA | CCCTTGCAGATGCCGGT | TGCTTCCGCGACCTCTGTTACCG |
| K4 | CCTGCTCGTCGCTGTGCT | GGAACGCGCCTCATACGA | ACATCACCACCCCACAGACCTGGAG |
| ORF8 | TCGCCGCACCAATACCATA | CCTGCGATCTACGTCGGG | CCCAGGCGCCGGTGAAGATG |
| ORF17 | CCCGCGAGTTATCCCAGAC | CCCAGGGCGCATAGTGATAC | CCCAGCGGTCCCGTGTTTCAAC |
| ORF25 | GCCACGCACGAATACGG | TTTGTGCACCCCTGTCCTG | AGGATGTGGCGCAGACCGTGC |
| ORF26 | CGAATCCAACGGATTTGACC | AGCGTGCCCCAGTTGCT | CGTGTTCCCCATGGTCGTGCC |
| ORF39 | AAAGACCAGAGCCCGCG | TCGGACTGGTATTGGGTCCTC | AAGGACATATCGACGCCCGCCC |
| ORF54 | TTGCGCCATAGGAAGCTAGC | TCGCGAAAATGCACTCGAG | CCTGGCGTCCGTTTCCGGG |
| ORF58 | TGTCATGCGTGGGCGTAT | GCCAAAGGCAGGAGAACAAA | TCGCCTCCCGACGAGCGC |
| ORF73 | CGCGAATACCGCTATGTACTCA | GGAACGCGCCTCATACGA | ACATCACCACCCCACAGACCTGGAG |
RESULTS
Purification of virion RNAs.
The goal of our study was to identify by DNA array analysis transcripts packaged into KSHV virions. In order to obtain a stock of pure virions, we isolated and purified KSHV particles from induced BCBL-1 cells on histodenz gradients. Fractions of the gradient were isolated and tested for the presence of the KSHV envelope glycoprotein K8.1 (by immunoblotting) and the viral DNA (by dot blotting). We then pooled the fractions that scored positive in both assays and used this as the source of our virion RNAs (5).
Gradient-purified virions were treated with micrococcal nuclease to remove any nucleic acids exterior to the particles. RNA was then extracted from the treated virions and subjected to further purification by treatment with RNase-free DNase. RNAs isolated from BJAB cells and KSHV virions were reverse transcribed and coupled to Cy3 and Cy5 dyes, respectively. The probes were then hybridized to an array representing the entire KSHV genome, tiled at 500-bp intervals; this array contains all KSHV sequences, coding and noncoding alike. We were able to identify a discrete number of KSHV spots that were labeled with the Cy5 probes, representing RNAs incorporated into the virions. A histogram showing the intensities of Cy5 signals for the KSHV tile array is shown in Fig. 1. There is a very intense signal observed for the K7/Pan region of the KSHV genome and smaller signals for several other regions, including the K2, K6, ORF17, ORF 54, ORF58, and ORF59 loci (Fig. 1). The y axis for this histogram was altered so that lower signal intensities could be visualized; the K7/Pan signal intensity exceeds the threshold for the graph, reaching a maximum at around 18,000.
FIG. 1.
Identification of KSHV virion RNAs by DNA array. RNAs isolated from BJAB cells and KSHV virions were reverse transcribed in the presence of amino-acyl dNTPs and then coupled to Cy3 and Cy5 dyes, respectively. The labeled cDNAs were then hybridized to the KSHV tile array as described in the text. The histogram displays the virion RNA signals across the KSHV tile array. The line across the histogram represents the arbitrary cutoff used in our analysis. The labeled peaks represent the RNAs consistently found in our KSHV virions.
We also employed another DNA microarray to examine the compositions of the virion RNAs. This array contained 20,000 human genes in addition to each of the KSHV and HCMV ORFs. In addition, two further experiments involved hybridizing 32P-radiolabeled virion cDNA to nylon membranes spotted with (i) plasmids containing individual cloned KSHV ORFs and (ii) uncloned, gel-purified PCR products corresponding to ORFs for which clones were not available. In all three experiments, the virion RNAs specifically hybridized to several discrete spots on the membrane or the microarray, indicating the presence of a limited number of transcripts in the virions (data not shown).
Comparisons between the two microarrays and the two nylon membranes allowed us to generate a list of specific viral RNAs that are incorporated in the KSHV virions (Table 1). We identified 11 transcripts present in the virions, and 10 of these were confirmed on all three types of arrays. The one discordance was the transcript labeled K5IR in Table 1; this transcript emanates from an intergenic region, and hence, its sequences were not represented on the arrays that corresponded to ORFs. (Recent work [39] indicates that this region generates a 1.4-kb RNA that has been postulated to play a role in the functioning of oriLyt, which also maps to this region.)
TABLE 1.
Expected sizes of RT-PCR products
| RNA | ORF | RT-PCR product size (bp) |
|---|---|---|
| Virion | K2 | 534 |
| K4 | 240 | |
| K5IR | 239 | |
| K6 | 209 | |
| K7 | 329 | |
| K8.1 | 720/540 | |
| K12 | 175 | |
| PAN | 334 | |
| ORF17 | 570 | |
| ORF54 | 189 | |
| ORF58 | 339 | |
| ORF59 | 320 | |
| Nonvirion | ORF8 | 432 |
| ORF50 | 351 | |
| ORF73 | 307 |
We consistently detected mRNAs for several of the so-called “K” genes unique to KSHV, including K2 (viral interleukin 6), K4 (vMIP-II), K6 (vMIP-I), K7 (vIAP), K8.1, and K12. Protein products from these transcripts have diverse functions (immunomodulatory, signaling, and antiapoptotic) and could significantly affect the phenotype of the newly infected cell and its microenvironment. We also found transcripts for genes involved in viral DNA replication. The ORF58/59 transcript encodes two proteins, one of which is uncharacterized while the other is an accessory factor for the viral DNA polymerase (11). ORF54 encodes the viral dUTPase, an enzyme responsible for hydrolyzing dUTP to dUMP, which can be further modified to dTMP. We also identified the RNA for ORF17, encoding the viral protease and assembly protein (35), which function to allow proper capsid maturation. In addition, we found the mRNA for PAN, a transcript expressed to high levels in lytic infection and whose function is presently unknown.
Confirmation of virion RNAs.
To confirm the specific packaging of these transcripts into the virion, we employed RT-PCR. KSHV virion RNA, as well as RNAs from BJAB and induced BCBL-1 cells, was reverse transcribed with an oligo(dT) primer and then amplified using gene-specific primers. KSHV BAC DNA was used as a positive control in the PCRs for the gene-specific primers. Table 1 lists the expected sizes of the RT-PCR products for the virion RNAs. As shown in Fig. 2, we were able to confirm the presence of all of the identified virion RNAs using RT-PCR. Interestingly, the RT-PCR for K8.1 produced two distinct products in the induced BCBl-1 cells and from the virion RNA (Fig. 2, bottom panel). K8.1 is expressed as several spliced transcripts, and the K8.1 primers were able to amplify two bands (720 and 540 bp) in the induced BCBL-1 lane, indicating the presence of both K8.1A and K8.1B mRNAs (28). Both of these bands are also present in the lane with the virion RNAs, suggesting that both mRNAs are incorporated into the virus particle. No signal was detectable in the BJAB RNA lane, and the PCR using the BAC DNA gave a single band of expected size, slightly higher than the transcripts since the transcripts are spliced. RT-PCR using primers for the remaining virion RNAs produced a single DNA product for both the induced BCBL-1 and virion RNA samples; these products were the same sizes as the products from the PCR using BAC DNA as a template for the gene-specific primers, confirming our array data. (It is important to note that K7 and PAN have overlapping sequences. We specifically employed primers that would amplify the 5′ untranslated region of the K7 ORF, which is unique to the K7 mRNA and not shared with PAN RNA.) We also examined the virion RNAs for the presence of several genes that did not score on our arrays, although they are present at high levels at late times of infection. The RT-PCRs for the virion RNA using primers for LANA, RTA, and ORF8 did not produce any specific products. Finally, control reactions performed on the induced BCBL-1 RNA produced bands of the expected sizes (Fig. 2, top), as did PCR from KSHV BAC DNA; no RT-PCR signal was generated from BJAB RNA. We conclude that the transcripts identified by our array analyses are indeed present in KSHV virions and that these transcripts are polyadenylated, since the RT-PCRs utilized an oligo(dT) primer for reverse transcription.
FIG. 2.
RT-PCR confirms the presence of KSHV virion RNAs. RNAs isolated from BJAB cells, uninduced and induced BCBL-1 cells, and KSHV virions were reverse transcribed using an oligo(dT) primer and then subjected to PCR with gene-specific primers. The PCR products were separated on 1.5 to 2% agarose gels. Lanes 1, KSHV BAC DNA; lanes 2, uninduced BCBL-1 cDNA; lanes 3, induced BCBL-1 cDNA; lanes 4, KSHV virion cDNA. Molecular size standards are the GeneRuler 100-bp DNA Ladder (Fermentas).
Confirmation that virion RNAs are full length.
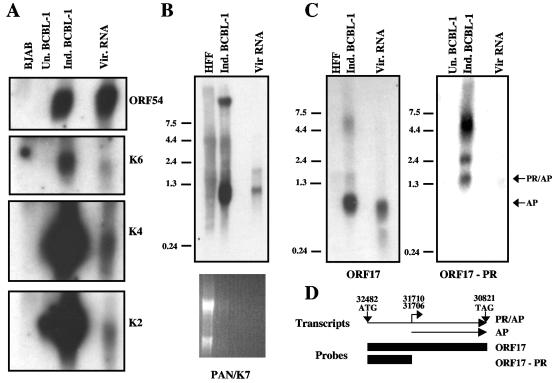
Next, we performed Northern blots on the virion RNA to verify for selected RNAs that the incorporated transcripts were full length. Poly(A)-purified RNAs isolated from BJAB and uninduced and induced BCBL-1 cells and KSHV virion RNA were separated on an agarose formaldehyde gel, transferred to nylon, and then probed for the specific transcripts. Strand-specific riboprobes were used for K2, K4, K6, K5IR, K12, and ORF 54 (Fig. 3A and data not shown). Although some of the blots are overexposed to visualize the virion RNA, it is clear that the ORF54, K2, K4, and K6 transcripts isolated from virus particles were the same size as the corresponding band in the induced BCBL-1 lanes. No specific signal was found in the lanes containing BJAB RNA, and occasionally there was a transcript of the appropriate size present in the uninduced BCBL-1 control lanes, suggesting that the cells used to isolate the RNA from uninduced BCBL-1 cells contained a small number of spontaneously reactivated cells. In addition, we probed for the presence of K7 and PAN mRNAs in the virion. The K7 and PAN genes exhibit substantial overlap, and a single DNA probe containing the entire PAN locus was used to probe for both K7 and PAN transcripts. As shown in Fig. 3B, the probe hybridized two bands: a 1.1-kb band representing the PAN transcript and a 1.8-kb transcript for the K7 mRNA (38). Since the blots are overexposed in order to detect signal from the virion RNAs, there is significant background in both the HFF and induced BCBL-1 lanes. On shorter exposures, there are specific bands corresponding to the two bands detected in the virion RNA lane and in the induced BCBL-1 lane, but not in the HFF lane (data not shown). It is important to note that the HFF RNA is present in significant molar excess above the induced BCBL-1 RNA, likely contributing to the background in that lane (ethidium bromide staining; Fig. 3B). K7 is expressed as three different transcripts that are differentially regulated during viral infection. The 1.8-kb band that we detected in our virions is the transcript detected at very early times of infection; in induced BC-P-1 cells, it is detected as early as 2 h postinduction. ORF17 encodes the viral protease (PR), as well as the assembly protein (AP), and several transcripts arise from the ORF17 locus. Only the transcriptional start site for the smaller AP transcript has been mapped; there are two start sites at genomic positions 31706 and 31710, leading to the production of a 950-bp transcript (Fig. 3D) (12). The longer transcripts arise from transcriptional start sites that are presently unmapped; the smallest transcript encompassing both PR and AP is 1.5 kb (35). Figure 3D depicts two of the transcripts from the locus, as well as the locations of the two riboprobes hybridized to Northern blots shown in Fig. 3C. When we probed RNAs isolated from KSHV virions, HFFs, and induced BCBL-1 cells with a riboprobe to the entire ORF17 locus, we observed a very strong signal for the 950-bp AP transcript in both the induced-BCBL-1 and virion RNA lanes, but not in the HFF lane (Fig. 3C). The ORF17 probe also hybridized to several less abundant transcripts; these are likely to represent the full-length PR/AP transcripts, since the AP transcript has been shown to accumulate to significantly higher levels during KSHV infection than the PR/AP transcript (35). To determine whether the full-length PR/AP transcript was also packaged into the virions, we probed KSHV virion RNA with a PR-specific riboprobe (Fig. 3C). We were able to observe three bands in induced BCBL-1 and KSHV virion RNAs (Fig. 3C). The PR/AP transcript migrates at 1.5 kb; this size correlated with the size of the fastest-migrating band observed in our Northern blot (Fig. 3C). We observed this band in both the induced BCBL-1 and KSHV virion RNAs, indicating that the full-length PR/AP transcript is packaged into virions. (The larger transcripts observed in both the induced BCBL-1 and KSHV virion RNA lanes have been suggested to be read-through of the ORF17 locus past the first canonical polyadenylation site [35].) As expected, our PR-specific riboprobe did not detect any of the 950-bp AP transcript (Fig. 3C). Since the transcripts detected were of the appropriate sizes and were the same sizes as the mRNAs detected in induced BCBL-1 cells, we concluded that the RNAs incorporated into the virion were full length.
FIG. 3.
Northern blots of KSHV virion RNA confirm that the transcripts are full length. (A) KSHV virion (Vir.) RNA and poly(A)-purified RNAs isolated from BJAB cells and uninduced (Un.) and induced (Ind.) BCBL-1 cells were separated on a 1% formaldehyde agarose gel and then blotted to nylon membranes and probed for ORF54, K6, K4, and K2 using strand-specific riboprobes. (B) RNAs isolated from HFFs, induced BCBL-1 cells, and KSHV virions were separated on a 1% formaldehyde agarose gel, transferred to nylon, and then probed for PAN (double-stranded DNA probe). The ethidium bromide staining of the Northern blot is shown below. (C) RNAs isolated from HFFs, induced BCBL-1 cells, and KSHV virions were separated on a 1% formaldehyde agarose gel, transferred to nylon, and then probed with a riboprobe to the entire ORF17 locus. Virion RNA and poly(A)-purified RNAs from uninduced and induced BCBL-1 cells were separated on a 1% agarose gel, transferred to nylon, and then hybridized with a riboprobe specific to the protease portion of ORF17. PR/AP and AP transcripts are marked with arrows. (D) Schematic diagram of two transcripts produced from the ORF17 locus. The start and stop codons for the longer PR/AP transcript are indicated, as are the two known transcriptional start sites for the AP transcript. The numbers refer to the genomic map positions. The two boxes represent the regions used as riboprobes in the Northern blots shown in Fig. 3C. The RNA ladders depicted in panels B and C are in kb.
In addition, we probed BJAB and uninduced and induced BCBL-1 RNAs with strand-specific primers for ORF58 and ORF59. Both ORF58 and ORF59 strand-specific probes hybridized to two transcripts (of 2.3 and 4.3 kb) present in induced BCBL-1 cells (data not shown). We were unable to detect any differences in the hybridization patterns of the two probes, and thus, we could not define an ORF58-specific monocistronic transcript. We conclude that the signals on our arrays and RT-PCRs arise from the bicistronic message.
ORF59 virion RNA is translated during infection.
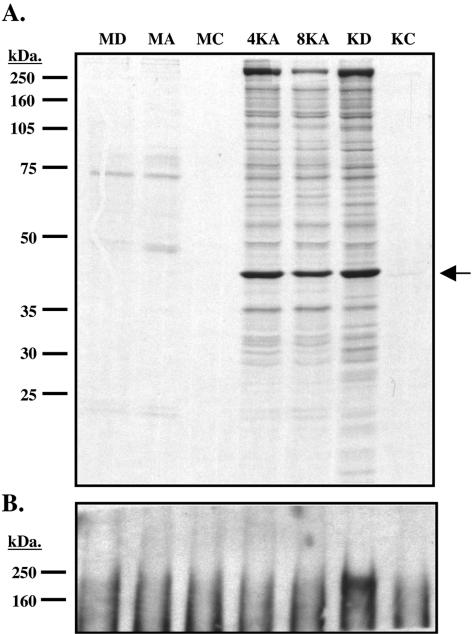
Next, we asked if the virion RNAs could be translated following de novo infection. We focused on the transcript for ORF59, since it is the only encapsidated mRNA for whose gene product a useful antibody is available. Mock- or KSHV-infected 293 cells were incubated with [35S]methionine and cysteine and then treated with either actinomycin D, cycloheximide, or DMSO. Lysates prepared from the cells at 4 or 8 h postinfection were immunoprecipitated by ORF59-specific antibodies, separated on a 10% SDS-PAGE gel, and examined by autoradiography (Fig. 4A). If imported ORF59 mRNA is translated, we would expect to detect labeled ORF59 protein in the cells infected in the presence of the transcription inhibitor actinomycin D, but such expression should be blocked by the translation inhibitor cycloheximide. As shown in Fig. 4A, we were able to detect labeled ORF59 protein in KSHV-infected cells at 4 and 8 h in the presence of actinomycin D at levels comparable to those in control cells untreated with inhibitors; expression was blocked efficiently by cycloheximide. As a control to demonstrate that actinomycin D was active under these conditions, lysates from each of these conditions were tested for expression of LANA. Since neither LANA nor its mRNA are incorporated into virions (5), LANA expression should be inhibited by both actinomycin D and cycloheximide. The lysates were immunoprecipitated with anti-LANA antibodies and then immunoblotted with the same antisera. As shown in Fig. 4B, LANA accumulation was blocked by both actinomycin and cycloheximide, as expected. We conclude that the encapsidated ORF59 transcript is indeed translated after de novo infection.
FIG. 4.
ORF59 mRNA packaged into virions is competent for translation. 293 cells were mock or KSHV infected in the presence of DMSO, actinomycin D (2 μg/ml), or cycloheximide (100 μg/ml) and labeled continuously with [35S]methionine and cysteine under these conditions for either 4 or 8 h. The cells were washed and lysed in 1% Ipegal buffer and then subjected to immunoprecipitation with antibodies specific for ORF59 (A) or LANA (B). (A) 35S-labeled ORF59 immunoprecitates were separated on a 10% SDS-PAGE gel, fixed, treated with Enhance (Perkin-Elmer), and then dried and exposed to film. The arrow indicates the ORF59 protein. (B) 35S-labeled LANA immunoprecipitates were separated on a 7.5% SDS-PAGE gel and then transferred to polyvinylidene difluoride and probed for LANA. MD, mock infected with DMSO; MA, mock infected with actinomycin D; MC, mock infected with cycloheximide; 4KA and 8KA, KSHV infected for 4 or 8 h with actinomycin D; KD, KSHV infected with DMSO; KC, KSHV infected with cycloheximide. Cycloheximide samples were harvested at 4 h postinduction. DMSO and actinomycin D samples were harvested at 8 h postinduction unless otherwise noted.
Selectivity of RNA encapsidation.
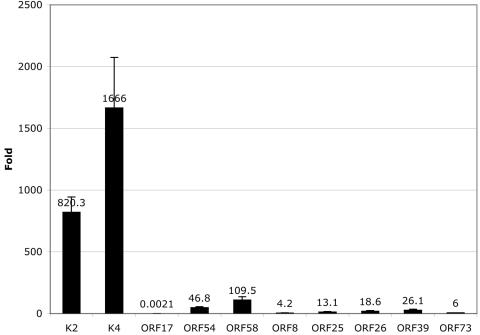
It has been controversial whether there exists a specific mechanism for herpesvirus RNA encapsidation versus the nonspecific packaging of the most abundant viral RNAs by simple inclusion with host cytoplasm during budding. To assess the specificity of incorporation of the viral transcripts into the KSHV virions, we employed quantitative, real-time RT-PCR to compare the levels of selected virion RNAs to those of viral genes that are not encapsidated into the virion. This analysis was performed late in infection, the period during which viral budding occurs. Specifically, we compared the levels of transcripts for K2, K4, ORF17, ORF54, and ORF58 (virion RNAs) to the levels of ORF8, ORF25, ORF26, ORF39, and ORF73 (unincorporated viral mRNAs). The unincorporated transcripts were selected to represent both latent (LANA) and late (ORF8 [gB], -25 [MCP], -26 [TRI-2], and -39 [gM]) temporal classes. We anticipated that the late mRNAs would be abundant, since they are made after the amplification of template viral DNA and encode abundant structural proteins. The primer pairs and probes for each gene are listed in Table 2. The abundance of each viral RNA in the infected cell is expressed as its concentration (n-fold) relative to host cell GAPDH mRNA; the results are displayed as a histogram to allow inspection of the levels of the viral mRNAs relative to one another (Fig. 5). We found that the majority of virion RNAs examined were more abundant at late times of infection than all of the transcripts examined that were not identified as virion components. The gM (ORF 39) transcript was the most abundant transcript analyzed that was not present in virions. K2 and K4 were present at levels 31 and 63 times higher than that of the gM (ORF39) transcript at late times of infection. ORF54 and ORF58 were also more abundant than gM, but only by two- and fourfold, respectively. These results are consistent with the notion that most of the encapsidated RNAs may be packaged simply as a result of their cytoplasmic abundance; if so, the threshold for such inclusion may be within two- to fourfold of the gM mRNA level.
FIG. 5.
Real-time PCR suggests that the ORF17 RNA is specifically incorporated into KSHV virions. RNA from induced BCBL-1 cells (3 days postinoculation) was reverse transcribed with an oligo(dT) primer and subjected to real-time PCR using gene-specific primers. Standard curves were generated for each primer/probe set (listed in Table 2) and then used to calculate the relative amounts of transcripts present in the sample. Levels of KSHV transcripts were normalized to levels of GAPDH transcript. The graph displays the normalized levels of each transcript (n-fold) over GAPDH. The data values are listed with the corresponding bars, and the error bars representing the standard deviations are shown.
In contrast, we found that the mRNA level for one virion RNA was far below that of gM or any of the other unincorporated transcripts. The real-time RT-PCR demonstrated that the ORF17 transcript is present at very low levels (12,000 times less than gM) at late times of infection yet is still incorporated into the virus particle. Its low abundance makes it unlikely to be incorporated into the virion via mass action and suggests that it may be specifically incorporated into the virus particle; the mechanism by which such incorporation proceeds, however, is unknown.
DISCUSSION
Our results show that several KSHV RNAs are packaged into virions. Using several different arrays, we were able to detect 11 virion RNAs incorporated into the virus particle. RT-PCR on the virion RNA samples demonstrated that all of the RNAs identified by our array analysis were present in virions, while transcripts not detected in the array experiments were not amplified by RT-PCR. We also established that these RNAs were polyadenylated and full length. Metabolic labeling of de novo infected cells allowed us to directly demonstrate that at least one virion mRNA, encoding ORF59, was translated following de novo infection. Although lack of available antisera precluded us from determining if this is true for the remaining transcripts, we have no reason to believe that this conclusion will not apply to all of the encapsidated mRNAs.
The majority of virion RNAs are present at very high levels at late times of infection. This suggests that these transcripts may be preferentially packaged simply due to their abundance and not by a specific mechanism. By contrast, a virion transcript encoding ORF17 is present at levels significantly lower than those of all of the unincorporated controls, suggesting that there may be a specific mechanism of incorporation for this mRNA. ORF17 encodes the viral protease (Pr) and assembly protein (AP); the genomic locus produces two transcripts, a minor mRNA encoding the Pr/AP polyprotein and a smaller, much more abundant transcript that produces only AP. Both transcripts are packaged (Fig. 3C). Our PCR primer/probe set was specific for the protease domain, and thus, the transcript we quantitated in Fig. 5 is that for the longer, more minor Pr/AP transcript. While the abundant AP transcript may be packaged nonspecifically, a specific mechanism likely operates for the Pr/AP message; if so, the relevant sequences for this packaging must reside in the portion of the mRNA that is specific to the protease. Tests of this possibility are now in progress.
Interestingly, 10 of the 11 virion RNAs correspond to lytic mRNAs detected at early times after infection by Krishnan and colleagues (21). These RNAs represent approximately 30% of the lytic RNAs detected by Krishnan et al. (21) and provide a satisfying, but only partial, explanation of their findings. Clearly, there must be de novo transcription of the remaining lytic genes during the first hours of infection. In fact, we have recently observed that even a gene whose mRNA is imported (K2) undergoes active transcription between 4 and 8 h after infection, since levels of K2 mRNA are higher in untreated cells than in those infected in the presence of actinomycin D (J. Bechtel and D. Ganem, unpublished observations). Possibly relevant to such de novo transcription is our recent observation that KSHV virions also encapsidate small quantities of the RTA protein, which could function to direct lytic transcription (5). Interestingly, virions also encapsidate the RAP (K8) protein, which has been shown to bind to RTA and impair its activation functions, at least on selected viral promoters (19, 23). These factors could, in the aggregate, result in an unusual pattern of transcript accumulation like that which characterizes these early time points. However, much remains to be learned about this phenomenon, not least how it is terminated in favor of traditional latency.
Functionally, the RNAs delivered by the virion could operate to make the cellular environment more conducive to the establishment of infection. For example, translation of some of these mRNAs will yield products that are antiapoptotic (K7 and K2) and immunomodulatory (K2, K4, and K6); others are involved in cellular signal transduction, both in infected cells (kaposin/K12) and in surrounding, uninfected cells (K2, K4, and K6). However, the rationale underlying the functions of other transcripts is not clear. The ORF17, ORF54, and ORF58/59 transcripts produce proteins whose known functions are involved in viral DNA replication (ORF54 and -59) and capsid assembly/DNA incorporation (ORF17 and Pr/AP). These activities seem unlikely to be relevant at very early times of infection, though it is possible that these proteins have evolved functions in addition to the ones presently ascribed to them. (KSHV already has one example of a protein conserved among all of the herpesviruses that has evolved a new function: the SOX [ORF37] protein is a canonical DNase that has also evolved an RNA degradation function [16].) Alternatively, their products may have no role in early viral replication and may simply be epiphenomena resulting from their nonspecific inclusion in virion RNA as a result of their abundance. It is harder, however, to explain away the presence of the viral protease mRNA in this fashion. We speculate that the protease may be expressed early in the cycle to allow cleavage of additional host or viral proteins. The recognition sequence for the viral protease has been characterized, and BLAST sequence analysis of the KSHV genome reveals that there are other proteins that contain the recognition sequences that could potentially serve as substrates (A. Marnett and J. Bechtel, unpublished observations). Additional experiments will be needed to explore this possibility.
Acknowledgments
We thank Dave Wang for the Illumina array and his assistance with the array hybridizations and scanning, Britt Glaunsinger for her generous provision of KSHV plasmids, Alan Marnett for his assistance in searching for proteins with potential protease cleavage sites, and Chris Sullivan for critical reading of the manuscript.
REFERENCES
- 1.Akula, S. M., P. P. Naranatt, N. S. Walia, F. Z. Wang, B. Fegley, and B. Chandran. 2003. Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) infection of human fibroblast cells occurs through endocytosis. J. Virol. 77:7978-7990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akula, S. M., N. P. Pramod, F. Z. Wang, and B. Chandran. 2002. Integrin α3β1 (CD 49c/29) is a cellular receptor for Kaposi's sarcoma-associated herpesvirus (KSHV/HHV-8) entry into the target cells. Cell 108:407-419. [DOI] [PubMed] [Google Scholar]
- 3.Ballestas, M. E., P. A. Chatis, and K. M. Kaye. 1999. Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science 284:641-644. [DOI] [PubMed] [Google Scholar]
- 4.Bechtel, J. T., Y. Liang, J. Hvidding, and D. Ganem. 2003. Host range of Kaposi's sarcoma-associated herpesvirus in cultured cells. J. Virol. 77:6474-6481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bechtel, J. T., R. C. Winant, and D. Ganem. 2005. Host and viral proteins in the virion of Kaposi's sarcoma-associated herpesvirus. J. Virol. 79:4952-4964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bozdech, Z., J. Zhu, M. P. Joachimiak, F. E. Cohen, B. Pulliam, and J. L. DeRisi. 2003. Expression profiling of the schizont and trophozoite stages of Plasmodium falciparum with a long-oligonucleotide microarray. Genome Biol. 4:R9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bresnahan, W. A., and T. Shenk. 2000. A subset of viral transcripts packaged within human cytomegalovirus particles. Science 288:2373-2376. [DOI] [PubMed] [Google Scholar]
- 8.Cannon, J. S., D. Ciufo, A. L. Hawkins, C. A. Griffin, M. J. Borowitz, G. S. Hayward, and R. F. Ambinder. 2000. A new primary effusion lymphoma-derived cell line yields a highly infectious Kaposi's sarcoma herpesvirus-containing supernatant. J. Virol. 74:10187-10193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cesarman, E., Y. Chang, P. S. Moore, J. W. Said, and D. M. Knowles. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N. Engl. J. Med. 332:1186-1191. [DOI] [PubMed] [Google Scholar]
- 10.Cesarman, E., P. S. Moore, P. H. Rao, G. Inghirami, D. M. Knowles, and Y. Chang. 1995. In vitro establishment and characterization of two acquired immunodeficiency syndrome-related lymphoma cell lines (BC-1 and BC-2) containing Kaposi's sarcoma-associated herpesvirus-like (KSHV) DNA sequences. Blood 86:2708-2714. [PubMed] [Google Scholar]
- 11.Chan, S. R., C. Bloomer, and B. Chandran. 1998. Identification and characterization of human herpesvirus-8 lytic cycle-associated ORF 59 protein and the encoding cDNA by monoclonal antibody. Virology 240:118-126. [DOI] [PubMed] [Google Scholar]
- 12.Chang, J., and D. Ganem. 2000. On the control of late gene expression in Kaposi's sarcoma-associated herpesvirus (human herpesvirus-8). J. Gen. Virol. 81:2039-2047. [DOI] [PubMed] [Google Scholar]
- 13.Chang, Y., E. Cesarman, M. S. Pessin, F. Lee, J. Culpepper, D. M. Knowles, and P. S. Moore. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266:1865-1869. [DOI] [PubMed] [Google Scholar]
- 14.Ciufo, D. M., J. S. Cannon, L. J. Poole, F. Y. Wu, P. Murray, R. F. Ambinder, and G. S. Hayward. 2001. Spindle cell conversion by Kaposi's sarcoma-associated herpesvirus: formation of colonies and plaques with mixed lytic and latent gene expression in infected primary dermal microvascular endothelial cell cultures. J. Virol. 75:5614-5626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao, S. J., J. H. Deng, and F. C. Zhou. 2003. Productive lytic replication of a recombinant Kaposi's sarcoma-associated herpesvirus in efficient primary infection of primary human endothelial cells. J. Virol. 77:9738-9749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Glaunsinger, B., and D. Ganem. 2004. Lytic KSHV infection inhibits host gene expression by accelerating global mRNA turnover. Mol. Cell 13:713-723. [DOI] [PubMed] [Google Scholar]
- 17.Gradoville, L., J. Gerlach, E. Grogan, D. Shedd, S. Nikiforow, C. Metroka, and G. Miller. 2000. Kaposi's sarcoma-associated herpesvirus open reading frame 50/Rta protein activates the entire viral lytic cycle in the HH-B2 primary effusion lymphoma cell line. J. Virol. 74:6207-6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greijer, A. E., C. A. Dekkers, and J. M. Middeldorp. 2000. Human cytomegalovirus virions differentially incorporate viral and host cell RNA during the assembly process. J. Virol. 74:9078-9082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Izumiya, Y., S. F. Lin, T. Ellison, L. Y. Chen, C. Izumiya, P. Luciw, and H. J. Kung. 2003. Kaposi's sarcoma-associated herpesvirus K-bZIP is a coregulator of K-Rta: physical association and promoter-dependent transcriptional repression. J. Virol. 77:1441-1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kedes, D. H., E. Operskalski, M. Busch, R. Kohn, J. Flood, and D. Ganem. 1996. The seroepidemiology of human herpesvirus 8 (Kaposi's sarcoma-associated herpesvirus): distribution of infection in KS risk groups and evidence for sexual transmission. Nat. Med. 2:918-924. [DOI] [PubMed] [Google Scholar]
- 21.Krishnan, H. H., P. P. Naranatt, M. S. Smith, L. Zeng, C. Bloomer, and B. Chandran. 2004. Concurrent expression of latent and a limited number of lytic genes with immune modulation and antiapoptotic function by Kaposi's sarcoma-associated herpesvirus early during infection of primary endothelial and fibroblast cells and subsequent decline of lytic gene expression. J. Virol. 78:3601-3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lagunoff, M., J. Bechtel, E. Venetsanakos, A. M. Roy, N. Abbey, B. Herndier, M. McMahon, and D. Ganem. 2002. De novo infection and serial transmission of Kaposi's sarcoma-associated herpesvirus in cultured endothelial cells. J. Virol. 76:2440-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liao, W., Y. Tang, S. F. Lin, H. J. Kung, and C. Z. Giam. 2003. K-bZIP of Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8 (KSHV/HHV-8) binds KSHV/HHV-8 Rta and represses Rta-mediated transactivation. J. Virol. 77:3809-3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mesri, E. A., E. Cesarman, L. Arvanitakis, S. Rafii, M. A. Moore, D. N. Posnett, D. M. Knowles, and A. S. Asch. 1996. Human herpesvirus-8/Kaposi's sarcoma-associated herpesvirus is a new transmissible virus that infects B cells. J. Exp. Med. 183:2385-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller, G., M. O. Rigsby, L. Heston, E. Grogan, R. Sun, C. Metroka, J. A. Levy, S. J. Gao, Y. Chang, and P. Moore. 1996. Antibodies to butyrate-inducible antigens of Kaposi's sarcoma-associated herpesvirus in patients with HIV-1 infection. N. Engl. J. Med. 334:1292-1297. [DOI] [PubMed] [Google Scholar]
- 26.Moses, A. V., K. N. Fish, R. Ruhl, P. P. Smith, J. G. Strussenberg, L. Zhu, B. Chandran, and J. A. Nelson. 1999. Long-term infection and transformation of dermal microvascular endothelial cells by human herpesvirus 8. J. Virol. 73:6892-6902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prichard, M. N., S. Jairath, M. E. Penfold, S. St Jeor, M. C. Bohlman, and G. S. Pari. 1998. Identification of persistent RNA-DNA hybrid structures within the origin of replication of human cytomegalovirus. J. Virol. 72:6997-7004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Raab, M. S., J. C. Albrecht, A. Birkmann, S. Yaguboglu, D. Lang, B. Fleckenstein, and F. Neipel. 1998. The immunogenic glycoprotein gp35-37 of human herpesvirus 8 is encoded by open reading frame K8.1. J. Virol. 72:6725-6731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Renne, R., M. Lagunoff, W. Zhong, and D. Ganem. 1996. The size and conformation of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) DNA in infected cells and virions. J. Virol. 70:8151-8154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sciortino, M. T., M. Suzuki, B. Taddeo, and B. Roizman. 2001. RNAs extracted from herpes simplex virus 1 virions: apparent selectivity of viral but not cellular RNAs packaged in virions. J. Virol. 75:8105-8116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sciortino, M. T., B. Taddeo, A. P. Poon, A. Mastino, and B. Roizman. 2002. Of the three tegument proteins that package mRNA in herpes simplex virions, one (VP22) transports the mRNA to uninfected cells for expression prior to viral infection. Proc. Natl. Acad. Sci. USA 99:8318-8323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun, R., S. F. Lin, L. Gradoville, Y. Yuan, F. Zhu, and G. Miller. 1998. A viral gene that activates lytic cycle expression of Kaposi's sarcoma-associated herpesvirus. Proc. Natl. Acad. Sci. USA 95:10866-10871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Terhune, S. S., J. Schroer, and T. Shenk. 2004. RNAs are packaged into human cytomegalovirus virions in proportion to their intracellular concentration. J. Virol. 78:10390-10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tomescu, C., W. K. Law, and D. H. Kedes. 2003. Surface downregulation of major histocompatibility complex class I, PE-CAM, and ICAM-1 following de novo infection of endothelial cells with Kaposi's sarcoma-associated herpesvirus. J. Virol. 77:9669-9684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Unal, A., T. R. Pray, M. Lagunoff, M. W. Pennington, D. Ganem, and C. S. Craik. 1997. The protease and the assembly protein of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8). J. Virol. 71:7030-7038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vieira, J., M. L. Huang, D. M. Koelle, and L. Corey. 1997. Transmissible Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) in saliva of men with a history of Kaposi's sarcoma. J. Virol. 71:7083-7087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vieira, J., P. O'Hearn, L. Kimball, B. Chandran, and L. Corey. 2001. Activation of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) lytic replication by human cytomegalovirus. J. Virol. 75:1378-1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang, H. W., T. V. Sharp, A. Koumi, G. Koentges, and C. Boshoff. 2002. Characterization of an anti-apoptotic glycoprotein encoded by Kaposi's sarcoma-associated herpesvirus which resembles a spliced variant of human survivin. EMBO J. 21:2602-2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang, Y., H. Li, M. Y. Chan, F. X. Zhu, D. M. Lukac, and Y. Yuan. 2004. Kaposi's sarcoma-associated herpesvirus ori-Lyt-dependent DNA replication: cis-acting requirements for replication and ori-Lyt-associated RNA transcription. J. Virol. 78:8615-8629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou, F. C., Y. J. Zhang, J. H. Deng, X. P. Wang, H. Y. Pan, E. Hettler, and S. J. Gao. 2002. Efficient infection by a recombinant Kaposi's sarcoma-associated herpesvirus cloned in a bacterial artificial chromosome: application for genetic analysis. J. Virol. 76:6185-6196. [DOI] [PMC free article] [PubMed] [Google Scholar]