Abstract
Helicase-like transcription factor (HLTF) is a member of the SWI/SNF (mating type switching/sucrose non-fermenting) family of ATPases/helicases and also has a RING-finger motif characteristic of ubiquitin ligase proteins. These features have led to suggestions that HLTF functions like yeast Rad5, which promotes replication through DNA lesions via a post-replication repair pathway. However, the function of HLTF in higher eukaryotes is still unknown. Herein, we found the overexpression of HLTF in radiation recurrent human uterine cervical carcinoma tissues when compared to disease free survived patients tissues. In this study, we used RNA interference techniques to investigate the potential function of HLTF in cervical cancer cell line HeLa and found that the cell proliferation was reduced by knockdown (KD) of HLTF. A host-cell reactivation assay showed that the capacity for repair to DNA damage induced by X-ray irradiation was reduced in HLTF KD cells. X-rays also increased apoptosis in HLTF KD cells. These results suggest that HLTF is involved in DNA repair and apoptosis in cancer cells, which might represent a target for gene therapies of human cancer.
Electronic supplementary material
The online version of this article (doi:10.1007/s00432-010-0925-5) contains supplementary material, which is available to authorized users.
Keywords: HLTF, Helicase-like transcription factor, Cervical cancer, Apoptosis, Radiation
Introduction
Cervical cancer is still the second most common cancer in women worldwide, despite the existence of effective screening methods (Waggoner 2003). Radiation therapy can be used to treat all stages of cervical cancer, but in early-stage disease, it is usually reserved for medically unfit patients. Including adjuvant radiotherapy following surgery, which is used in approximately 1/3 of patients with stage IB disease, radiotherapy is the most commonly used treatment modality for 460% of cases of cervical cancer (Chung et al. 2005). The most significant change in the standard radiation treatment of cervical cancer has been cisplatin-containing concurrent chemotherapy with radiation for patients with loco-regionally advanced disease (Keys et al. 1999; Morris et al. 1999; Rose et al. 1999). Despite the fact that concurrent chemoradiation demonstrated a marked improvement in pelvic disease control and survival, the resistance of tumor cells to radiation remains a major therapeutic problem.
Due to the essential nature of DNA for genetic inheritance, all organisms have evolved mechanisms to recognize and respond to DNA damage. Following radiation-induced DNA damage, cells either undergo cell cycle arrest, to facilitate DNA damage repair, or apoptosis (Shiloh 2003). The efficiency of DNA repair is one of the critical determinants of cell fate following radiotherapy (Polischouk et al. 2001). Base and nucleotide excision repair mechanisms are particularly important in the repair of DNA strand breaks caused by radiotherapy. The DNA repair capacity varies between individuals as a result of inheritance, environmental factors and physiological factors (Scully et al. 2000).
Human HLTF has recently been shown to share several functional and structural similarities with yeast Rad5. Reduction in HLTF expression enhances DNA damage sensitivity and promotes GCR upon DNA damage, and HLTF is able to partially complement for yeast Rad5 function in a sensitized genetic background (Gangavarapu et al. 2006; Unk et al. 2008). Moreover, HLTF has a yeast Rad5-like domain structure with a C3HC4 RING domain embedded into a SWI/SNF2 helicase motif. Similarly to other RING domain-containing proteins, HLTF is a ubiquitin ligase which, together with Rad6-Rad18 and Mms2-Ubc13 ubiquitin-conjugating complexes, carries out PCNA polyubiquitylation (Gangavarapu et al. 2006; Lee and Myung 2008; Unk et al. 2008). Traditionally, HLTF considered to be a tumor suppressor gene, and the concept was supported by the detection of HLTF promoter hypermethylation detected in various types of cancer tissues and cell lines (Moinova et al. 2002); however, recent observations of increased expression of HLTF in transformed cells and cancer tissues suggest that HLTF could be associated with carcinogenesis and may act like an oncogene (Capouillez et al. 2009; Debauve et al. 2008). More recently, Blastyak et al. reported the double-stranded DNA translocase activity of human HLTF in replication of damaged DNA. However, there is no evidence about the function of HLTF in radioresistance of human cancer. In this study, we examined the expression of HLTF in DF and recurrence uterine cervical carcinoma tissues using immunohistochemistry. We also analyzed the role of HLTF by knockdown (KD) using small interfering RNA (siRNA) in cell proliferation, apoptosis and DNA repair after X-ray irradiation in human cancer cells.
Our study was performed on cervical carcinomas, because this type of cancer is most commonly treated with radiotherapy alone, at least in advanced stages, which cannot be cured by surgery. A previous study provided evidence that in vitro intrinsic radioresponse of cervical carcinoma cells may be predictive of both local control of the disease and survival Rose et al. 1999. Apoptosis, intrinsic radiosensitivity and prediction of radiotherapy response in cervical carcinoma supporting a role for the activation of the DNA repair/G1 arrest pathway in the therapeutic potential of ionizing radiations (IR).
Materials and methods
Cells and culture conditions
A cervix cancer cell line, HeLa, was purchased from the Korean Cell Line Bank, Korea. HeLa cells were grown in DMEM supplemented with 10% fetal bovine serum (FBS), 100 IU/ml penicillin and 100 μg/ml streptomycin at 37°C in 5% CO2. Trypsin–EDTA was used for cell harvesting and passage. All standard culture reagents were obtained from Invitrogen (Carlsbad, CA, USA).
Patients sample
Formalin-fixed paraffin-embedded specimens of human cervical carcinoma tissues of patients received curative radiation therapy from May 2003 to July 2005 at Chungbuk National University Hospital were used. All experiments were performed under the approval of the Institutional Review Board, Chungbuk National University Hospital.
Immunohistochemistry
Immunohistochemical staining was performed on paraffin-embedded tissue sections using Immunohistochemistry Accessory Kit (Bethyl Laboratories, Montgomery, TX, USA) as per manufacturer’s instructions after antigen retrieval. In brief, de-paraffinized sections were antigen retrieved using Citrate buffer PH 6.0 in a microwave oven for 6 min. Endogenous peroxidase was blocked using 0.6% hydrogen peroxide in methanol for 20 min. Tissue sections were treated with protein block solution for 30 min. Primary antibody (dilution for HLTF 1:100 (Aviva Systems San Diego, CA, USA)) was added to slides and incubated at 4°C for overnight. After washing in PBST, the sections were incubated with appropriate secondary antibodies for 60 min. The color reaction was developed using Liquid DAB substrate Pack (Bethyl Laboratory, Montgomery, TX, USA). Sections were counterstained with hematoxylin.
siRNA treatment
The siRNA oligos of HLTF were synthesized in Ambion (Austin, TX, USA) as (s13138)5′GGAAUUUUAGCUGAUGAUATT3′. For transient transfection, 5 × 104 HeLa cells were seeded in six-well plates and cultured to 70% confluency. After removing the culture media, 0.4 nM siRNA of HLTF dissolved in 2 μl of NeoFX transfection reagent (Ambion) was added to each well according to the manufacturer’s recommendation. After 48 h of siRNA transfection, the culture medium was changed to one containing 10% FBS, and the cells were irradiated with 4 Gy at room temperature. The effectiveness of RNA inhibition was measured by quantitative real-time PCR and Western blotting.
Real-time RT–PCR analysis
Total RNA was isolated from HeLa cells using Trizol (Invitrogen) and further purified with the RNeasy kit (Qiagen, Germantown, MD, USA) according to the manufacturer’s protocol. RNA was treated with iScript (Bio-Rad, Hercules, CA, USA) for cDNA synthesis. Briefly, 1 μg of RNA was reverse transcribed with reverse transcriptase at 42°C for 30 min. Real-time PCR was performed (iCycler iQ, Bio-Rad) with a PCR master mix comprising 10 μl of master mix (iQ SYBR Green Supermix, Bio-Rad), 1 μl of cDNA template, 1 μl of each HLTF primer pair (20 nM; forward, TGAAGGACATGCCATACGAA; reverse, TCCTCCTTCATCTCCCATTG) and 7 μl of RNase-free water. The reaction involved denaturation at 95°C for 2 min and 40 cycles of amplification at 95°C for 15 s, 60°C for 15 s and 72°C for 15 s. Real-time quantification of HLTF gene was normalized to the threshold number of cycles (C
T) of glyceraldehyde-3-phosphate dehydrogenase (GAPDH), where C
T equals the PCR cycle number at which the amount of amplified sample product reached 100 relative fluorescence units. The difference in HLTF mRNA expression relative to KD expression was calculated by comparing C
T
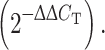 A melting curve for all products was obtained immediately after amplification by increasing the temperature in 0.4°C increments from 65°C for 85 cycles of 10 s each. Each experiment was performed independently three times.
A melting curve for all products was obtained immediately after amplification by increasing the temperature in 0.4°C increments from 65°C for 85 cycles of 10 s each. Each experiment was performed independently three times.
Cell proliferation assay
Cell proliferation was assessed using WST-1 (Roche, Indianapolis, IN, USA), which is an assay based on cleavage of the tetrazolium salt to formazan by cellular mitochondrial dehydrogenases. HeLa cells were plated in 96-well microplates at a density of 1 × 105 cells per well. After culturing to 70% confluency, the cells were transfected with siRNA for HLTF or with control siRNA using NeoFX reagent (Ambion) for 48 h, and control cells were left untreated. The WST-1 reagent was applied for 1 h at 0, 24 and 48 h, and the formazan dye formed was quantified using a microplate reader (SpectraMax M5, Molecular Devices, Sunnyvale, CA, USA) at 450 nm. The cell proliferation was assessed in triplicate. The data are presented as percentages relative to negative-control proliferation, with P values of <0.05 considered significant. Each experiment was performed three times.
DNA fragmentation assay
A DNA fragmentation assay was performed using the Quick apoptotic DNA ladder detection kit (BioVision, Mountain View, CA, USA) according to the manufacturer’s recommendations. Briefly, 5 × 104 HeLa cells were cultured in six-well culture plates to 70% confluency. After 48 h of siRNA transfection, the culture medium was changed to one containing 10% FBS, and the cells were irradiated at 4 Gy at room temperature. DNA was isolated from the cells and electrophoresed in 1.5% agarose gel at 100 V for 20 min, and an image of the gel was obtained (SL-20 Image Visualizer, Seolin Scientific, Seoul, Korea).
Western blotting
The cells were washed with PBS and dissolved in 50 μl of ice-cold lysis buffer (Cell Signaling Technology, Danvers, MA, USA) consisting of 20 mM Tris HCl (pH 7.5), 150 mM NaCl, 1 mM Na2EDTA, 2.5 mM sodium pyrophosphate, 1% Triton, 1 mM EGTA and protease inhibitors. After sonication on ice, protein lysates were obtained by centrifugation at 19,000g at 4°C for 10 min. The protein concentration was determined with Bradford assay reagent (Bio-Rad). Fifteen microliters of protein was mixed with 3 μl of 6× sample buffer (360 mM Tris HCl [pH 6.7], 60% glycerol, and 12% SDS). The protein samples were then heated at 100°C for 5 min and subjected to 10% SDS–PAGE (sodium dodecyl sulfate–polyacrylamide gel electrophoresis) and transferred to a PVDF membrane (Bio-Rad). The membrane was blocked with 0.5% skim milk and probed with monoclonal antibodies against HLTF, survivin, endonuclease G (EndoG), X-linked inhibitor of apoptosis (XIAP) and poly ADP ribose polymerase (PARP), and then polyclonal antibodies against apoptosis-inducing factor (AIF). The antibody for HLTF was a product of Aviva Systems (San Diego, CA, USA); those for survivin, PARP, cytochrome c and AIF were products of Santa Cruz Biotechnology (Santa Cruz, CA, USA); and those for EndoG and XIAP were products of Cell Signaling Technology. The filter was then incubated with a horseradish-peroxidase-conjugated secondary antibody and developed with WEST-ZOL Plus (iNtRON Biotechnology, Gyeonggi-do, Korea). To determine whether the amounts of proteins in each lane were comparable, the filter was stripped and probed again with a goat polyclonal antibody against β-actin (Santa Cruz Biotechnology).
Host-cell reactivation (HCR) assay
The plasmid expression vector pGL-3 (Promega, Madison, WI, USA) containing the luciferase (LUC) reporter gene was used for the LUC assay. One milliliter of pGL-3 (500 μg/ml) was either not irradiated or irradiated with X-rays (4 Gy). After incubation at 37°C for 3 h, the plasmid DNA was precipitated with 99% ethanol, washed with 75% ethanol, dissolved in 0.1 M Tris–EDTA buffer at a final concentration of 0.3 mg/ml and stored at –20°C until being used for transfection. HeLa cells were seeded in six-well plates at 2 × 105 cells/well. Perfectin (Bio-Rad) was used to transfect cells with undamaged or X-ray-damaged pGL-3. After transfection for 48 h, the cells were washed with PBS and harvested by scraping and centrifugation at 14,000g at 4°C. The cell pellets were suspended in 50 μl of reporter lysis buffer (Promega), frozen, thawed in an ethanol–dry-ice water bath at 37°C and then centrifuged at 1,400g for 15 s. For each LUC assay, 20 μl of cell extract supernatant was mixed with 20 μl of LUC assay substrate (Promega) in 96-well plates at room temperature. LUC activity was measured with a luminometer (Lumat LB 9507, Berthold Technologies, Bad Wildbad, Germany) and quantified in arbitrary LUC light-intensity units that were recorded for cells with undamaged plasmids (control reading) and X-ray-damaged plasmids (repair reading). The DNA repair capacity was calculated as the percentage LUC activity of the damaged plasmid relative to that of the undamaged plasmid.
Statistical analysis
All experiments were performed in triplicate and repeated at least three times. All data are expressed as mean and SD values. Statistical differences were assessed by Student’s t test (two-way ANOVA), with a P value of <0.05 considered significant.
Results
HLTF expression in human DF and recurrent cervical carcinoma
We analyzed the expression of HLTF in 27 human cervical carcinoma tissues which include 16 diseases free (DF) survival and 11 recurrence samples after curative radiotherapy using immunohistochemistry. The expression and localization of HLTF varies greatly between DF and recurrent samples. The expression of HLTF in DF samples mostly cytoplasmic and with the moderate strength associated with the tumor cell groups (Fig 1a–d and Table 1). HLTF Immunohistochemical staining of recurrent tissue sample revealed the interesting pattern with the over expression associated with the nucleocytoplasmic localization. The staining intensity was higher in stromal invading malignant cell groups. The expression was observed around the perinuclear region of some malignant cells with large hollow nucleus (Fig. 1e–h).Tumor adjacent cervical intra neoplastic lesions show notable staining pattern of HLTF. The expression of HLTF with moderate strength observed at CIN I and CIN II, the early cervical neoplastic grades, whereas the higher grade CIN III shows no expression of HLTF expression (Supplementary Fig. 1).
Fig. 1.
Representative pictures of HLTF immunohistochemistry. a–d Moderate staining pattern of HLTF in DF human cervical carcinoma. e–h Strong overexpression and nucleocytoplasmic localization of HLTF in recurrent human cervical carcinoma (bar = 200 μm)
Table 1.
Immunohistochemical staining intensity of HLTF expression in human cervical carcinoma recurrent and DF patients’ tissues
| Sample | Number | HLTF expression | |||
|---|---|---|---|---|---|
| − | + | ++ | +++ | ||
| DF | 16 | − | 2 | 10 | 4 |
| Recurrence | 11 | − | − | 2 | 9 |
HLTF siRNA transfection decreases HLTF expression
We assessed the role of HLTF by transiently knocking down HLTF expression in HeLa cells using RNAi technology (Fig. 2) and examining the effects on cell growth (Fig. 3). HLTF siRNA transfection decreased HLTF protein expression compared with controls (Fig. 1b). To determine whether the decreased production of HLTF was due to a decrease in gene transcription, HLTF transcripts were assessed using real-time RT–PCR. As shown in Fig. 1a, the transcript levels of HLTF were significantly lower in siRNA-transfected cells than in controls (P < 0.05).
Fig. 2.
Inhibition of HLTF expression using RNA interference. a RT–PCR was employed to detect the transcription level of HLTF after transfection with HLTF siRNA. Transcript levels of HLTF were significantly lower in HLTF siRNA-transfected cells than in cells transfected with NeoFX reagent (P < 0.05). GAPDH was used as an internal control to normalize mRNA loading. The relative mRNA level of HLTF is depicted as the relative change in HLTF to GAPDH at the same time point. b Western blotting was used to analyze the HLTF protein level in HLTF siRNA-transfected HeLa cells. HLTF protein expression was lower in HLTF siRNA transfection than in controls (P < 0.05). Beta-actin was used as an internal control to normalize protein loading. The result shown is representative of triplicate experiments
Fig. 3.
Inhibition of cell proliferation by siHLTF. The cell proliferation assay was performed using WST-1. HeLa cells were transiently transfected with siRNA for 48 h, and either not irradiated (a) or irradiated with 4 Gy (b). In both cases, the optical density was measured at 0, 24 and 48 h. The lines represent no-treatment controls (NT, diamonds), controls treated with transfection reagent only (Rg, squares) and siRNA treatment (triangles)
Effect of HLTF siRNA on the proliferation of HeLa cells
The effect of HLTF KD on HeLa cell proliferation was assessed using the WST-1 assay. Proliferation was reduced in cells with HLTF functional loss induced by siRNA transfection (Fig. 3a), and it was increased by irradiation (Fig. 3b) compared with controls. The number of proliferating cells was significantly lower after siRNA transfection and irradiation than in the two control groups (P < 0.05).
Down-regulation of HLTF induces apoptosis
The effect of KD of HLTF was analyzed by treating HeLa cells with 0.4 nM siRNA in six-well plates for 48 h and then irradiating them with X-rays. The siRNA-treated HeLa cells showed morphological characteristics typical of apoptosis, such as cell shrinkage (data not shown). DNA fragmentation analysis was used to further validate the induction of apoptosis by siRNA. The typical laddering pattern—which is believed to occur at the later stage of apoptosis–was more profound in siRNA-treated cells (Fig. 4a).
Fig. 4.
Characterization of the apoptotic process. a DNA fragmentation assay for apoptosis. Cells were treated or not treated with siRNA for 48 h and irradiated at 4 Gy. The fragmented DNA was extracted 0, 24 and 48 h post-irradiation and resolved in a 1.5% agarose gel. b Expression of apoptosis-related proteins. HeLa cells were treated or not treated with siHLTF for 48 h, irradiated at 4 Gy and subjected to Western blotting at 0, 24 and 48 h post-irradiation. Beta-actin was used as an internal control to ensure equal loading
Western blotting was used to examine the expression of the proapoptotic proteins AIF and EndoG (caspase-independent) (Daugas et al. 2000; Suzuki et al. 1999), PARP (caspase-dependent) and the antiapoptotic genes XIAP and survivin. The expression of proapoptotic EndoG and PARP was increased after irradiation, the expressions of the antiapoptotic genes XIAP and survivin were decreased, and AIF was not affected. This suggests that the inhibition of cell growth and increased apoptosis induced by siRNA were correlated with the changes in the expressions of EndoG, XIAP, PARP and survivin proteins (Fig. 4b).
Down-regulation of HLTF expression leads to defective repair of exogenously damaged plasmids
The host-cell reactivation (HCR) assay is used to investigate the DNA-repair capacity of cells by quantifying the function of repaired exogenous DNA that had been damaged before being introduced into host cells (Mallya and Sikpi 1998; Yang et al. 1998). We therefore used the HCR assay to detect the functional recovery of pGL3 DNA damaged by X-rays and introduced into HeLa cells. PGL3 was undamaged or damaged by X-rays (4 Gy) and transfected into host HeLa cells, and the relative LUC units were determined 24, 48 and 72 h after transfection.
In the cells transfected with non-irradiated (undamaged) plasmids, the LUC activities were reduced to 24.9, 21.9 and 9.37% of control (non-siRNA-treated) cells by HLTF KD at 24, 48 and 72 h, respectively (Fig. 5a). In the cells transfected with irradiated (damaged) plasmids, the LUC activities were reduced to 9.97, 15.1 and 3.37% of control cells by HLTF KD (Fig. 5b). Fig. 5c shows the LUC activities in siRNA-treated host cells as percentages relative to those in control cells. The percentages at 48 and 72 h were lower in irradiated plasmids than in non-irradiated plasmids, which suggest a reduction in the repair capacity of X-ray damage by HLTF KD.
Fig. 5.
Host-cell reactivation assay. The HCR assay was performed to assess the DNA repair capacity of HeLa cells. HeLa cells were transiently transfected or not transfected with siRNA for 48 h and then transfected with either undamaged (a) or damaged (irradiated) (b) pGL3. The LUC activity of the HeLa cells was compared at 24, 48 and 72-h post-transfection between controls and siRNA treatment. The percentage LUC activity of undamaged and damaged (irradiated) pGL3 in siRNA-treated HeLa cells is also shown (c). Data are expressed mean and SD values for three independent experiments
Discussion
HLTF is a member of the SWI/SNF family that is reportedly hypermethylated in various cancer cells (Hamai et al. 2003; Jo et al. 2007; Kim et al. 2006; Leung et al. 2003; Wallner et al. 2006). There is accumulating evidence that HLTF plays an important role in DNA double-strand break repair (Wang et al. 2005). Down-regulation of HLTF can not only induce growth inhibition and apoptosis in cancer cells, but can also increase the sensitivity of cancer cells to chemotherapy and ionizing radiation. At present, there are no studies evaluating DNA repair protein HLTF expression in radioresistant cervical cancer. We report that the over expression of HLTF correlates with radiotherapy outcome in cervical cancer. Our immunohistochemical data show high expression of HLTF associated with radio therapy recurrence. This antibody detects the 95 kDa variant of HLTF in HeLa cells and in tissues (SMARCA-3 Antibody from Aviva Biosystems, San Diego, CA, USA) indicates that the 95 kDa variant HLTF is highly expressed in the cervical cancer DFS and recurrent patients tissues. Our results were in consistent with the recent observation using hypopharyngeal carcinoma, and they reported the HLTF is a strong predictor of recurrence (Capouillez et al. 2009). Our in vitro analysis also provide evidence that down-regulation of HLTF induces apoptosis in HeLa cells. The RNA KD and WST-1 assay clearly showed that down-regulation of HLTF can induce apoptosis in radiation-damaged HeLa cells. Apoptotic processes are usually controlled by proapoptotic and antiapoptotic genes. The increased expression of the proapoptotic gene EndoG and the decreased expressions of the antiapoptotic genes XIAP and survivin might explain the apoptotic process induced by HLTF KD. It is commonly believed that the IAP (inhibitor of apoptosis) family protects against apoptosis via a caspase-dependent pathway. In contrast, the expression of the caspase-independent gene EndoG was increased and that of survivin was decreased. This suggests that a caspase-independent pathway is also involved in the HLTF-mediated apoptotic process.
The Rad5–Mms2–Ubc13-dependent PRR pathway promotes error-free lesion bypass in yeast (Gangavarapu et al. 2006; Kannouche et al. 2004). For UV-induced lesions, in addition to this pathway, Polη and Polζ also contribute to error-free bypass via the promotion of error-free TLS using cyclobutane pyrimidine dimers. This increases the frequency of UV-induced mutations when both the Rad5-dependent PRR and Polη-dependent TLS pathways are inactivated (Gangavarapu et al. 2006; Kannouche et al. 2004). The SWI/SNF ATPase activity of Rad5 is also critical for PRR, because inactivation of this function causes the same high degree of PRR defect as conferred upon inactivating its function as a ubiquitin ligase (Lorick et al. 1999). Recent studies show that RING-finger ATPase contributes to DNA double-strand break repair in yeast Rad5 (Chai et al. 2005; Chen et al. 2005). These results suggest that both the ligase and ATPase domains also play an important role in responses to DNA damage.
In human, SHPRH and HLTF are ubiquitin ligases for mediating Mms2–Ubc13-dependent PCNA polyubiquitylation, and the promotion of error-free replication in DNA lesions by this pathway raises the strong possibility that SHPRH and HLTF represent important obstacles to mutagenesis and carcinogenesis in human cells (Motegi et al. 2006; Unk et al. 2006, 2008). In this line, our study has shown that HLTF KD enhances radiosensitivity in a human cervical cancer cell line. In addition, we provide evidence that HLTF is associated with the repair of radiation-induced DNA damage in human cells. Because the host cells (HeLa) are not irradiated (i.e., DNA is not damaged) in the HCR assay, the observed DNA repair capacity reflects that of the host cells. Further studies are necessary to define the exact role of HLTF in radiation-induced DNA repair. Taken together with our tissue staining data and in vitro experimental results clearly suggest that the HLTF is an important molecule determines the outcome of the radiotherapy.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
This research was supported by the program of Basic Atomic Energy Research Institute (BAERI), which is a part of the Nuclear R & D Programs funded by the Ministry of Science and Technology (MOST) of Korea in 2008.
Conflict of interest
We declare that we have no conflict of interest.
Footnotes
S. Cho and S. Cinghu have contributed equally to the work.
References
- Capouillez A, Debauve G, Decaestecker C, Filleul O, Chevalier D, Mortuaire G, Coppee F, Leroy X, Belayew A, Saussez S (2009) The helicase-like transcription factor is a strong predictor of recurrence in hypopharyngeal but not in laryngeal squamous cell carcinomas. Histopathology 55:77–90 [DOI] [PubMed] [Google Scholar]
- Chai B, Huang J, Cairns BR, Laurent BC (2005) Distinct roles for the RSC and Swi/Snf ATP-dependent chromatin remodelers in DNA double-strand break repair. Genes Dev 19:1656–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Davies AA, Sagan D, Ulrich HD (2005) The RING finger ATPase Rad5p of Saccharomyces cerevisiae contributes to DNA double-strand break repair in a ubiquitin-independent manner. Nucleic Acids Res 33:5878–5886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung YM, Kim BG, Park CS, Huh SJ, Kim J, Park JK, Cho SM, Kim BS, Kim JS, Yoo YD, Bae DS (2005) Increased expression of ICAM-3 is associated with radiation resistance in cervical cancer. Int J Cancer 117:194–201 [DOI] [PubMed] [Google Scholar]
- Daugas E, Susin SA, Zamzami N, Ferri KF, Irinopoulou T, Larochette N, Prevost MC, Leber B, Andrews D, Penninger J, Kroemer G (2000) Mitochondrio-nuclear translocation of AIF in apoptosis and necrosis. FASEB J 14:729–739 [PubMed] [Google Scholar]
- Debauve G, Capouillez A, Belayew A, Saussez S (2008) The helicase-like transcription factor and its implication in cancer progression. Cell Mol Life Sci 65:591–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangavarapu V, Haracska L, Unk I, Johnson RE, Prakash S, Prakash L (2006) Mms2-Ubc13-dependent and -independent roles of Rad5 ubiquitin ligase in postreplication repair and translesion DNA synthesis in Saccharomyces cerevisiae. Mol Cell Biol 26:7783–7790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamai Y, Oue N, Mitani Y, Nakayama H, Ito R, Matsusaki K, Yoshida K, Toge T, Yasui W (2003) DNA hypermethylation and histone hypoacetylation of the HLTF gene are associated with reduced expression in gastric carcinoma. Cancer Sci 94:692–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo H, Kang S, Kim JW, Kang GH, Park NH, Song YS, Park SY, Kang SB, Lee HP (2007) Hypermethylation of the COX-2 gene is a potential prognostic marker for cervical cancer. J Obstet Gynaecol Res 33:236–241 [DOI] [PubMed] [Google Scholar]
- Kannouche PL, Wing J, Lehmann AR (2004) Interaction of human DNA polymerase eta with monoubiquitinated PCNA: a possible mechanism for the polymerase switch in response to DNA damage. Mol Cell 14:491–500 [DOI] [PubMed] [Google Scholar]
- Keys HM, Bundy BN, Stehman FB, Muderspach LI, Chafe WE, Suggs C L 3rd, Walker JL, Gersell D (1999) Cisplatin, radiation, and adjuvant hysterectomy compared with radiation and adjuvant hysterectomy for bulky stage IB cervical carcinoma. N Engl J Med 340:1154–1161 [DOI] [PubMed] [Google Scholar]
- Kim JJ, Chung SW, Kim JH, Kim JW, Oh JS, Kim S, Song SY, Park J, Kim DH (2006) Promoter methylation of helicase-like transcription factor is associated with the early stages of gastric cancer with family history. Ann Oncol 17:657–662 [DOI] [PubMed] [Google Scholar]
- Lee KY, Myung K (2008) PCNA modifications for regulation of post-replication repair pathways. Mol Cells 26:5–11 [PMC free article] [PubMed] [Google Scholar]
- Leung WK, Yu J, Bai AH, Chan MW, Chan KK, To KF, Chan FK, Ng EK, Chung SC, Sung JJ (2003) Inactivation of helicase-like transcription factor by promoter hypermethylation in human gastric cancer. Mol Carcinog 37:91–97 [DOI] [PubMed] [Google Scholar]
- Lorick KL, Jensen JP, Fang S, Ong AM, Hatakeyama S, Weissman AM (1999) RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc Natl Acad Sci USA 96:11364–11369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallya SM, Sikpi MO (1998) Evidence of the involvement of p53 in gamma-radiation-induced DNA repair in human lymphoblasts. Int J Radiat Biol 74:231–238 [DOI] [PubMed] [Google Scholar]
- Moinova HR, Chen WD, Shen L, Smiraglia D, Olechnowicz J, Ravi L, Kasturi L, Myeroff L, Plass C, Parsons R et al (2002) HLTF gene silencing in human colon cancer. Proc Natl Acad Sci USA 99:4562–4567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris M, Eifel PJ, Lu J, Grigsby PW, Levenback C, Stevens RE, Rotman M, Gershenson DM, Mutch DG (1999) Pelvic radiation with concurrent chemotherapy compared with pelvic and para-aortic radiation for high-risk cervical cancer. N Engl J Med 340:1137–1143 [DOI] [PubMed] [Google Scholar]
- Motegi A, Sood R, Moinova H, Markowitz SD, Liu PP, Myung K (2006) Human SHPRH suppresses genomic instability through proliferating cell nuclear antigen polyubiquitination. J Cell Biol 175:703–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polischouk AG, Grenman R, Granath F, Lewensohn R (2001) Radiosensitivity of human squamous carcinoma cell lines is associated with amount of spontaneous DNA strand breaks. Int J Cancer 96(Suppl):43–53 [DOI] [PubMed] [Google Scholar]
- Rose PG, Bundy BN, Watkins EB, Thigpen JT, Deppe G, Maiman MA, Clarke-Pearson DL, Insalaco S (1999) Concurrent cisplatin-based radiotherapy and chemotherapy for locally advanced cervical cancer. N Engl J Med 340:1144–1153 [DOI] [PubMed] [Google Scholar]
- Scully C, Field JK, Tanzawa H (2000) Genetic aberrations in oral or head and neck squamous cell carcinoma (SCCHN): 1. Carcinogen metabolism, DNA repair and cell cycle control. Oral Oncol 36:256–263 [DOI] [PubMed] [Google Scholar]
- Shiloh Y (2003) ATM and related protein kinases: safeguarding genome integrity. Nat Rev Cancer 3:155–168 [DOI] [PubMed] [Google Scholar]
- Suzuki A, Tsutomi Y, Miura M, Akahane K (1999) Caspase 3 inactivation to suppress Fas-mediated apoptosis: identification of binding domain with p21 and ILP and inactivation machinery by p21. Oncogene 18:1239–1244 [DOI] [PubMed] [Google Scholar]
- Unk I, Hajdu I, Fatyol K, Szakal B, Blastyak A, Bermudez V, Hurwitz J, Prakash L, Prakash S, Haracska L (2006) Human SHPRH is a ubiquitin ligase for Mms2-Ubc13-dependent polyubiquitylation of proliferating cell nuclear antigen. Proc Natl Acad Sci USA 103:18107–18112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unk I, Hajdu I, Fatyol K, Hurwitz J, Yoon JH, Prakash L, Prakash S, Haracska L (2008) Human HLTF functions as a ubiquitin ligase for proliferating cell nuclear antigen polyubiquitination. Proc Natl Acad Sci USA 105:3768–3773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waggoner SE (2003) Cervical cancer. Lancet 361:2217–2225 [DOI] [PubMed] [Google Scholar]
- Wallner M, Herbst A, Behrens A, Crispin A, Stieber P, Goke B, Lamerz R, Kolligs FT (2006) Methylation of serum DNA is an independent prognostic marker in colorectal cancer. Clin Cancer Res 12:7347–7352 [DOI] [PubMed] [Google Scholar]
- Wang L, Baiocchi RA, Pal S, Mosialos G, Caligiuri M, Sif S (2005) The BRG1- and hBRM-associated factor BAF57 induces apoptosis by stimulating expression of the cylindromatosis tumor suppressor gene. Mol Cell Biol 25:7953–7965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang WL, Cvijic ME, Ishii K, Chin KV (1998) The requirement of yeast Ssl2 (Rad25) for the repair of cisplatin-damaged DNA. Biochem Biophys Res Commun 250:593–597 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.







