Abstract
A novel high-throughput screening method that overcame product inhibition was used to isolate a mutant ω-transaminase from Vibrio fluvialis JS17. An enzyme library was generated using error-prone PCR mutagenesis and then enriched on minimal medium containing 2-aminoheptane as the sole nitrogen source and 2-butanone as an inhibitory ketone. An identified mutant enzyme, ω-TAmla, showed significantly reduced product inhibition by aliphatic ketone. The product inhibition constants of the mutant with 2-butanone and 2-heptanone were 6- and 4.5-fold higher than those of the wild type, respectively. Using ω-TAmla (50 U/ml) overexpressed in Escherichia coli BL21, 150 mM 2-aminoheptane was successfully resolved to (R)-2-aminoheptane (enantiomeric excess, >99%) with 53% conversion with an enantioselectivity of >100.
Transaminases (TAs) have been studied extensively due to their use in the production of amino acids and chiral amines (5, 18, 19). TAs play an important role in amino acid metabolism and are ubiquitous in both microorganisms and eukaryotic cells (6, 18). Despite their advantages, including broad substrate specificity and high stability (19), the industrial use of these enzymes has been limited due to the low equilibrium constants of the TA reactions. However, by removing one of the products via coupling to other enzymes or whole-cell biotransformation, d and l amino acids have been successfully produced (3, 5, 17, 19).
TAs can be classified as α- or ω-TA according to the position of the amino group transferred with respect to the carboxyl group of the substrate (8, 9, 21). Among the TAs, only ω-TA has been reported to show catalytic activity towards primary amines (6, 21). Therefore, the chemical properties of the starting amino donor and acceptor substrates in the ω-TA reaction are often different from those of the resulting products. For example, when (S)-α-methylbenzylamine [(S)-α-MBA] is deaminated with pyruvate, the amine and the keto acid are the substrates, whereas an amino acid (alanine) and a ketone (acetophenone) are the products. Unlike α-TA reactions, the equilibrium constant of the ω-TA reaction is high enough (Keq = 88,100) to yield highly enantiopure (R)-α-MBA via kinetic resolution. This makes the ω-TA an attractive biocatalyst for preparing chiral amines (12).
The ω-TA from Vibrio fluvialis JS17 screened from soil microorganism shows high enantioselectivity (E) for the (S) enantiomers of various chiral amines, such as α-MBA and sec-butylamine, with remarkable stability and a high reaction rate (11-16). However, severe inhibition by the ketone product prevented the enzyme from maintaining the maximum activity. A limited solution to this problem was an extractive biphasic reaction system that reduced the inhibitory acetophenone concentration in the aqueous phase (11). However, like most two-phase systems, problems with the solvent toxicity of the enzyme and mass transfer limitation due to the restricted interfacial area were encountered (15).
Removing the inhibitory effect of the product through protein mutagenesis would also allow higher yields. Of the two methods site-directed mutagenesis and random mutagenesis, the first requires a detailed understanding of both the enzyme structure and the reaction mechanism and yields inconsistent results. However, random mutagenesis is a relatively simple approach, requiring only a selection or rapid screening procedure to obtain the proteins of interest (1, 10). Here we report an enrichment culture method combined with random mutagenesis for the high-throughput production and isolation of mutants overcoming product inhibition. This method is based on using the amine donor substrate 2-aminoheptane as the sole nitrogen source in minimal medium. The concomitantly generated inhibitory ketone compound 2-heptanone reduces the growth rate of the host cells and exerts product inhibition on the enzyme. Thus, the higher growth rates of mutants resistant to inhibition allow them to be enriched in culture, greatly reducing the number of colonies that would otherwise have to be screened. We employed this strategy to identify a mutant enzyme, ω-TAmla, which exhibited significantly reduced product inhibition by aliphatic ketone, and we used it for kinetic resolution of racemic 2-aminoheptane.
MATERIALS AND METHODS
Chemicals.
(R)-, (S)-, and (R,S)-2-aminoheptanes, 1,2-dimethylpropylamine, 1,5-dimetylhexylamine, 2-butanone, 2,3,4,6-tetra-o-acetyl-α-d-glucopyranosyl isothiocyanate (GITC), and antibiotics were purchased from Sigma-Aldrich (St. Louis, MO). All other chemicals were analytical or reagent grade.
Random mutagenesis.
The ω-TA gene was cloned into the isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible expression vector pET24ma using PCR amplification as previously reported (16). Plasmid pET24ma::ω-TA DNA was prepared using a Miniprep kit (QIAGEN, Valencia, CA) and was used as a template for PCR mutagenesis. A GeneAmp PCR 2400 kit (Perkin-Elmer, United States) and thin-wall PCR tubes were used in all PCRs. The total reaction volume was 50 μl. Random mutants were generated using a Diversify PCR random mutagenesis kit purchased from Clontech (Palo Alto, CA) (94°C for 30 s for one cycle; 94°C for 30 s and 68°C for 1 min for 25 cycles; 68°C for 1 min for one cycle) with 0.5 pmol of primer 5′-GGAGATATACATATGAACAAA-CCGCAA-3′, 0.5 pmol of primer (5′-CTCGAATTCGGATCCTCAGGCAACCTCGGC-3′), 0.32 mM MnSO4, 0.04 mM dGTP, 1× Diversify deoxynucleoside triphosphate mixture, 5 U TITANIUM Taq polymerase, and 1 μl plasmid DNA (pET24ma::ω-TA). The fragments were digested with the NdeI/BamHI restriction enzymes and inserted into pET24ma. The plasmids were transformed into Escherichia coli BL21, and the clones were grown in Luria-Bertani (LB) broth containing 50 μg/ml of kanamycin at 37°C.
Saturation mutagenesis.
Saturation mutagenesis was performed at selected positions using the following PCR conditions: 94°C for 4 min for one cycle; 94°C for 30 s, 60°C for 30 s, and 68°C for 12 min for 16 cycles; and 68°C for 20 min for one cycle. For the PCR 2.5 U Pfu polymerase (Stratagene), each deoxynucleoside triphosphate at a concentration of 0.2 mM, 1 μl plasmid DNA (pET24ma::ω-TA), and 1 pmol of each primer were used. The following primers were used: for amino acid position 233, (5′-GTGATTNNNCCGGCCAAGGGGTATTTCCAG-3′ and 5′-GGCCGGNNNAATCACGCCGCCCGCGCCCAT-3′); and for amino acid position 297, (5′-GGGGCGNNNATCCTTGGCCCGGAACTTTCC-3′ and 5′-AAGGATNNNCGCCCCCATGGGGAAAAAGCC-3′). After DpnI treatment of the PCR products, E. coli transformations were carried out by using the standard method, as mentioned above. In both cases, about 3,000 colonies were obtained. Double point saturation mutagenesis at amino acids 233 and 297 was performed by the following procedure. The amino acid position 297 saturation mutagenesis was carried out using the plasmid extracted from the amino acid position 233 saturation mutant library. The amino acid position 233 saturation mutagenesis was carried out using the plasmid extracted from the amino acid position 297 saturation mutant library and the standard method, as mentioned above. Each PCR product was transformed into E. coli BL21, about 3,000 colonies were obtained, and the two libraries were mixed.
Enrichment of ω-TA mutants resistant to product inhibition.
To isolate a mutant ω-TA less affected by inhibitory aliphatic ketone, the random mutant library was incubated overnight in 50 ml of LB broth. After incubation, 0.05 ml of the culture broth was transferred to 50 ml of minimal medium (100 mM glucose, 10 mM pyruvate, 50 mM potassium phosphate buffer [pH 7.0], 1.0 g/liter MgSO4 · 7H2O, 0.2 mM CaCl2, 0.1 mg/liter ZnCl2, 0.1 mg/liter MnSO4 · 4H2O, 0.02 mg/liter H3BO3, 0.1 mg/liter CuSO4 · 5H2O, 0.05 mg/liter CoCl2, 0.1 mg/liter NiSO4 · 6H2O, 2.0 mg/liter NaMoO4, 4.0 mg/liter FeSO4 · 7H2O, 0.2 mM IPTG, 50 μg/ml of kanamycin, 2 mM 2-aminoheptane as a sole nitrogen source, and 20 mM 2-butanone as an inhibitory ketone) (22). Enrichment of the recombinant E. coli BL21 (random mutant library) was carried out by repeated dilution (1,000-fold) of culture broth with fresh minimal medium at 37°C whenever the optical density at 600 nm of the culture broth reached about 0.3. The concentration of 2-butanone was serially increased up to 120 mM in steps of 20 mM. Finally, when the optical density reached about 0.3 in 50 ml of minimal medium containing 120 mM 2-butanone, the diluted culture broth was spread on LB agar plates containing 50 μg/ml of kanamycin. Six mutants were randomly selected from the agar plates to be assayed.
Preparation of enzyme.
To express the mutant enzyme, the plasmid prepared from a mutant colony selected as described above was introduced into fresh E. coli BL21. The recombinant E. coil BL21 strain harboring the ω-TA gene or the ω-TAmla gene was grown in 1 liter of LB broth containing 50 μg/ml of kanamycin at 37°C. When the optical density reached 0.75, 1 mM IPTG was added to the cell broth. After 5 h of cultivation, the cells were harvested, washed twice with 200 ml of ice-cold 200 mM phosphate buffer (pH 7.0), suspended in 50 ml of buffer, and kept in ice for further study. To obtain a cell extract, the harvested cells were suspended in 50 ml of 200 mM phosphate buffer (pH 7.0) containing 0.2 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 20 μM pyridoxal 5′-phosphate (PLP), and 0.01% (vol/vol) 2-mercaptoethanol and then subjected to ultrasonic disruption for 25 min. The supernatant solution was obtained after centrifugation (17,000 × g, 20 min), and it was dialyzed overnight against 50 mM phosphate buffer (pH 7.0) at 4°C. The purified enzyme was obtained as described elsewhere (16).
Enzyme assays.
One unit of ω-TA activity was defined as the amount of enzyme that catalyzed the formation of 1 μmol acetophenone from 50 mM (S)-α-MBA and 50 mM pyruvate in 1 min (11). The kinetic parameters were determined at 37°C (23) in a 1-ml assay solution (200 mM phosphate buffer, pH 7.0) containing 0.5 U of the purified enzyme and 20 mM pyruvate. The concentration of racemic sec-butylamine was varied from 5 to 400 mM, and the concentration of of 2-butanone was varied from 0 to 20 mM. The initial reaction rate was measured by analysis of pyruvate. The values for Km, Vmax, and the competitive product inhibition constant (Ki) were calculated using a series of Lineweaver-Burk plots determined at different concentrations of 2-butanone. For determination of the kinetic parameters for 2-aminoheptane, all the reaction conditions were kept constant except that 0.1 U/ml of enzyme was used. The relative activities of the enzyme (wild-type ω-TA or ω-TAmla) with various substrates were determined by incubating the enzyme (0.2 U/ml) in 0.2 ml of 100 mM phosphate buffer containing 10 mM target racemic amine [or 5 mM enantiopure (S)-α-MBA] and 5 mM pyruvate at 37°C for 30 min. The amount of pyruvate consumed was determined by high-performance liquid chromatography (HPLC).
Enzyme stability.
The crude cell extracts or whole cells of recombinant E. coli BL21 overexpressing ω-TAmla (5 U/ml) were incubated in 200 mM borate buffer (pH 9.0) containing various concentrations of 2-aminoheptane (0 to 250 mM) and 40 mM pyruvate at 37°C for 1 h. The residual enzyme activity was assayed by determining the reaction of 50 mM (S)-α-MBA and 50 mM pyruvate in 100 mM phosphate buffer (pH 7.0). An aliquot of the reaction mixture (100 μl) was sampled and incubated with 0.9 ml of the assay solution at 37°C. The reaction rate was measured by analyzing acetophenone.
Analytical methods.
Quantitative and chiral analyses of 2-aminoheptane were performed using a C18 Symmetry column (Waters, Milford, MA) with a Waters HPLC system after derivatization of a sample with GITC (7). For determination of enantiomeric purity, the analysis conditions were as follows: mobile phase, 35% acetonitrile-water containing 0.1% trifluoroacetic acid; flow rate, 1.0 ml/min; and detection at 254 nm. The retention times were 27.4 min for (S)-amine and 28.8 min for (R)-amine. For quantitative analysis of 2-aminoheptane, ethylamine derivatized with GITC was used as the internal standard (23). Pyruvate was analyzed using an Aminex HPX-87H HPLC column (Bio-Rad, Richmond, CA) with elution with a 5 mM sulfuric acid solution at a wavelength at 210 nm. Analysis of acetophenone was performed with a Symmetry HPLC column (Waters) with isocratic elution with acetonitrile-water (50/50, vol/vol) at a flow rate of 1 ml/min (23).
RESULTS AND DISCUSSION
Development of enrichment method.
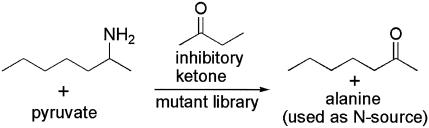
The ω-TA reaction produces alanine, which is utilized as both a nitrogen source and an energy source (Fig. 1). This fact can be exploited for rapid isolation of microbes possessing ω-TA activity. Selection in minimal media containing 2 mM 2-aminobutane or 2-aminoheptane was used to isolate cells carrying ω-TA. In E. coli BL21 cells harboring pET24ma with and without the ω-TA gene, only the cells with the insert grew. Compared to the activity with (S)-α-MBA, the apparent activities of ω-TA with 2-aminoheptane and 2-aminobutane are 43 and 7%, respectively (14), indicating that 2-aminoheptane is a better amino group donor than 2-aminobutane. This was reflected in the growth pattern of the recombinant E. coli possessing ω-TA activity, which grew three times faster in the minimal medium containing 2 mM 2-aminoheptane than in the minimal medium containing 2 mM 2-aminobutane. To reach an optical density at 600 nm of 0.3, growth in the presence of 2 mM 2-aminoheptane took 2 days, whereas growth in the presence of 2 mM 2-aminobutane took 6 days.
FIG. 1.
Schematic diagram of the enrichment procedure using a minimal medium containing 2-aminoheptane as the sole N source. In the presence of the inhibitory product 2-butanone, the mutants most resistant to product inhibition can be selected.
To isolate ω-TA mutants more resistant to product inhibition by ketones, a similar approach was used to selectively enrich for strains which grew better under product-inhibitory conditions. The less toxic (but still inhibitory) compound 2-butanone was added to the medium as the inhibitory ketone, while the more active compound 2-aminoheptane was used as the sole nitrogen source (Fig. 1).
Mutant library construction and selection.
Random mutations were introduced into the ω-TA gene from V. fluvialis coding for 453 amino acids under PCR conditions designed to generate an average of one or two amino acid substitutions per sequence. About 85,000 clones were obtained, and the library of mutants was incubated overnight in 50 ml of LB broth. Fifty microliters of each overnight culture was subcultured in 50 ml of fresh minimal medium containing 2 mM 2-aminoheptane and 20 mM 2-butanone. In a series of subcultures, the concentration of 2-butanone was increased in steps of 20 mM up to 120 mM. Finally, six colonies were randomly selected from the culture broth containing 120 mM 2-butanone and 2 mM 2-aminoheptane, and the ω-TA genes were sequenced. The sensitivities of the mutated ω-TAs to inhibition were compared by measuring the enzyme activities of IPTG-induced cells in the presence of 100 mM 2-butanone, 50 mM pyruvate, and 50 mM (S)-α-MBA. The activity of cells containing the wild-type ω-TA was reduced to 30% of the activity without 100 mM 2-butanone, whereas the activity of the mutants was reduced to 60% of this activity, suggesting that the mutant enzymes showed greater resistance to the inhibition by 2-butanone.
DNA sequencing analysis of the mutants revealed that all the mutants had the same nucleotide sequence with substitutions at the proline at position 233 (CCC), which was changed to leucine (CTC), and at the valine at position 297 (GTG), which was changed to alanine (GCG). The mutant enzyme was designated ω-TAmla. Previous studies have shown that saturation mutagenesis performed at sites identified by error-prone PCR often generates further improvements (1, 4, 20, 24). Saturation mutagenesis was thus attempted at both position 233 and position 297. About 3,000 mutant colonies were collected for each position, and 6,000 colonies were collected for the double point mutations. All the saturation mutants were pooled and subjected to enrichment culture screening as described above. The degrees of product inhibition for the enzyme activities of the selected mutants were similar to that of ω-TAmla, but the activities with 2-aminoheptane and 2-aminobutane were about 10 and 20% lower than those of ω-TAmla, respectively (data not shown). The double point saturation mutation did not yield improved mutants, suggesting that ω-TAmla (i.e., P233L and V297A double mutant) was isolated serendipitously during the primary enrichment culture without further optimization.
Comparison of wild-type ω-TA and ω-TAmla.
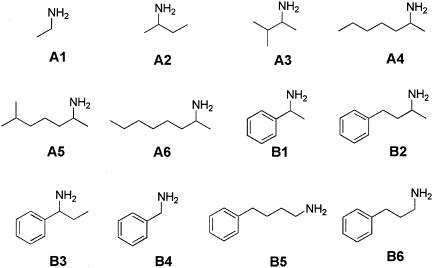
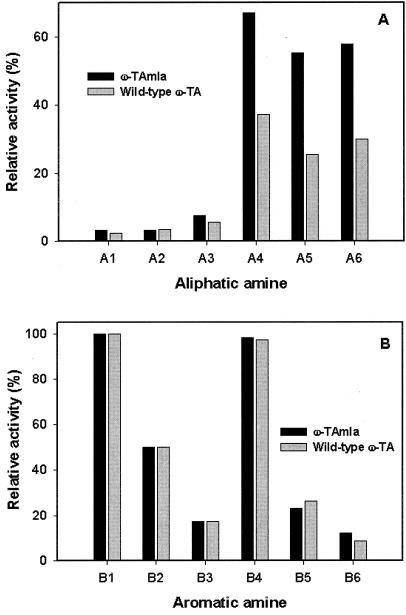
For a comparison of the two enzymes, (S)-α-MBA was used as a model substrate. The specific activities of ω-TA from crude extracts of recombinant E. coli BL21 cells harboring the pET24ma::ω-TA gene and recombinant E. coli BL21 cells harboring the pET24ma::ω-TAmla gene were 19.2 and 20.4 U/mg, respectively. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis also revealed that the two strains produced similar amounts of the enzyme, suggesting that the two enzymes have similar specific enzyme activities. However, the mutant ω-TAmla enzyme was less susceptible to product inhibition by aliphatic ketones. The product Kis of wild type ω-TA and ω-TAmla for 2-butanone were 5.1 and 30.6 mM, respectively, and the Kis for 2-heptanone were 4.1 and 18.5 mM, respectively (Table 1). Aromatic ketones, such as acetophenone and phenylacetone, inhibited both wild-type ω-TA and ω-TAmla to the same extent up to a concentration of 20 mM. The initial reaction rates in the presence of aromatic ketones (0 to 20 mM) were similar for ω-TAmla and wild-type ω-TA (data not shown). Substrate specificities were examined with various aliphatic and aromatic amines by comparing the initial rates under similar reaction conditions (Fig. 2). The relative apparent activities of the crude cell extract were always in good agreement with previous results obtained for the purified enzyme (14). Compared with wild-type ω-TA, the ω-TAmla crude extract displayed twofold-higher activity for long-chain aliphatic amines, such as 2-aminoheptane (A4), 1,5-dimetylhexylamine (A5), and 1-methylheptylamine (A6) in the reaction conditions used (Fig. 3). However, its activities with short-chain aliphatic amines (A1 to A3) and aromatic amines (B1 to B6) were similar to those of wild-type ω-TA.
TABLE 1.
Kinetic parameters of wild-type ω-TA and ω-TAmlaa
| Enzyme | Substrate | Km (mM) | Vmax (U/mg) | Ki (mM) |
|---|---|---|---|---|
| Wild-type ω-TA | Racemic 2-aminoheptane | 21.5 ± 1.0 | 231 ± 10 | 4.1 ± 0.58 |
| Racemic sec-butylamineb | 18.1 ± 0.45 | 25.4 ± 0.80 | 5.1 ± 0.41 | |
| ω-TAmla | Racemic 2-aminoheptane | 12.1 ± 0.87 | 283 ± 15 | 18.5 ± 0.52 |
| Racemic sec-butylamine | 11.6 ± 0.50 | 18.4 ± 0.93 | 30.6 ± 1.1 |
Kinetic parameters were determined for the purified enzymes in the presence of 20 mM pyruvate at 37°C.
See reference 23.
FIG. 2.
Amino donors used to determine substrate specificities of the enzyme.
FIG. 3.
Substrate specificities of wild-type ω-TA and ω-TAmla (0.2 U/ml). Pyruvate (10 mM) and each amino donor (racemic form) at a concentration of 10 mM were used, and the initial reaction rate was measured by analyzing the pyruvate consumed from the reaction mixture. (A) Aliphatic amine. (B) Aromatic amine. The activity of each enzyme for 5 mM (S)-α-MBA (enantiopure form) was defined as 100%.
Enzyme stability.
The effects of the cofactor PLP and the substrate 2-aminoheptane on the stability of the ω-TAmla enzyme in the crude cell extract and the recombinant E. coli BL21 whole cells were studied by incubating the reaction mixtures in 200 mM borate buffer (pH 9.0) containing various concentrations of 2-aminoheptane (0 to 250 mM) and 40 mM pyruvate at 37°C for 1 h. It was observed that in the presence of 10 mM 2-aminoheptane the enzyme lost 60% of its activity in 1 h and that it was completely deactivated in the presence of 80 mM 2-aminoheptane. Enzyme stability was notably improved in the presence of 1 mM PLP, and only 5% of the activity was lost in the presence of 80 mM 2-aminoheptane. It is well known that addition of PLP to reaction mixtures helps maintain the stability of PLP-containing enzymes (15, 22). In comparison, whole cells tolerated up to 100 mM 2-aminoheptane with no loss of activity. The residual activity of the whole cells in the presence of 1 mM PLP with 150 mM 2-aminoheptane was 44.4% of the original activity, and the residual activity with 150 mM 2-aminoheptane without added PLP was 15.6% of the original activity. Almost all activity was lost both from the cell extract and whole cells when the 2-aminoheptane concentration was increased to 200 mM. Similar results were obtained with wild-type ω-TA.
Kinetic resolution of racemic 2-aminoheptane using ω-TAmla.
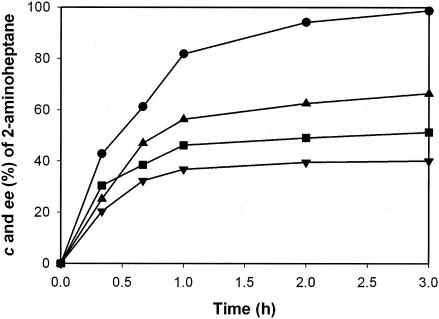
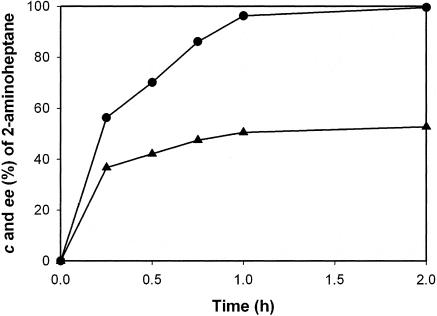
Both wild-type ω-TA and ω-TAmla displayed optimum activity at pH 9.0 for kinetic resolution of 2-aminoheptane. Crude cell extracts (20 U/ml) containing wild-type ω-TA or ω-TAmla were added to 5 ml of 200 mM borate buffer (pH 9.0) containing 100 mM 2-aminoheptane, 100 mM pyruvate, and 0.5 mM PLP at 37°C. After 3 h, the enatiomeric excess (ee) of (R)-2-aminoheptane was 99% in the ω-TAmla reaction, whereas the ee of (R)-2-aminoheptane was 66% in the wild-type ω-TA reaction (Fig. 4). The E was >100 for both enzymes. Since the apparent activity of ω-TAmla is twofold higher than that of wild ω-TA (Fig. 3), kinetic resolution was carried out with 40 U/ml of wild-type ω-TA. The ee of (R)-2-aminoheptane obtained after 3 h was only marginally increased to 70%, suggesting that the greater conversion and higher ee using ω-TAmla were mainly due to the decreased product inhibition. The kinetic resolution observed with the whole cells also yielded similar results; 150 mM racemic 2-aminoheptane was successfully resolved to (R)-2-aminoheptane (>99% ee) at 53% conversion (Fig. 5). Kinetic resolution of the solution at 2-aminoheptane concentrations above 150 mM could not be performed efficiently because of the toxicity of 2-aminoheptane to the enzyme. In order to perform kinetic resolution at higher concentrations of 2-aminoheptane, perhaps another round of directed evolution to improve the enzyme resistance to 2-aminoheptane or to develop a new reaction system working at high concentrations of 2-aminoheptane is required.
FIG. 4.
Reaction profiles of the kinetic resolution of 2-aminoheptane using the crude cell extracts containing wild-type ω-TA and ω-TAmla. The crude cell extracts (20 U/ml) containing wild-type ω-TA and ω-TAmla were added to 5 ml of 200 mM borate buffer (pH 9.0) containing 100 mM 2-aminoheptane, 100 mM pyruvate, and 0.5 mM PLP. The conversion (c) (▪) and ee (•) of 2-aminoheptane in the ω-TAmla reaction and the conversion (▾) and ee (▴) of 2-aminoheptane in the wild-type ω-TA reaction were determined.
FIG. 5.
Reaction profiles of the kinetic resolution of 2-aminoheptane using recombinant E. coli BL21 overexpressing ω-TAmla. Whole cells (50 U/ml) were added to 5 ml of 200 mM borate buffer (pH 9.0) containing 150 mM 2-aminoheptane, 100 mM pyruvate, and 1 mM PLP. The conversion (c) (▴) and ee (•) of 2-aminoheptane were determined.
In conclusion, we developed an efficient high-throughput selection method for isolation of ω-TA resistant to product inhibition by a ketone. The mutant ω-TAmla enzyme showing significantly reduced product inhibition by aliphatic ketones was identified from the ω-TA library generated by error-prone PCR mutagenesis. We identified an improved ω-TA mutant from 85,000 mutants with simple enrichment subcultures within 2 weeks. However, this method could not be applied to enrichment of mutants resistant to product inhibition by aromatic ketones because of their toxicity to E. coli. If cells of more solvent-resistant organisms, such as Pseudomonas putida, were used as host cells, this simple selection method could be extended to identify ω-TA mutants resistant to product inhibition by aromatic ketones. In addition, this enrichment method is applicable to other enzymes as long as one can correlate the activity of the enzyme reaction with an increased growth rate of the host cells. Specifically, this enrichment selection method would be useful to broaden the substrate spectrum of ω-TA for less-reactive unnatural amino acids and amines. For example, ω-amino acid:pyruvate transaminase from Alcaligenes denitrificans Y2k-2 can transfer an amine group from various aliphatic β-amino acids, such as l-β-amino-n-butyric acid, to pyruvate with high E (>80), whereas aromatic β-amino acids were inert (22). If an enzyme library was generated using error-prone PCR mutagenesis and then enriched on minimal medium containing β-phenylalanine as a sole nitrogen source, the mutant reactive for β-phenylalanine could be selected.
To understand why the replacement of two amino acids conferred resistance to product inhibition by an aliphatic ketone, comparative homology modeling was attempted (2). According to the model structure constructed based on dialkylglycine decarboxylase (PDB ID code 1DGE) as a template, Pro233 is located in the loop region exposed to the reaction media and Val297 is involved in constructing the active site binding pocket (unpublished data). However, more kinetic studies and computer modeling are required to understand how replacement of the two amino acid residues increased the resistance to product inhibition by an aliphatic ketone.
Acknowledgments
This study was supported in part by Korea Energy Management Corporation and 21C Frontier R&D Programs (Microbial Genomics & Applications Center, Korea).
REFERENCES
- 1.Arnold, F. H., and A. A. Volkov. 1999. Directed evolution of biocatalysts. Curr. Opin. Chem. Biol. 3:54-59. [DOI] [PubMed] [Google Scholar]
- 2.Baker, D., and A. Sali. 2001. Protein structure prediction and structural genomics. Science 294:93-96. [DOI] [PubMed] [Google Scholar]
- 3.Bartsch, K., R. Schneider, and A. Schulz. 1996. Stereospecific production of the herbicide phosphinothricin: purification of aspartate transaminase from Bacillus stearothermophilus, cloning of the corresponding gene, aspC, and application in a coupled transaminase process. Appl. Environ. Microbiol. 62:3794-3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bornscheuer, U. T., J. Altenbuchner, and H. H. Meyer. 1998. Directed evolution of an esterase for the stereoselective resolution of a key intermediate in the synthesis of epothilones. Biotechnol. Bioeng. 58:554-559. [DOI] [PubMed] [Google Scholar]
- 5.Chao, Y.-P., Z. J. Lai, P. Chen, and J.-T. Chern. 1999. Enhanced conversion rate of l-phenylalanine by coupling reactions of aminotransferases and phosphoenolpyruvate carboxykinase in Escherichia coli K-12. Biotechnol. Prog. 15:453-458. [DOI] [PubMed] [Google Scholar]
- 6.Christen, P., and D. E. Metzler. 1985. Transaminases. Wiley, New York, N.Y.
- 7.Ito, S., A. Ota, K. Yamamoto, and Y. Kawashima. 1992. Resolution of the enantiomers of thiol compounds by reversed-phase liquid chromatography using chiral derivatization with 2,3,4-tetra-O-acetyl-β-d-glucopyranosyl isothiocyanate. J. Chromatogr. 626:187-196. [Google Scholar]
- 8.Mehta, P. K., and P. Christen. 1994. Homology of 1-aminocyclopropane-1-carboxylate synthase, d-amino-7-oxononanoate synthase, 2-amino-6-carprolactam racemase, 2,2-dialkylglycine decarboxylase, glutamate-1-semialdehyde 1,2-aminomutase and isopenicilin-N-epimerase with aminotransferases. Biochem. Biophy. Res. Commun. 198:138-143. [DOI] [PubMed] [Google Scholar]
- 9.Mehta, P. K., T. I. Hale, and P. Christen. 1993. Aminotransferases: demonstration of homology and division into evolutionary subgroups. Eur. J. Biochem. 214:549-561. [DOI] [PubMed] [Google Scholar]
- 10.Salazar, O., and L. Sun. 2003. Evaluating a screen and analysis of mutant libraries, p. 85-97. In F. H. Arnold and G. Georgiou (ed.), Methods in molecular biology. Directed enzyme evolution. Humana Press, Totowa, N.J. [DOI] [PubMed]
- 11.Shin, J.-S., and B.-G. Kim. 1997. Kinetic resolution of α-methylbenzylamine with ω-transaminase screened from soil microorganisms: application of a biphasic system to overcome product inhibition. Biotechnol. Bioeng. 55:348-358. [DOI] [PubMed] [Google Scholar]
- 12.Shin, J.-S., and B.-G. Kim. 1998. Kinetic modeling of ω-transamination for enzymatic kinetic resolution of α-methylbenzylamine. Biotechnol. Bioeng. 60:534-540. [DOI] [PubMed] [Google Scholar]
- 13.Shin, J.-S., and B.-G. Kim. 1999. Asymmetric synthesis of chiral amines with ω-transaminase. Biotechnol. Bioeng. 65:206-211. [PubMed] [Google Scholar]
- 14.Shin, J.-S., and B.-G. Kim. 2002. Exploring the active site of amine:pyruvate aminotransferase on the basis of the substrate; structure-reactivity relationship: how the enzyme controls substrate specificity and stereoselectivity. J. Org. Chem. 67:2848-2853. [DOI] [PubMed] [Google Scholar]
- 15.Shin, J.-S., B.-G. Kim, A. Liese, and C. Wandrey. 2001. Kinetic resolution of chiral amines using enzyme-membrane reactor. Biotechnol. Bioeng. 73:179-187. [DOI] [PubMed] [Google Scholar]
- 16.Shin, J.-S., H. Yun, J.-W. Jang, I. Park, and B.-G. Kim. 2003. Purification, characterization, and molecular cloning of a novel amine:pyruvate transaminase from Vibrio fluvialis JS17. Appl. Microbiol. Biotechnol. 61:463-471. [DOI] [PubMed] [Google Scholar]
- 17.Stinson, S. C. 1997. Chiral drug market shows signs of maturity. Chem. Eng. News 20(10):38-70. [Google Scholar]
- 18.Stirling, D. I. 1992. The use of aminotransferases for the production of chiral amino acids and amines, p. 209-235. In A. N. Collins, G. N. Sheldrake, and J. Crosby (ed.), Chirality in industry. Wiley, New York, N.Y.
- 19.Taylor, P. P., D. P. Pantaleone, R. F. Senkpeil, and I. G. Fotheringham. 1998. Novel biosynthetic approaches to the production of unnatural amino acids using transaminases. Trends Biotechnol. 16:412-418. [DOI] [PubMed] [Google Scholar]
- 20.Van Kampen, M. D., N. Dekker, M. R. Egmond, and H. M. Verheij. 1998. Substrate specificity of Staphylococcus hyicus lipase and Staphylococcus aureus lipase as studied by in vivo chimeragenesis. Biochemistry 37:3459-3466. [DOI] [PubMed] [Google Scholar]
- 21.Yonaha, K., S. Toyama, and K. Soda. 1987. ω-Amino acid-pyruvate aminotransferase. Methods Enzymol. 143:500-505. [DOI] [PubMed] [Google Scholar]
- 22.Yun, H., S. Lim, B.-K. Cho, and B.-G. Kim. 2004. ω-Amino acid:pyruvate transaminase from Alcaligenes denitrificans Y2k-2: a new catalyst for kinetic resolution of β-amino acids and amines. Appl. Environ. Microbiol. 70:2529-2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yun, H., B.-K. Cho, and B.-G. Kim. 2004. Kinetic resolution of (R,S)-sec-butylamine using omega-transaminase from Vibrio fluvialis JS17 under reduced pressure. Biotechnol. Bioeng. 87:772-777. [DOI] [PubMed] [Google Scholar]
- 24.Zhao, H., and F. H. Arnold. 1999. Directed evolution converts subtilisin E into a functional equivalent of thermitase. Protein Eng. 12:47-52. [DOI] [PubMed] [Google Scholar]