Abstract
Despite viral contamination of recreational waters, only bacterial, not viral, indicators are monitored routinely, due to a lack of rapid and cost-effective assays. We used negatively charged filters to capture enteroviruses from seawater and freshwater. Viral RNA was extracted using a commercial kit, and the viruses were quantified by real-time quantitative reverse transcriptase PCR (qRT-PCR). Poliovirus (6.6 to 330,000 virus particles/ml) was added to samples from watersheds in Los Angeles, California, and analysis showed that with 50-ml samples, a cellulose acetate/nitrate (HA) filter yielded final recovery of 51% (r2 = 0.99) in fresh water and 23% (r2 = 0.90) in seawater. However, for additions of low levels of virus (more likely to represent field samples; <104 enterovirus particles/ml), the recovery was lower and more variable, with HA being best in freshwater (17%, r2 = 0.97) and the type GF/F glass filter having higher average recovery in seawater (GF/F, 17%; r2 = 0.93; HA 12%, r2 = 0.87). The optimized method was used with 1-liter field samples from two very different freshwater “creeks” that drain into Santa Monica Bay, California: Topanga Creek (TC), a relatively pristine mountain creek, and Ballona Creek (BC), a concrete-lined urban storm drain. One TC site out of 10 and 2 BC sites out of 7 tested significantly positive for enteroviruses, with higher enterovirus concentrations in BC than in TC (ca. 10 to 25 versus 1 equivalent enterovirus particle/ml). The presented filtration-qRT-PCR approach is fast (<8 h from sampling to results), sensitive, and cost efficient and is promising for monitoring viral contamination in environmental water samples.
About 37% of the United States population resides in coastal areas, with about 1.0 × 1010 gallons of wastewater released into rivers and seas from outfalls and storm drains per day (8). Before their release, wastewaters are not always treated to remove viruses, and storm water is usually not treated at all (24). Epidemiology studies have shown that recreational exposure from swimming or surfing in locations impacted by contaminated storm water leads to a significant increase in a variety of illnesses (8, 10). There can be even greater potential exposure through the consumption of contaminated shellfish due to concentration of contaminants by the filter feeders (6, 8, 25, 29). The illnesses of concern include (but are not limited to) diarrhea, ocular and respiratory infection, gastroenteritis, hepatitis, myocarditis, meningitis, paralysis, and severe chronic disease. Bacteria and viruses are both important etiological agents of these illnesses. Important waterborne pathogenic viruses include many enteric viruses that are transmitted via the fecal-oral route. These include Picornaviridae (which includes the enteroviruses, e.g., poliovirus, coxsackievirus, and echovirus), Adenoviridae, Caliciviridae (norovirus, calicivirus), Astroviridae, and Reoviridae (reovirus, rotavirus) (8).
In the United States, Escherichia coli and enterococci are currently used as indicators of microbial water quality, serving as proxies for the potential presence of pathogenic bacteria and viruses. There are no currently mandated tests for viral indicators in recreational and shellfish harvesting waters, in part because of the lack of suitable available assays. However, bacterial indicators alone are inadequate predictors of viral pathogens, which have been shown to sometimes be present even in waters with low bacterial indicator concentrations; reports from numerous locations, including the study area reported here (11, 12, 22), show an absence of a significant quantitative relationship between indicator bacteria and viral pathogens, as reviewed by Griffin et al. (8).
The traditional “gold standard” method of human pathogenic viral detection is cell culture assay. It is costly, time-consuming, and impractical for monitoring, because it requires large sample volumes and days to weeks for results. Also, some viruses do not grow in tissue culture. This is unsuitable for management of recreational or shellfish harvesting waters (and many clinical and environmental applications), and rapid genetic techniques as potential alternatives have recently been developed (8, 12, 14, 17-21). Although genetic methods may also detect inactive viruses, such methods have been shown to correlate with detection of viable viruses by cell culture when used with samples in natural waters (5). Some recent studies have used integrated cell culture and PCR, with a cell culture step of several hours to a few days followed by PCR, showing viruses capable of infecting the cell culture much more rapidly than cell culture alone (7, 15, 26, 27). While this approach has some advantages and may be suitable for research studies, it also is more costly and time-consuming than direct reverse transcriptase PCR (RT-PCR). For this study, we tested and applied an approach with features that may be particularly suitable for routine monitoring and management of beaches and shellfish harvesting waters: small sample volume (1 liter), minimized turnaround time, and no costs associated with cell culture.
In order to apply genetic detection methods to detect viruses from natural waters, it is necessary to concentrate the viruses into a very small volume and extract the RNA so that it is free of impurities. Although it was shown several years ago that small charged filters could be used to capture viruses with reasonably good efficiency (2, 30), there were no established methods to recover the viral RNA from the filters. Most previously published approaches for PCR analysis of viruses from seawater samples used ultrafiltration and subsequent RNA extraction of this concentrate. The concentrate was then subjected to conventional RT-PCR (9, 11, 22) or quantitative RT-PCR (qRT-PCR) analysis (4, 13). Katayama et al. (13) reported that negatively charged filters were efficient (>61%) in capturing viruses from relatively small volumes of seawater for qRT-PCR and for virus cultivation. This appeared to be a promising protocol for routine virus assays. However, our attempt to reproduce that method in our laboratory was prevented by the lack of availability (in the United States) of the proprietary extraction kit described in that report; we also noted that the concentrations of poliovirus they used in their seeding experiments (300 to 770 PFU/ml) were very high compared to what one might see in field samples, leading to questions about recovery at realistic field concentrations. We modified the Katayama et al. (13) protocol to include a readily available extraction kit, different qRT-PCR parameters that we tested in a previous study to avoid inhibition (1), and a range of test virus concentrations at levels more realistic of real-world field samples. We also tested this modified protocol with both freshwater and seawater samples and compared the cellulose mixed-ester filter retention with the retention of more porous and freely flowing glass fiber filters used in a previous enterovirus field study (1). Overall, we found that our modified filter/qRT-PCR method was rapid and reasonably efficient for both fresh- and seawater samples, with the best efficiency found with the cellulose acetate/nitrate filters in freshwater and fine-pore glass filters in seawater. We had reduced and somewhat variable, but still acceptable, recovery even at very low virus concentrations that had not been tested in previous studies. Finally, we applied the protocol to successfully detect and quantify enteroviruses in two creek or storm drain systems whose outlets are near popular bathing beaches of Santa Monica Bay (SMB) in Los Angeles, Calif.
MATERIALS AND METHODS
Seeding experiment.
We performed two sets of seeding experiments with both seawater and freshwater, one set with higher seeding concentrations, and the other set with lower seeding concentrations. For both experiments, freshwater was sampled from lower Topanga Creek (TC; 34°03′N, 118°35′W), and seawater was sampled from Topanga Beach, SMB (34°02′N, 118°34′W) in acid-washed polypropylene bottles. Two liters of both fresh water and seawater were collected on 23 September 2003 for high-level virus additions and on 24 March 2004 for low-level virus additions. Each water sample was divided into 200-ml subsamples. A Sabin strain type 1 poliovirus stock was used to seed these subsamples, and the poliovirus concentration in the stock was measured by direct counts with SYBR Green II (23) at 6.6 × 106 ± 0.7 × 106 viral particles/μl (mean ± standard errors, n = 3). That stock was initially titered by a 50% tissue culture infective dose (TCID50) approach (16) as 1.43 × 105 per μl, which is equivalent to an estimated 105 PFU per μl as calculated by 0.7× TCID50 (3); it was then divided into several tubes and stored at −80°C until thawed for these experiments. Amounts of poliovirus added in the high concentration range experiment were 0 (blank), 1.32 × 106, 2.64 × 106, 6.6 × 106, 1.32 × 107, 2.64 × 107, 6.6 × 107 virus particles (corresponding to 0.2 to 10 μl of stock). In the low-concentration additions, stock was serially diluted to add calculated amounts of 1.32 × 103, 3.96 × 103, 1.32 × 104, 3.96 × 104, 1.32 × 105, 3.96 × 105, and 1.32 × 106 (corresponding to 0.0002 to 0.2 μl of undiluted stock). Although we had no way to independently verify the poliovirus concentrations in the serial dilutions, similar dilutions prepared for the qRT-PCR standard curve (below) had the expected log-linear response. The poliovirus and water samples were allowed to interact for 1 h at room temperature. The recovery of the following filters was tested (all 47 mm in diameter): Millipore cellulose acetate/nitrate filter (Type HAWP, denoted as HA; Millipore Corp.), with a nominal pore size of 0.45 μm; Gelman glass borosilicate filter (type A/E; Pall Gelman Corp.), with a nominal pore size of 1.2 μm; and Whatman glass borosilicate filter (type GF/F; Whatman International Ltd.), with a nominal pore size of 0.7 μm. Three 50-ml subsamples were taken from each of the 200-ml samples and filtered through the HA, A/E, and GF/F filters, respectively, held in plastic Millipore filter holders. With the HA and GF/F filters, a gentle vacuum was used (<150 mm Hg); gravity filtration was used for the A/E filters. One filter of each type was used per seeding level.
Filters were collected and stored flat in polyethylene Whirl-Pak bags (18 oz.) at −80°C prior to RNA extraction. Preliminary tests in which the filters were instead folded into quarters and put into 2-ml microcentrifuge tubes yielded much lower recovery. Filters were extracted using the RNeasy mini kit (QIAGEN category no. 74106) and QIAvac 24 vacuum manifold (QIAGEN category no. 19403). The extraction protocol was modified from the manufacturer's instructions as follows: 1 ml lysis buffer RLT (with 10 μl β-mercaptoethanol) was added directly into each Whirl-Pak bag and allowed to soak the filter for 10 min, and the resulting extracts (lysates) were carefully removed by pipette into 2-ml microcentrifuge tubes (droplets hanging in the bag and water clinging to the filter were first squeezed to the bottom corner of the bag by manually applying pressure to the outside of the bag). If there was visible filter or sample debris, the particulate matter was removed by brief centrifugation. One volume of 70% ethanol (usually 1 ml) was then added to the extract and mixed by pipetting. Samples were transferred to the RNeasy spin columns filtered through with the QIAvac at approximately 500 mm Hg vacuum and were washed on the manifold, once with 700 μl RW1 solution and twice with 500 μl RPE solution to remove contaminants. The columns were cleared of remaining droplets of buffer by centrifugation into a 2-ml collection tube (14,000 rpm, Eppendorf 5415 microfuge, 2 min), and the buffer was discarded. The RNA was eluted from the columns into a 1.5-ml collection tube with 50-μl volumes of RNase-free water by centrifugation (10,000 rpm, 2 min; Eppendorf) after allowing the water to stay in the column 1 min. This filter extraction step typically took up to 2 h for 15 samples. In this study, we routinely froze the extracts at −80°C for qRT-PCR analysis the next day.
For each PCR, 5 μl of the 50-μl RNA was analyzed by qRT-PCR on a Mx3000P Thermal Cycler (Stratagene, Inc.). The PCR protocol was modified from the single-tube RT-PCR method previously developed for sludge samples by Monpoeho et al. (19, 20). Primers and probe, unchanged from the original published method (except for the BHQ quencher), were reverse primer Ev1 [5′-GATTGTCACCATAAGCAGC-3′] and forward primer Ev2 [5′-CCCCTGAATGCGGCTAATC-3′], synthesized by QIAGEN, and Ev-probe (5′-FAM-CGGAACCGACTACTTTGGGTGTCCGT-BHQ-Phosphor-3′), synthesized by Sigma Genosys. A GenBank BLAST search done on 3 June 2004 revealed that only human (not other animal) enteroviruses matched all three primer and probe sequences. Each PCR contained 5 μl RNA extract and 20 μl master mix; each 20 μl master mix contained 1× Taq gold buffer (ABI), 5.5 mM MgCl2 (ABI), 500 μM deoxynucleoside triphosphates (ABI), 6% glycerol (Sigma Chemical Co.), 2% PVP 40(polyvinylpyrrolidone; average molecular weight of 40,000; Sigma Chemical Co.), 500 nM Ev1, 400 nM Ev2, 120 nM Ev-probe, 1.5 μg T4 gene 32 protein (Ambion), 10 U of RNAsin (ABI), 2.5 U of AmpliTaq gold (ABI), and 5 U of murine leukemia virus RT (ABI). Each RNA extract was analyzed in duplicate. Enterovirus RNA was transcribed into cDNA at 50°C for 45 min, the cDNA was amplified by PCR after a 95°C 10-min hot start for 50 cycles at 94°C for 15 s and 60°C for 1 min. Fluorescence measurements were made during the extension step, every cycle at 60°C. Calculations for quantification were done by the Stratagene QPCR software in real time, with raw data saved for possible reanalysis. Parameters (e.g., fluorescence threshold) were set manually after PCR was completed to generate a standard curve with optimal statistics (usually r2 > 0.95; slope around 3.3), and unknowns were calculated based on that standard curve. Standards were prepared using the poliovirus stock described above. Standards used in the high concentration set were 10-fold dilutions ranging from 330 viral particles/well to 3.3 × 105 viral particles/well, while the seven standards used in the low-concentration set were 33, 99, 330, 990, 3,300, 9,900, and 33,000 viral particles/well. The lower range of standards was used with lower seeding concentrations. qRT-PCR results were available 3 h after the start of analysis, making the total PCR preparation and analysis time less than 5 h for 15 samples (or <8 h including filtration and reasonable setup and transfer times; somewhat less for single samples). Results are reported as equivalent virus particles per unit of sample volume, meaning that this is where the qRT-PCR calculation indicated the sample appeared relative to the standard curve prepared from poliovirus standards. For various reasons (see Discussion) these should not be considered necessarily to be linearly proportional to the viral titer in the field but instead are treated as relative units.
To estimate the loss of recovery efficiency due to inhibition of the qRT-PCR assay by contaminants present in the RNA extracts and for losses during extraction, filters from unspiked field samples (i.e., with no seeded poliovirus) were cut in half before extraction, with half of the filter being “spiked” with 0.1 μl (6.6 × 105 particles) poliovirus and the other half untreated. The filters went through the extraction and qRT-PCR assays together with other samples. Comparison of the recovered virus in the spiked field sample (i.e., spiked minus unspiked field sample) with the normal standard curve (i.e., poliovirus added directly to a PCR with no sample extract) permitted estimation of the extent of RT-PCR inhibition by the extracts plus extraction losses.
Field study.
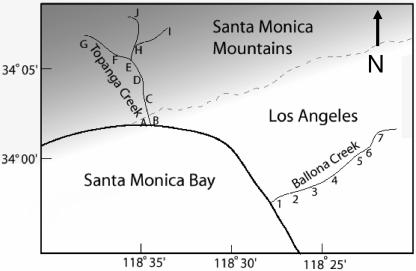
HA filters were used for the investigation of enterovirus contamination along Ballona Creek (BC) of Los Angeles on 16 July 2003 and investigation of TC on 13 April 2004. BC originates from the center of Los Angeles, encompassing an urban watershed ending at the Pacific Ocean, receiving urban wastewaters from streets, lawns, and potentially sewage leaks along the way. Two 1-liter water samples were obtained from seven different locations along BC, representing locations where bridges cross the concrete-lined channel (Fig. 1). TC is a small mountain creek that runs through a canyon and mountain community with low population density and no centralized sewer system (homes are on individual septic systems). Two 1-liter water samples were obtained from 10 different locations along Topanga Creek (Fig. 1). One liter of each water sample was filtered through the HA filter, and extraction and analysis in duplicate was performed as described for the seeding experiments above. Other parameters measured included temperature and salinity by a handheld probe from Yellow Springs Instruments, Inc.
FIG. 1.
Map of field sample locations. The light zone marked “Los Angeles” is largely urbanized, and the darker “Santa Monica Mountains” has greatly reduced population density.
RESULTS
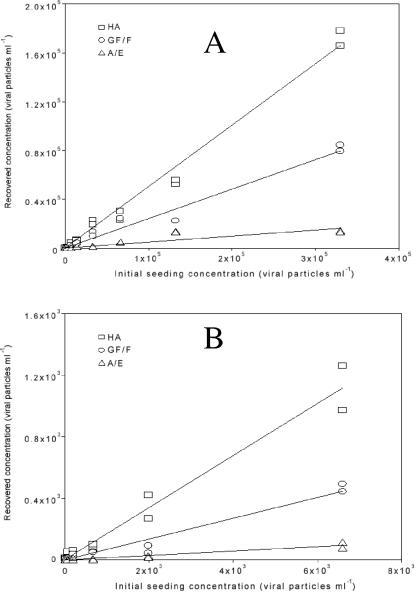
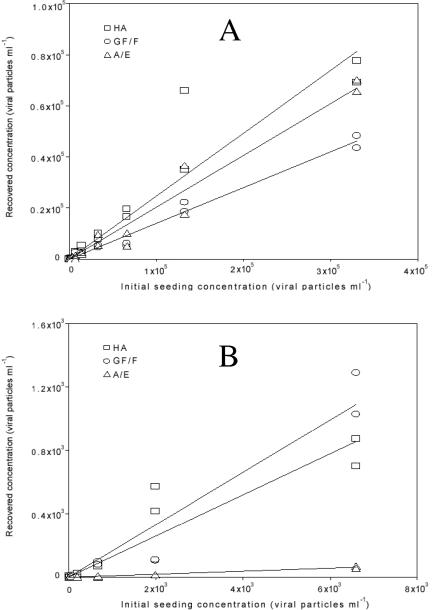
The seeding experiment showed that over the full range of initial seeding concentrations (6.6 × 103 to 3.3 × 105 poliovirus particles/ml in the first experiment and 6.6 to 6,600 poliovirus particles/ml in the second, 50 ml total volume), the HA filter demonstrated better overall recovery than the GF/F or A/E and the GF/F better than A/E filters in freshwater (Table 1, Fig. 2). Lower seeding concentrations demonstrated less recovery in general (Table 1). In seawater, the comparison was more complicated, with the HA best at high concentrations, followed by the A/E and GF/F in that order, but the GF/F was similar to or even better than HA in recovery at low concentrations (up to 6,600 enterovirus particles/ml). Note that at some concentrations in seawater, such as 2 × 104 enterovirus particles/ml, the HA recovery was substantially above the regression line even for the GF/F, demonstrating that although the results are semiquantitative, at low concentrations the recovery percentage is fairly “noisy” and not always close to a linear response (Fig. 3). The A/E was consistently much lower in recovery at those low levels (Table 1, Fig. 3). At an initial seeding concentration of 6.6 poliovirus particles/ml or below, recovery became unpredictable in that one out of two duplicates gave no detection.
TABLE 1.
Average recovery efficiencies (combination of filtration, extraction and PCR efficiencies from 50 ml samples) of seawater and freshwater over a poliovirus seeding range from 6.6 to 330,000 enterovirus particles/ml, as determined by linear regression (see Fig. 2 and 3)a
| Filter | Enterovirus particles/mlb | Seawater
|
Freshwater
|
||
|---|---|---|---|---|---|
| % Recovery efficiencyc | r2 | % Recovery efficiencyc | r2 | ||
| HA | 6.6-330,000 | 22.6 ± 2.8 | 0.90 | 51.2 ± 1.9 | 0.99 |
| GF/F | 6.6-330,000 | 13.9 ± 0.6 | 0.98 | 23.5 ± 1.5 | 0.97 |
| A/E | 6.6-330,000 | 20.7 ± 1.3 | 0.97 | 4.4 ± 0.7 | 0.82 |
| HA | 6.6-6,600 | 12.3 ± 1.3 | 0.87 | 16.7 ± 0.8 | 0.97 |
| GF/F | 6.6-6,600 | 17.3 ± 1.2 | 0.93 | 6.9 ± 0.4 | 0.96 |
| A/E | 6.6-6,600 | 0.9 ± 0.05 | 0.97 | 1.4 ± 0.1 | 0.92 |
Efficiencies are the slope of the regressions, and we also report the standard errors of the calculated slopes to indicate their variance. Boldface text indicates the low-concentration range up to 6,600 enterovirus particles/ml, that we believe is most appropriate to compare to actual field samples from natural waters.
In the poliovirus stock used, there was approximately 1 PFU per 66 enterovirus particles.
Mean ± standard error.
FIG. 2.
qRT-PCR-estimated recovery of seeded polioviruses from freshwater samples with three filter types. The y axis indicates where the sample results occurred on the standard curve prepared from poliovirus. Lines are linear regressions forced through the origin. Panel A shows data for the entire concentration range, and panel B shows only the lower range. All data points from duplicate measurements (two assays from each extracted filter) are shown. The same poliovirus stock was used for the seeding (x axis) and for the standard curve used to calibrate the y axis, hence the slope reflects the recovery. In the poliovirus stock used there was approximately 1 PFU per 66 enterovirus particles.
FIG. 3.
qRT-PCR-estimated recovery of seeded polioviruses from seawater samples with three filter types. This figure is the same as Fig. 2, but it is for seawater samples.
Tests for extraction loss and inhibition during the PCR step indicated that our seawater samples had more extraction loss and PCR inhibition than our freshwater samples. With the samples from 23 September 2003, the sum of extraction loss and PCR inhibition observed in seawater samples was 62% ± 5% and 23% ± 2% in freshwater samples at the spike level of 0.1 μl poliovirus stock, which was equivalent to 1.3 × 104 enterovirus particles/ml. The overall recovery efficiency observed (which was the combination of filtration, extraction, and PCR recovery efficiencies) at this seeding concentration was 30% ± 11% in seawater and 51% ± 1% in freshwater. If one uses the inverse of this inhibition and extraction loss as an average correction factor, the calculated corrected recovery is 92% ± 30% in seawater and 67% ± 1% in freshwater (note this used only the average correction factor and did not include the variation). However, we found in numerous preliminary tests, and with low spike additions of 19.8 poliovirus particles/ml, that variability in the apparent extraction loss and PCR inhibition was often high between field replicates. Had we used these to calculate “final corrected” efficiencies, they would sometimes exceed 100%.
In the field study, samples were freshwater with the exception of Pacific (salinity of 30 practical salinity units [PSU] compared to seawater at about 33 PSU) and Lincoln (9.5 PSU); temperatures in BC ranged 23 to 26°C and 12 to 15°C in TC. Field results of enterovirus measurements (Table 2) showed that from TC, the negative controls showed no detectable enteroviruses, and 2 out of 10 samples were detectably positive in both replicates: Falls and Backbone. A third sample, from Paradise, appeared positive in only one of the two replicates. Only the Falls sample was statistically significant above controls. In BC (Table 2), two out of seven samples were clearly positive for enteroviruses: Pacific and Higuera. These are samples taken from nonadjacent areas. There was a very low but detectable background level seen in one out of three negative controls from BC, and some of the field samples from BC had one replicate undetectable and one slightly detectable. We interpret the detectable negative control as probably a small contamination of RNA or PCR products in a reagent or tube used in that study. It did not interfere with detection and quantification of enteroviruses in the clearly contaminated samples from the Pacific and Higuera locations.
TABLE 2.
Ballona Creek (16 July 2003) and Topanga Creek (13 April 2004) field study enterovirus concentrations, in units of equivalent enterovirus particles per milliliterb
| Location | Fig. 1 symbol | Enterovirus particles/ml (mean ± SE)a |
|---|---|---|
| Ballona Creek | ||
| Pacific | 1 | 23.0 ± 2.3* |
| Lincoln | 2 | 0.007 ± 0.007 |
| Centinela | 3 | 0.11 ± 0.07 |
| Sepulveda | 4 | 0.45 ± 0.11 |
| Higuera | 5 | 9.8 ± 0.4* |
| National | 6 | 0.61 ± 0.54 |
| Fairfax | 7 | 0.35 ± 0.04 |
| No template control | 0.04 ± 0.04 | |
| Topanga Creek | ||
| Lagoon | A | 0 ± 0 |
| Rodeo Grounds | B | 0 ± 0 |
| Bridge | C | 0 ± 0 |
| Falls | D | 1.1 ± 0.37* |
| Abuelitas | E | 0 ± 0 |
| Backbone | F | 1.06 ± 0.73 |
| Zuniga | G | 0 ± 0 |
| Highvale | H | 0 ± 0 |
| Paradise | I | 0.33 ± 0.33 |
| Entrado | J | 0 ± 0 |
| No template control | 0 ± 0 |
*, significant (P < 0.05) versus control.
In the poliovirus stock used to make the standard curve, there was approximately 1 PFU per 66 enterovirus particles.
Freeze-thaw losses.
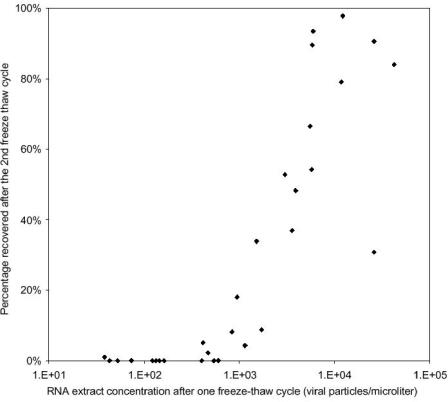
In the process of developing the method, analyzing data, and reanalyzing some stored (frozen) RNA samples, we noticed a rapid loss of enteroviruses in some RNA extracts after two freeze-thaw cycles, notably in samples with small amounts of RNA (note that one freeze-thaw cycle was normal in our analysis, as we froze the RNA extracts and analyzed them on a subsequent day). A plot of such results shows that when the concentration of RNA in samples (extracts) measured after one freeze-thaw cycle was below about 700 equivalent enterovirus particles/μl, the signal was completely lost after an additional freeze-thaw cycle. When the initial concentration of RNA extracts was higher than about 104 virus particles/μl, almost none was lost, with recovery usually from 80% to 100% (Fig. 4).
FIG. 4.
Amount of RNA detected in an extract after it was subjected to two freeze-thaw cycles as a percentage of the amount of RNA detected after one freeze-thaw cycle.
DISCUSSION
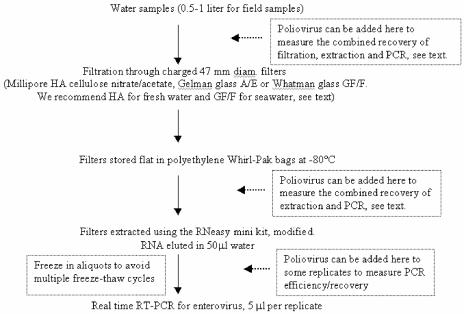
The results confirm the data of Katayama et al. (13) and Boehm et al. (1) showing that small charged filters can be used to collect enteroviruses from small volumes (∼1 liter) of field samples and that simple extraction of those filters and qRT-PCR can be used to detect viruses within hours. Our protocol differed from that of Katayama et al. (13) in a few ways that we think are more appropriate for our laboratory. First, we did not rinse the filters with acid and elute with base as done by Katayama et al. Instead, we extracted the filters directly into a small volume suitable for direct qRT-PCR analysis. We found in preliminary experiments that the acid-base treatment did not significantly improve overall recovery but that it did require extraction from a much larger liquid volume and thus made the subsequent extraction steps more difficult and time-consuming (we were also not using the extracts to grow viruses in cell culture, so this part of the Katayama procedure was deemed unnecessary). Second, we used an RNA extraction kit readily available in the United States and Europe (QIAGEN, Inc.) rather than the kit used by Katayama et al. (13), which is not available in the United States. This allows the procedure to be used in many more laboratories around the world. Third, we used a modified real-time PCR protocol originally developed for sewage sludge by Monpoeho et al. (19), which includes components to reduce interference common in many field samples (especially in “dirty” samples where human fecal-borne pathogens are suspected). These components include T4 gene 32 protein (a single-stranded nucleic acid binding protein), polyvinylpyrrolidone, and RNase inhibitor. We found in preliminary experiments that these components improved overall recovery by twofold or more in some field samples, hence we used these qRT-PCR reagents instead of a commercially available kit. Our overall recommended protocol is shown as a flowchart in Fig. 5.
FIG. 5.
Flow chart of protocol for measuring enteroviruses in natural waters, with dashed lines indicating possible locations of tests for recovery and efficiency of various steps. diam., diameter.
Of the three filter types we tested, the best overall recovery in freshwater was with the HA filters, and the GF/F filter appeared similar or even slightly better in seawater at the low concentrations one is likely to see in field samples. The A/E filter had generally lower recovery. Note that even in freshwater, the GF/F filters collected viruses with reasonable efficiency (all factors considered), and they have much more rapid flow and reduced likelihood of clogging than the HA type. Faster filtration permits testing of significantly more water with these filters, and filtration of highly turbid water samples through glass filters is much easier. In a previous study (1) we used the A/E filters to detect enteroviruses in saline groundwater because it is very difficult to filter turbid samples through fine-pore-size filters such as the HA filters.
In our tests with added polioviruses, the overall recovery percentage was highest at high additions and was reduced at low additions. This suggests that low but detectable virus amounts in field samples are probably underestimates. It also means that attempts to use internal standards to correct for all losses would require multiple parallel additions (different concentrations) to each sample to cover the proper internal standard range. Owing to the variability in losses and detection, we report results as “equivalent enterovirus particles,” but they should not necessarily be considered to be proportional to an actual virus titer. The reduced efficiency at low titers may be due to difficulty in capturing viruses, extraction losses, or storage losses. Regarding storage losses, we found that refreezing the extract for later analysis led to significant losses at low virus concentrations (Fig. 4). Thus, refreezing might result in false negatives. We speculate that this loss may be due to adsorption of viruses on the container walls.
While our recovery values from seeded field samples at low virus abundance may seem low in an absolute sense, it is important to consider we tested under realistic conditions highly relevant to actual field studies, not under ideal lab conditions or by spiking artificial seawater. At high enterovirus concentrations typical of other studies (see, e.g., reference 13), our recovery was comparably high, but we found such results to be unrealistic for field studies. We are not aware of any comparable small-volume and rapid method that has been shown to be better at the low (but still potentially hazardous) virus concentrations expected from contaminated recreational or shellfish harvesting waters.
We noted clear differences in uncorrected recovery in fresh- and seawater samples, and our inhibition experiments suggested that the differences may be in large part due to differences in inhibition of the PCR due to materials in the field extracts. Inhibition has been reported to be common and variable in extracts from field samples (28). Internal standards may be used to make some corrections for inhibition (Fig. 5), but as we noted, the concentration-dependent recovery would require careful choice of a range of internal standard additions, and the additional cost and effort may not be suitable for all studies.
For the two sets of field results, we saw a much higher level of enterovirus contamination (>10 times greater at “hotspots”) in BC than in TC. This may be because BC is an urban storm drain that passes through a highly urban watershed, while TC passes through a residential but relatively pristine mountain area with a much smaller potential for human fecal contamination. Nevertheless, it shows that even relatively pristine waterways in inhabited areas can have detectable enterovirus contamination.
In summary, the approach used here represents a readily usable means to detect human enteroviruses in field samples from freshwater and marine environments. It has the potential to be applied in routine monitoring.
Acknowledgments
This work was supported by USC Sea Grant and the State of California Water Quality Control Board, Proposition 13 funds.
We thank Ximena Hernandez, Rosi Dagit, Steve Williams, Ian Hewson, Mike Schwalbach, Anand Patel, Josh Steele, Denene Blackwood, and Jason Gregory for assistance in sample collection and preparation and Douglas Moore for providing the titered poliovirus stocks.
REFERENCES
- 1.Boehm, A. B., J. A. Fuhrman, R. D. Mrše, and S. B. Grant. 2003. A tiered approach for identification of a human fecal pollution source at a recreational beach: case study at Avalon Bay, Catalina Island, California, USA. Env. Sci. Technol. 37:673-680. [DOI] [PubMed] [Google Scholar]
- 2.Borrego, J. J., R. Cornax, D. R. Preston, S. R. Farrah, R. McElhaney, and G. Bitton. 1991. Development and application of new positively charged filters for recovery of bacteriophages from water. Appl. Environ. Microbiol. 57:1218-1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davis, B. D., R. Dulbecco, H. N. Eisen, and H. S. Ginsberg. 1990. Microbiology, 4th ed. J. B. Lippincott, Philadelphia, Pa.
- 4.Donaldson, K. A., D. W. Griffin, and J. H. Paul. 2002. Detection, quantitation and identification of enteroviruses from surface waters and sponge tissue from the Florida Keys using real-time RT-PCR. Water Res. 36:2505-2514. [DOI] [PubMed] [Google Scholar]
- 5.Enriquez, C. E., M. Abbaszadegan, I. L. Pepper, K. J. Richardson, and C. P. Gerba. 1993. Poliovirus detection in water by cell culture and nucleic-acid hybridization. Water Res. 27:1113-1118. [Google Scholar]
- 6.Gerba, C. P. 1988. Viral disease transmission by seafoods. Food Technol. 42:99-103. [Google Scholar]
- 7.Greening, G. E., J. Hewitt, and G. D. Lewis. 2002. Evaluation of integrated cell culture-PCR (C-PCR) for virological analysis of environmental samples. J. Appl. Microbiol. 93:745-750. [DOI] [PubMed] [Google Scholar]
- 8.Griffin, D. W., K. A. Donaldson, J. H. Paul, and J. B. Rose. 2003. Pathogenic human viruses in coastal waters. Clin. Microbiol. Rev. 16:129-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Griffin, D. W., C. J. Gibson, E. K. Lipp, K. Riley, J. H. Paul, and J. B. Rose. 1999. Detection of viral pathogens by reverse transcriptase PCR and of microbial indicators by standard methods in the canals of the Florida Keys. Appl. Environ. Microbiol. 65:4118-4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haile, R. W., J. S. Witte, M. Gold, R. Cressey, C. McGee, R. C. Millikan, A. Glasser, N. Harawa, C. Ervin, P. Harmon, J. Harper, J. Dermand, J. Alamillo, K. Barrett, M. Nides, and G. Y. Wang. 1999. The health effects of swimming in ocean water contaminated by storm drain runoff. Epidemiology 10:355-363. [PubMed] [Google Scholar]
- 11.Jiang, S., R. Noble, and W. P. Chui. 2001. Human adenoviruses and coliphages in urban runoff-impacted coastal waters of Southern California. Appl. Environ. Microbiol. 67:179-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang, S. C., and W. Chu. 2004. PCR detection of pathogenic viruses in southern California urban rivers. J. Appl. Microbiol. 97:17-28. [DOI] [PubMed] [Google Scholar]
- 13.Katayama, H., A. Shimasaki, and S. Ohgaki. 2002. Development of a virus concentration method and its application to detection of enterovirus and Norwalk virus from coastal seawater. Appl. Environ. Microbiol. 68:1033-1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Le Cann, P., S. Ranarijaona, S. Monpoeho, F. Le Guyader, and V. Ferre. 2004. Quantification of human astroviruses in sewage using real-time RT-PCR. Res. Microbiol. 155:11-15. [DOI] [PubMed] [Google Scholar]
- 15.Lee, H. K., and Y. S. Jeong. 2004. Comparison of total culturable virus assay and multiplex integrated cell culture-PCR for reliability of waterborne virus detection. Appl. Environ. Microbiol. 70:3632-3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lennette, E. H., D. A. Lennette, and E. T. Lennette. 1995. Diagnostic procedures for viral, rickettsial, and chlamydial infections, 7th edition. American Public Health Association, Washington, D.C.
- 17.Li, J. W., X. W. Wang, C. Q. Yuan, J. L. Zheng, M. Jin, N. Song, X. Q. Shi, and F. H. Chao. 2002. Detection of enteroviruses and hepatitis A virus in water by consensus primer multiplex RT-PCR. World J. Gastroenterol. 8:699-702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mohamed, N., A. Elfaitouri, J. Fohlman, G. Friman, and J. Blomberg. 2004. A sensitive and quantitative single-tube real-time reverse transcriptase-PCR for detection of enteroviral RNA. J. Clin. Virol. 30:150-156. [DOI] [PubMed] [Google Scholar]
- 19.Monpoeho, S., A. Dehee, B. Mignotte, L. Schwartzbrod, V. Marechal, J. C. Nicolas, S. Billaudel, and V. Ferre. 2000. Quantification of enterovirus RNA in sludge samples using single tube real-time RT-PCR. BioTechniques 29:88-93. [DOI] [PubMed] [Google Scholar]
- 20.Monpoeho, S., A. Maul, B. Mignotte-Cadiergues, L. Schwartzbrod, S. Billaudel, and V. Ferre. 2001. Best viral elution method available for quantification of enteroviruses in sludge by both cell culture and reverse transcription-PCR. Appl. Environ. Microbiol. 67:2484-2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nijhuis, M., N. van Maarseveen, R. Schuurman, S. Verkuijlen, M. de Vos, K. Hendriksen, and A. M. van Loon. 2002. Rapid and sensitive routine detection of all members of the genus enterovirus in different clinical specimens by real-time PCR. J. Clin. Microbiol. 40:3666-3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Noble, R. T., and J. A. Fuhrman. 2001. Enteroviruses detected by reverse transcriptase polymerase chain reaction from the coastal waters of Santa Monica Bay, California: low correlation to bacterial indicator levels. Hydrobiologia 460:175-184. [Google Scholar]
- 23.Noble, R. T., and J. A. Fuhrman. 1998. Use of SYBR Green I for rapid epifluorescence counts of marine viruses and bacteria. Aquat. Microb. Ecol. 14:113-118. [Google Scholar]
- 24.Paul, J. H., J. B. Rose, S. C. Jiang, X. T. Zhou, P. Cochran, C. Kellogg, J. B. Kang, D. Griffin, S. Farrah, and J. Lukasik. 1997. Evidence for groundwater and surface marine water contamination by waste disposal wells in the Florida Keys. Water Res. 31:1448-1454. [Google Scholar]
- 25.Pina, S., M. Puig, F. Lucena, J. Jofre, and R. Girones. 1998. Viral pollution in the environment and in shellfish: human adenovirus detection by PCR as an index of human viruses. Appl. Environ. Microbiol. 64:3376-3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reynolds, K. A., C. P. Gerba, M. Abbaszadegan, and I. L. Pepper. 2001. ICC/PCR detection of enteroviruses and hepatitis A virus in environmental samples. Can. J. Microbiol. 47:153-157. [PubMed] [Google Scholar]
- 27.Reynolds, K. A., C. P. Gerba, and I. L. Pepper. 1996. Detection of infectious enteroviruses by an integrated cell culture PCR procedure. Appl. Environ. Microbiol. 62:1424-1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reynolds, K. A., K. Roll, R. S. Fujioka, C. P. Gerba, and I. L. Pepper. 1998. Incidence of enteroviruses in Mamala Bay, Hawaii using cell culture and direct polymerase chain reaction methodologies. Can. J. Microbiol. 44:598-604. [PubMed] [Google Scholar]
- 29.Schwab, K. J., F. H. Neill, M. K. Estes, T. G. Metcalf, and R. L. Atmar. 1998. Distribution of Norwalk virus within shellfish following bioaccumulation and subsequent depuration by detection using RT-PCR. J. Food Prot. 61:1674-1680. [DOI] [PubMed] [Google Scholar]
- 30.Sobsey, M. D., K. J. Schwab, and T. R. Handzel. 1990. A simple membrane-filter method to concentrate and enumerate male-specific RNA coliphages. J. Am. Water Works Assoc. 82:52-59. [Google Scholar]