Abstract
Toluene o-xylene monooxygenase (ToMO) and phenol hydroxylase (PH) of Pseudomonas stutzeri OX1 act sequentially in a recombinant upper pathway for the degradation of aromatic hydrocarbons. The catalytic efficiency and regioselectivity of these enzymes optimize the degradation of growth substrates like toluene and o-xylene. For example, the sequential monooxygenation of o-xylene by ToMO and PH leads to almost exclusive production of 3,4-dimethylcatechol (3,4-DMC), the only isomer that can be further metabolized by the P. stutzeri meta pathway. We investigated the possibility of producing ToMO mutants with modified regioselectivity compared with the regioselectivity of the wild-type protein in order to alter the ability of the recombinant upper pathway to produce methylcatechol isomers from toluene and to produce 3,4-DMC from o-xylene. The combination of mutant (E103G)-ToMO and PH increased the production of 4-methylcatechol from toluene and increased the formation of 3,4-DMC from o-xylene. These data strongly support the idea that the products and efficiency of the metabolic pathway can be controlled not only through mutations that increase the catalytic efficiency of the enzymes involved but also through tuning the substrate specificity and regioselectivity of the enzymes. These findings are crucial for the development of future metabolic engineering strategies.
Bacterial multicomponent monooxygenases (BMMs) catalyze the chemically difficult hydroxylation of aliphatic and aromatic hydrocarbons, which is crucial for the ability of several bacterial strains to grow on aliphatic and aromatic hydrocarbons (5, 14, 17, 21).
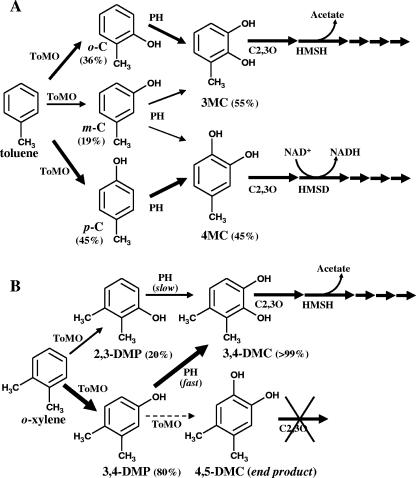
The regioselectivity of these enzymes may be the molecular basis for explaining (i) the ability of microorganisms to grow on selected substrates or (ii) the metabolic efficiency of the use of growth substrates (12, 20, 21). For example, in the case of Pseudomonas mendocina KR1, the restricted regioselectivity (10) on toluene of toluene 4-monooxygenase (T4MO), which is a para directing enzyme, is essential because only p-cresol can be further metabolized through progressive oxidation of the p-methyl group (20, 21). Moreover, BMMs from other organisms, such as Pseudomonas stutzeri and Burkohlderia sp. strain JS150, are able to further hydroxylate cresols to 3-methylcatechol (3-MC) and/or 4-methylcatechol (4-MC) (Fig. 1A), which are metabolized through the ortho or meta pathway (1, 2, 6). At least in the case of the meta pathway, 3-MC and 4-MC are not metabolically equivalent because they are metabolized through different branches of the pathway (Fig. 1A) (12). Thus, the metabolic fate of a growth substrate like toluene is determined by the ability of the enzymes involved in toluene hydroxylation to produce one of the two methylcatechol isomers and hence by their regioselectivity.
FIG. 1.
Proposed transformations of toluene (A) and o-xylene (B) catalyzed by ToMO and PH. The thickness of the arrows is roughly proportional to the relative abundance of each species. The possible catabolic fate of the hydroxylation products through the meta pathway is also shown. HMSD, 2-hydroxymuconic semialdehyde dehydrogenase which produces NADH; HMSH, 2-hydroxymuconic semialdehyde hydrolase which produces acetate (12); o-C, o-cresol; m-C, m-cresol; p-C, p-cresol.
P. stutzeri OX1, like a few other bacteria that are able to grow on aromatic molecules, possesses two BMMs, toluene o-xylene monooxygenase (ToMO) and phenol hydroxylase (PH). Both of these enzymes are capable of oxidizing substituted benzenes to catechols, which are subsequently metabolized through the meta pathway (1, 2). In the accompanying paper (4) we show that ToMO and PH act sequentially on toluene, catalyzing its oxidation to an equimolar mixture of 3-MC and 4-MC (Fig. 1A). This is mainly due to the ability of ToMO to produce similar amounts of o- and p-cresols, which are converted by PH to 3-MC and 4-MC, respectively (4). Both methylcatechols can be cleaved by P. stutzeri catechol 2,3-dioxygenase (C2,3O) and enter the two branches of the meta pathway (1). Moreover, we also show in the accompanying paper (4) that the combined action of ToMO and PH converts o-xylene almost exclusively to 3,4-dimethylcatechol (3,4-DMC) (Fig. 1B), the sole dimethylcatechol that can be cleaved by P. stutzeri C2,3O, thus allowing P. stutzeri OX1 to grow on the most recalcitrant xylene isomer (1). This depends on the regioselectivity of ToMO, which converts o-xylene predominantly into 3,4-dimethylphenol (3,4-DMP), the optimal substrate for PH, and on the regioselectivity of PH, which transforms 3,4-DMP mainly into 3,4-DMC (Fig. 1B).
In this paper we report the use of protein engineering techniques to produce ToMO variants with altered regioselectivity compared with the regioselectivity of the wild-type protein. This was done by searching for ToMO mutants which could alter the ability of the metabolic chain consisting of ToMO and PH to produce methylcatechol isomers from toluene and to produce 3,4-DMC from o-xylene. We discovered that the use of mutant (E103G)-ToMO and PH together increased the production of 4-MC from toluene and, more interestingly, improved the conversion of o-xylene to 3,4-DMC, the only dimethylcatechol which can be metabolized further by the meta pathway of P. stutzeri. Thus, our data indicate that rational design of ToMO mutants with altered regioselectivity can be used to tune the relative abundance of the products of toluene and o-xylene hydroxylation. These findings are crucial for the development of future metabolic engineering strategies.
MATERIALS AND METHODS
Materials.
Bacterial culturing, plasmid purification, and transformation were performed as described by Sambrook et al. (13). Escherichia coli strains JM109 and CJ236 and vector pET22b(+) were obtained from Novagen. Plasmid pBZ1260 (2), which was used to express the ToMO cluster, was kindly supplied by P. Barbieri (Dipartimento di Biologia Strutturale e Funzionale, Università dell'Insubria, Varese, Italy). Plasmid pGEM-3Z/PH, which was used to express the PH complex, was prepared as described in the accompanying paper (4). E. coli strain JM101 was purchased from Boehringer. The pGEM-3Z expression vector and Wizard SV gel and PCR clean-up system for elution of DNA fragments from the agarose gel were obtained from Promega. Enzymes and other reagents used for DNA manipulation were obtained from New England Biolabs. The oligonucleotides were synthesized at the Stazione Zoologica ‘A. Dohrn’ (Naples, Italy). All other chemicals were obtained from Sigma. The methods used for expression and purification of recombinant C2,3O from P. stutzeri OX1 are described elsewhere (19).
Analysis of ToMO structure and docking of (di)methylphenols.
The ToMO H structure (PDB code 1T0Q) (15) was analyzed by using Swiss PDB viewer (http://www.expasy.org/spdbv/) and Pymol (DeLano Scientific LLC). Residues of the active site pocket were identified using the program Pymol. Phenol, o-, m-, and p-cresols, and 2,3-, 2,4-, and 3,4-DMPs were manually docked using Pymol. (Di)methylphenols were assumed to bind to ToMO in the anionic form. The phenolate oxygen was placed at 2.19 Å from Fe(1) and at 2.09 Å from Fe(2) (see Fig. 2) in the position that is occupied by the bridging oxygen of the thioglycolate molecule in the crystallographic complex. The aromatic ring was rotated using the phenolate oxygen as the rotation center in order to search for conformations which fit the van der Waals volume of the substrate in the active site cavity.
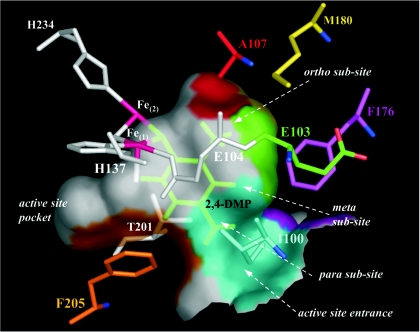
FIG. 2.
Active site of ToMO A with a docked 2,4-DMP molecule (green). The carbon atoms of the residues that take part in the formation of the hypothetical subsites for methyl groups are red, yellow, green, magenta, orange, and cyan, whereas nitrogen and oxygen atoms of these residues are blue and light red, respectively. Iron ions are purple. All other atoms are white. The numbers of the residues are also shown. The color of the surface of each active site pocket corresponds to the color of the residues that form the cavity. Hydrogen atoms are shown only on the 2,4-DMP molecule.
Coordinates for the (di)methylphenols were generated by using the programs CS ChemDraw Pro and Chem3D Pro (Cambridge Soft Corporation) and energy minimized before docking of the compounds in the active site.
Sequence alignments of the multicomponent monooxygenase α-subunits.
A multiple-sequence alignment was prepared and analyzed as described previously using the same set of 33 α-subunit sequences reported by Notomista et al. (9). The pairwise alignment of ToMO A and T4MO A was extracted from the multiple alignment of α-subunits and was used to identify active site residues which are different in the two proteins.
ToMO A mutagenesis.
ToMO A mutants were produced by site-directed mutagenesis by the method of Kunkel (7). Initially, a pGEM-3Z derivative lacking the unique SalI site (pGEM-3Z-Sal−) which would have interfered with the subsequent cloning procedures described below was prepared. The commercial vector pGEM-3Z was digested with the SalI restriction endonuclease. The protruding ends were filled in with Klenow DNA polymerase, and the linearized plasmid was ligated with DNA ligase. The ToMO coding sequence was excised from plasmid pBZ1260 by digestion with XbaI and KpnI. The resulting fragment was ligated to plasmid pGEM3Z-Sal− digested with the same enzymes. The resulting vector, designated pTou, contains a single SalI site located 138 bp upstream of the stop codon of the open reading frame coding for the ToMO A subunit (touA).
pTou was digested with MluI and SalI to extract a 1,200-bp internal fragment from the touA open reading frame. This fragment lacked the first 36 bp and the last 138 bp of the coding sequence and coded for amino acids 14 to 451 of the ToMO A subunit. The MluI/SalI fragment was cloned into the pET22b(+) commercial vector, and the resulting plasmid was designated pET22b(+)/touA. This plasmid was used to obtain mutants (E103G)-, (E103L)-, and (E103M)-ToMO A. The following mutagenic oligonucleotides were used: 5′-TGCGGCGTATTCTCCAAGTGCGATCGC-3′ for the E103G mutation, 5′-TGCGGCGTATTCAAGAAGTGCGATCGC-3′ for the E103L mutation, and 5′-TGCGGCGTATTCCATAAGTGCGATCGC-3′ for the E103M mutation. The MluI/SalI fragments of the three mutants were completely sequenced (MWG-Biotech), digested with MluI and SalI, and cloned back into the pTou plasmid previously digested with the same enzymes. The resulting plasmids were designated pTou/touA-(E103G), -(E103L), and -(E103M).
Identification of reaction products and determination of the apparent kinetic parameters.
All the assays were performed using whole E. coli JM109 cells transformed with plasmids pGEM-3Z/PH, pBZ1260, pTou/touA-(E103G), pTou/touA-(E103L), and pTou/touA-(E103 M), which expressed PH, ToMO, (E103G)-ToMO, (E103L)-ToMO, and (E103M)-ToMO, respectively. Details are described in the accompanying paper (4). The specific activities of the cells ranged from 10 to 14 mU/optical density at 600 nm for cells expressing PH and from 10 to 20 mU/optical density at 600 nm for cells expressing wild-type ToMO and ToMO mutants. One milliunit was defined as the amount of catalyst that oxidized one nmol of phenol per min at 25°C. The levels of expression of ToMO mutants, determined as described in the accompanying paper (4), were similar to the levels of expression of wild-type ToMO. The reaction products and the apparent kinetic parameters of the ToMO mutants were determined as reported for wild-type ToMO in the accompanying paper (4).
Time course of toluene and o-xylene oxidation.
The rate of product formation by E. coli cells expressing ToMO mutants or a mixture of ToMO (either wild-type or mutant enzyme) and wild-type PH was measured by the high-performance liquid chromatography (HPLC) discontinuous assay described in the accompanying paper (4). Each assay mixture contained either 30 μM toluene or 20 μM o-xylene. All cells were used at a concentration of 1 mU/ml when toluene was used as the substrate and at a concentration of 1.5 mU/ml when o-xylene was used.
Rate of (di)methylcatechol production as a function of ToMO concentration.
The rate of 3,4-DMC production from 40 μM o-xylene was measured as a function of the ToMO or (E103G)-ToMO concentration at concentrations ranging from 0.3 to 6 mU/ml with a constant concentration of cells expressing PH (0.5 mU/ml). 3,4-DMC was measured by the continuous coupled assay with C2,3O (3 U/ml) described in the accompanying paper (4). Experimental data were fitted to the following equation: rate = (Vmax × [ToMO])/(K + [ToMO]), where Vmax is the maximum rate and K is a constant equal to the concentration of ToMO which gives a rate of Vmax/2. Data were linearized by plotting rates as a function of the rate/[ToMO] ratio.
RESULTS
Design of ToMO mutants.
The recently determined X-ray crystal structure of the hydroxylase component of ToMO (ToMO H) (15) allows analysis of the active site pocket, which may provide a better understanding of the substrate regiospecificity and assist in the design of mutants that improve the catalytic efficiency with aromatic substrates. The active site cavity, which is in the A subunit of ToMO H, has an irregular, round, flat shape with a radius greater than that of a single benzene ring, and it is in close proximity to the catalytic diiron center (Fig. 2). Manual docking of alcohol products like phenol, cresols, and dimethylphenols into the active site such that the aromatic hydroxyl group was positioned at the bridging position ∼2.0 Å from each of the iron atoms provided insight into how the toluene and o-xylene substrates may be oriented before hydroxylation. In all cases, van der Waals contacts were minimized when the aromatic ring was positioned orthogonal to the plane defined by the iron atoms and the bridging hydroxide.
When cresols are docked in this cavity, the methyl group of o-cresol can be accommodated in a putative subsite between the methylene groups of the E103 side chain and the side chains of A107 and M180. The methyl group of m-cresol is accommodated in a subsite between the methylene groups of the E103 side chain and the side chain of F176 (Fig. 2). When p-cresol is docked in the same cavity, its methyl group is accommodated in a subsite between the side chains of F205 and I100, which are positioned at the interface between the active site pocket and the long channel that provides access to the diiron center from the surface of the protein (15). The positions of the methyl groups from the docked 2,3-, 2,4-, and 3,4-DMP products are also in agreement with the defined ortho, meta, and para subsites described above. Previous mutation studies of position 103 of T4MO A (8, 16) and of position 100 of ToMO A (18) indicated that changes at these positions alter the distribution of cresol isomers produced from toluene. In the case of the A subunit of T4MO, the G103L mutation increases the percentage of o-cresol from less than 1 to 55%, whereas in the case of the A subunit of ToMO the I100Q mutation increases the percentage of m-cresol from 19 to 44%.
To experimentally check whether changes in these subsites affect the regioselectivity of the enzyme, we changed residue E103 of ToMO A to glycine, the residue present at the homologous position of T4MO A, and to leucine and methionine, which are the only two residues found at homologous positions in 15 monooxygenases belonging to the subfamily containing the toluene 2-monooxygenases/phenol hydroxylases (9). Three ToMO variants, (E103G)-ToMO, (E103M)-ToMO, and (E103L)-ToMO, were produced by site-directed mutagenesis and expressed.
Apparent kinetic parameters and regioselectivity of ToMO mutants.
Tables 1 and 2 show the apparent kinetic constants of the three mutants for phenol, cresols, benzene, toluene, and o-xylene. Mutations E103M and E103L caused a threefold decrease in the kcat for phenol and a fivefold increase in the Km for phenol, respectively. All the other apparent kinetic parameters are identical or very similar to those measured for wild-type ToMO. Apparent kinetic constants for cresols were measured only for the E103G mutant (Table 2), and they were found to be identical to those measured for wild-type ToMO.
TABLE 1.
Apparent catalytic parameters of ToMO and ToMO mutants with benzene, toluene, and o-xylene
| Enzyme |
kcat(s−1) witha:
|
||
|---|---|---|---|
| Benzene | Toluene | o-Xylene | |
| ToMO | 0.36 | 0.42 | 0.25 |
| (E103G)-ToMO | 0.43 | 0.42 | 0.42 |
| (E103L)-ToMO | 0.29 | 0.32 | 0.36 |
| (E103M)-ToMO | 0.26 | 0.30 | 0.2 |
The errors for kcat values were about 26%.
TABLE 2.
Apparent kinetic constants of ToMO and ToMO mutants for phenol and cresols
| Compound | Parameter | ToMO | (E103G)-ToMO | (E103L)-ToMO | (E103M)-ToMO |
|---|---|---|---|---|---|
| Phenol | Km(μM)a | 2.18 | 2.7 | 15 | 2.5 |
| kcat(s−1)b | 1.0 | 1.0 | 0.89 | 0.29 | |
| kcat/Km(s−1μM−1) | 0.46 | 0.37 | 0.064 | 0.1 | |
| o-Cresol | Km(μM)a | 6 | 11.2 | ||
| kcat(s−1)b | 0.9 | 1.5 | |||
| kcat/Km(s−1μM−1) | 0.15 | 0.13 | |||
| m-Cresol | Km(μM)a | 9.38 | 13.2 | ||
| kcat(s−1)b | 0.44 | 1 | |||
| kcat/Km(s−1μM−1) | 0.047 | 0.076 | |||
| p-Cresol | Km(μM)a | 13.3 | 11 | ||
| kcat(s−1)b | 0.63 | 0.59 | |||
| kcat/Km(s−1μM−1) | 0.047 | 0.054 |
The errors for Km values were less than 10%.
The errors for kcat values were about 26%. The errors for the kcat values for phenol were about 20%.
In contrast, large variations were observed in the regioselectivity of the three mutants on toluene, o-xylene, m-cresol, and 3,4-DMP (Table 3). Mutation E103G increased the frequency of hydroxylation at positions para to methyl groups. The percentage of p-cresol obtained from toluene was about doubled, whereas the percentage of 3,4-DMP increased up to 99%. The same increase could be measured, although it was less evident, when hydroxylated substrates such as m-cresol and 3,4-DMP were used. Also, mutation E103L caused a similar, more limited increase in the frequency of para hydroxylation of toluene and o-xylene. Finally, mutation E103M caused inversion of the percentages of o- and p-cresols produced from toluene but no change in the distribution of dimethylphenols produced from o-xylene.
TABLE 3.
Regioselectivities of PH and ToMO (wild type and mutants) for toluene, m-cresol, o-xylene, and 3,4-DMP
| Substrate | Product | % witha:
|
|||
|---|---|---|---|---|---|
| ToMO | (E103G)-ToMO | (E103L)-ToMO | (E103M)-ToMO | ||
| Toluene | o-Cresol | 36 | 9 | 20 | 47 |
| m-Cresol | 19 | 6 | 11 | 19 | |
| p-Cresol | 45 | 85 | 69 | 34 | |
| o-Xylene | 2,3-DMP | 20 | 1 | 6 | 18 |
| 3,4-DMP | 80 | 99 | 94 | 82 | |
| m-Cresol | 3-MC | 5 | 2 | NDb | ND |
| 4-MC | 95 | 98 | ND | ND | |
| 3,4-DMP | 3,4-DMC | 10 | 1 | ND | ND |
| 4,5-DMC | 90 | 99 | ND | ND | |
The errors were ∼1%.
ND, not determined.
Toluene and o-xylene metabolism by ToMO mutant/PH mixtures.
Mixtures of E. coli cells expressing each ToMO mutant and cells expressing wild-type PH were incubated with toluene, and the formation of methylcatechols was monitored by HPLC as described above for wild-type ToMO and PH. The results shown in Table 4 indicate that the E103M mutation slightly increased 3-MC production, whereas the E103L and E103G mutations reduced the relative production of 3-MC 1.8- and 4-fold, respectively.
TABLE 4.
Regioselectivities of PH/ToMO (wild type and mutant) mixtures on toluene
| Product | % witha:
|
|||
|---|---|---|---|---|
| Wild-type ToMO/PH | (E103G)-ToMO/PH | (E103L)-ToMO/PH | (E103M)-ToMO/PH | |
| 3-MC | 55 (54)b | 14 (14) | 29 (30) | 67 (65) |
| 4-MC | 45 (46) | 86 (86) | 71 (70) | 33 (35) |
The errors were ∼1.5%.
The values in parentheses are theoretical percentages of 3- and 4-MC calculated by assuming that ToMO (wild type and mutants) converts toluene to cresols and PH converts cresol isomers to MC. Theoretical values were calculated with the following equation: 3-MC% = o-C% + 0.96 × m-C%, where 3-MC% is the percentage of 3-MC compared with the total amount of methylcatechols produced and o-C% and m-C% are the percentages of o-cresol and m-cresol, respectively, compared with the total amount of cresols produced by ToMO. For details see the accompanying paper (4).
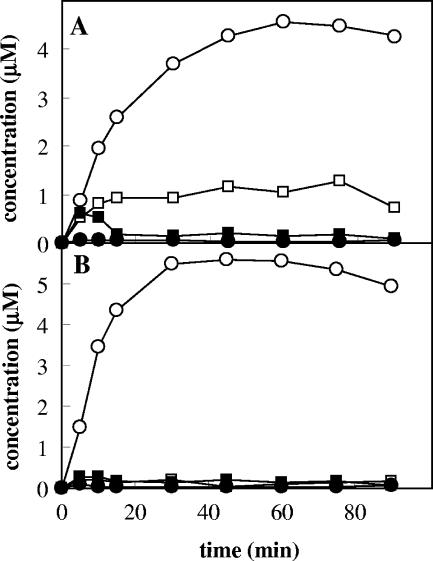
For o-xylene, the substrate at a concentration of 20 μM was incubated with a mixture of E. coli cells expressing (E103G)-ToMO (1.5 mU/ml) and cells expressing PH (1.5 mU/ml). The concentrations of 2,3- and 3,4-DMPs and 3,4- and 4,5-DMCs were measured by HPLC at different times (Fig. 3B). In contrast to the results obtained with wild-type ToMO and PH (Fig. 3A) (4), no 2,3-DMP accumulation was detected, and the rate of formation of 3,4-DMC was doubled.
FIG. 3.
Kinetics of 2,3-DMP (□), 3,4-DMP (▪), 3,4-DMC (○), and 4,5-DMC (•) production by a mixture of cells expressing ToMO (1.5 mU/ml) and PH (1.5 mU/ml) (A) and a mixture of cells expressing (E103G)-ToMO (1.5 mU/ml) and PH (1.5 mU/ml) (B) in the presence of 20 μM o-xylene.
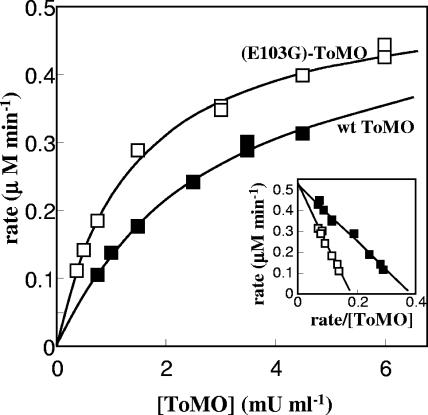
To confirm these results, mixtures of a constant amount of E. coli cells expressing PH (0.5 mU/ml) and different concentrations of E. coli cells expressing (E103G)-ToMO or wild-type ToMO (0.3 to 6 mU/ml) were incubated with 40 μM o-xylene. The rate of 3,4-dimethylcatecol formation was measured spectrophotometrically using the coupled assay with C2,3O. In both cases a hyperbolic dependence of the rate of 3,4-DMC formation was observed (Fig. 4). However, the rate of 3,4-DMC formation was higher when (E103G)-ToMO and PH were used than when wild-type ToMO and PH were used. The same maximum rate (Fig. 4, inset) was obtained in both cases, but the maximum rate was reached with lower concentrations of (E103G)-ToMO. Larger differences in the reaction rates with (E103G)-ToMO and ToMO were observed at low (E103G)-ToMO concentrations. The difference decreased as the ToMO concentration increased, likely because at higher ToMO concentrations the concentration of PH becomes the rate-limiting factor (4).
FIG. 4.
Kinetics of 3,4-DMC formation. Cells expressing PH were used at a constant concentration (0.5 mU/ml), and the rate of 3,4-DMC formation was measured as a function of the concentration of cells expressing ToMO (▪) or (E103G)-ToMO (□) in the presence of 40 μM o-xylene. The rate of 3,4-DMC production was measured by the continuous coupled assay with C2,3O as described in Materials and Methods. The inset shows linearization of the data obtained by plotting rates (μM min−1) as a function of the rate/[ToMO] ratio (μM min−1/mU ml−1). wt, wild type.
DISCUSSION
In the accompanying paper we show that ToMO and PH act sequentially to convert (di)methylbenzenes to (di)methylcatechols. In the case of toluene, ToMO produces similar amounts of o- and p-cresols and a smaller amount of m-cresol. On the other hand, PH converts p-cresol to 4-MC and o- and m-cresols to 3-MC. Thus, the combined action of the two enzymes yields an equimolar mixture of 3-MC and 4-MC from toluene. For the hydroxylation of o-xylene, the sequential action of the two enzymes mainly produces 3,4-DMC, which is the only dimethylcatechol isomer which can be further transformed by C2,3O. However, hydroxylation of o-xylene by ToMO yields 3,4-DMP, the optimal substrate for PH, and also small amounts of 2,3-DMP, which transiently accumulates during o-xylene degradation.
Protein engineering was performed to explore the possibility of obtaining ToMO mutants capable of producing only 3-MC or 4-MC from toluene and of more efficiently producing 3,4-DMC from o-xylene when the enzyme was coupled to wild-type PH. ToMO was chosen as the target for protein engineering studies because it is more efficient with nonhydroxylated molecules and is the primary agent in the channeling of aromatic molecules into the lower pathway of P. stutzeri. The E103G, E103M, and E103L mutations in the ToMO A subunit did not significantly change the efficiency of the enzyme in the first hydroxylation step; the kcat values for benzene, toluene, and o-xylene were very similar to those measured for wild-type ToMO (Table 1). The apparent kinetic constants for cresols were measured only for the E103G mutant (Table 2), and they are identical to those measured for wild-type ToMO.
Although the mutations in the ToMO Α subunit did not significantly influence the catalytic parameters, they caused major alterations in ToMO regioselectivity (Table 3). The E103G mutation caused the greatest change compared with wild-type ToMO. The regioselectivity of this mutant resembled that of T4MO, which is highly para directing (11). This finding was not unexpected since residue 103 is one of the two active site residues that are different in the sequences of ToMO, T4MO, and toluene para-monooxygenase (formerly toluene 3-monooxygenase); the other different residue is at position 180 and is methionine, isoleucine, and phenylalanine, respectively (15). The regioselectivity of the (E103L)-ToMO mutant was intermediate between those of (E103G)-ToMO and wild-type ToMO. In contrast, the similar mutation E103M caused a slight increase in the o-cresol relative abundance compared with the amount obtained with ToMO.
The regiospecific effects of the ToMO A mutations at position 103 on toluene hydroxylation (Table 3) may be explained by the docking of (di)methylphenols in the active site pocket of ToMO. The relaxed regiospecificity of ToMO indicates that at least three different positions in the active site pocket can accommodate the methyl group of toluene. Our docking results suggest that there are three subsites that could orient the methyl groups of the different substrates such that the ortho, meta, or para carbon is presented to the diiron center (Fig. 2). Thus, it may be hypothesized that it is the difference in the affinity of each subsite for the binding of the methyl groups that determines the relative abundance of the three enzyme-substrate complexes and hence the observed distribution of cresols produced by wild-type ToMO. Consequently, the low percentage of m-cresol produced by ToMO may suggest that the meta subsite has a lower affinity for methyl groups than the ortho and para subsites.
Thus, the reduced hydroxylation at the ortho and meta positions of toluene that we observed with mutant E103G (Table 3) may be explained on the basis of destabilization of the interaction between the methyl group of the substrate and the ortho and meta subsites given the reduced nonpolar portion of the residue 103 side chain (Fig. 2).
On the other hand, only minor changes in regioselectivity, such as a small increase in o-cresol production, were observed when E103 was changed to methionine (Table 3). This may be attributed to stabilization of the interaction at the ortho subsite, given the presence of a quasi-isosteric but more hydrophobic residue at position 103. For mutation E103L, the reduction of o- and m-cresols suggests that there is destabilization of the binding at the ortho and meta subsites. This finding is surprising because leucine has a van der Waals volume and hydrophobicity similar to those of methionine and is the residue that is most frequently found at homologous positions in the members of the toluene 2-monooxygenase subfamily (14 of 15 sequences). Moreover, it has been reported that the T4MO G103L mutation causes an increase in the percentage of o-cresol produced from toluene (8). Therefore, the G103L and E103L mutations cause opposite effects in T4MO and ToMO in spite of the fact that the A subunits of the two enzymes exhibit about 67% identity (3) and the fact that the residues at positions 103 and 180 are the only different active site residues in the two enzymes. The substitution of methionine for isoleucine at position 180 could explain the observed differences between (G103L)-T4MO and (E103L)-ToMO because the side chain of this residue also contributes to the ortho subsite (Fig. 2).
The changes in the regioselectivity of ToMO mutants have remarkable effects on toluene metabolism when these mutants are used in conjunction with wild-type PH. We found that mutation E103M results in a slight increase in 3-MC production, whereas mutations E103L and E103G reduce the formation of 3-MC. It is worth noting that the (E103G)-ToMO/PH combination produces essentially 4-MC.
Thus, the regulation of ToMO regioselectivity alone allows fine-tuning of the relative abundance of 3-MC and 4-MC. This understanding is crucial for the development of future metabolic engineering strategies.
The production of 2,3-DMP by the E103L and E103G mutants from o-xylene was 3.3- and 20-fold less, respectively, than the production of 2,3-DMP by the wild-type enzyme (Table 3). Formation of 2,3-DMP requires the binding of o-xylene methyl groups to the ortho and meta subsites of ToMO A. However, as shown above, mutations E103L and E103G destabilize the interactions of methyl substituents with these two subsites.
The changes in regioselectivity in o-xylene transformations by the E103L and E103G mutants are metabolically relevant because they lead to an increase in the production of 3,4-DMP (Table 3), which is efficiently transformed by PH in the second step of the reaction into 3,4-DMC, the only dimethylcatechol which can be further transformed by C2,3O, as described in the accompanying paper (4). The results in Fig. 3 and 4 clearly show that use of the (E103G)-ToMO mutant coupled with PH improved the efficiency of conversion of o-xylene to 3,4-DMC. It is worth noting that the concentrations of o-xylene used (20 to 40 μM) are saturating for both ToMO and (E103G)-ToMO (data not shown). Therefore, the results that we obtained cannot be attributed to variation in the Km of (E103G)-ToMO for o-xylene. Moreover, as (E103G)-ToMO has a kcat for o-xylene that is slightly higher than that of ToMO, the improvement in the conversion of o-xylene to 3,4-DMC can be at least in part attributed to the fact that the main product of (E103G)-ToMO is 3,4-DMP (99%), the best substrate for PH, which converts it to 3,4-DMC. Therefore, the efficiency of the metabolic pathway can be controlled not only through mutations that increase the catalytic efficiency of the enzymes involved but also through fine-tuning their substrate specificity and their regioselectivity.
In conclusion, the data that we collected strongly support the idea that the spectrum of products that can be obtained from substituted aromatic compound by the combined action of ToMO and PH can be predicted with reasonable confidence based on knowledge of the structure of the enzymes. Such predictability may be useful for future industrial biocatalysis applications.
Acknowledgments
We are indebted to Giuseppe D'Alessio, Department of Biological Chemistry, University of Naples Federico II, and to Gennaro Marino, Department of Organic Chemistry and Biochemistry, University of Naples Federico II, for critically reading the manuscript.
This work was supported by grants PRIN/2000 and PRIN/2002 from the Ministry of University and Research.
REFERENCES
- 1.Arenghi, F. L., D. Berlanda, E. Galli, G. Sello, and P. Barbieri. 2001. Organization and regulation of meta cleavage pathway gene for toluene and o-xylene derivative degradation in Pseudomonas stutzeri OX1. Appl. Environ. Microbiol. 67:3304-3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bertoni, G., F. Bolognesi, E. Galli, and P. Barbieri. 1996. Cloning of the genes for and characterization of the early stages of toluene catabolism in Pseudomonas stutzeri OX1. Appl. Environ. Microbiol. 62:3704-3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bertoni, G., M. Martino, E. Galli, and P. Barbieri. 1998. Analysis of the gene cluster encoding toluene/o-xylene monooxygenase from Pseudomonas stutzeri OX1. Appl. Environ. Microbiol. 64:3626-3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cafaro, V., E. Notomista, P. Capasso, and A. Di Donato. 2005. Regiospecificity of two multicomponent monooxygenases from Pseudomonas stutzeri OX1: molecular basis for catabolic adaptation of this microorganism to methylated aromatic compounds. Appl. Environ. Microbiol. 71:4736-4743. [DOI] [PMC free article] [PubMed]
- 5.Fetzner, S., and J. R. van der Meer. 2000. Enzymes involved in the aerobic bacterial degradation of N-heteroaromatic compounds: molybdenum hydroxylases and ring-opening 2,4-dioxygenases. Naturwissenschaften 87:59-69. [DOI] [PubMed] [Google Scholar]
- 6.Johnson, G. R., and R. H. Olsen. 1997. Multiple pathways for toluene degradation in Burkholderia sp. strain JS150. Appl. Environ. Microbiol. 63:4047-4052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kunkel, T. A. 1987. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc. Natl. Acad. Sci. USA 82:488-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mitchell, K. H., J. M. Studts, and B. G. Fox. 2002. Combined participation of hydroxylase active site residues and effector protein binding in a para to ortho modulation of toluene 4-monooxygenase regiospecificity. Biochemistry 41:3176-3188. [DOI] [PubMed] [Google Scholar]
- 9.Notomista, E., A. Lahm, A. Di Donato, and A. Tramontano. 2003. Evolution of bacterial and archaeal multicomponent monooxygenases. J. Mol. Evol. 56:435-445. [DOI] [PubMed] [Google Scholar]
- 10.Pikus, J. D., J. M. Studts, C. Achim, K. E. Kauffmann, E. Munck, R. J. Steffan, K. McClay, and B. G. Fox. 1996. Recombinant toluene-4-monooxygenase: catalytic and Mossbauer studies of the purified diiron and Rieske components of a four-protein complex. Biochemistry 35:9106-9119. [DOI] [PubMed] [Google Scholar]
- 11.Pikus, J. D., J. M. Studts, K. McClay, R. J. Steffan, and B. G. Fox. 1997. Changes in the regiospecificity of aromatic hydroxylation produced by active site engineering in the diiron toluene 4-monooxygenase. Biochemistry 36:9283-9289. [DOI] [PubMed] [Google Scholar]
- 12.Powlowski, J., and V. Shingler. 1994. Genetics and biochemistry of phenol degradation by Pseudomonas sp. CF600. Biodegradation 5:219-236. [DOI] [PubMed] [Google Scholar]
- 13.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 14.Sanseverino, J., B. M. Applegate, J. M. King, and G. S. Sayler. 1993. Plasmid-mediated mineralization of naphthalene, phenanthrene, and anthracene. Appl. Environ. Microbiol. 59:1931-1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sazinsky, M. H., J. Bard, A. Di Donato, and S. J. Lippard. 2004. Crystal structure of the toluene/o-xylene monooxygenase hydroxylase from Pseudomonas stutzeri OX1. Insight into the substrate specificity, substrate channeling, and active site tuning of multicomponent monooxygenases. J. Biol. Chem. 279:30600-30610. [DOI] [PubMed] [Google Scholar]
- 16.Tao, Y., A. Fishman, W. E. Bentley, and T. K. Wood. 2004. Altering toluene 4-monooxygenase by active-site engineering for the synthesis of 3-methoxycatechol, methoxyhydroquinone, and methylhydroquinone. J. Bacteriol. 186:4705-4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van der Meer, J. R. 1997. Evolution of novel metabolic pathways for the degradation of chloroaromatic compounds. Antonie Leeuwenhoek 71:159-178. [DOI] [PubMed] [Google Scholar]
- 18.Vardar, G., and T. K. Wood. 2004. Protein engineering of toluene-o-xylene monooxygenase from Pseudomonas stutzeri OX1 for synthesizing 4-methylresorcinol, methylhydroquinone, and pyrogallol. Appl. Environ. Microbiol. 70:3253-3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Viggiani, A., L. Siani, E. Notomista, L. Birolo, P. Pucci, and A. Di Donato. 2004. The role of conserved residues H246, H199 and Y255 in the catalysis of catechol 2,3-dioxygenase from Pseudomonas stutzeri OX1. J. Biol. Chem. 279:48630-48639. [DOI] [PubMed] [Google Scholar]
- 20.Whited, G. M., and D. T. Gibson. 1991. Separation and partial characterization of the enzymes of the toluene-4-monooxygenase catabolic pathway in Pseudomonas mendocina KR1. J. Bacteriol. 173:3017-3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Whited, G. M., and D. T. Gibson. 1991. Toluene-4-monooxygenase, a three component enzyme system that catalyzes the oxidation of toluene to p-cresol in Pseudomonas mendocina KR1. J. Bacteriol. 173:3010-3016. [DOI] [PMC free article] [PubMed] [Google Scholar]