Abstract
The human immunodeficiency virus type 1 nucleocapsid protein (NC) is a nucleic acid chaperone that catalyzes the rearrangement of nucleic acids into their thermodynamically most stable structures. In the present study, a combination of optical and thermodynamic techniques were used to characterize the influence of NC on the secondary structure, thermal stability and energetics of monomolecular DNA quadruplexes formed by the sequence d(GGTTGGTGTGGTTGG) in the presence of K+ or Sr2+. Circular dichroism studies demonstrate that NC effectively unfolds the quadruplexes. Studies carried out with NC variants suggest that destabilization is mediated by the zinc fingers of NC. Calorimetric studies reveal that NC destabilization is enthalpic in origin, probably owing to unstacking of the G-quartets upon protein binding. In contrast, parallel studies performed on a related DNA duplex reveal that under conditions where NC readily destabilizes and unfolds the quadruplexes, its effect on the DNA duplex is much less pronounced. The differences in NC's ability to destabilize quadruplex versus duplex is in accordance with the higher ΔG of melting for the latter, and with the inverse correlation between nucleic acid stability and the destabilizing activity of NC.
INTRODUCTION
The human immunodeficiency virus type 1 (HIV-1) carries its genetic information on a single-stranded RNA that is reverse transcribed into double-stranded DNA before integration into the host cell's genome. Reverse transcription involves a complicated series of biochemical reactions and strand transfer events. It has been shown that plus-strand DNA synthesis terminates by a strand displacement synthesis at the center of the DNA duplex, which generates a ‘flap’ of 99 nt (1,2). As a result, there exists at the center of the DNA genome a three-stranded region with two identical plus-strands and the complementary minus-strand segment, which appears to be important for proper replication of HIV-1 (2–4). Both plus-strands contain two adjacent guanine tracts consisting of four and six guanines in a row, separated by 9 nt (5). It has been shown that the G-tracts can be involved in the formation of G-quartets (6); these are formed by four guanine residues associated in a square planar configuration, where each guanine interacts with its two neighbors through Hoogsteen hydrogen bonds (8 per quartet). The formation of G-quartets requires the presence of cations that bind specifically to guanine O6 carbonyl groups between the planes of the G-quartets. Owing to the cation coordination and stacking interactions between G-quartets, quadruplexes are remarkably stable. The stability depends strongly on the size of the cation, with the most favorable ionic radius between 1.3 and 1.4 Å (e.g. K+ or Sr2+) (7–9).
The HIV-1 nucleocapsid protein (NC) is a small, highly basic protein containing 15 positively charged amino acids out of a total of 55 (10–13). Owing to its high charge density, NC strongly binds and aggregates nucleic acids. NC also contains two zinc finger motifs of the CCHC type, which are involved in sequence-specific binding to DNA or RNA single-stranded regions (14–16). It has been reported that NC facilitates reverse transcription by accelerating tRNA primer annealing (17–19), minus- and plus-strand transfer reactions (20–22) and strand displacement synthesis that produces the central DNA flap (1). In the latter work, a full-length DNA flap was investigated using an in vitro assay that mimics HIV-1 strand displacement synthesis. Optimal conditions that enable synthesis of the complete central flap in vitro in the presence of NC have been achieved. Based on this experiment, it was suggested that NC facilitates structural reorganization within the flap, which in turn accelerates the displacement synthesis (1). Recently, it was shown that NC preferentially recognizes the intermolecular G-quadruplex structures formed by the sequences derived from the DNA flap (6).
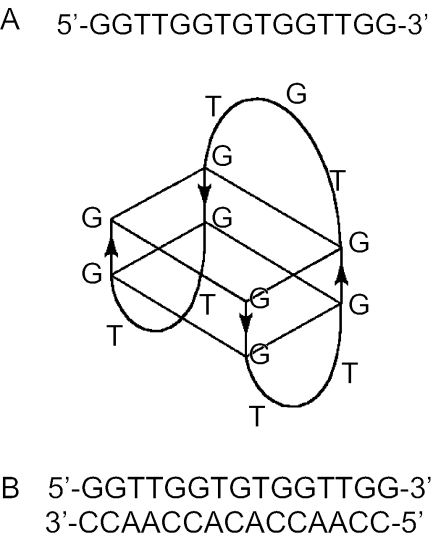
Here, we further characterize the interaction between NC and G-quartets. The oligonucleotide d(GGTTGGTGTGGTTGG), which folds into a monomolecular quadruplex with two G-quartets connected by a TGT and two TT loops, was used in this work (Figure 1A) (23,24). This quadruplex was shown previously to bind thrombin with high affinity and is therefore referred to as the thrombin binding aptamer (TBA) (25). There are several reasons for using this particular molecule in our study: (i) the quadruplex forms readily in solution (9); (ii) the sequence contains only guanines and thymidines, which are the preferred binding sites for NC (14,26,27); (iii) depending on the identity of the counterion, TBA forms quadruplexes of different stability (9); and (iv) by simple counterion exchange (for instance, Cs+ for K+), the entire folding process of the quadruplex may be monitored at ambient temperature (9).
Figure 1.
Sequences of (A) the TBA and (B) the DNA duplex studied in this work. The scheme showing the folded TBA molecule is based on its known 3D structure (23,24).
Here, we study the effect of NC on the thermodynamic properties of the TBA quadruplex and a related DNA duplex (Figure 1B) using circular dichroism (CD) spectroscopy, ultraviolet (UV) melting experiments and isothermal titration calorimetry (ITC). CD experiments allowed us to evaluate the influence of NC on the secondary structure of the DNA molecules, while UV melting studies and ITC measured the effect of NC on the stabilization/destabilization of the duplex or the quadruplex formed in the presence of different cations. Taken together, these studies show that NC destabilizes various monomolecular quadruplex structures, even at low concentrations of the protein (1 NC per 15 nt). Addition of a second equivalent of NC results in further quadruplex destabilization. Removal of Zn2+ ions from NC results in a complete loss of the quadruplex destabilization activity of the protein, whereas deletion of the N-terminal basic domain while retaining the Zn finger structures does not affect the destabilization activity. Interestingly, under conditions where we observe quadruplex unfolding, NC has only moderate, if any, effects on the thermal stability and the secondary structure of the DNA duplex.
MATERIALS AND METHODS
Proteins and nucleic acids
NC was prepared by solid-phase synthesis, cleaved, purified and activated with Zn2+, as will be described elsewhere. The truncated NC(11–55), with its C-terminal a carboxylic acid and its N-terminal a free amine, was prepared similarly; the structure was confirmed by ESI-MS: m/z for [M + 7H]calc.: 783.4; m/z for [M + 7H]found: 783.4.
DNA oligonucleotides were purchased from Eppley Research Institute at the University of Nebraska Medical Center; they were high-performance liquid chromatography purified and desalted by dialysis against water at 4°C using Spectrum dialysis tubing with a molecular weight cut-off of 500 Da. The concentrations of the oligonucleotides were determined at 260 nm and 80°C, using the following molar extinction coefficients (given in mM−1 cm−1 of strands): 145 for d(GGTTGGTGTGGTTGG) and 141 for d(CCAACCACACCAACC). These values were calculated using procedures reported earlier (28). All solutions contained 10 mM Cs–HEPES at pH 7.5. UV unfolding and CD studies on the TBA quadruplexes were performed in the presence of either 50 mM KCl or 10 mM SrCl2, as indicated in the figure legends.
CD spectroscopy
CD spectra were obtained with a JASCO J710 spectropolarimeter equipped with a water-jacketed cell holder using 10 mm path length cells. Typically, the solutions of DNA oligonucleotides in the desired buffer were titrated by stepwise addition of 3–5 μl aliquots of NC, from 0.8 to 1 mM stock solution, directly into optical cells. NC alone does not contribute to the CD signal between 230 and 350 nm.
UV melting experiments
UV absorption readings were taken as a function of temperature, using a GBC 918 spectrophotometer equipped with thermoelectrically controlled cell holders. The melting curves were recorded at either 260 or 300 nm. In a typical experiment, DNA and/or NC samples were mixed and diluted into the desired degassed buffers in optical cuvettes. The NC-free solutions were incubated at 80°C for ∼20 min in the cell holder. The temperature was then ramped to the desired starting temperature, NC was added and the melting experiments were performed at a heating rate of 0.5°C/min. Melting curves allowed an estimation of melting temperature, Tm, the midpoint temperature of the unfolding process. Then, van't Hoff enthalpies, ΔHvH, were calculated using the equation (29).
ITC measurements
A MicroCal MCS calorimeter was used to measure the heat evolved during quadruplex or duplex formation. In the case of quadruplex formation, the oligonucleotide solution without or with NC protein was placed in the reaction cell and titrated with K+ or Sr2+. Typically, 5 or 10 μl aliquots of salt solution (1–5 mM) were injected into the oligonucleotide solution (5–10 μM) by a syringe spinning at 400 r.p.m. The heat of duplex formation was followed by titrating TBA (∼30 μM) into its complementary strand, d(CCAACCACACCAACC) (2–3 μM). In the experiments with NC, both oligonucleotides in the cell and in the syringe were premixed with the desired amounts of NC. The resulting curves were corrected by subtracting the signal obtained from control experiments in which the cation or oligonucleotide solutions were injected into the buffer. The data analysis program provided by MicroCal was used to obtain the number of binding sites, n, the binding constants, K, and the enthalpies of binding, ΔH. Free energies were calculated using the equation ΔG = −RTlnK and the entropic term was calculated using the equation TΔS = ΔH − ΔG.
RESULTS AND DISCUSSION
CD measurements
CD spectroscopy is a sensitive technique for probing the secondary structure of nucleic acids, particularly of quadruplexes. Upon folding of the TBA quadruplex, the absolute value of the CD signal between 220 and 320 nm increases several fold (9). For the present work, we have used CD spectroscopy to determine the influence of NC on the secondary structures of both the monomolecular TBA quadruplex and of a DNA duplex formed by annealing the TBA sequence to a perfectly complementary oligonucleotide. In addition to wild-type synthetic NC, we studied two derivatives of NC: (i) a truncated polypeptide retaining Zn2+, but lacking the N-terminal basic domain, i.e. NC(11–55) and (ii) an unstructured polypeptide formed by removal of Zn2+ ions with EDTA treatment.
Quadruplexes
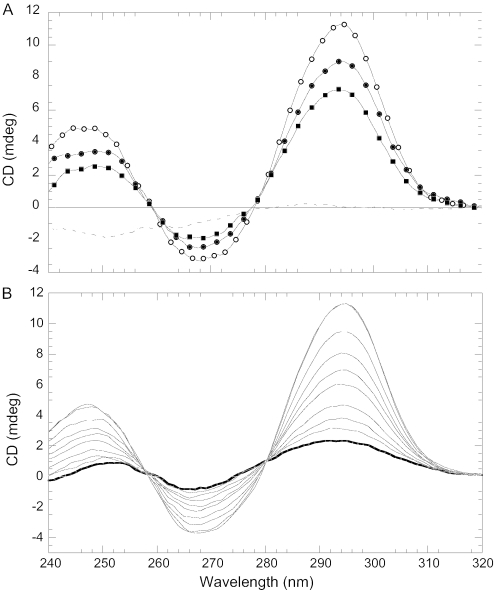
Figure 2A shows the CD spectra of K-TBA in 50 mM KCl, 10 mM Cs–HEPES at pH 7.5 and 20°C. The spectrum before adding NC is characteristic for the potassium salt of TBA: positive bands with maxima ∼245 and 295 nm and a negative peak ∼268 nm (9) (Figure 2A, open circles). NC protein significantly changes the CD profile of the quadruplex: drops by ∼18% ellipticity at all bands upon the addition of 1 NC per TBA (Figure 2A, closed circles). Addition of a second equivalent of NC has a similar effect (Figure 2A, closed squares). Although the addition of a third NC per TBA also shows an effect, parallel measurements of optical absorption at 350 nm reveals some aggregation; therefore, this result is not shown here. The CD profiles in Figure 2A also reveal isosbestic points ∼258 and 280 nm, which indicate the two-state nature of the structural transition of the quadruplex upon NC binding. It is interesting that the same isosbestic points are observed during folding of the TBA quadruplex by K+ ions (Figure 2B). This observation shows that NC cooperatively unfolds the quadruplex structure and suggests that each equivalent of NC added results in complete unfolding of a certain fraction of quadruplex molecules, while the remaining molecules stay folded. These conclusions are also supported by UV melting and ITC experiments as described below. CD characterization of Sr-TBA, which is significantly more stable than K-TBA based on an 18°C difference in Tm (9), also reveals similarly strong destabilization of the quadruplex by NC (Supplementary Material, Figure S1).
Figure 2.
CD spectra showing the effects of NC and KCl on the secondary structure of TBA. (A) CD spectra of K-TBA in 50 mM KCl and 10 mM Cs–HEPES, pH 7.5 at 20°C before (open circles) and after addition of NC: [NC]/[TBA] = 1 (closed circles) and [NC]/[TBA] = 2 (closed squares). The dashed line corresponds to unfolded TBA in water at 80°C. (B) CD spectra of TBA in 10 mM Cs–HEPES, pH 7.5 at 20°C in the absence (bold line) and in the presence of various concentrations of KCl. As the concentration of KCl was increased from 0 to 3 mM, the absolute value of the CD signal increased at all three bands. The concentration of TBA is ∼2 μM in both spectra.
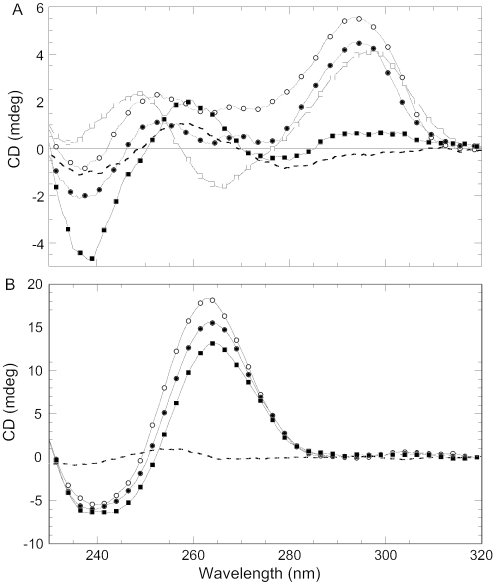
We also studied the effect of NC on two other monomolecular quadruplexes: K+ salts of (GGGTTA)3GGG and (GGGT)4. The first quadruplex is derived from the human telomere sequence and its Na+ form was reported previously to be stabilized by NC (6). The latter quadruplex was chosen based on its significantly higher stability [Tm increased by 42°C compared to K-TBA (30)], which is attributed to the presence of highly ordered loops in the presence of either Na+ or K+ (30,31). Figure 3A reveals that NC effectively unfolds the (GGGTTA)3GGG quadruplex; in the presence of two equivalents of NC, the CD signal at the positive bands drops by 20% (K+) and 60% (Na+), respectively. Similarly, a 30% decrease in the CD signal was observed upon the addition of two equivalents of NC to (GGGT)4 (Figure 3B). The discrepancy between the present study, which shows almost complete unfolding of the human telomere repeat in the presence of Na+, and earlier work (6) that reported NC-induced stabilization of (GGGTTA)3GGG, likely reflects differences in total DNA and NC concentrations, which are much higher in our experiments.
Figure 3.
(A) CD spectra of (GGGTTA)3GGG under different experimental conditions. Circles correspond to 50 mM KCl and 10 mM Cs–HEPES, pH 7.5 at 20°C before (open circles) and after addition of 2 NCs per strand (closed circles). Squares correspond to 10 mM NaCl, 100 mM LiCl and 10 mM Cs–HEPES, pH 7.5 at 20°C before (open squares) and after addition of 2 NCs per strand (closed squares). (B) CD spectra of (GGGT)4 in 50 mM KCl, 10 mM Cs–HEPES, pH 7.5 at 20°C before (open circles) and after addition of NC: [NC]/[strand] = 1 (closed circles) and [NC]/[strand] = 2 (closed squares). The dashed lines in both spectra correspond to unfolded oligonucleotides in water at 80°C. Concentrations of oligonucleotides are ∼2 μM.
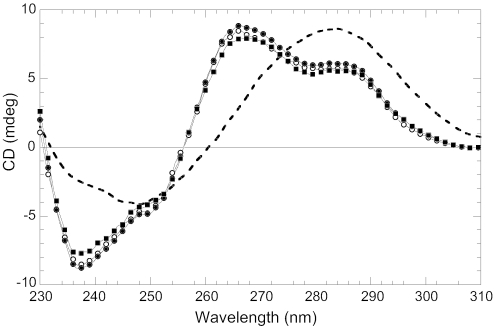
When Zn2+ is removed from NC by EDTA treatment, protein binding results in essentially no effect on K-TBA stability (Figure 4). These data reveal the important role of the Zn fingers in quadruplex destabilization, and are in agreement with earlier studies which showed that Zn finger mutations decrease or abolish NC chaperone activities (32–37).
Figure 4.
CD spectra showing the effect of NC variants on the secondary structure of K-TBA. Spectra were measured in 50 mM KCl and 10 mM Cs–HEPES, pH 7.5 at 20°C before (solid line) and after addition of protein: two equivalents of wild-type NC (dashed line), two equivalents of NC treated with EDTA to remove the Zn2+ ions (open circles), two equivalents of NC(11–55) (closed circles) and four equivalents of NC(11–55) (closed squares).
In contrast to results obtained with EDTA-treated NC, the deletion of 10 amino acids from the N-terminus did not result in a significant decrease in the destabilizing activity of NC (Figure 4). This result supports the conclusion that destabilization of TBA quadruplexes is induced primarily by the Zn finger domains of NC. As expected based on previous studies with NC lacking the N-terminal domain (38,39), we did not observe any aggregation, even upon the addition of four equivalents of NC(11–55) per oligonucleotide. Moreover, as shown in Figure 4, the addition of 4 NC(11–55) molecules resulted in further destabilization of the TBA quadruplex.
DNA duplex
The CD profile of the DNA duplex shown in Figure 1B, taken in 20 mM CsCl and 10 mM Cs–HEPES, corresponds to the expected B-DNA secondary structure of a guanine-rich duplex. The CD spectrum contains two positive bands at 266 and 283 nm, and a negative band ∼240 nm (Figure 5). In contrast to the result with TBA quadruplexes, the addition of NC does not significantly affect the CD spectrum; we observed only a slight increase of the CD signal upon the addition of one equivalent of NC and a slight decrease after the addition of second equivalent (Figure 5). The CD spectrum of a completely unfolded/melted duplex (Figure 5, dashed line) clearly indicates that the small CD effects observed upon NC binding do not correspond to any measurable duplex unfolding. Similarly, no measurable effects were observed in the case of NC(11–55), even at four proteins per strand (data not shown). Thus, we observe dramatically different effects of NC on the secondary structures of the DNA molecules investigated here. Binding of two equivalents of NC results in significant destabilization of monomolecular quadruplexes, with essentially no influence on the secondary structure of the DNA duplex derived from the same sequence.
Figure 5.
CD spectra of the DNA duplex, shown in Figure 1B, taken in 20 mM CsCl and 10 mM Cs–HEPES, pH 7.5 at 20°C before (open circles) and after addition of NC protein: [NC]/[Duplex] = 2 (closed circles) and [NC]/[Duplex] = 4 (closed squares). The dashed line corresponds to unfolded duplex in water at 20°C. The concentration of the duplex is ∼3 μM.
Salt concentration dependence
The CD studies discussed above were performed under a variety of solution conditions. All experiments with TBA contained 10 mM Cs–HEPES, pH 7.5, whereas the duplex was studied in the presence of 20 mM CsCl. To fold TBA into a stable quadruplex, 50 mM KCl or 10 mM SrCl2 were added. These salt concentrations (i) ensured formation of the desired secondary structures at the experimental temperature of 20°C, (ii) allowed a comparison of K-TBA and the DNA duplex under conditions of similar thermal stability (Figure 7) and (iii) facilitated NC binding to oligonucleotides (40,41). Binding is expected to be strong under the low salt conditions, but becomes weaker in high salt.
Figure 7.
(A) UV unfolding of K-TBA at 300 nm in the absence (open circles) and in the presence of NC protein: [NC]/[TBA] = 1 (closed circles) and [NC]/[TBA] = 2 (closed squares). Experimental conditions are as indicated in Figure 2A. The concentration of TBA is ∼6 μM. (B) UV unfolding of the duplex shown in Figure 1B at 260 nm, in the absence (open circles) and in the presence of NC protein: [NC]/[Duplex] = 2 (closed circles) and [NC]/[Duplex] = 4 (closed squares). Experimental conditions are as indicated in Figure 5.
To rule out the possibility that the different effects of NC on the quadruplex and duplex structures were induced by differences in solution conditions, we performed CD studies of the duplex in the presence of 50 mM KCl. However, NC did not significantly affect the CD spectrum of K-TBA under these conditions (Supplementary Material, Figure S2).
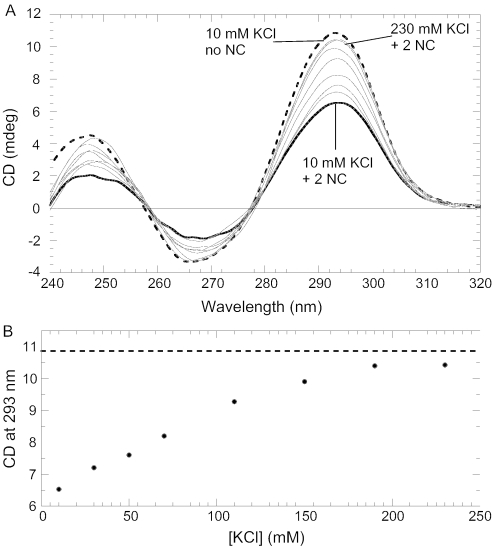
Non-specific electrostatic forces are major contributors to NC–nucleic acid interactions (42). To determine whether NC–quadruplex interactions are salt sensitive, we performed CD studies of quadruplex•NC complexes as a function of different monovalent cations. Since certain cations favor TBA quadruplex formation (e.g. K+, and Rb+) while others do not (e.g. Li+, Na+ and Cs+) (9), increasing the cation concentration in the former case not only contributes to the ionic strength but also directly affects the secondary structure of TBA. Figure 6A shows CD spectra of the TBA•2NC as a function of KCl concentration. The bold line corresponds to TBA•2NC in 10 mM KCl; increasing the K+ concentration results in an increase in the CD signal at all bands. Figure 6B shows the dependence of the CD signal at 293 nm on KCl concentration. At higher concentrations of KCl (i.e. ∼200 mM) the CD spectrum of the TBA•2NC complex resembles the spectrum obtained at low salt (i.e. 10 mM KCl) in the absence of NC (Figure 6A and B, dashed lines). This result suggests that NC is displaced from the quadruplex by high salt, similar to dissociation from other single- and double-stranded nucleic acids. This strong salt dependence of binding is a signature of the important contribution of non-specific ionic interactions to NC-TBA binding.
Figure 6.
(A) CD spectra of the TBA•2NC complex (solid curves) at different concentrations of K+ ions. The concentration of KCl ranges from 10 (solid, bold curve) to 230 mM with an increase in the CD signal at all three bands with increasing K+. The dashed curve corresponds to the CD spectrum in 10 mM KCl in the absence of NC. (B) Graph showing dependence of the CD signal at 293 nm on the KCl concentration. The dashed line corresponds to the CD signal of a fully folded quadruplex at 293 nm. The concentration of TBA is ∼2 μM.
UV melting experiments
We next used optical melting techniques to characterize thermodynamic parameters of TBA and DNA duplex unfolding, and to study the impact of NC on the unfolding process.
Quadruplexes
Figure 7A shows differential UV thermal unfolding profiles of the K-TBA quadruplex in the absence and in the presence of NC protein. As expected, K-TBA in 50 mM KCl melts at ∼50°C (9). The transition peak is negative, owing to the hypochromic nature of quadruplex unfolding at ∼300 nm (9,43). The unfolding is monophasic, corresponding to a two-state transition. As in the case of the CD measurements, UV thermal unfolding experiments were performed in the presence of 1 and 2 NC molecules per TBA strand. In the presence of NC, TBA is destabilized and the melting transition spans the entire temperature range. Owing to such broad melting transition, it was not possible to determine accurately the Tm values in the presence of NC.
DNA duplex
The differential unfolding curve of the duplex in 20 mM CsCl, 10 mM Cs–HEPES buffer (Figure 7B, open circles) is also characterized by a single sharp peak, corresponding to a two-state transition. Adding one equivalent of NC per strand does not induce any change in Tm value (Figure 7B and Table 1). However, the presence of NC broadens the transition, indicating a decrease in the transition enthalpy (Table 1). In the presence of 2 NC proteins per strand, we observe an ∼4°C decrease in Tm, and a further reduction in the cooperativity of the helix–coil transition, resulting in a ΔHvH value of 77 kcal/mol (Table 1). Thus, in contrast to the results obtained with the quadruplexes, 1 NC per DNA strand does not induce significant destabilization of the duplex, but the addition of 2 NC proteins per strand (i.e. 1 NC per 7–8 nt) slightly destabilizes the structure. We also performed UV melting experiments on the DNA duplex in the presence of NC(11–55); results were almost identical to wild-type NC (data not shown). Our melting studies are in general agreement with previous studies on DNA duplexes of varying length and sequence (44). The literature experiments showed that 1 NC per 8 nt destabilizes 21mer and 28mer DNA duplexes, while also broadening the melting transition.
Table 1.
Melting temperatures and van't Hoff enthalpies derived from UV unfolding profiles measured for the DNA duplex in the absence and presence of NCa
| Molecule/complex | Tm (°C) | ΔHvH (kcal/mol) |
|---|---|---|
| Duplex alone | 51.5 | 120 |
| Duplex•1NC | 51.5 | 109 |
| Duplex•2NC | 47.5 | 77 |
aThe DNA duplex sequence is shown in Figure 1B. Tm and ΔHvH were derived from the shapes of UV melting curves at a concentration of ∼3 μM per strand; buffer: 20 mM CsCl and 10 mM Cs–HEPES, pH 7.5. Experimental errors are: Tm (±0.5°C) and ΔHvH (±10%).
ITC experiments
ITC is a sensitive tool to study biomolecular interactions and can provide direct information on binding stoichiometry, affinity and enthalpy. We have used ITC to study the influence of NC on the folding of DNA quadruplexes and the DNA duplex at constant temperature. In the case of quadruplexes, aliquots of K+ or Sr2+ ions were injected into the Cs-TBA (unfolded quadruplex) solution in the absence and in the presence of NC, and the heat changes accompanying quadruplex formation and TBA-cation binding were measured. Results of a typical experiment are shown in Figure 8A. The apparent discontinuity in peaks is due to an increase in volume of titrant during the experiments. This allowed us to obtain binding isotherms with evenly distributed experimental points (Figure 8B) and to reliably estimate the binding parameters: number of binding sites, n, binding constants, K, and enthalpy, ΔH. A simple model, which assumes a single non-cooperative binding site, accurately describes the experimental titration curves (45). The solid line in Figure 8B is a result of nonlinear fitting based on this model. The thermodynamic parameters obtained from the fits are reported in Tables 2 and 3.
Figure 8.
ITC measurement of the interaction of K+ ions with TBA in 10 mM Cs–HEPES, pH 7.5 at 20°C. (A) The upper curve represents heats of KCl dilution in the same buffer. The lower curve shows the heat released upon titration of K+ into a 10 μM TBA solution by injection of various volumes (2, 4, 8 and 16 μl) of 7 mM KCl solution. (B) Plot showing the cumulative heats of reaction as a function of [K+]/[TBA], based on the data shown in A. The data are fit to a model that assumes a single non-cooperative binding site (45).
Table 2.
Thermodynamic parameters derived from ITC experiments measured for TBA quadruplex folding in 10 mM Cs–HEPES, pH 7.5 at 20°C
| Molecule/complex | n | ΔG (kcal/mol) | ΔH (kcal/mol) | TΔS(kcal/mol) |
|---|---|---|---|---|
| K-TBA | 3.1 ± 0.5 | −5.5 ± 0.1 | −21.6 ± 0.6 | −16.1 |
| K-TBA•1NC | 3.2 ± 1.3 | −4.5 ± 0.1 | −6.4 ± 1.5 | −1.9 |
| Sr-TBA | 1.0 ± 0.1 | −8.5 ± 0.1 | −14.3 ± 0.3 | −5.8 |
| Sr-TBA•1NC | 1.2 ± 0.3 | −7.4 ± 0.1 | −5.3 ± 0.3 | 2.1 |
Table 3.
Thermodynamic parameters derived from ITC experiments measured for duplex folding at 20°C
| Molecule/complex | n | ΔG (kcal/mol) | ΔH (kcal/mol) | TΔS (kcal/mol) |
|---|---|---|---|---|
| Low salt a | 1.0 ± 0.1 | −9.1 ± 0.1 | −64.1 ± 1.2 | −55.0 |
| High saltb | 1.1 ± 0.1 | n.d.c | −66.6 ± 1.8 | n.d.c |
| NC in low saltd | 1.1 ± 0.1 | −9.2 ± 0.1 | −69.8 ± 1.7 | −60.6 |
aLow salt corresponds to 10 mM Cs–HEPES, pH 7.5. The DNA duplex sequence is shown in Figure 1B.
bHigh salt corresponds to 100 mM NaCl and 10 mM Cs–HEPES, pH 7.5.
cNot determined.
dNC in low salt corresponds to duplex formation from the DNA single strands pre-equilibrated with equimolar amounts of NC in 10 mM Cs–HEPES, pH 7.5.
K-TBA
A binding stoichiometry of 3:1 K+ cations per quadruplex was determined for solutions of TBA both alone and in the presence of NC. Earlier NMR studies detected only two potassium binding sites (46). The present stoichiometry of 3:1 is, however, in good agreement with earlier acoustic experiments of TBA in the presence of K+ (9). As shown in Table 2, NC binding results in a strong decrease in quadruplex formation enthalpy, which is reduced to ∼70% of the ΔH value measured in the absence of NC. Addition of NC also results in a 14.2 kcal/mol increase in TΔS, which suggests that NC entropically stabilizes quadruplex formation. Thus, NC-induced enthalpic destabilization (15.2 kcal/mol) is almost compensated by entropic stabilization, resulting in only 1 kcal/mol net reduction in overall free energy of folding (Table 2). This result is in agreement with the NC-induced decrease in melting enthalpy and increase in melting entropy of both the TBA quadruplex and the DNA duplex observed in the UV thermal unfolding studies.
Sr-TBA
Binding of Sr2+ ions to TBA reveals a stoichiometry of 1:1 (Table 2), which agrees well with previous hydration (9) and NMR (48) studies. As in the case of K-TBA, the presence of NC does not affect binding stoichiometry, within experimental error (Table 2). The enthalpy of Sr-TBA formation, which is also in good agreement with an earlier determination (9), is strongly affected by the presence of NC. Thus, NC enthalpically destabilizes (by 9 kcal/mol) and entropically stabilizes (by 7.9 kcal/mol) the Sr-TBA quadruplex, resulting in a destabilization free energy of 1.1 kcal/mol, which is similar to the value determined for K-TBA. The thermodynamic data summarized in Table 2 reveal an enthalpically driven destabilization of both quadruplexes upon NC binding.
Duplex
In the ITC studies carried out with the DNA duplex, 30 μl aliquots of TBA were added to the complementary DNA strand, and the heat of duplex formation was measured. The influence of NC on duplex formation was studied by incubating equimolar amounts of NC with each oligonucleotide, prior to mixing. A strand stoichiometry of 1:1 was measured both in the absence and in the presence of NC (Table 3). The enthalpy of duplex formation in 10 mM Cs–HEPES buffer is 64 kcal/mol, which is significantly smaller than the unfolding value obtained from the UV melting experiments (Table 1). A similar discrepancy between ITC and UV unfolding enthalpies has been observed for other oligonucleotides (B. I. Kankia, unpublished data) and can be explained by the different nature of the experiments. In particular, thermal unfolding enthalpies contain heats of unstacking of nucleic acid bases, which are absent in ITC experiments. As in the case of TBA quadruplex unfolding, the absolute values of the enthalpies and entropies of duplex unfolding are significantly decreased in the presence of NC, according to both our ITC and UV data. The molecular basis for this phenomenon and its implication for the kinetics of duplex opening/closing by NC, will be discussed in detail elsewhere (I. Rouzina, manuscript in preparation).
ITC measurements performed at low ionic strength allow an estimation of the affinity between complementary strands, as well as of the free energy of duplex formation. As shown in Figure 9 (upper curve), titration experiments in 10 mM Cs–HEPES reveal gradually decreasing heats of reaction, which allowed us to estimate a value of 9.1 kcal/mol for the free energy of duplex formation in the absence of NC (Table 3). At higher ionic strength (100 mM NaCl) the binding affinity between complementary strands increases dramatically, which results in the ITC titration profile shown in the middle curve of Figure 9. Under these conditions, the affinity between the complementary strands is too high to be estimated from ITC (Table 3). However, the n and ΔH values determined under these conditions are similar to the values determined at low ionic strength (Table 3). The kinetics of duplex formation is highly dependent on solution ionic strength. At low ionic strength, duplex formation is slow, with a waiting time of 10 min between each injection. At high ionic strength, standard 3 min waiting periods were sufficient (Figure 9). Interestingly, in the presence of NC at low ionic strength, duplex formation is accelerated (Figure 9, bottom curve), while binding affinity and the free energy of duplex formation stay the same (Table 3).
Figure 9.
ITC measurement of duplex formation in 10 mM Cs–HEPES, pH 7.5 in the absence (upper curve) or in the presence of 100 mM NaCl (middle curve), or in the presence of NC (lower curve). In the latter case, each oligonucleotide in 10 mM Cs–HEPES was premixed with an equimolar amount of NC before loading into the calorimetric cell and the syringe. For clarity, the middle and bottom titration curves are offset by −0.4 and −1.8 μcal/s, respectively.
CONCLUSIONS
As reported in Table 2, favorable free energies of folding for both K-TBA and Sr-TBA quadruplexes result from the compensation of a favorable folding enthalpy by an unfavorable entropic term. Sr-TBA forms a significantly more stable quadruplex than K-TBA (by 3 kcal/mol); its enthalpy of folding is less favorable (by 7.3 kcal/mol), while its folding entropy is more favorable (by 10.3 kcal/mol). Thus, the more favorable folding ΔG of Sr-TBA is due to the entropic term. Based on the similarity of the K-TBA and Sr-TBA structures (47), it is reasonable to assume that hydrogen bonding and stacking interactions contribute similarly to the thermodynamic parameters. Thus, differences in the stability of the two quadruplexes may be explained by differences in ionic interactions and hydration effects resulting from binding of three K+ ions to K-TBA versus one Sr2+ cation to Sr-TBA (9).
In the presence of NC, quadruplex folding is destabilized enthalpically and stabilized entropically relative to the NC-free quadruplex systems. We observe a similar decrease (∼15%) in the folding free energy for both quadruplexes as a result of strong enthalpy–entropy compensation (Table 2). This result agrees well with the established notion that the free energy of folding is the result of a delicate balance between mutually compensatory enthalpic and entropic terms (48–50). The decrease in enthalpy of quadruplex folding, most likely, comes from enthalpic stabilization of the unfolded DNA state by NC. The latter effect is due to partial stacking of aromatic residues, F16 and W37, within the zinc fingers of NC with otherwise unstacked DNA bases (51).
Our UV data show that in the presence of NC, the melting profile of TBA molecules is significantly altered, with the transition becoming more uncooperative. In addition, both ITC and CD analyses show that at 20°C, NC effectively unfolds and increases the disorder of the quadruplex system. However, even at high NC concentrations, quadruplex unfolding is incomplete, in accordance with the limited capability of NC to melt nucleic acid secondary structures. The fact that we only observe partial quadruplex unfolding in the presence of NC does not imply that each quadruplex molecule is only partially unfolded. Indeed, the presence of isosbestic points in the CD spectra shown in Figure 2 provides evidence that NC induces a two-state transition between the fully folded and the fully unfolded quadruplex.
Taken together, ITC, UV melting and CD spectroscopy studies reveal that NC destabilizes and unfolds quadruplexes. The unfolding activity of NC does not require ATP and does not, therefore, reflect G4-helicase activity. The slight preference of NC for single-stranded nucleic acids is, in part, responsible for its destabilizing activity (43). Experiments with EDTA-treated NC and with truncated NC(11–55) show clearly that this effect is due to the zinc finger structures. Although a previous study showed that a zinc finger peptide derived from the DNA-binding domain of Zif268 binds to a quadruplex derived from the human telomeric sequence in a structure-specific manner (52), the Zif268-like zinc finger domains do not resemble the CCHC type retroviral zinc knuckles of NC.
In contrast, under conditions where we observe NC-induced quadruplex unfolding, the effect of NC on the related DNA duplex is much less pronounced. A reasonable explanation for the much weaker NC-induced DNA duplex versus quadruplex destabilization is the higher overall free energy of duplex melting by comparison to the ΔG of quadruplex melting (Tables 2 and 3). While the duplex contains 15 bp, there are only four base pairing interactions within the quadruplex. Therefore, assuming that NC has a similar destabilization effect per base pair on both DNA structures, the modest destabilization by NC within the quadruplex results in a major overall structural perturbation. In contrast, the same destabilization activity of NC (per base pair) on the duplex results in a very little change in duplex structure. This phenomenon is similar to the previously observed negative correlation between the destabilizing effect of NC and the stability of transactivation response element-derived DNA hairpins (53–55). As the stability of hairpin structures increased, the capability of NC to destabilize these structures decreased. We conclude, based on the recent work with DNA quadruplexes reported here, that NC has the capability to function as a relatively weak destabilizer of a variety of nucleic acid secondary and tertiary structures. This conclusion is in good agreement with NC's function in the HIV-1 lifecycle as a non-specific chaperone of a wide variety of nucleic acid restructuring events (42).
Acknowledgments
We thank Dr Ioulia Rouzina for critical reading of the manuscript and helpful discussions, and Dr Daniel G. Mullen and Ms Brandie J. Kovaleski for chemical synthesis of NC and NC(11–55). This work was supported by NIH grant GM65056. Funding to pay the Open Access publication charges for this article was provided by NIH grant GM65056.
Conflict of interest statement. None declared.
REFERENCES
- 1.Hameau L., Jeusset J., Lafosse S., Coulaud D., Delain E., Unge T., Restle T., Le Cam E., Mirambeau G. Human immunodeficiency virus type 1 central DNA flap: dynamic terminal product of plus-strand displacement DNA synthesis catalyzed by reverse transcriptase assisted by nucleocapsid protein. J. Virol. 2001;75:3301–3313. doi: 10.1128/JVI.75.7.3301-3313.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Charneau P., Mirambeau G., Roux P., Paulous S., Buc H., Clavel F. HIV-1 reverse transcription. A termination step at the center of the genome. J. Mol. Biol. 1994;241:651–662. doi: 10.1006/jmbi.1994.1542. [DOI] [PubMed] [Google Scholar]
- 3.Charneau P., Alizon M., Clavel F. A second origin of DNA plus-strand synthesis is required for optimal human immunodeficiency virus replication. J. Virol. 1992;66:2814–2820. doi: 10.1128/jvi.66.5.2814-2820.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hungnes O., Tjotta E., Grinde B. Mutations in the central polypurine tract of HIV-1 result in delayed replication. Virology. 1992;190:440–442. doi: 10.1016/0042-6822(92)91230-r. [DOI] [PubMed] [Google Scholar]
- 5.Lyonnais S., Hounsou C., Teulade-Fichou M.P., Jeusset J., Le Cam E., Mirambeau G. G-quartets assembly within a G-rich DNA flap. A possible event at the center of the HIV-1 genome. Nucleic Acids Res. 2002;30:5276–5283. doi: 10.1093/nar/gkf644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lyonnais S., Gorelick R.J., Mergny J.L., Le Cam E., Mirambeau G. G-quartets direct assembly of HIV-1 nucleocapsid protein along single-stranded DNA. Nucleic Acids Res. 2003;31:5754–5763. doi: 10.1093/nar/gkg716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen F.M. Sr2+ facilitates intermolecular G-quadruplex formation of telomeric sequences. Biochemistry. 1992;31:3769–3776. doi: 10.1021/bi00130a006. [DOI] [PubMed] [Google Scholar]
- 8.Smirnov I., Shafer R.H. Lead is unusually effective in sequence-specific folding of DNA. J. Mol. Biol. 2000;296:1–5. doi: 10.1006/jmbi.1999.3441. [DOI] [PubMed] [Google Scholar]
- 9.Kankia B.I., Marky L.A. Folding of the thrombin aptamer into a G-quadruplex with Sr2+: stability, heat, and hydration. J. Am. Chem. Soc. 2001;123:10799–10804. doi: 10.1021/ja010008o. [DOI] [PubMed] [Google Scholar]
- 10.Green L.M., Berg J.M. Retroviral nucleocapsid protein-metal ion interactions: folding and sequence variants. Proc. Natl Acad. Sci. USA. 1990;87:6403–6407. doi: 10.1073/pnas.87.16.6403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Henderson L.E., Copeland T.D., Sowder R.C., Smythers G.W., Oroszlan S. Primary structure of the low molecular weight nucleic acid-binding proteins of murine leukemia viruses. J. Biol. Chem. 1981;256:8400–8406. [PubMed] [Google Scholar]
- 12.Berg J.M. Potential metal-binding domains in nucleic acid binding proteins. Science. 1986;232:485–487. doi: 10.1126/science.2421409. [DOI] [PubMed] [Google Scholar]
- 13.Covey S.N. Amino acid sequence homology in gag region of reverse transcribing elements and the coat protein gene of cauliflower mosaic virus. Nucleic Acids Res. 1986;14:623–633. doi: 10.1093/nar/14.2.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fisher R.J., Rein A., Fivash M., Urbaneja M.A., Casas-Finet J.R., Medaglia M., Henderson L.E. Sequence-specific binding of human immunodeficiency virus type 1 nucleocapsid protein to short oligonucleotides. J. Virol. 1998;72:1902–1909. doi: 10.1128/jvi.72.3.1902-1909.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vuilleumier C., Bombarda E., Morellet N., Gérard D., Roques B.P., Mély Y. Nucleic acid sequence discrimination by the HIV-1 nucleocapsid protein NCp7: a fluorescence study. Biochemistry. 1999;38:16816–16825. doi: 10.1021/bi991145p. [DOI] [PubMed] [Google Scholar]
- 16.Amarasinghe G.K., De Guzman R.N., Turner R.B., Chancellor K.J., Wu Z.R., Summers M.F. NMR structure of the HIV-1 nucleocapsid protein bound to stem–loop SL2 of the psi-RNA packaging signal. Implications for genome recognition. J. Mol. Biol. 2000;301:491–511. doi: 10.1006/jmbi.2000.3979. [DOI] [PubMed] [Google Scholar]
- 17.Hargittai M.R., Gorelick R.J., Rouzina I., Musier-Forsyth K. Mechanistic insights into the kinetics of HIV-1 nucleocapsid protein-facilitated tRNA annealing to the primer binding site. J. Mol. Biol. 2004;337:951–968. doi: 10.1016/j.jmb.2004.01.054. [DOI] [PubMed] [Google Scholar]
- 18.De Rocquigny H., Gabus C., Vincent A., Fournié-Zaluski M.C., Roques B., Darlix J.L. Viral RNA annealing activities of human immunodeficiency virus type 1 nucleocapsid protein require only peptide domains outside the zinc fingers. Proc. Natl Acad. Sci. USA. 1992;89:6472–6476. doi: 10.1073/pnas.89.14.6472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Darlix J.-L., Lapadat-Tapolsky M., de Rocquigny H., Roques B.P. First glimpses at structure–function relationships of the nucleocapsid protein of retroviruses. J. Mol. Biol. 1995;254:523–537. doi: 10.1006/jmbi.1995.0635. [DOI] [PubMed] [Google Scholar]
- 20.Guo J., Henderson L.E., Bess J., Kane B., Levin J.G. Human immunodeficiency virus type 1 nucleocapsid protein promotes efficient strand transfer and specific viral DNA synthesis by inhibiting TAR-dependent self-priming from minus-strand strong-stop DNA. J. Virol. 1997;71:5178–5188. doi: 10.1128/jvi.71.7.5178-5188.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Auxilien S., Keith G., Le Grice S.F.J., Darlix J.-L. Role of post-transcriptional modifications of primer tRNALys,3 in the fidelity and efficacy of plus strand DNA transfer during HIV-1 reverse transcription. J. Biol. Chem. 1999;274:4412–4420. doi: 10.1074/jbc.274.7.4412. [DOI] [PubMed] [Google Scholar]
- 22.Wu T., Guo J., Bess J., Henderson L.E., Levin J.G. Molecular requirements for human immunodeficiency virus type 1 plus-strand transfer: analysis in reconstituted and endogenous reverse transcription systems. J. Virol. 1999;73:4794–4805. doi: 10.1128/jvi.73.6.4794-4805.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Macaya R.F., Schultze P., Smith F.W., Roe J.A., Feigon J. Thrombin-binding DNA aptamer forms a unimolecular quadruplex structure in solution. Proc. Natl Acad. Sci. USA. 1993;90:3745–3749. doi: 10.1073/pnas.90.8.3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang K.Y., Krawczyk S.H., Bischofberger N., Swaminathan S., Bolton P.H. The tertiary structure of a DNA aptamer which binds to and inhibits thrombin determines activity. Biochemistry. 1992;32:11285–11292. doi: 10.1021/bi00093a004. [DOI] [PubMed] [Google Scholar]
- 25.Bock L.C., Griffin L.C., Latham J.A., Vermaas E.H., Toole J.J. Selection of single-stranded DNA molecules that bind and inhibit human thrombin. Nature. 1992;355:564–566. doi: 10.1038/355564a0. [DOI] [PubMed] [Google Scholar]
- 26.Berglund J.A., Charpentier B., Rosbash M. A high affinity binding site for the HIV-1 nucleocapsid protein. Nucleic Acids Res. 1997;25:1042–1049. doi: 10.1093/nar/25.5.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.D'Souza V., Summers M.F. Structural basis for packaging the dimeric genome of Moloney murine leukemia virus. Nature. 2004;431:586–590. doi: 10.1038/nature02944. [DOI] [PubMed] [Google Scholar]
- 28.Kankia B.I. Optical absorption assay for strand-exchange reactions in unlabeled nucleic acids. Nucleic Acids Res. 2004;32:e154. doi: 10.1093/nar/gnh152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marky L.A., Breslauer K.J. Calculating thermodynamic data for transitions of any molecularity from equilibrium melting curves. Biopolymers. 1987;26:1601–1620. doi: 10.1002/bip.360260911. [DOI] [PubMed] [Google Scholar]
- 30.Jing N., Rando R.F., Pommier Y., Hogan M.E. Ion selective folding of loop domains in a potent anti-HIV oligonucleotide. Biochemistry. 1997;36:12498–12505. doi: 10.1021/bi962798y. [DOI] [PubMed] [Google Scholar]
- 31.Jing N., Hogan M.E. Structure-activity of tetrad-forming oligonucleotides as a potent anti-HIV therapeutic drug. J. Biol. Chem. 1998;273:34992–34999. doi: 10.1074/jbc.273.52.34992. [DOI] [PubMed] [Google Scholar]
- 32.Guo J., Wu T., Anderson J., Kane B.F., Johnson D.G., Gorelick R.J., Henderson L.E., Levin J.G. Zinc finger structures in the human immunodeficiency virus type 1 nucleocapsid protein facilitate efficient minus- and plus-strand transfer. J. Virol. 2000;74:8980–8988. doi: 10.1128/jvi.74.19.8980-8988.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo J., Wu T., Kane B.F., Johnson D.G., Henderson L.E., Gorelick R.J., Levin J.G. Subtle alterations of the native zinc finger structures have dramatic effects on the nucleic acid chaperone activity of human immunodeficiency virus type 1 nucleocapsid protein. J. Virol. 2002;76:4370–4378. doi: 10.1128/JVI.76.9.4370-4378.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Williams M.C., Rouzina I., Wenner J.R., Gorelick R.J., Musier-Forsyth K., Bloomfield V.A. Mechanism for nucleic acid chaperone activity of HIV-1 nucleocapsid protein revealed by single molecule stretching. Proc. Natl Acad. Sci. USA. 2001;98:6121–6126. doi: 10.1073/pnas.101033198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Williams M.C., Gorelick R.J., Musier-Forsyth K. Specific zinc-finger architecture required for HIV-1 nucleocapsid protein's nucleic acid chaperone function. Proc. Natl Acad. Sci. USA. 2002;99:8614–8619. doi: 10.1073/pnas.132128999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heath M.J., Derebail S.S., Gorelick R.J., DeStefano J.J. Differing roles of the N- and C-terminal zinc fingers in human immunodeficiency virus nucleocapsid protein-enhanced nucleic acid annealing. J. Biol. Chem. 2003;278:30755–30763. doi: 10.1074/jbc.M303819200. [DOI] [PubMed] [Google Scholar]
- 37.Bampi C., Jacquenet S., Lener D., Decimo D., Darlix J.L. The chaperoning and assistance roles of the HIV-1 nucleocapsid protein in proviral DNA synthesis and maintenance. Curr. HIV Res. 2004;2:79–92. doi: 10.2174/1570162043485022. [DOI] [PubMed] [Google Scholar]
- 38.Stoylov S.P., Vuilleumier C., Stoylova E., De Rocquigny H., Roques B.P., Gerard D., Mély Y. Ordered aggregation of ribonucleic acids by the human immunodeficiency virus type 1 nucleocapsid protein. Biopolymers. 1997;41:301–312. doi: 10.1002/(SICI)1097-0282(199703)41:3<301::AID-BIP5>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 39.Bernacchi S., Stoylov S., Piémont E., Ficheux D., Roques B.P., Darlix J.L., Mély Y. HIV-1 nucleocapsid protein activates transient melting of least stable parts of the secondary structure of TAR and its complementary sequence. J. Mol. Biol. 2002;317:385–399. doi: 10.1006/jmbi.2002.5429. [DOI] [PubMed] [Google Scholar]
- 40.Lapadat-Tapolsky M., Pernelle C., Borie C., Darlix J.-L. Analysis of the nucleic acid annealing activities of nucleocapsid protein from HIV-1. Nucleic Acids Res. 1995;23:2434–2441. doi: 10.1093/nar/23.13.2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mély Y., de Rocquigny H., Sorinas-Jimeno M., Keith G., Roques B.P., Marquet R., Gerard D. Binding of the HIV-1 nucleocapsid protein to the primer tRNA(3Lys), in vitro, is essentially not specific. J. Biol. Chem. 1995;270:1650–1656. doi: 10.1074/jbc.270.4.1650. [DOI] [PubMed] [Google Scholar]
- 42.Levin J.G., Guo J., Rouzina I., Musier-Forsyth K. Nucleic acid chaperone activity of HIV-1 nucleocapsid protein: critical role in reverse transcription and molecular mechanism. Prog. Nucleic Acid Res. Mol. Biol. 2005.;280:217–286. doi: 10.1016/S0079-6603(05)80006-6. [DOI] [PubMed] [Google Scholar]
- 43.Mergny J.L., Phan A.T., Lacroix L. Following G-quartet formation by UV-spectroxcopy. FEBS Lett. 1998;435:74–78. doi: 10.1016/s0014-5793(98)01043-6. [DOI] [PubMed] [Google Scholar]
- 44.Urbaneja M.A., Wu M., Casas-Finet J.R., Karpel R.L. HIV-1 nucleocapsid protein as a nucleic acid chaperone: spectroscopic study of its helix-destabilizing properties, structural binding specificity, and annealing activity. J. Mol. Biol. 2002;318:749–764. doi: 10.1016/S0022-2836(02)00043-8. [DOI] [PubMed] [Google Scholar]
- 45.Kankia B.I. Hydration effects of Ni2+ binding to synthetic polynucleotides with regularly alternating purine–pyrimidine sequences. Nucleic Acids Res. 2000;28:911–916. doi: 10.1093/nar/28.4.911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marathias V.M., Bolton P.H. Structures of the potassium-saturated, 2:1, and, 1:1, forms of a quadruplex DNA. Nucleic Acids Res. 2000;28:1969–1977. doi: 10.1093/nar/28.9.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mao X., Marky L.A., Gmeiner W.H. NMR structure of the thrombin-binding DNA aptamer stabilized by Sr2+ J. Biomol. Struct. Dyn. 2004;22:25–33. doi: 10.1080/07391102.2004.10506977. [DOI] [PubMed] [Google Scholar]
- 48.Lumry R., Rajender S. Enthalpy–entropy compensation phenomena in water solutions of proteins and small molecules: a ubiquitous property of water. Biopolymers. 1970;9:1125–1227. doi: 10.1002/bip.1970.360091002. [DOI] [PubMed] [Google Scholar]
- 49.Breslauer K.J., Remeta D.P., Chou W.-Y., Ferrante R., Curry J., Zaunczkowski D., Snyder J., Marky L.A. Enthalpy–entropy compensations in drug–DNA binding studies. Proc. Natl Acad. Sci. USA. 1987;84:8922–8926. doi: 10.1073/pnas.84.24.8922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gallicchio E., Kubo M.M., Levy R.M. Entropy–enthalpy compensation in solvation and ligand binding revisited. J. Am. Chem. Soc. 1998;120:4526–4527. [Google Scholar]
- 51.Mély Y., Piemont E., Sorinas-Jimeno M., de Rocquigny H., Jullian N., Morellet N., Roques B.P., Gerard D. Structural and dynamic characterization of the aromatic amino acids of the human immunodeficiency virus type I nucleocapsid protein zinc fingers and their involvement in heterologous tRNA(Phe) binding: a steady-state and time-resolved fluorescence study. Biophys. J. 1993;65:1513–1522. doi: 10.1016/S0006-3495(93)81222-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Isalan M., Patel S.D., Balasubramanian S., Choo Y. Selection of zinc fingers that bind single-stranded telomeric DNA in the G-quadruplex conformation. Biochemistry. 2001;40:830–836. doi: 10.1021/bi001728v. [DOI] [PubMed] [Google Scholar]
- 53.Cosa G., Harbron E.J., Zeng Y., Liu H.W., O'Connor D.B., Eta-Hosokawa C., Musier-Forsyth K., Barbara P.F. Secondary structure and secondary structure dynamics of DNA hairpins complexed with HIV-1 NC protein. Biophys. J. 2004;87:2759–2767. doi: 10.1529/biophysj.104.043083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Beltz H., Azoulay J., Bernacchi S., Clamme J.P., Ficheux D., Roques B., Darlix J.L., Mély Y. Impact of the terminal bulges of HIV-1 cTAR DNA on its stability and the destabilizing activity of the nucleocapsid protein NCp7. J. Mol. Biol. 2003;328:95–108. doi: 10.1016/s0022-2836(03)00244-4. [DOI] [PubMed] [Google Scholar]
- 55.Beltz H., Piemont E., Schaub E., Ficheux D., Roques B., Darlix J.L., Mély Y. Role of the structure of the top half of HIV-1 cTAR DNA on the nucleic acid destabilizing activity of the nucleocapsid protein NCp7. J. Mol. Biol. 2004;338:711–723. doi: 10.1016/j.jmb.2004.03.019. [DOI] [PubMed] [Google Scholar]