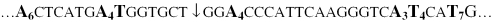Abstract
The electrophoretic mobility of a curved DNA restriction fragment taken from the VP1 gene in the SV40 minichromosome has been measured in polyacrylamide gels and free solution, using capillary electrophoresis. The 199 bp restriction fragment has an apparent bend angle of 46 ± 2° located at SV40 sequence position 1922 ± 2 bp [Lu Y.J., Weers B.D. and Stellwagen N. C. (2005) Biophys. J., 88, 1191–1206]. The ‘curvature module’ surrounding the apparent bend center contains five unevenly spaced A- and T-tracts, which are responsible for the observed curvature. The parent 199 bp fragment and sequence mutants containing at least one A-tract in the curvature module migrate anomalously slowly in free solution, as well as in polyacrylamide gels. Hence, the anomalously slow mobilities observed for curved DNA molecules in polyacrylamide gels are due in part to their anomalously slow mobilities in free solution. Analysis of the gel and free solution mobility decrements indicates that each A- or T-tract contributes independently, but not equally, to the curvature of the 199 bp fragment and its A-tract mutants. The relative contribution of each A- or T-tract to the observed curvature depends on its spacing with respect to the first A-tract in the curvature module.
INTRODUCTION
It has been known for many years that curved DNA molecules migrate anomalously slowly in polyacrylamide gels, compared with normal DNA molecules containing the same number of base pairs (1,2). The anomalously slow mobilities have been attributed to the curvature of the helix backbone because curved DNA molecules have larger cross-sectional areas than normal DNA molecules of the same size and therefore require larger pores to migrate through the gel matrix, behaving electrophoretically as though they were larger than their true sizes (1,3,4). Several empirical equations have been developed to relate the extent of DNA curvature to the magnitude of the mobility anomalies observed under specific gel-running conditions (5–9).
Attributing the anomalously slow mobilities observed for curved DNA molecules in polyacrylamide gels to the restrictive pore size of the gel matrix is intuitively reasonable, and is consistent with the fact that the mobility anomalies increase with increasing polyacrylamide gel concentration (1–3), i.e. decreasing gel pore size (10). However, if this explanation is correct, it should be possible to eliminate the gel mobility anomalies by increasing the gel pore size sufficiently. There are two methods of increasing the pore size of a polyacrylamide gel—decreasing the acrylamide concentration at constant crosslinker concentration, the usual method of increasing the gel pore size, or decreasing the percentage of the crosslinker in gels with a constant acrylamide concentration (10–12).
Previous studies (12,13), using DNA molecules containing multiple copies of curved and normal 147 bp restriction fragments taken from plasmid pBR322, showed that a curved fragment (called 12A) and its multimers migrated more slowly than the corresponding normal fragment (called 12B) and its multimers, even when the acrylamide concentration was reduced to the limit at which a gel can be formed. If the gel pore size was increased by decreasing the crosslinker concentration at constant acrylamide concentration, anomalously slow mobilities were observed for fragment 12A and its multimers if the effective gel pore radius was smaller than the radius of gyration of the multimer. When the effective gel pore radius was larger than the radius of gyration, the mobility anomalies leveled off and became constant with increasing gel pore size (13).
One explanation of these results is that curved DNA molecules are transiently trapped by the polyacrylamide gel fibers during electrophoresis, causing curved DNAs to be retarded more than normal DNA molecules containing the same number of base pairs. However, another possibility is that curved and normal DNA molecules of the same size have different mobilities in free solution and therefore exhibit different mobilities in large-pore polyacrylamide gels.
Here, we have addressed this question directly by measuring the gel and free solution mobilities of curved and normal DNA molecules that have the same molecular weight but are characterized by different degrees of curvature. The target fragment is a 199 bp restriction fragment taken from the VP1 gene of the SV40 minichromosome. Recent transient electric birefringence studies have shown that this restriction fragment contains an apparent bend of 46 ± 2° located at SV40 sequence position 1922 ± 2 bp (14). This sequence position is defined as the apparent bend center in the following discussion. The apparent bend angle is nearly independent of ionic strength, indicating that the 199 bp fragment is stably curved, rather than anisotropically flexible, as discussed in detail elsewhere (14). The apparent bend angle is also independent of temperature from 4 to 37°C (14), suggesting that the curvature of the VP1 gene at this sequence location is apt to be biologically important.
The 60 bp sequence surrounding the apparent bend center contains four A- and T-tracts of various lengths plus a mixed A3T4 sequence motif, as shown in Scheme 1. For brevity, this 60 bp sequence element is called the ‘curvature module.’ Replacing the curvature module with a same-sized fragment of normal DNA taken from an uncurved region of plasmid pUC19 (15) causes the modified SV40 construct to adopt a normal conformation in free solution (14). Hence, the 60 bp curvature module is necessary and sufficient to cause stable curvature of the DNA helix backbone.
Scheme 1.
SV40 sequence from 1905 to 1965 bp, with the location of the apparent bend center denoted by a vertical arrow. The individual A- and T-tracts in the curvature module are designated in bold by the compact notation, A6, A4T, A4, A3T4 and T7, that will be used in the subsequent text. Note that the second A-tract is designated A4T, both to distinguish it from the following A4-tract and in recognition of the fact that thymine residues following an A-tract exhibit many of the properties of the preceding adenine residues (16–19).
Scheme 1 shows SV40 sequence from 1905 to 1965 bp, with the location of the apparent bend center denoted by a vertical arrow. The individual A- and T-tracts in the curvature module are designated in bold by the compact notation, A6, A4T, A4, A3T4 and T7 that will be used in the subsequent text. Note that the second A-tract is designated A4T, both to distinguish it from the following A4-tract and in recognition of the fact that thymine residues following an A-tract exhibit many of the properties of the preceding adenine residues (16–19).
Since the A- and T-tracts in the SV40 curvature module are of different lengths, have different sequences and are not regularly spaced with respect to the helix repeat, it is not clear whether they contribute equally to the curvature of the 199 bp fragment. To investigate this question, each of the A- and T-tracts in the curvature module was modified by site-directed mutagenesis. The electrophoretic mobilities of the parent 199 bp fragment and its A-tract mutants were then measured in polyacrylamide gels and free solution, using capillary electrophoresis.
The results indicate that curved DNA molecules that migrate anomalously slowly in polyacrylamide gels also migrate anomalously slowly in free solution. Hence, the anomalously slow mobilities observed for curved DNA molecules in polyacrylamide gels are due, at least in part, to their intrinsically lower free solution mobilities. Analysis of the mobility decrements observed in polyacrylamide gels and free solution indicates that each of the A-tracts in the curvature module contributes additively, but not equally, to the curvature of the parent 199 bp fragment. The relative importance of each A- or T-tract appears to be determined by its phasing with respect to the first A-tract in the curvature module.
MATERIALS AND METHODS
DNA samples
The parent 199 bp restriction fragment used for the present studies was prepared by adding BamHI linkers to an SV40 fragment with coordinates 1826–2017 and subcloning the construct into plasmid pUC19, using methods described previously (14,15). After large-scale plasmid preparation, the desired fragment was isolated by BamHI digestion and agarose gel electrophoresis. The agarose was dissolved with a chaotropic salt (Qiagen QIAquick Gel Extraction Kit) and the DNA was concentrated and desalted by adsorption on small DEAE columns. After elution from the column and ethanol precipitation, the DNA was redissolved in T0.1E (10 mM Tris–HCl buffer, pH 8.0 and 0.1 mM EDTA) and stored at −20°C until needed. The integrity of the 199 bp fragment was verified by sequencing.
The A- and T-tracts in the parent 199 bp fragment were modified by site-directed mutagenesis, using standard methods (20). The forward primers contained 35–45 nt with 1–3 single residue mismatches near their centers; the reverse primers were the reverse complements of the forward primers. The DNAs were amplified with Pfu polymerase for 24 cycles using temperatures of 95°C for 45 s, 56–62°C (5°C below the melting temperature) for 45 s and 72°C for 1 min, with a final extension step of 72°C for 1 min. The PCR-amplified mutants were purified by digesting the template with DpnI, which digests only methylated cytosine residues. The PCR products were then transformed into One Shot MAX Efficiency DH5α-T1 cells and were spread on Luria–Bertani-ampicillin plates. Plasmid DNAs were isolated using the Qiaprep Spin Miniprep Kit (Qiagen) and were analyzed by restriction enzyme digestion and agarose gel electrophoresis. Desirable mutants were sequenced to verify their identities, subcloned into pUC19 and purified as described above. Subcloning into pUC19 was necessary to eliminate small molecular weight heterogeneities in the PCR-amplified fragments (data not shown).
The normal DNA fragments used in the gel electrophoresis experiments were the fragments of a 50 bp ladder (Invitrogen, Carlsbad, CA). The normal DNA fragments used as controls in the capillary electrophoresis experiments were isolated from plasmids pUC19, pBR322 or Litmus 28, contain 79–789 bp, have no or only a few isolated An- or Tn-tracts (n ≥ 4), have normal transient electric birefringence relaxation times and exhibit normal electrophoretic mobilities in polyacrylamide gels, indicating that they have normal conformations in free solution (15). The preparation and characterization of these fragments is described elsewhere (15).
PAGE
The parent 199 bp fragment and its A-tract derivatives were characterized by electrophoresis in polyacrylamide gels containing 6.9% T (total acrylamide, w/v acrylamide and N,N′-methylenebisacrylamide (bis)] and 3% C (crosslinker, w/w bis/total acrylamide), using methods described in detail previously (10,11). The gels were cast and run in 40 mM TAE buffer (40 mM Tris and 1 mM EDTA, brought to pH 8.0 with glacial acetic acid) at 4°C, using an electric field strength of 5.0 V/cm. All gels were aged overnight to allow the polymerization reaction to go to completion and pre-electrophoresed for 2 h before the samples were loaded, to eliminate polar impurities. The 50 bp ladder (Invitrogen) was run in each gel as a normal mobility marker. The mobilities of all DNA fragments were calculated from the equation below:
| 1 |
where μobs is the observed mobility, d is the distance migrated in the gel in cm, E is the applied electric field strength in V/cm and t is the migration time in seconds. All gels were run in triplicate; the average standard deviation of the mobilities was ±3%.
The anomalous mobilities of the parent 199 bp fragment and its A-tract derivatives were quantitated by calculating mobility ratios, μR = μcurved/μnormal, for each of the target fragments, taking the mobility of a normal 199 bp fragment from the equation of the line describing the mobility of fragments in the 50 bp ladder run in the same gel. The mobility ratios calculated for the 199 bp fragment and its A-tract derivatives from three gels run in triplicate were then averaged and used to calculate mobility decrements, μdecr, which are defined by
| 2 |
The mobility decrements may be thought of as shape factors characterizing the apparent curvature of a DNA fragment as determined by electrophoresis.
Capillary electrophoresis
The free solution mobilities of the various DNA samples were measured in a Beckman Coulter P/ACE MDQ Capillary Electrophoresis System, operated in the anodic migration mode (anode on the detector side) with UV detection at 254 nm. Migration times and peak profiles were analyzed using the 32 Karat™ software, using methods described previously (21,22). Coated capillaries, 40 cm in length (30 cm to the detector) and 75 or 100 μm in internal diameter, were used to eliminate the electroosmotic flow (EOF) of the solvent. Previous studies have shown that DNA electrophoretic mobilities measured in coated capillaries are independent of the particular polymer used to coat the capillary wall, and that the free solution mobilities measured by capillary electrophoresis are equal to those observed by other methods (21). Hence, the DNA molecules do not interact with the capillary wall during electrophoresis.
The running buffer used for all experiments was 40 mM TAE buffer (defined above). The capillary was filled with buffer alone; no entangled linear polymers were present. The DNA samples, diluted to a concentration of ∼50 ng/μl, were pressure injected for 3 s at 20 psi (0.0035 MPa); the sample zone comprised 0.9% of the capillary volume. The applied electric field was 200 V/cm; the temperature of electrophoresis was 20 ± 0.1°C. Control experiments showed that the mobilities were independent of electric field strength and DNA concentration under the chosen conditions.
Each DNA sample was analyzed separately, to prevent the overlap of peaks with similar mobilities. As a control, two samples with widely differing mobilities were occasionally electrophoresed together to verify the mobility differences measured separately. The migration times of all samples were measured at least 2–3 times under a given set of conditions; the average standard deviation of the measurements was ±0.04%, with a maximum day-to-day variation of ±0.2%.
The free solution mobilities of the various members of the 199 bp ‘family’ and the corresponding mobility decrements were calculated from Equations 1 and 2. No corrections were needed for the EOF of the solvent, which was negligible. However, the mobilities of three of the A-tract mutants were measured in capillaries that were different from the one used for the majority of the measurements. To correct for the mobility differences caused by differences in the residual EOF in different capillaries, the mobility of the parent 199 bp fragment was measured in all three capillaries and corrected to the value measured in the original capillary, using the following equation:
| 3 |
where μparent,new cap is the mobility of the parent fragment measured in the new capillary, μparent,first cap is the mobility of the parent fragment measured in the first capillary and Δμ is the difference between the mobilities measured in the two capillaries. The mobilities of derivatives measured in the new capillary, μderivative,new cap, were then corrected to the value that would have been observed in the first capillary, μderivative,first cap, using the following equation:
| 4 |
The mobility correction was ∼2% for one capillary and ∼8% for the other capillary. As a control, the mobilities of two of the sequence mutants that had been measured in the original capillary were remeasured in one of the new capillaries and were corrected using Equation 4. The corrected mobilities were independent of the capillary in which the mobilities were measured.
Fitting equation
To test the additivity of the mobility decrements observed for each of the A- and T-tracts in the curvature module, mobilities were calculated for the parent 199 bp fragment and each of its A-tract mutants, using the following equation:
| 5 |
where (Δμ/μ)i is the mobility decrement calculated for the i-th mutant, the (Δμ/μ)j are the mobility decrements of the five derivatives, j, containing single A- or T-tracts, and the ai are structural parameters equal to 1 if a particular A-tract is present in the ith mutant and 0 otherwise. No other weighting factors were used. Mobilities were calculated for the 199 bp fragment and each of its A-tract mutants, assuming initially that the five (Δμ/μ)j were equal to those observed experimentally and that the mobility decrements of the parent 199 bp fragment and fragment 199K, with no A- or T-tracts, were equal to those observed experimentally. The calculated mobilities were then summed and compared with the sum of the experimental mobility decrements. The calculation was repeated iteratively, choosing different values for the five initial (Δμ/μ)j, until the mean square difference between the calculated and measured mobility decrements, , was minimized.
RESULTS AND DISCUSSION
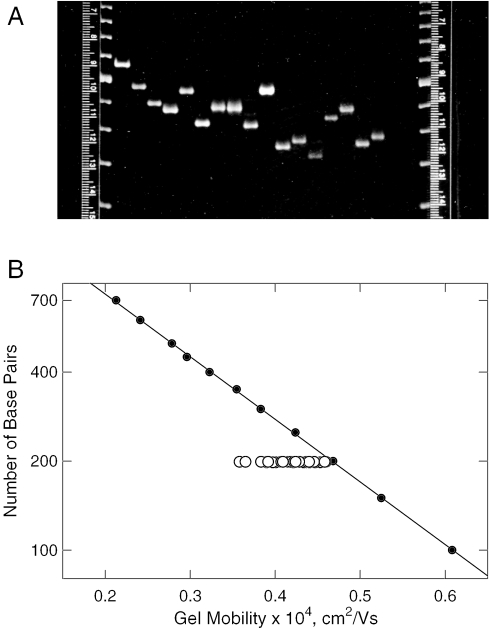
The parent 199 bp SV40 restriction fragment was modified by site-directed mutagenesis, generating 31 derivatives with all possible combinations of the A- and T-tracts in the curvature module (Scheme 1). Efforts were made to minimize the sequence changes in the various mutants: the A6-tract in the curvature module was modified to TCTAGA or AACGAA; the A4-tracts were mutated to ACGA or CTAG; the A3T4-tract was mutated to ATCTAGA or AACGTTT; and the T7-tract was modified to TCTAGAT or TTCGTTT. The names of the various derivatives, each of which contained 198 or 199 bp, are given in the first column of Table 1. The A- and T-tracts present in each derivative are indicated in the next five columns of this table. In addition, an extra base pair was added to the curvature module of fragment 198D, containing the A6 and T7 sequence motifs, to test the effect of small changes in phasing on the observed mobility anomalies. No significant differences in the gel mobility decrements were observed for 198 and 199 bp derivatives containing the A6 and T7 sequence motifs. The parent 199 bp fragment and its A-tract mutants were first characterized by PAGE. A photograph of a typical gel is shown in Figure 1A, where it can be seen that the various members of the 199 bp family exhibited a wide range of mobilities, even though their molecular weights were virtually identical. The mobilities calculated for the parent 199 bp fragment and its A-tract mutants are compared with the mobilities observed for the normal fragments in the 50 bp ladder in Figure 1B. The slowest mobility was observed for the parent 199 bp fragment. The various A-tract derivatives migrated progressively more rapidly as the number of A-tracts in the curvature module was decreased, until fragment 199K, with no A- or T-tracts in the curvature module, migrated with a mobility approximately equal to that of the 200 bp marker fragment. Since fragment 199K also exhibits normal transient birefringence relaxation times (14), the conformation of this fragment is that of normal DNA. Hence, the A- and T-tracts in the curvature module shown in Scheme 1 are responsible for the curvature of the parent 199 bp fragment, rather than the intervening sequences between the A-tracts. The gel mobility decrements calculated for the 199 bp fragment and its A-tract mutants using Equation 2 are compiled in column 7 of Table 1. The free solution mobilities of the parent 199 bp fragment and its A-tract mutants were measured by capillary electrophoresis; a typical electropherogram observed for the 199 bp fragment is illustrated in the inset of Figure 2. The free solution mobilities of normal DNA molecules of different sizes are plotted as the closed circles in the main part of Figure 2. As observed previously (21), the free solution mobilities of small DNA molecules increase with increasing molecular weight, before leveling off and becoming constant at molecular weights above ∼400 bp. The increase in mobility with increasing DNA molecular weight can be attributed to the greater solvent friction experienced by small, relatively rigid DNA oligomers (23). This effect gradually becomes less important with the onset of coiling and free draining effects at higher molecular weights (23–25).
Table 1.
The parent 199 bp SV40 fragment and derivatives missing one or more of the A- and T-tracts in the curvature module
| DNA derivative | A- and T-tracts present | Gel mobility decrement | Free solution mobility decrement | ||||
|---|---|---|---|---|---|---|---|
| 199 parent | A6 | A4T | A4 | A3T4 | T7 | 23.5 | 1.63 |
| 199F | — | A4T | A4 | A3T4 | T7 | 18.1 | 1.16 |
| 199B | A6 | — | A4 | A3T4 | T7 | 16.6 | 1.30 |
| 199C | A6 | A4T | — | A3T4 | T7 | 22.0 | 1.51 |
| 198A | A6 | A4T | A4 | — | T7 | 14.4 | 1.26 |
| 199E | A6 | A4T | A4 | A3T4 | — | 18.1 | 1.24 |
| 199Q | — | — | A4 | A3T4 | T7 | 9.8 | 0.85 |
| 199R | — | A4T | — | A3T4 | T7 | 13.0 | 1.19 |
| 198X | — | A4T | A4 | — | T7 | 7.4 | 1.06 |
| 199G | — | A4T | A4 | A3T4 | — | 11.4 | 0.93 |
| 199D | A6 | — | — | A3T4 | T7 | 12.9 | 1.10 |
| 198C | A6 | — | A4 | — | T7 | 6.7 | 0.71 |
| 199P | A6 | — | A4 | A3T4 | — | 10.6 | 1.02 |
| 198B | A6 | A4T | — | — | T7 | 16.7 | 1.12 |
| 199V | A6 | A4T | — | A3T4 | — | 16.3 | 1.24 |
| 199U | A6 | A4T | A4 | — | — | 12.6 | 0.77 |
| 199S | — | — | — | A3T4 | T7 | 8.5 | 0.56 |
| 198J | — | — | A4 | — | T7 | 4.5 | 0.24 |
| 199L | — | — | A4 | A3T4 | — | 2.1 | 0.63 |
| 198I | — | A4T | — | — | T7 | 10.0 | 0.77 |
| 199H | — | A4T | — | A3T4 | — | 11.2 | 0.82 |
| 199O | — | A4T | A4 | — | — | 6.0 | 0.50 |
| 198D | A6 | — | — | — | T7 | 10.0 | 0.65 |
| 199T | A6 | — | — | A3T4 | — | 9.4 | 0.54 |
| 198H | A6 | — | A4 | — | — | 6.9 | 0.53 |
| 198G | A6 | A4T | — | — | — | 15.2 | 1.06 |
| 198E | — | — | — | — | T7 | 3.2 | 0.35 |
| 199I | — | — | — | A3T4 | — | 4.8 | 0.53 |
| 199N | — | — | A4 | — | — | 2.1 | 0.36 |
| 199J | — | A4T | — | — | — | 6.6 | 0.74 |
| 198F | A6 | — | — | — | — | 5.9 | 0.69 |
| 199K | — | — | — | — | — | 1.8 | 0.50 |
Figure 1.
PAGE of the parent 199 bp fragment and its A-tract mutants. (A) Typical polyacrylamide gel. The two lanes on the edges of the gel contain the 50 bp ladder (fragments with 150–550 bp). From left to right, the fragments in the other lanes are: 222D (containing two tandem copies of the curvature module), the parent 199 bp fragment, 199A, 199B, 199C, 199D, 199E, 199F, 199G, 199H, 199I, 199J, 199K, 198A, 198B, 198C and 198D (Table 1). (B) Dependence of the mobility of the parent 199 bp fragment and its A-tract mutants on the number of base pairs in the fragment. The symbols correspond to: closed circle, fragments in the standard marker ladder; open circle, 199 bp family.
Figure 2.
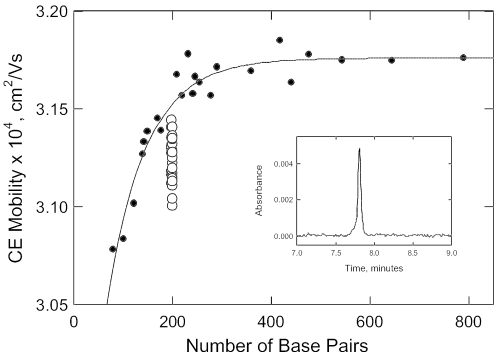
Free solution mobility of curved (open circles) and normal (closed circles) DNA molecules, measured by capillary electrophoresis. Inset: typical electropherogram observed for the parent 199 bp restriction fragment.
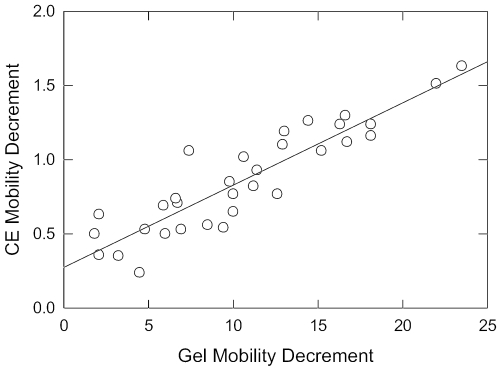
Surprisingly, the free solution mobilities observed for the parent 199 bp fragment and its A-tract mutants (except fragment 199K, with no A- or T-tracts) fall below the curve describing the mobilities of normal DNA molecules of the same size, as shown by the open circles in Figure 2. Hence, curved DNA molecules migrate anomalously slowly in free solution, just as they do in polyacrylamide gels. The free solution mobility decrements observed for the parent 199 bp fragment and its A-tract mutants are compiled in the last column of Table 1. Although the absolute values of the mobility decrements observed in polyacrylamide gels and free solution differ by more than an order of magnitude, the mobility decrements are highly correlated, as shown in Figure 3. Hence, both electrophoretic techniques are sensitive to DNA curvature.
Figure 3.
Correlation between the mobility decrements observed in free solution (ordinate) and in polyacrylamide gels (abscissa). The drawn line is a least squares regression line with a correlation coefficient, r2 = 0.82.
In polyacrylamide gels, DNA mobilities are determined primarily by sieving effects, owing to the fact that only a fraction of the pores in the gel matrix are large enough to accommodate the migrating DNA molecules (13,21). To obtain the intrinsic mobilities of curved and normal DNA molecules of the same size, the observed gel mobilities must be extrapolated to zero gel concentration (N. C. Stellwagen, manuscript in preparation). The extrapolated mobilities of curved DNA molecules are lower than those of normal DNAs containing the same number of base pairs, as observed here for the free solution mobilities of the 199 bp fragment and its A-tract derivatives. Molecular weight sieving effects in the polyacrylamide gel matrix thus amplify the intrinsic mobility differences between curved and normal DNAs.
In free solution, gel matrix sieving effects do not exist, and the intrinsic mobilities of curved and normal DNA fragments can be measured directly, using capillary electrophoresis. It is important to realize that the anomalously slow mobilities observed for curved DNA molecules in free solution can be compared only with the mobilities of normal DNAs containing the same number of base pairs, because the relative importance of solvent friction varies with molecular weight (23). Therefore, it is not possible to calculate an ‘effective size’ of curved DNA molecules in free solution based on their anomalously slow electrophoretic mobilities.
To test the additivity of the mobility decrements observed for the various A- and T-tracts in the curvature module, mobility decrements were calculated for the parent 199 bp fragment and its A-tract derivatives by assuming that each A- or T-tract contributed the same amount of curvature to each derivative in which it was present. The mobility decrements of mutants containing single A- or T-tracts were taken to be the optimized mobility decrements obtained from the fitting procedure described above. The fitted mobility decrements of the five mutants containing single A- or T-tracts are compared with the experimentally measured values in Table 2. The fitted and measured gel mobility decrements are very similar for all five mutants. The agreement is not as good as that for the fitted and measured free solution mobility decrements, most likely because of greater experimental error in measuring the very small mobility decrements of derivatives containing only a single A- or T-tract. Nevertheless, the fitted and measured free solution mobility decrements exhibit the same trends.
Table 2.
Comparison of the fitted and measured mobility decrements observed for derivatives containing a single A- or T-tract in the curvature module
| Derivative name | A-tract present | Gel mobility decrement | Free solution mobility decrement | ||
|---|---|---|---|---|---|
| Fitted | Measured | Fitted | Measured | ||
| 198F | A6 | 5.6 | 5.9 | 0.36 | 0.69 |
| 199J | A4T | 6.8 | 6.6 | 0.48 | 0.74 |
| 199N | A4 | 0.5 | 2.1 | 0.16 | 0.36 |
| 199I | A3T4 | 4.9 | 4.8 | 0.34 | 0.53 |
| 198E | T7 | 3.4 | 3.4 | 0.29 | 0.35 |
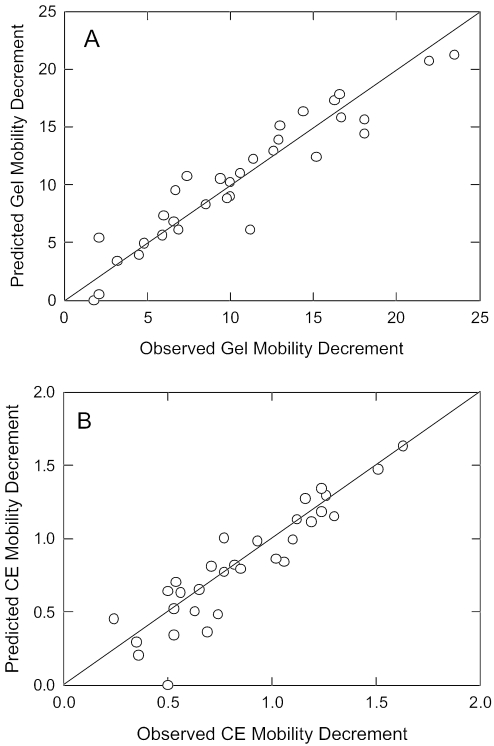
The fitted mobility decrements of the five mutants containing single A- or T- tracts were used to predict the mobilities of mutants containing two or more A-tracts. The predicted gel and free solution mobility decrements are compared with the measured mobility decrements in Figure 4. In both polyacrylamide gels and free solution, there is a strong correlation between the measured mobility decrements and those predicted by assuming that each A- or T-tract contributes a constant degree of bending to the curvature module. The average standard deviation of the predicted and measured mobility decrements was ±1.2 for the gel mobility decrements (6.8%) and ±0.12 for the free solution mobility decrements (7.6%). Very similar results were obtained by using the measured mobility decrements of mutants containing single A- or T-tracts to predict the mobility decrements of the other A-tract mutants (data not shown). Hence, the individual A- and T-tracts in the curvature module contribute in an additive manner to the total curvature of the parent 199 bp DNA fragment, even though the A- and T-tracts are of variable lengths and have a variable spacing with respect to each other.
Figure 4.
Comparison of the measured and predicted mobility decrements. (A) Polyacrylamide gel mobility decrements; (B) free solution mobility decrements. The drawn lines are least squares regression lines with correlation coefficients of r2 = 0.89 for (A) and 0.84 for (B).
The linear dependence of the mobility decrements on curvature differs from the dependence of relative length on the square of DNA curvature observed for oligomer ladders containing phased A-tracts (7), and the dependence of relative mobility on the cosine of one-half of the bend angle, observed for DNA fragments containing phased A-tracts in their centers (6). The latter relationship is close to the linear dependence observed here, since, for small values of the bend angle, α, cos α/2 ≈ α/2. The type of DNA sequence in the latter study is also closer to the A-tract mutants studied here since, in both cases, a region of curvature in the center of the fragment is flanked by long stretches of normal DNA on either side.
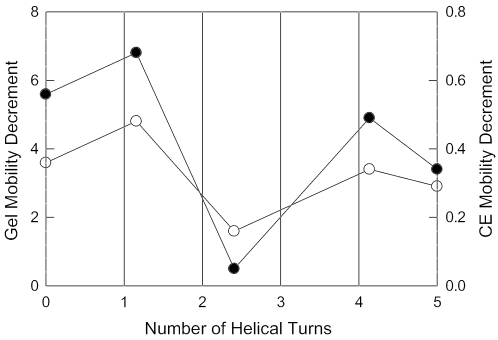
The gel and free solution mobility decrements obtained for derivatives with a single A- or T-tract are plotted in Figure 5 as a function of the number of helical turns (10.4 bp) between the particular A- or T-tract and the A6-tract at the beginning of the curvature module. The mobility decrements observed for the A4T, A3T4 and T7 sequence motifs, which are approximately in phase with each other and with the initial A6-tract in the curvature module, decrease progressively with the increasing number of helical turns between the particular A- or T-tract and the initial A6-tract. A relatively small mobility decrement is observed for the A4-tract nearest to the apparent bend center, apparently because this A-tract is nearly out-of-phase with the initial A6-tract and with the adjacent A4T- and A3T4-tracts. Hence, the phasing of the A- and T-tracts that follow the first A-tract in the curvature module, and the distance between the initial A-tract and subsequent A- and T-tracts, determine the relative contribution of each A- or T-tract to the curvature of the parent 199 bp fragment.
Figure 5.
Dependence of the gel and free solution mobility decrements of mutants containing single A- or T-tracts (fitted values) on the number of 10.4 bp helical turns between the initial A6-tract in the curvature module and subsequent A- and T-tracts. The symbols correspond to: closed circles, gel mobility decrements (left ordinate); open circles, free solution mobility decrements (right ordinate). Similar results are observed if the measured mobility decrements are plotted instead of the fitted values.
CONCLUDING REMARKS
The parent 199 bp SV40 restriction fragment and derivatives containing one or more A- or T-tracts in the curvature module migrate anomalously slowly in free solution, as well as in polyacrylamide gels. Hence, the anomalously slow mobilities observed for curved DNA molecules in polyacrylamide gels are due, in part, to the fact that curved DNA molecules have intrinsically lower electrophoretic mobilities than normal DNAs containing the same number of base pairs. The good correlation between the mobility decrements observed in polyacrylamide gels and in free solution indicates that polyacrylamide gels do not introduce artifacts into the mobility measurements. Rather, the intrinsic free solution mobility differences between curved and normal DNA molecules of the same size are enhanced by the sieving effect of the polyacrylamide gel matrix, which preferentially retards the migration of curved DNA molecules.
The curvature module in the VP1 gene of SV40 contains five unevenly spaced A- and T-tracts that contribute in an additive manner to the total curvature of the helix backbone. However, the various A- and T-tracts are not equally important. In particular, the A4-tract near the apparent bend center contributes very little to the observed curvature, most likely because it is nearly out of phase with the initial A6-tract in the curvature module and with the adjacent A4T- and A3T4-tracts.
The biological function of curvature in the middle of the VP1 gene in SV40 is not clear. The apparent bend center forms the boundary of a strong nucleosome positioning signal in vitro (26). However, nucleosomes are excluded from this site in vivo (27), leading to speculation that it might be involved in the formation of higher order nucleosome structures (27).
The anomalously slow electrophoretic mobilities observed for the 199 bp fragment and its A-tract mutants suggest that the A- and T-tracts in the curvature module bind additional monovalent counterions, decreasing the effective net charge of the various members of the 199 bp family and consequently decreasing their free solution mobilities. Similar decreases in mobility upon monovalent counterion binding have been observed for small DNA oligomers containing A-tracts (28). These results are consistent with recent studies (29) indicating that DNA curvature results in the net crowding of the charged phosphate residues, leading to a net increase in counterion condensation and a corresponding increase in charge neutralization. The additivity of the mobility decrements observed for the 199 bp fragment and its A-tract mutants indicates that each A- or T-tract in the curvature module binds counterions independently, as expected for an electrostatic interaction between DNA and its counterions. However, phasing effects also appear to be important, since the magnitude of the mobility decrements observed for derivatives containing a single A or T-tract depends on the spacing between that A- or T-tract and the first A-tract in the curvature module.
Acknowledgments
The expert technical assistance of Brock Weers in preparing the various DNA samples is gratefully acknowledged. Financial support from the National Institute of General Medical Sciences, grants GM071464 and GM61009 (to N.C.S.), is also acknowledged. Funding to pay the Open Access publication charges for this article was provided by the National Institute of General Medical Sciences, grants GM071464 and GM61009.
Conflict of interest statement. None declared.
REFERENCES
- 1.Marini J.D., Levene S.D., Crothers D.M., Englund P.T. Bent helical structure in kinetoplast DNA. Proc. Natl Acad. Sci. USA. 1982;79:7664–7668. doi: 10.1073/pnas.79.24.7664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stellwagen N.C. Anomalous electrophoresis of deoxyribonucleic acid restriction fragments on polyacrylamide gels. Biochemistry. 1983;22:6186–6193. doi: 10.1021/bi00295a023. [DOI] [PubMed] [Google Scholar]
- 3.Calladine C.R., Drew H.R., McCall M.J. The intrinsic curvature of DNA in solution. J. Mol. Biol. 1988;210:127–137. doi: 10.1016/0022-2836(88)90444-5. [DOI] [PubMed] [Google Scholar]
- 4.Wu H.-M., Crothers D.M. The locus of sequence-directed and protein-induced DNA bending. Nature. 1984;308:509–513. doi: 10.1038/308509a0. [DOI] [PubMed] [Google Scholar]
- 5.Eckdahl T.T., Anderson J.N. Computer modeling of DNA structures involved in chromosome maintenance. Nucleic Acids Res. 1987;15:8531–8545. doi: 10.1093/nar/15.20.8531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thompson J.F., Landy A. Empirical estimation of protein-induced bending angles: applications to lambda site-specific recombination complexes. Nucleic Acids Res. 1988;16:9687–9705. doi: 10.1093/nar/16.20.9687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koo H.-S., Crothers D.M. Calibration of DNA curvature and a unified description of sequence-directed bending. Proc. Natl Acad. Sci. USA. 1988;85:1763–1767. doi: 10.1073/pnas.85.6.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bolshoy A., McNamara P., Harrington R.E., Trifonov E.N. Curved DNA without A-A: experimental estimation of all 16 DNA wedge angles. Proc. Natl Acad. Sci. USA. 1991;88:2312–2316. doi: 10.1073/pnas.88.6.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ross E.D., Den R.B., Hardwidge P.R., Maher L.J. Improved quantitation of DNA curvature using ligation ladders. Nucleic Acids Res. 1999;27:4135–4142. doi: 10.1093/nar/27.21.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holmes D.L., Stellwagen N.C. Estimation of polyacrylamide gel pore size from Ferguson plots of normal and anomalously migrating DNA fragments. I. Gels containing 3% N,N′-methylenebisacrylamide. Electrophoresis. 1991;12:253–263. doi: 10.1002/elps.1150120405. [DOI] [PubMed] [Google Scholar]
- 11.Holmes D.L., Stellwagen N.C. Estimation of polyacrylamide gel pore size from Ferguson plots of linear DNA fragments. II. Comparison of gels with different crosslinker concentrations, added agarose and added linear polyacrylamide. Electrophoresis. 1991;12:612–619. doi: 10.1002/elps.1150120903. [DOI] [PubMed] [Google Scholar]
- 12.Stellwagen A., Stellwagen N.C. Anomalously slow electrophretic mobilities of DNA restriction fragments in polyacrylamide gels are not eliminated by increasing the gel pore size. Biopolymers. 1990;30:309–324. doi: 10.1002/bip.360300309. [DOI] [PubMed] [Google Scholar]
- 13.Stellwagen N.C. DNA mobility anomalies are determined primarily by polyacrylamide gel concentration, not gel pore size. Electrophoresis. 1997;18:34–44. doi: 10.1002/elps.1150180108. [DOI] [PubMed] [Google Scholar]
- 14.Lu Y.J., Weers B.D., Stellwagen N.C. Intrinsic curvature in the VP1 gene of SV40: comparison of solution and gel results. Biophys. J. 2005;88:1191–1206. doi: 10.1529/biophysj.104.039834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu Y.J., Weers B., Stellwagen N.C. DNA persistence length revisited. Biopolymers. 2002;61:261–275. doi: 10.1002/bip.10151. [DOI] [PubMed] [Google Scholar]
- 16.Hagerman P.J. Sequence-directed curvature of DNA. Nature. 1986;321:449–450. doi: 10.1038/321449a0. [DOI] [PubMed] [Google Scholar]
- 17.Burkhoff A.M., Tullius T.D. The unusual conformation adopted by the adenine tracts in kinetoplast DNA. Cell. 1987;48:935–943. doi: 10.1016/0092-8674(87)90702-1. [DOI] [PubMed] [Google Scholar]
- 18.Leroy J.-L., Charretier E., Kochoyan M., Gueron M. Evidence from base-pair kinetics for two types of adenine tract structures in solution: their relation to DNA curvature. Biochemistry. 1988;27:8894–8898. doi: 10.1021/bi00425a004. [DOI] [PubMed] [Google Scholar]
- 19.Mollegaard N.E., Bailly C., Waring M.J., Nielsen P.E. Effects of diaminopurine and inosine substitutions on A-tract induced DNA curvature. Importance of the 3′-A-tract junction. Nucleic Acids Res. 1997;25:3497–3502. doi: 10.1093/nar/25.17.3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sambrook J., Russell D.W. Molecular Cloning. 3rd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 21.Stellwagen N.C., Gelfi C., Righetti P.G. The free solution mobility of DNA. Biopolymers. 1997;42:687–703. doi: 10.1002/(SICI)1097-0282(199711)42:6<687::AID-BIP7>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 22.Dong Q., Stellwagen E., Dagle J.M., Stellwagen N.C. Free solution mobility of small single-stranded oligonucleotides with variable charge densities. Electrophoresis. 2003;24:3323–3329. doi: 10.1002/elps.200305589. [DOI] [PubMed] [Google Scholar]
- 23.Mohanty U., Stellwagen N.C. Free solution mobility of oligomeric DNA. Biopolymers. 1999;49:209–214. [Google Scholar]
- 24.Cottet H., Gareil P., Theodoly O., Williams C.E. A semi-empirical approach to the modeling of the electrophoretic mobility in free solution: application to polystyrenesulfonates of various sulfonation rates. Electrophoresis. 2000;21:3529–3540. doi: 10.1002/1522-2683(200011)21:17<3529::AID-ELPS3529>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 25.Stellwagen E., Lu Y.J., Stellwagen N.C. Unified description of electrophoresis and diffusion for DNA and other polyions. Biochemistry. 2003;42:11745–11750. doi: 10.1021/bi035203p. [DOI] [PubMed] [Google Scholar]
- 26.Stein A. Unique positioning of reconstituted nucleosomes occurs in one region of simian virus 40 DNA. J. Biol. Chem. 1987;262:3872–3879. [PubMed] [Google Scholar]
- 27.Ambrose C., Lowman H., Rajadhyaksha A., Blasquez V., Bina M. Location of nucleosomes in simian virus 40 chromatin. J. Mol. Biol. 1990;214:875–884. doi: 10.1016/0022-2836(90)90342-J. [DOI] [PubMed] [Google Scholar]
- 28.Stellwagen N.C., Magnusdóttir S., Gelfi C., Righetti P.G. Preferential counterion binding to A-tract DNA oligomers. J. Mol. Biol. 2001;305:1025–1033. doi: 10.1006/jmbi.2000.4362. [DOI] [PubMed] [Google Scholar]
- 29.Range K., Mayaan E., Maher L.J., III, York D.M. The contribution of phosphate-phosphate repulsions to the free energy of DNA bending. Nucleic Acids Res. 2005;33:1257–1268. doi: 10.1093/nar/gki272. [DOI] [PMC free article] [PubMed] [Google Scholar]