Abstract
Long non-coding RNAs (lncRNAs) and microRNAs (miRNAs) are closely related to the treatment of human diseases. Traditional biological experiments often require time-consuming and labor-intensive in their search for mechanisms of disease. Computational methods are regarded as an effective way to predict unknown lncRNA-miRNA interactions (LMIs). However, most of them complete their tasks by mainly focusing on a single lncRNA-miRNA network without considering the complex mechanism between biomolecular in life activities, which are believed to be useful for improving the accuracy of LMI prediction. To address this, a heterogeneous information network (HIN) learning model with neighborhood-level structural representation, called HINLMI, to precisely identify LMIs. In particular, HINLMI first constructs a HIN by integrating nine interactions of five biomolecules. After that, different representation learning strategies are applied to learn the biological and network representations of lncRNAs and miRNAs in the HIN from different perspectives. Finally, HINLMI incorporates the XGBoost classifier to predict unknown LMIs using final embeddings of lncRNAs and miRNAs. Experimental results show that HINLMI yields a best performance on the real dataset when compared with state-of-the-art computational models. Moreover, several analysis experiments indicate that the simultaneous consideration of biological knowledge and network topology of lncRNAs and miRNAs allows HINLMI to accurately predict LMIs from a more comprehensive perspective. The promising performance of HINLMI also reveals that the utilization of rich heterogeneous information can provide an alternative insight for HINLMI to identify novel interactions between lncRNAs and miRNAs.
Keywords: Network structural representation, Heterogeneous information networks, Biological and network representations, LMIs
Graphical abstract

1. Introduction
Non-coding RNA (ncRNA) has been demonstrated to be associated with mammalian and human genomes, and further study of non-coding RNA can improve the understand of gene regulatory networks for researchers [16], [31], [20], [5], [39]. As researchers delve deeper into the study of non-coding RNAs (ncRNAs), it becomes evident that long non-coding RNAs (lncRNAs) and microRNAs (miRNAs) play crucial roles in regulating various cellular processes. These ncRNAs are found to have significant impacts on the control of life activities. In particular, lncRNAs have been extensively investigated and have been shown to possess diverse and intricate mechanisms for governing gene expression. They exert their influence at both the transcriptional and post-transcriptional levels, adding to their importance in cellular regulation [41]. Moreover, lncRNAs are closely interested with miRNAs in terms of stability, which makes miRNAs also indispensable in process of physiological and pathological. MicroRNAs (miRNAs) are single-stranded (<22nt) ncRNA sequences, which is the most attractive and most clearly characterized regulatory ncRNAs [22]. Therefore, studying unknown interactions between lncRNA and miRNA can help us better understand the functional expression of lncRNAs and miRNAs, and further gain new insights for medical research.
Traditional biological experiments, the acquisition of unknown lnRNA-miRNA interactions (LMIs) is time consuming and expensive, resulting in inefficiency and unpopularity. Artificial intelligence techniques have been successfully applied in bioinformatics, including LMIs prediction, has attracted much attention as an alternative yet complementary strategy to discover new interactions for lncRNAs and miRNAs, thus offering significant advantages to accelerate the process of biological research.
In recent years, there has been growing interest in the representation learning of ncRNAs within Heterogeneous Information Networks (HINs), owing to the rich feature information of lncRNAs and miRNAs contained in these networks. While this method enhances precision, it overlooks the complex relationships and interactions that biomolecules exhibit within rich molecular association networks, making it challenging to understand the molecular mechanisms of disease occurrence from a holistic biological systems perspective. For example, in the treatment of gastric cancer, the post-transcriptional regulatory roles played by lncRNAs and miRNAs, particularly in regulating protein stability, are crucial [17], [15]. Therefore, incorporating a variety of significant biomolecules such as proteins and diseases to construct a systematic and enriched biomolecular association network to explore potential interactions between lncRNAs and miRNAs holds significant biomedical importance [4], [14].
To address the issues mentioned, we develop a novel computational model for learning in Heterogeneous Information Networks (HINs) with neighborhood structures, named HINLMI, which predicts new lncRNA-miRNA interactions (LMIs) by leveraging the biological knowledge and network topology of lncRNAs and miRNAs. Specifically, HINLMI first integrates nine types of biomolecular networks, including lncRNA-miRNA, lncRNA-disease, lncRNA-protein, mi-RNA-disease, miRNA-protein, protein-disease, protein-protein, protein-drug, and drug-disease, into a HIN, where the nucleotide sequences of lncRNAs and miRNAs serve as their biological knowledge. Subsequently, the popular sequence representation learning algorithm k-mer [32] is employed to represent the biological knowledge of lncRNAs and miRNAs. Then, to more precisely capture the complex relationships of lncRNAs and miRNAs within the HIN, HINLMI calculates the neighborhood structural similarity between molecules, including both first-order and higher-order similarities, to obtain a comprehensive network representation. Finally, the ultimate representation of lncRNAs and miRNAs, composed of both biological and network topology representations derived from the given HIN, is fed into an XGBoost classifier to perform the LMI prediction task. Experimental results demonstrate that HINLMI outperforms the latest computational models used for predicting novel LMIs on several independent metrics in real datasets. Furthermore, our case studies indicate that the rich biological information gains new insight into lncRNA-miRNA interaction prediction with improved accuracy. The main contributions of this work are summarized as:
(1) An effective computational algorithm, namely HINL-MI, is proposed to precisely identify novel DDAs by calculating neighborhood-level structural representation of lncRNAs and miRNAs through a XGBoost classifier.
(2) Different representation learning strategies are developed to learn the biological and network representations of lncRNAs and miRNAs from different perspectives.
(3) First-order and high-order similarity structure information are taken into account simultaneously to better learn the feature representations of lncRNAs and miRNAs on heterogeneous information networks.
(4) Experimental results demonstrate that HINLMI performs better than several state-of-the-art computational models under 5-fold cross-validation. The promising performance of HINLMI also reveals that the utilization of rich heterogeneous information allows HINLMI to identify novel relationships between lncRNAs and miRNAs.
2. Related works
Existing LMI prediction methods can be divided into two categories based on feature extraction strategies: similarity network-based computation and graph representation learning. Similarity network-based methods rely on phenotypic data of lncRNAs and miRNAs, predicting interactions by calculating their similarities. These methods are computationally simple and easy to implement, but their performance may decline with sparse data due to high dependency on available data. For instance, Huang et al. [21] propose a computational-based model, called EPLMI, for predicting unknown LMIs. EPLMI assumes that highly similar lncRNAs have similar patterns of interaction or non-interaction with miRNAs, and then different similarity matrices are constructed to identify unknown LMIs. Hu et al. [19] present a matrix factorization-based model, namely LMNLMI, to complete the LMIs prediction task. First, LMNLMI obtains patterns according to the sequences expression and functional of lncRNAs and miRNAs, and then constructs several similarity networks, and finally the interaction scores of lncRNAs and miRNAs are calculated. GNMFLMI [36] combines known interaction information, sequence data, and graph regularization to predict potential lncRNA-miRNA interactions, which leverages both biological knowledge and network structure to enhance the accuracy of LMI prediction. LMI-INGI [42] constructs two graphs according to the similarity of lncRNAs and miRNAs, and then calculates the scores for lncRNA-miRNA pairs by these two graphs and the known interactions.
In contrast, graph representation learning methods transform lncRNA-miRNA relationships into low-dimensional, dense vector representations while preserving the original graph properties. For example, GCNCRF [37] initially constructs a heterogeneous network using known interactions, similarity networks, and feature matrices of lncRNAs and miRNAs, then employs a graph convolutional neural network equipped with an attention mechanism to obtain representations of the nodes. Subsequently, these representations are decoded through a decoding layer to yield scores. MGCAT [26] adopts a multi-view graph neural network with cascaded attention to learn informative node representations by leveraging view-level, node-level, and layer-level attentions for LMI prediction.
3. Materials and methods
3.1. Dataset
To construct a HIN for performance evaluation, we have based on a heterogeneous dataset, namely MAN dataset, composed of five kinds of biomolecules, i.e., lncRNA, miRNA, protein, drug, and disease, and their interactions. To systematically and comprehensively establish a biomolecular relationship network, we followed the methodology described by Guo et al. [13], downloading known associations between small biological molecules (miRNA, lncRNA, and proteins), diseases, and drugs from multiple databases. These databases include lncRNASNP2, lncRNADisease, lncRNA2Target, HMDDv3.0, miRTarBase, DisGeNET, and etc. Initially, we extracted all lncRNA and miRNA interaction pairs from the lncRNASNP2 database and performed the following processing steps: unifying identifiers to ensure data consistency across different databases; removing duplicates to retain unique interaction pairs; simplifying the data structure by keeping key fields such as lncRNA ID, miRNA ID, and interaction type; and eliminating irrelevant items to ensure data relevance and accuracy. We applied the same preprocessing steps to other types of data to maintain consistency and relevance. After completing these steps, we obtained a non-redundant and simplified molecular association network (MAN) dataset. The detailed data statistics are shown in Table 1, Table 2, where lncRNA-miRNA interactions are regarded as the benchmark dataset.
Table 1.
Nine biological molecules association.
| Associations | Number | Database |
|---|---|---|
| lncRNA-miRNA | 8,374 | lncRNASNP2 [28] |
| lncRNA-disease | 1,264 | lncRNADisease [2], lncRNASNP2 |
| lncRNA-protein | 690 | lncRNA2Target [24] |
| miRNA-disease | 16,427 | HMDDv3.0 [23] |
| miRNA-protein | 4,944 | miRTarBase [7] |
| protein-disease | 25,087 | DisGeNET [30] |
| protein-protein | 19,237 | STRING [33] |
| protein-drug | 11,107 | DrugBank5.0 [38] |
| drug-disease | 18,416 | CTD [8] |
| Total | 105,546 | MAN [14] |
Table 2.
The number of five biomolecular types.
| Name | lncRNA | miRNA | protein | drug | disease | Total |
|---|---|---|---|---|---|---|
| Number | 769 | 1023 | 1649 | 1025 | 2062 | 6528 |
3.2. Construction of HIN
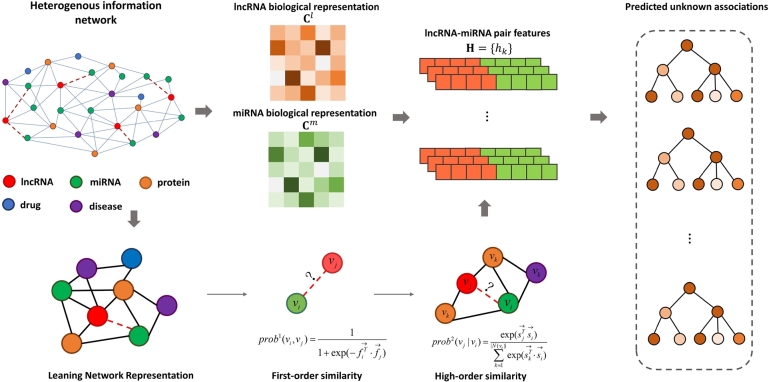
As mentioned before, a HIN of interest is consisted of nine different types associations [10], [1], [43], [11], and then two kinds of biological information are contained, i.e., the biological and network representations of lncRNAs and miRNAs. The framework overview of HINLMI is shown in Fig. 1. To model a HIN, we develop a classical three-element tuple, i.e., , where is a set of all biological molecules, is a set of biological representation of lncRNAs and miRNAs. is composed of all lncRNA-miRNA associations , lncRNA-disease associations , lncRNA-protein associations , miRNA-disease associations , miRNA-protein associations , protein-disease associations , protein-protein associations , drug-protein associations and drug-disease associations . Besides, is the size of V, L and M are assumed as the number of lncRNAs and miRNAs, respectively.
Fig. 1.
The framework overview of HINLMI.
3.3. Learning biological representation
In this paper, we collect the sequences of lncRNAs and miRNAs are derived from the miRbase [25] and NONCODE [9], respectively. For the sake of convenience, a gene sequence which can be made up of A (Adenine), G (Guanine), C (Cytosine), and U (Uracil) is divided a series of subsequences by k-mer algorithm (k=3) [32]. For example, “” first can be divided into “AAC, ACT, CTG, TGA, GAC, ACT, CTG, TGA”. Second, the representation vectors of lncRNAs and miRNAs are obtained by counting sub-sequences occurrence frequency. At last, these representation vectors are normalized as final biological representation of lncRNAs and miRNAs.
3.4. Learning network representation
Since the HIN involves many molecular attributes, which is more complex network [18]. To fully capture the network representation of lncRNAs and miRNAs, we take into account the features of nodes in given HIN from first-order and high-order similarity structures. Among which, the first-order similarity indicates known associations between biological molecules, and high-order similarity regards biological molecules are similar if they have shared neighborhood structures. Inspired by graph representation learning algorithm [34], first-order similarity is constructed by the joint probability among node and () as follows:
| (1) |
where and denote the feature vector of the node and respectively, as a probability distribution in the space . Subsequently, defines the empirical probability of Eq. (1), as follows:
| (2) |
where W is a trainable weight matrix and represents the first-order similarity between node i and j. To preserve , we optimize Eq. (1) by minimizing the following objective functions.
| (3) |
where the function D denotes the distance between probability and . For the sake of calculation, the KL-divergence is introduced to replace the function D by following:
| (4) |
Hence, the first-order representations of all nodes in are represented as by minimize the Eq. (4). In the same way, we calculate the second-order representations in HIN by following:
| (5) |
where denotes a set of neighbor of nodes, defines a conditional probability function. and are the second-order representations of node and in , respectively. To preserve , we optimize Eq. (5) by minimize the following objective function.
| (6) |
where denotes the degree of node , and is empirical probability of Eq. (5).
| (7) |
where . For the sake of calculation, the KL-divergence is introduced to replace the function D. The KL-divergence is used in the HINLMI model to minimize the distance between two probability distributions: the empirical distribution and the joint distribution for both first-order and second-order proximities. The advantages of using KL-divergence include three folds, fidelity to the data distribution, effectiveness in sparse data, and non-symmetry. Its calculation process by following:
| (8) |
Therefore, the second-order representations of all nodes in are represented as by minimize the Eq. (8). In the end, first-order and second-order representations are spliced together, and we have a matrix to collect the network representations of lncRNAs and miRNAs, where is a splice function.
There are three primary reasons why third-order proximities are not deemed necessary in the HINLMI model. Firstly, the combination of first- and second-order proximities effectively captures a substantial amount of the structural information inherent in the HIN. This comprehensive capture of network structure minimizes the need for higher-order proximities. Secondly, the inclusion of second-order proximity already significantly enhances the performance of HINLMI on various prediction tasks compared to using only first-order proximity, suggesting a robust improvement in embedding quality without necessitating further complexity. Lastly, while higher-order proximities may potentially yield even more representative embeddings, the increased computational complexity will compromise the model's efficiency, particularly when scaling to very large networks such as molecular association networks.
3.5. Predicting novel LMIs
According to the above steps, HINLMI have been obtained two kinds of representations for lncRNAs and miRNAs for a given , i.e., the biological representation C and network representation P. The lncRNA-miRNA interaction prediction issue usually is regarded as a binary classification, which is positive samples consist of known LMI and negative samples consist of unknown LMI are fed to a machine learning classifier to train the model, and then predictive samples are input to the train model to obtain their scores as predicted results. Hence, we connect them by a connection matrix to represent the final representations of lncRNAs and miRNAs. To accurately predict novel LMIs, HINLMI predict novel LMIs by incorporating the XGBoost classifier [3]. In particular, we prepare a training dataset, denoted as , to build the XGBoost classifier based on the feature representations of lncRNAs and miRNAs. contains kinds of feature vectors , and each feature vector is composed of and , where , . Similarly, is also constructed by N lncRNA-miRNA pairs with unknown interactions.
For the XGBoost classifier consist of K decision trees, each decision tree can be regarded as a projection function , and its calculation process as follows:
| (9) |
where is the prediction value of k-th training samples in , is t-th decision tree, T is the number of all training trees. The objective function of the XGBoost classifier contains two steps: training loss and complexity regularization. Given a training sample , its objective function can be defined as:
| (10) |
where is the loss function that measure the error between the predicted value and the real value . is a regularization of the complexity for t-th decision tree, usually defined as:
| (11) |
where is the number of leaf nodes in k-th decision tree, is the weight of the j-th leaf node, and γ and λ are the regularization parameters. Subsequently, we calculate candidate lncRNA-miRNA pairs by following a logical function:
| (12) |
where σ is sigmoid function, which converts logarithmic odds to probabilities.
| (13) |
where denotes the final classification of the candidate sample . Through the above calculation process, the XGBoost classifier can be used to give the probability for . Regarding the predicted results, a N-dimension vector r is adopted to collect the prediction score of each lncRNA-miRNA pair in . One should note that the range of each element in r is within [0, 1]. The main symbols used in this paper are summarized in Table 3.
Table 3.
Main symbols used.
| Symbol | Description |
|---|---|
| HIN | Heterogeneous Information Network |
| MAN | Molecular Association Network |
| LMI | LncRNA-miRNA Interaction |
| The HIN graph | |
| V | The set of all biological molecules |
| C | The set of biological representation of lncRNAs and miRNAs |
| E | The set of all molecular associations |
| |V| | The number of all molecules |
| L | The number of lncRNAs |
| M | The number of miRNAs |
| C | The matrix of the final biological representations |
| F | The set of the first-order representations |
| S | The set of second-order representations |
| P | The matrix of the network representations |
| X | The matrix of the final train representations |
| Etrain | The train set of LMIs |
| Etest | The test set of LMIs |
| H | The feature vector set of Etrain |
4. Results and discussion
4.1. Evolution metrics
To evaluate the predictive performance of HINLMI from different perspectives, several evaluation metrics are introduced, including Accuracy, Matthew's correlation coefficient (MCC), Precision, Recall, and F1-score. The definition of them is defined as:
| (14) |
| (15) |
| (16) |
| (17) |
| (18) |
where TP, TN, FP and FN represent the numbers of true positives, true negatives, false positives and false negatives respectively for predicted LMIs.
Besides, AUC and AUPR are also used as important evaluation metrics, they are the areas under the Receiver Operating Characteristic (ROC) curve and the Precision-Recall (PR) curve respectively.
4.2. Evaluate performance
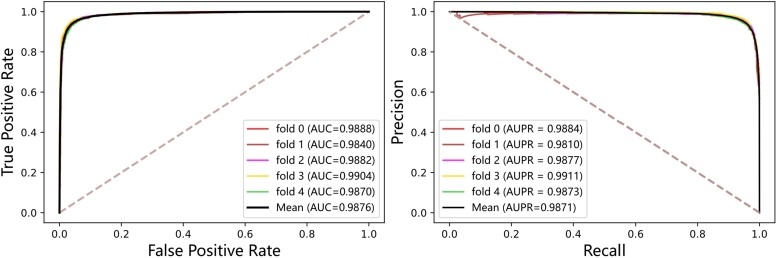
In the experiments, the performance of HINLMI is evaluated by following a 5-fold cross-validation (CV) scheme. Specifically, the benchmark dataset is first divided into 5-folds, and then each fold is alternatively taken as a testing dataset while the rest compose the training dataset. The experiment results of 5-fold CV on the MAN dataset are shown in Table 4 and Fig. 2. We note that the average results achieved by HINLMI in term of AUC, AUPR, Accuracy, MCC and F1-score are 98.76%, 98.71%, 95.02%, 90.04%, and 95.01%, respectively. More importantly, HINLMI yields a promising performance in terms of robustness and stability, as it obtains relatively small variances for these independent evaluation metrics.
Table 4.
The performance of HINLMI using 5-fold CV on the MAN dataset.
| Fold | Accuracy | MCC | Precision | Recall | F1-score |
|---|---|---|---|---|---|
| 1 | 0.9484 | 0.8969 | 0.9563 | 0.9379 | 0.9479 |
| 2 | 0.9472 | 0.8944 | 0.9534 | 0.9403 | 0.9468 |
| 3 | 0.9534 | 0.9069 | 0.9529 | 0.9540 | 0.9535 |
| 4 | 0.9558 | 0.9116 | 0.9564 | 0.9552 | 0.9558 |
| 5 | 0.9460 | 0.8921 | 0.9374 | 0.9558 | 0.9465 |
| Mean | 0.9502±0.0042 | 0.9004±0.0084 | 0.9513±0.0079 | 0.9490±0.0082 | 0.9501±0.0043 |
Fig. 2.
The ROC and PR curves of HINLMI using 5-fold CV on the MAN dataset.
Regarding the hardware environment, all experiments have been conducted on an AMD Ryzen machine with an 8-core CPU running at 3.9 GHz, 128 GB of RAM, and an NVIDIA GeForce GTX 2080 Ti GPU. The Python code was executed in an Anaconda3 environment.
4.3. Comparison with state-of-the-art models
To demonstrate the prediction ability of HINLMI, we compare it with several state-of-the-art baseline models, i.e., EPLMI [21], GNMFLMI [36], LMI-INGI [42], GCNCRF [37]. The work details of these comparing models are presented in Introduction. Regarding the parameter settings used for training, we explicitly adhere to the default values recommended in the original work to ensure a fair comparison. The experimental results of all competing models, evaluated under 5-fold cross-validation on the benchmark dataset, are presented in Table 5. We note that HINLMI yields the best performance across the benchmark dataset, as on average it gives 1.69%, 1.07%, 32.65%, and 5.94% relative improvement in AUC, respectively, over all baseline models.
Table 5.
The performance of all comparing models using 5-fold CV.
| Model | Accuracy | MCC | Precision | Recall | F1-score | AUC | AUPR |
|---|---|---|---|---|---|---|---|
| EPLMI | N/A | N/A | N/A | N/A | N/A | 0.9707±0.0051 | N/A |
| GNMFLMI | 0.9209±0.0052 | N/A | 0.9038±0.0063 | 0.9420±0.0049 | 0.9225±0.0048 | 0.9769±0.0037 | N/A |
| LMI-INGI | 0.5794±0.0061 | 0.2023±0.0074 | 0.6163±0.0057 | 0.5082±0.0046 | 0.5780±0.0049 | 0.6611±0.0062 | 0.6913±0.0053 |
| GCNCRF | 0.9316±0.0049 | 0.1050±0.0067 | 0.8729±0.0043 | 0.2727±0.0058 | 0.0945±0.0061 | 0.9282±0.0045 | 0.1710±0.0064 |
| MGCAT | 0.9038±0.0054 | 0.8147±0.0063 | 0.9087±0.0049 | 0.8979±0.0058 | 0.9033±0.0062 | 0.9414±0.0051 | 0.9059±0.0048 |
| HINLMI | 0.9502±0.0042 | 0.9004±0.0084 | 0.9513±0.0079 | 0.9490±0.0082 | 0.9501±0.0043 | 0.9876±0.0024 | 0.9871±0.0037 |
In addition to its superior accuracy, HINLMI also demonstrates greater robustness compared to other algorithms, as evidenced by its evaluation scores. For instance, when comparing HINLMI with GCNCRF, the scores for Accuracy and AUC are significantly higher, while the scores for MCC, F1-score, and AUPR are comparatively lower for GCNCRF. This phenomenon can be attributed to the fact that the number of positive samples correctly predicted by comparing models is much lower than that of HINLMI. In other words, HINLMI outperforms the comparing models, particularly in terms of its ability to discover novel LMIs, as indicated by its superior performance in Recall. Furthermore, the predictive power of GCNCRF is limited due to the inherent issue of over-smoothing in graph neural networks. In contrast, HINLMI demonstrates significantly lower performance fluctuations across all evaluation metrics compared to other prediction models. The robustness of HINLMI can be attributed to two key factors. Firstly, the integration of multiple molecular associations enables HINLMI to learn node features from both first-order and high-order similarity structures. This comprehensive perspective allows it to effectively predict unknown lncRNA-miRNA associations, benefiting from the rich biological and topological information available in the heterogeneous information network. Secondly, HINLMI functions as an effective ensemble model by incorporating the XGBoost classifier to perform the binary classification task. This ensemble approach contributes to the robustness and generalization ability of HINLMI, enhancing its performance across diverse scenarios.
Regarding the unsatisfactory performance of LMI-INGI for the LMIs prediction task, its operations conducted in the lncRNA-miRNA network have a two-fold effect. First, LMI-INGI requires more biological attributes in term of biological knowledge, such as sequence similarity, expression profiles and functional similarity of lncRNAs and miRNAs, but many biological attributes are lacking in practical application. In this regard, HINLMI have flexibility and robustness against feature representations of nodes by mining the first-order and high-order similar structure in given HIN can be enhanced as indicated by the experimental results. Second, the features obtained only in the light of the lncRNA-miRNAs association network, its expression power is weak, and it is difficult to capture outstanding feature representations.
To conduct the experiments with MGCAT, we first download the source codes from the GitHub repositories provided in their original work and compile these codes to run the prediction models for performance comparison. For parameter settings, we explicitly adopt the default values recommended in their original work to ensure a fair comparison. The experimental results of 5-fold cross-validation on the benchmark dataset are presented in Table 5. MGCAT achieves 90.38%, 81.47%, 90.87%, 89.79%, 90.33%, 94.14%, and 90.59% in terms of Accuracy, MCC, Precision, Recall, F1-score, AUC, and AUPR, respectively. On average, HINLMI performs better by 4.62% and 8.12% than MGCAT in terms of AUC and AUPR, respectively. We analyze that the possible reason why MGCAT performs lower than HINLMI is that the graph neural network mechanism adopted by MGCAT has difficulty capturing expressive features due to its inherent limitations.
In summary, the multiple molecular associations and their biological knowledge of HIN are benefited for HINLMI to correctly capture its full complexity and structural richness, and the applied graph representation learning algorithm allows HINLMI to seamlessly incorporate such features for learning high-quality network representations of lncRNAs and miRNAs. Thus, HINLMI yields a promising performance for identifying novel LMIs.
4.4. Ablation study
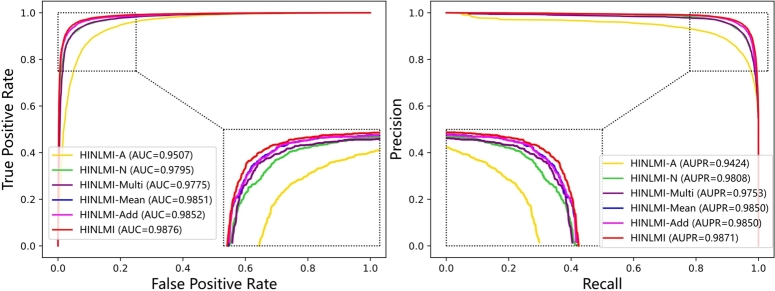
To investigate the impact of different feature representations in HINLMI, we conducted an ablation study with two variants: HINLMI-A and HINLMI-N. The primary difference between them lies in how they capture the feature representations of lncRNAs and miRNAs in HIN. In HINLMI-A, we only consider the biological knowledge of lncRNAs and miRNAs for representation learning. On the other hand, HINLMI-N solely incorporates the network representations of lncRNAs and miRNAs, omitting their biological knowledge. The experimental results of 5-fold cross-validation are presented in Table 4 and Fig. 3. Several observations can be made from these results. The performance of HINLMI-A is influenced by focusing exclusively on biological knowledge, while HINLMI-N relies solely on the network structure. By comparing their results with the original HINLMI, we gain insights into the individual contributions of biological knowledge and network topology in predicting lncRNA-miRNA interactions. This ablation study highlights the importance of integrating both biological knowledge and network structure in HINLMI, showcasing how diverse feature representations enhance its predictive capabilities for identifying potential lncRNA-miRNA interactions, where several things can be noted.
Fig. 3.
The ROC and PR curves for several variants of HINLMI were generated using 5-fold CV on the MAN dataset.
Firstly, HINLMI-A exhibits the lowest performance among HINLMI and its variants. This suggests that relying solely on the biological knowledge of lncRNAs and miRNAs may not be sufficient to achieve the desired prediction performance. Secondly, HINLMI-N demonstrates a considerable performance advantage over HINLMI-A in each evaluation metric. Specifically, HINLMI-N outperforms HINLMI-A by 2.88%, 3.84%, 4.42%, 8.76%, and 4.20% in terms of AUC, AUPR, Accuracy, MCC, and F1-score, respectively. This significant margin indicates that the network topology information represented by the HIN enables HINLMI-N to better capture the characteristics of lncRNAs and miRNAs during the training of the XGBoost classifier. Lastly, an additional improvement is observed in HINLMI by combining the strengths of both HINLMI-A and HINLMI-N. Comparing the performance of HINLMI-A with that of HINLMI-N, we deduce that it is the integration of more heterogeneous association information that contributes the most to the performance enhancement of HINLMI. In other words, HINLMI can comprehensively leverage the feature representations of lncRNAs and miRNAs, thus enhancing the predictive capability of the LMIs prediction task from different perspectives.
To effectively harness diverse features from various projection spaces, we analyze the optimal aggregation schemes for both biological and network representations of lncRNA and miRNAs. Specifically, we design four types of feature aggregation methods: HINLMI, HINLMI-Multi, HINLMI-Mean, and HINLMI-Add. These methods perform multiplication, mean, and addition operations on two types of representation vectors. Experimental trials employing different variants within a 5-fold CV framework demonstrate that HINLMI achieves optimal performance by concatenating the biological and network representations, as indicated in Table 6 and Fig. 3. Notably, the AUC value of HINLMI-Multi decreased by 1.01% and the AUPR value decreased by 1.18% compared to HINLMI, suggesting that the multiplication of biological and network representations may introduce incompatibilities and amplify noise, thereby leading to a loss of meaningful information and overall performance degradation.
Table 6.
The performance of several variants of HINLMI using 5-fold CV on the MAN dataset.
| Model | Accuracy | MCC | Precision | Recall | F1-score |
|---|---|---|---|---|---|
| HINLMI-A | 0.8885±0.0044 | 0.7778±0.0088 | 0.8725±0.0067 | 0.9101±0.0081 | 0.8909±0.0044 |
| HINLMI-N | 0.9327±0.0073 | 0.8654±0.0145 | 0.9358±0.0093 | 0.9291±0.0092 | 0.9324±0.0073 |
| HINLMI-Multi | 0.9319±0.0025 | 0.8639±0.0051 | 0.9255±0.0046 | 0.9395±0.0069 | 0.9324±0.0027 |
| HINLMI-Mean | 0.9442±0.0049 | 0.8884±0.0098 | 0.9473±0.0092 | 0.9408±0.0059 | 0.9440±0.0048 |
| HINLMI-Add | 0.9442±0.0056 | 0.8884±0.0112 | 0.9472±0.0099 | 0.9409±0.0056 | 0.9440±0.0054 |
| HINLMI | 0.9502±0.0042 | 0.9004±0.0084 | 0.9513±0.0079 | 0.9490±0.0082 | 0.9501±0.0043 |
In summary, the ablation study highlights the importance of incorporating both biological knowledge and network topology information for accurate prediction of lncRNA-miRNA interactions. The findings underscore the significance of leveraging diverse feature representations in HINLMI, leading to improved performance in identifying potential regulatory interactions between lncRNAs and miRNAs.
4.5. Heterogeneous network performance analysis
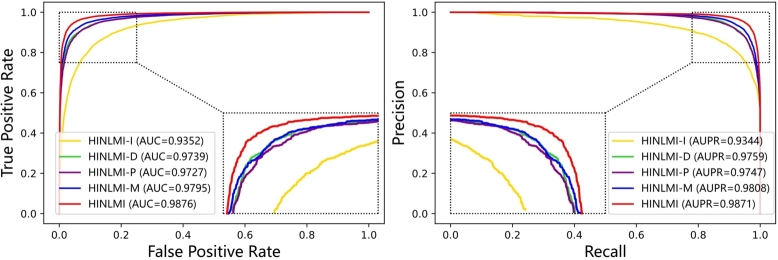
To better analysis the contribution of the given HIN for the performance of LMIs prediction, we construct this experiment to evaluate the importance of biological molecules. In particular, several sub-heterogeneous networks, i.e., HINLMI-I, HINLMI-P, HINLMI-D, and HINLMI-N, are designed by biological knowledge. HINLMI-I merely contains associations between lncRNAs and miRNAs, whereas HINLMI-P additionally integrates protein-related networks on the basis of HINLMI-I. Similar, HINLMI-D can enrich the information of HINLMI-I by incorporating drug-related associations. HINLMI-N consists of five types of nodes and nine association networks. Experimental results are also presented in Table 7 and Fig. 4, and several things are worth noting.
Table 7.
The performance of HINLMI on different heterogeneous network structure.
| Model | Accuracy | MCC | Precision | Recall | F1-score |
|---|---|---|---|---|---|
| HINLMI-I | 0.8636±0.0061 | 0.7273±0.0121 | 0.8633±0.0110 | 0.8641±0.0091 | 0.8637±0.0057 |
| HINLMI-P | 0.9198±0.0065 | 0.8380±0.0130 | 0.9246±0.0094 | 0.9124±0.0091 | 0.9184±0.0065 |
| HINLMI-D | 0.9197±0.0040 | 0.8395±0.0079 | 0.9247±0.0022 | 0.9138±0.0087 | 0.9192±0.0044 |
| HINLMI-N | 0.9327±0.0073 | 0.8654±0.0145 | 0.9358±0.0093 | 0.9291±0.0092 | 0.9324±0.0073 |
| HINLMI | 0.9502±0.0042 | 0.9004±0.0084 | 0.9513±0.0079 | 0.9490±0.0082 | 0.9501±0.0043 |
Fig. 4.
The ROC and PR curves of different heterogeneous network structure by HINLMI.
On the one hand, HINLMI-P and HINLMI-D have obvious improvement against the performance of LMIs predictors. On average, they perform better by 7.62%, 8.18%, 11.23%, 22.29%, 12.27%, 9.80%, 11.02% than HINLMI-I in term of AUC, AUPR, Accuracy, MCC and F1-score, respectively. Obviously, after the addition of protein or drug associations, rich network association information can provide additional pathways for lncRNAs and miRNAs. On the other hand, multi-molecular association network contributes to LMIs prediction most as its highest evaluation metrics. The reason is that due to HINLMI-N can integrate the advantages of HINLMI-P and HINLMI-D to enhance the expressive power of features of lncRNAs and miRNAs.
4.6. Parameter analysis
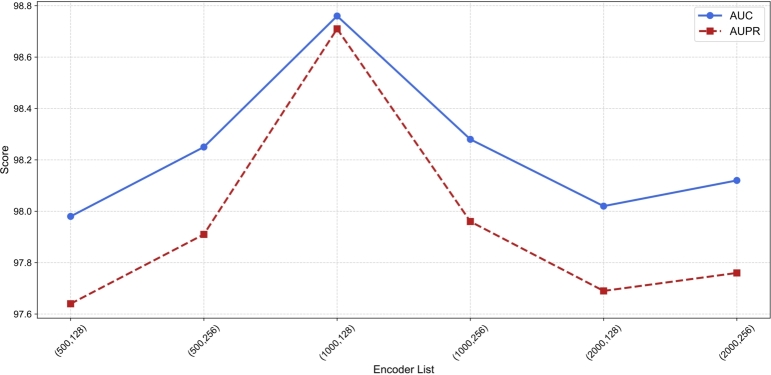
We perform an optical choice experiment for crucial parameters to achieve the optimal performance of HINLMI. As has been pointed out by [35], the encoder-list is considered the most critical hyperparameter because it directly determines the structure and representation of HINLMI. By fine-tuning the encoder-list, the performance of HINLMI can be significantly enhanced. Typically, the encoder-list specifies the number of neurons in each encoder layer and the dimension of the output node representation. Therefore, we construct a set (500, 128), (500, 256), (1000, 128), (1000, 256), (2000, 128), (2000, 256) to represent different combinations of two variables for the encoder-list. Fig. 5 illustrates the performance of HINLMI, showing that the highest AUC score of 98.76% and the highest AUPR score of 98.71% are achieved with the encoder-list configuration of (1000,128). This indicates that this configuration provides the most balanced and effective structure for HINLMI, optimizing its ability to capture complex relationships in the data. Additionally, the performance across different configurations shows significant variability, highlighting the importance of choosing the right encoder structure. The results demonstrate the sensitivity of HINLMI to the encoder-list configuration, underscoring the critical role of hyperparameter tuning in developing effective neural network models for complex tasks.
Fig. 5.
The AUC and AUPR values for different combinations of two variables in the encoder list.
4.7. Classifier selection
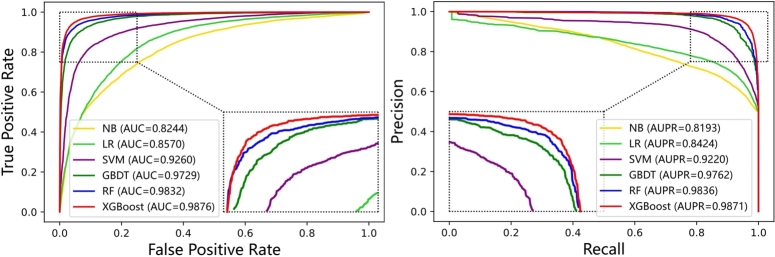
Given the availability of well-established classifiers, such as Naïve Bayes (NB), Logistic Regression (LR), Support Vector Machine (SVM), Gradient Boosting Decision Tree (GBDT), Random Forest (RF), and XGBoost, the selection of an appropriate classifier is crucial to achieve the best performance for HINLMI. To address this, we conducted experiments comparing the performance of HINLMI with different classifiers. The experimental results of 5-fold cross-validation are presented in Table 8 and Fig. 6. Overall, HINLMI demonstrates the best performance when employing XGBoost as its classifier. The decision to incorporate XGBoost into HINLMI for predicting novel LMIs is based on its superior performance compared to other classifiers. XGBoost's ability to handle complex data and exploit the rich features from heterogeneous information networks makes it well-suited for enhancing the predictive capability of HINLMI. Furthermore, several noteworthy points are worth mentioning. The consistent superiority of HINLMI with XGBoost as the classifier indicates the effectiveness of this combination in accurately predicting lncRNA-miRNA interactions. This suggests that the integration of a powerful classifier like XGBoost plays a crucial role in improving the performance of HINLMI. In summary, the experimental comparison of different classifiers highlights the significance of selecting XGBoost as the preferred classifier for HINLMI. Its strong performance, coupled with the inherent advantages of HINLMI, makes it a powerful and reliable method for predicting novel lncRNA-miRNA interactions in heterogeneous biological networks. Besides, there are several points worth further commentary.
Table 8.
The performance of HINLMI by using different classifiers.
| Model | Accuracy | MCC | Precision | Recall | F1-score |
|---|---|---|---|---|---|
| NB | 0.7459±0.0098 | 0.4920±0.0197 | 0.7464±0.0160 | 0.7457±0.0103 | 0.7459±0.0075 |
| LR | 0.7801±0.0044 | 0.5603±0.0086 | 0.7821±0.0111 | 0.7770±0.0128 | 0.7794±0.0039 |
| SVM | 0.8600±0.0084 | 0.7202±0.0168 | 0.8657±0.0133 | 0.8524±0.0125 | 0.8589±0.0082 |
| GBDT | 0.9187±0.0031 | 0.8376±0.0062 | 0.9240±0.0070 | 0.9126±0.0076 | 0.9182±0.0032 |
| RF | 0.9371±0.0044 | 0.8742±0.0087 | 0.9321±0.0083 | 0.9429±0.0047 | 0.9375±0.0041 |
| XGBoost | 0.9502±0.0042 | 0.9004±0.0084 | 0.9513±0.0079 | 0.9490±0.0082 | 0.9501±0.0043 |
Fig. 6.
The ROC and PR curves of HINLMI by using different classifiers.
First, among all classifiers, we note that the performance of NB and LR is the worst in terms of Accuracy, MCC, F1-score and AUC. The main reason for its unsatisfactory performance is that NB and LR have low fitting ability to heterogeneous information features and are not applicable for the LMIs prediction task. Second, the performances of SVM and GBDT are only worse than RF, but better than NB and LR. This could be a strong indicator that high-dimension features require more complex classifiers to fit it. Last, although RF is the second-best classifier, its ability of fault tolerance tends to become less efficient when the number of features increases.
4.8. Case studies
In this experiment, we have conducted additional experiments on the MAN Dataset to demonstrate the effectiveness of HINLMI in practical prediction task. Firstly, we select a lncRNA, i.e., nonhsat022132.2, which is closely associated with mainstream diseases. As a kind of malignant tumor, colon cancer usually tends to be found at the borders of sigmoid colon and rectum [29]. According to statistics, colon cancer has been the third common cancer in the United States, and it also becomes the third leading cause of cancer death for human [27]. All the time, patients with early stage colon cancer usually present subtle symptoms so that colon cancer is difficult to be detected. However, an upward trend for its incidence is reported [6]. Thus, computing-based methods are an effective measure by predicting miRNAs-lncRNAs interactions for colon cancer. In biological experiments, the available lncRNAs have been shown to have a significant regulatory effect on colon tumors. For instance, nonhsat022132.2 has been shown to be associated with colon cancer and to affect the growth of colon cancer cells [2]. Moreover, breast cancer and prostate cancer also are proved to be associated with ncRNAs molecules [40], [12].
Secondly, miRNAs with associated (a total 109 LMIs) the lncRNA (nonhsat022132.2) are removed from the MAN dataset, and then the remaining 8256 LMIs as a training set. In doing so, the generalization ability of HINLMI is proved by predicting unknown associations for new nodes. Finally, the associations between lncRNA (nonhsat022132.2) and all miRNAs are predicted by HINLMI, where the predicted results are present in Table 9. Obviously, HINLMI successfully predict 22 kinds of unknown LMIs for top-30 of predicted results. In this regard, HINLMI is a useful tool for the implementation of LMIs prediction task.
Table 9.
Top-30 predicted results for nonhsat022132.2.
| miRNAs | Confirmed | miRNAs | Confirmed |
|---|---|---|---|
| hsa-mir-3167 | yes | hsa-mir-367-3p | yes |
| hsa-mir-873-5p | yes | hsa-mir-25-3p | yes |
| hsa-mir-23c | yes | hsa-mir-4262 | yes |
| hsa-mir-19b-3p | no | hsa-mir-425-5p | yes |
| hsa-mir-301a-3p | no | hsa-mir-146a-5p | yes |
| hsa-mir-361-5p | no | hsa-mir-224-5p | yes |
| hsa-mir-329-3p | no | hsa-mir-346 | yes |
| hsa-mir-4465 | yes | hsa-mir-370-3p | yes |
| hsa-mir-144-3p | yes | hsa-mir-454-3p | no |
| hsa-mir-519a-5p | no | hsa-mir-149-5p | yes |
| hsa-mir-23b-3p | yes | hsa-mir-4725-5p | no |
| hsa-mir-374a-5p | yes | hsa-mir-202-3p | yes |
| hsa-mir-135b-5p | yes | hsa-mir-378b | yes |
| hsa-mir-4306 | yes | hsa-mir-378e | yes |
| hsa-mir-92b-3p | yes | hsa-mir-378e | no |
5. Conclusion
In this work, a novel computational method, namely HINLMI, is presented for lncRNA-miRNA interactions (LMIs) prediction based on neighborhood-level structural representation. To better capture the feature representations of lncRNAs and miRNAs from a more comprehensive perspective. HINLMI first integrates five kinds of biomolecules and their interaction and biological knowledge, thus composing a complicated HIN. After that, the biological and network representations of lncRNAs and miRNAs are obtained by different representation learning techniques from the perspectives of biological knowledge and network topology, respectively. Last, HINLMI combines the XGBoost classifier to predict new LMIs by jointly considering these two kinds of feature representations. Experimental results indicate that HINLMI yields the best performance than state-of-the-art computational models in terms of several evaluation metrics. Case studies further demonstrate that HINLMI a useful tool in the practical LMIs prediction task.
The importance of integrating biological information and network topology lies in our study's ability to enhance the accuracy of LMI predictions by combining these two elements. This approach demonstrates the significance of utilizing multiple information sources in biomedical predictions, offering new perspectives and methodologies for LMI predictions. The HINLMI model constructs a complex heterogeneous information network by integrating various biomolecules and their interactions. Compared to traditional single-network methods, HINLMI captures the intricate relationships between lncRNAs and miRNAs more effectively, exhibiting superior predictive performance and stability. In future work, we first plan to develop an end-to-end model to further improve the precision and efficiency of LMI predictions. Second, we will incorporate a wider range of biomolecular data and their associated information in future research. This will help to more vividly reconstruct complex biological regulatory networks and, consequently, develop richer datasets. By diversifying the datasets, we aim to better demonstrate the versatility and effectiveness of the proposed method. Last, we will consider viewing associations as semantic relationships based on biological information, further enhancing the model's applicability.
CRediT authorship contribution statement
Bo-Wei Zhao: Conceptualization, Methodology, Writing – original draft, Writing – review & editing. Xiao-Rui Su: Methodology, Software. Yue Yang: Resources, Software. Dong-Xu Li: Data curation, Investigation. Guo-Dong Li: Software, Validation. Peng-Wei Hu: Data curation, Software, Funding acquisition. Xin Luo: Conceptualization, Writing – original draft, Funding acquisition. Lun Hu: Conceptualization, Writing – original draft, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors would like to thank all anonymous reviewers for their constructive advice. This work was supported in part by the National Natural Science Foundation of China under grants 62373348, 62302495 and 62272078, in part by the Natural Science Foundation of Xinjiang Uygur Autonomous Region under grants 2021D01D05 and 2023D01E15, in part by the Tianshan Talent Training Program under grant 2023TSYCLJ0021, in part by the Xinjiang Tianchi Talents Program under grant E33B9401, in part by the Chonqing Natural Science Foundation under grant CSTB2023NSCQ-LZX0069, and in part by the Pioneer Hundred Talents Program of Chinese Academy of Sciences.
Contributor Information
Bo-Wei Zhao, Email: zhaobowei19@mails.ucas.ac.cn.
Xiao-Rui Su, Email: suxiaorui19@mails.ucas.ac.cn.
Yue Yang, Email: yangyue233@mails.ucas.ac.cn.
Dong-Xu Li, Email: lidongxu22@mails.ucas.ac.cn.
Guo-Dong Li, Email: liguodong22@mails.ucas.ac.cn.
Peng-Wei Hu, Email: hpw@ms.xjb.ac.cn.
Xin Luo, Email: luoxin@swu.edu.cn.
Lun Hu, Email: hulun@ms.xjb.ac.cn.
References
- 1.Berahmand K., Mohammadi M., Sheikhpour R., Li Y., Xu Y. Wsnmf: weighted symmetric nonnegative matrix factorization for attributed graph clustering. Neurocomputing. 2024;566 [Google Scholar]
- 2.Chen G., Wang Z., Wang D., Qiu C., Liu M., Chen X., et al. Lncrnadisease: a database for long-non-coding rna-associated diseases. Nucleic Acids Res. 2012;41:D983–D986. doi: 10.1093/nar/gks1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen T., Guestrin C. Proceedings of the 22nd ACM SIGKDD international conference on knowledge discovery and data mining. 2016. Xgboost: a scalable tree boosting system; pp. 785–794. [Google Scholar]
- 4.Chen X., Yan G.Y. Novel human lncRNA-disease association inference based on lncRNA expression profiles. Bioinformatics. 2013;29:2617–2624. doi: 10.1093/bioinformatics/btt426. [DOI] [PubMed] [Google Scholar]
- 5.Chen X., Zhu C.C., Yin J. Ensemble of decision tree reveals potential miRNA-disease associations. PLoS Comput Biol. 2019;15:1–23. doi: 10.1371/journal.pcbi.1007209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chong V., Abdullah M., Telisinghe P., Jalihal A. 2009. Colorectal cancer: incidence and trend in Brunei Darussalam. [PubMed] [Google Scholar]
- 7.Chou C.H., Shrestha S., Yang C.D., Chang N.W., Lin Y.L., Liao K.W., et al. miRTarBase update 2018: a resource for experimentally validated microRNA-target interactions. Nucleic Acids Res. 2018;46:D296–D302. doi: 10.1093/nar/gkx1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davis A.P., Grondin C.J., Johnson R.J., Sciaky D., King B.L., McMorran R., et al. The comparative toxicogenomics database: update 2017. Nucleic Acids Res. 2017;45:D972–D978. doi: 10.1093/nar/gkw838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fang S., Zhang L., Guo J., Niu Y., Wu Y., Li H., et al. Noncodev5: a comprehensive annotation database for long non-coding rnas. Nucleic Acids Res. 2018;46:D308–D314. doi: 10.1093/nar/gkx1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Forouzandeh S., Berahmand K., Sheikhpour R., Li Y. A new method for recommendation based on embedding spectral clustering in heterogeneous networks (reschet) Expert Syst Appl. 2023;231 [Google Scholar]
- 11.Forouzandeh S., Rostami M., Berahmand K., Sheikhpour R. Health-aware food recommendation system with dual attention in heterogeneous graphs. Comput Biol Med. 2024;169 doi: 10.1016/j.compbiomed.2023.107882. [DOI] [PubMed] [Google Scholar]
- 12.Gmyrek G.A., Walburg M., Webb C.P., Yu H.M., You X., Vaughan E.D., et al. Normal and malignant prostate epithelial cells differ in their response to hepatocyte growth factor/scatter factor. Am J Pathol. 2001;159:579–590. doi: 10.1016/S0002-9440(10)61729-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo Z.H., Yi H.C., You Z.H. Construction and comprehensive analysis of a molecular association network via lncrna–mirna–disease–drug–protein graph. Cells. 2019;8:866. doi: 10.3390/cells8080866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo Z.H., Yi H.C., You Z.H. Construction and comprehensive analysis of a molecular association network via lncRNA-miRNA -disease-drug-protein graph. Cells. 2019;8:866. doi: 10.3390/cells8080866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hao N.B., He Y.F., Li X.Q., Wang K., Wang R.L. The role of mirna and lncrna in gastric cancer. Oncotarget. 2017;8 doi: 10.18632/oncotarget.19197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holley C.L., Topkara V.K. An introduction to small non-coding rnas: mirna and snorna. Cardiovasc Drugs Ther. 2011;25:151–159. doi: 10.1007/s10557-011-6290-z. [DOI] [PubMed] [Google Scholar]
- 17.Hu L., Wang X., Huang Y.A., Hu P., You Z.H. A survey on computational models for predicting protein–protein interactions. Brief Bioinform. 2021;22 doi: 10.1093/bib/bbab036. [DOI] [PubMed] [Google Scholar]
- 18.Hu L., Zhang J., Pan X., Yan H., You Z.H. HiSCF: leveraging higher-order structures for clustering analysis in biological networks. Bioinformatics. 2020 doi: 10.1093/bioinformatics/btaa775. [DOI] [PubMed] [Google Scholar]
- 19.Hu P., Huang Y.A., Chan K.C.C., You Z.H. Learning multimodal networks from heterogeneous data for prediction of lncRNA-miRNA interactions. IEEE/ACM Trans Comput Biol Bioinform. 2019;5963:1. doi: 10.1109/tcbb.2019.2957094. [DOI] [PubMed] [Google Scholar]
- 20.Huang Y.A., Chan K.C., You Z.H. Constructing prediction models from expression profiles for large scale lncrna–mirna interaction profiling. Bioinformatics. 2018;34:812–819. doi: 10.1093/bioinformatics/btx672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang Y.A., Chan K.C., You Z.H. Constructing prediction models from expression profiles for large scale lncRNA-miRNA interaction profiling. Bioinformatics. 2018;34:812–819. doi: 10.1093/bioinformatics/btx672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang Y.A., Chan K.C., You Z.H., Hu P., Wang L., Huang Z.A. Predicting microrna–disease associations from lncrna–microrna interactions via multiview multitask learning. Brief Bioinform. 2021;22 doi: 10.1093/bib/bbaa133. [DOI] [PubMed] [Google Scholar]
- 23.Huang Z., Shi J., Gao Y., Cui C., Zhang S., Li J., et al. Hmdd v3. 0: a database for experimentally supported human microrna–disease associations. Nucleic Acids Res. 2019;47:D1013–D1017. doi: 10.1093/nar/gky1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang Q., Wang J., Wu X., Ma R., Zhang T., Jin S., et al. Lncrna2target: a database for differentially expressed genes after lncrna knockdown or overexpression. Nucleic Acids Res. 2015;43:D193–D196. doi: 10.1093/nar/gku1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kozomara A., Birgaoanu M., Griffiths-Jones S. miRBase: from microrna sequences to function. Nucleic Acids Res. 2019;47:D155–D162. doi: 10.1093/nar/gky1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li H., Wu B., Sun M., Ye Y., Zhu Z., Chen K. Multi-view graph neural network with cascaded attention for lncrna-mirna interaction prediction. Knowl-Based Syst. 2023;268 [Google Scholar]
- 27.Liu F., Yuan D., Wei Y., Wang W., Yan L., Wen T., et al. 2012. Systematic review and meta-analysis of the relationship between ephx1 polymorphisms and colorectal cancer risk. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miao Y.R., Liu W., Zhang Q., Guo A.Y. lncRNASNP2: an updated database of functional SNPs and mutations in human and mouse lncRNAs. Nucleic Acids Res. 2018;46:D276–D280. doi: 10.1093/nar/gkx1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Phipps A.I., Lindor N.M., Jenkins M.A., Baron J.A., Win A.K., Gallinger S., et al. Colon and rectal cancer survival by tumor location and microsatellite instability: the colon cancer family registry. Dis Colon Rectum. 2013;56:937. doi: 10.1097/DCR.0b013e31828f9a57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Piñero J., Bravo À., Queralt-Rosinach N., Gutiérrez-Sacristán A., Deu-Pons J., Centeno E., et al. Disgenet: a comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res. 2016 doi: 10.1093/nar/gkw943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quinn J.J., Chang H.Y. Unique features of long non-coding rna biogenesis and function. Nat Rev Genet. 2016;17:47–62. doi: 10.1038/nrg.2015.10. [DOI] [PubMed] [Google Scholar]
- 32.Steinegger M., Söding J. Mmseqs2 enables sensitive protein sequence searching for the analysis of massive data sets. Nat Biotechnol. 2017;35:1026–1028. doi: 10.1038/nbt.3988. [DOI] [PubMed] [Google Scholar]
- 33.Szklarczyk D., Morris J.H., Cook H., Kuhn M., Wyder S., Simonovic M., et al. The string database in 2017: quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Res. 2016 doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang J., Qu M., Wang M., Zhang M., Yan J., Mei Q. Proceedings of the 24th international conference on world wide web. 2015. Line: large-scale information network embedding; pp. 1067–1077. [Google Scholar]
- 35.Wang D., Cui P., Zhu W. Proceedings of the 22nd ACM SIGKDD international conference on knowledge discovery and data mining. 2016. Structural deep network embedding; pp. 1225–1234. [Google Scholar]
- 36.Wang M.N., You Z.H., Li L.P., Wong L., Chen Z.H., Gan C.Z. Gnmflmi: graph regularized nonnegative matrix factorization for predicting lncrna-mirna interactions. IEEE Access. 2020;8:37578–37588. [Google Scholar]
- 37.Wang W., Zhang L., Sun J., Zhao Q., Shuai J. Predicting the potential human lncrna–mirna interactions based on graph convolution network with conditional random field. Brief Bioinform. 2022;23 doi: 10.1093/bib/bbac463. [DOI] [PubMed] [Google Scholar]
- 38.Wishart D.S., Feunang Y.D., Guo A.C., Lo E.J., Marcu A., Grant J.R., et al. Drugbank 5.0: a major update to the drugbank database for 2018. Nucleic Acids Res. 2018;46:D1074–D1082. doi: 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wong L., Huang Y.A., You Z.H., Chen Z.H., Cao M.Y. Lnrlmi: linear neighbour representation for predicting lncrna-mirna interactions. J Cell Mol Med. 2020;24:79–87. doi: 10.1111/jcmm.14583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu N., Wang F., Lv M., Cheng L. Microarray expression profile analysis of long non-coding rnas in human breast cancer: a study of Chinese women. Biomed Pharmacother. 2015;69:221–227. doi: 10.1016/j.biopha.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 41.Yoon J.H., Abdelmohsen K., Gorospe M. Seminars in cell & developmental biology. Elsevier; 2014. Functional interactions among micrornas and long noncoding rnas; pp. 9–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang L., Liu T., Chen H., Zhao Q., Liu H. Predicting lncrna–mirna interactions based on interactome network and graphlet interaction. Genomics. 2021;113:874–880. doi: 10.1016/j.ygeno.2021.02.002. [DOI] [PubMed] [Google Scholar]
- 43.Zhao B.W., Hu L., You Z.H., Wang L., Su X.R. HINGRL: predicting drug–disease associations with graph representation learning on heterogeneous information networks. Brief Bioinform. 2022;23:1–15. doi: 10.1093/bib/bbab515. [DOI] [PubMed] [Google Scholar]