Abstract
Analyses for the presence of indicator organisms provide information on the microbiological quality of water. Indicator organisms recommended by the United States Environmental Protection Agency for monitoring the microbiological quality of water include Escherichia coli, a thermotolerant coliform found in the feces of warm-blooded animals. These bacteria can also be isolated from environmental sources such as the recreational and pristine waters of tropical rain forests in the absence of fecal contamination. In the present study, E. coli isolates were compared to E. coli K12 (ATCC 29425) by restriction fragment length polymorphism using pulsed-field gel electrophoresis. Theoretically, genomic DNA patterns generated by PFGE are highly specific for the different isolates of an organism and can be used to identify variability between environmental and fecal isolates. Our results indicate a different band pattern for almost every one of the E. coli isolates analyzed. Cluster analysis did not show any relations between isolates and their source of origin. Only the discriminant function analysis grouped the samples with the source of origin. The discrepancy observed between the cluster analysis and discriminant function analysis relies on their mathematical basis. Our validation analyses indicate the presence of an artifact (i.e., grouping of environmental versus fecal samples as a product of the statistical analyses used and not as a result of separation in terms of source of origin) in the classification results; therefore, the large genetic heterogeneity observed in these E. coli populations makes the grouping of isolates by source rather difficult, if not impossible.
Fecal pollution of water resources is an environmental problem of increasing importance as demographic densities increase. Fecal indicator bacteria are used to assess the microbial quality of water because they are not typically disease causing but may be correlated with the presence of several waterborne disease-causing organisms. An indicator of recent fecal contamination recommended universally to be used for monitoring the microbiological quality of water is Escherichia coli, a thermotolerant coliform found in the feces of warm-blooded animals (1). The use of E. coli as an indicator of fecal contamination relies on the assumption that its presence in water is a direct evidence of fecal contamination and indicates the possible presence of pathogens. However, several studies have shown that E. coli can be isolated from the pristine areas of a tropical rain forest in Puerto Rico (2, 3, 17, 20) and also from tropical soils and waters in Hawaii and subtropical areas such as Florida (11, 22). This continuous detection in nonhuman disturbance areas suggests that E. coli is a natural inhabitant in these environments and that it may be part of a previously established community.
For over a decade, the source of fecal indicator bacteria (such as thermotolerant coliforms, Escherichia coli, and enterococci) has been a pressing question in water quality assessment. Accurate risk analysis, effective remediation efforts, and valid total maximum daily load assessments all depend upon knowledge of the source of contamination (i.e., failing septic systems, overloads at sewage treatment facilities, wildlife, domestic pets, and runoff from nonpoint sources). The standard methods of measuring fecal pollution do not distinguish between human and animal sources (7). For this reason, there are efforts to develop methods to identify sources of fecal pollution in surface waters and groundwater. The methods used for tracking the source of contamination rely on some assumptions: geographical structure of the bacterial population, host specificity, and a stable clonal composition through time (10).
Molecular (DNA) fingerprinting methods have been described as promising for discriminating fecal-origin bacteria from humans versus animals. There are a number of genetic fingerprinting methods, pulsed-field gel electrophoresis (PFGE), restriction fragment length polymorphism, ribotyping, repetitive sequence-based PCR (including BOX-PCR, enterobacterial repetitive intergenic consensus [ERIC]-PCR, and repetitive extragenic palindromic [REP]-PCR), and amplified fragment length polymorphism (AFLP), that are being developed for discriminating between sources of indicator bacteria in natural waters. The data obtained with the different fingerprinting approaches are commonly complex, and thus multivariate analyses such as cluster analysis are used to group the isolates by similarities and therefore identify the major source of bacterial contamination. This approach has been useful in closed environments and especially so when dealing with clonal populations. As different possible sources of contamination are identified and a highly diverse population exists, it becomes more difficult to identify the possible source of contamination by cluster analysis. In these cases, methods such as discriminant analysis can be used to classify the isolates and group them by source (6, 21, 28). However because the discriminant analysis maximizes differences between groups, the analyst needs to be careful with the interpretation of the results.
Since 1996, the Centers for Disease Control and Prevention (CDC) have developed a national network of public health laboratories, called PulseNet, that permits access to a large database of the DNA fingerprints of isolated foodborne pathogens. The network identifies and labels each fingerprint pattern and permits rapid comparison of these patterns through the electronic database at the CDC to identify related strains. The fingerprinting uses PFGE, which can distinguish strains of pathogenic organisms such as Escherichia coli, Salmonella, Shigella, or Listeria at the genome level. PulseNet works well when dealing with clonal populations, as would be expected with pathogens.
In this study, we used PFGE to attempt to differentiate between fecal-origin bacteria (animal and human) and environmental-origin isolates. PFGE involves embedding organisms in agarose, lysing the organisms in situ, and digesting the chromosomal DNA with the restriction endonuclease XbaI that cleaves infrequently (8). We also analyzed the genetic heterogeneity of different E. coli populations and the use of PFGE conjointly with multivariate statistical analyses to classify the isolates.
MATERIALS AND METHODS
E. coli isolation.
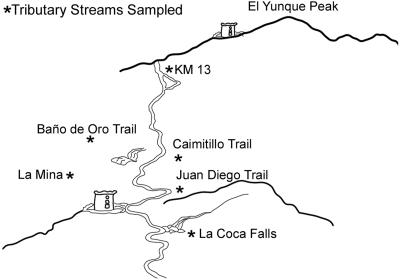
E. coli isolates were collected from six tributary streams (Fig. 1) and two different forests at different points in El Yunque and from soils in Bolivia. The surface water was divided into two areas: contaminated-recreational and pristine waters. Four tributary streams were used for recreational purposes and 21 samples were taken at these sites, and 17 samples were taken from two different pristine tributary streams. The pristine samples consisted of samples taken from isolated low-human-impact environments. These environments were located upstream from the recreational samples and had previously been shown not to be impacted by human or animal wastes. Soil samples were taken randomly at a distance of 5 m from a stream at a depth of 0 to 10 cm. A total of 23 soil isolates were analyzed. A total of 21 isolates from human feces and 4 from animal feces were also included in the analyses. All samples were collected in sterile bottles and kept at 4 to 7°C until processed, within 24 h. E. coli was isolated using standard membrane filtration on mFC agar (Difco Laboratories, MI) and incubated at 44.5°C for 18 to 24 h. All dark blue colonies on mFC agar were first subcultured onto eosin methylene blue agar (EMB; Difco Laboratories, MI) and then onto methylumbelliferyl-β-d-glucopyranoside (MUG)-containing media to test for uidA activity. Fecal E. coli isolates were obtained from humans and warm-blooded animals using rectal swabs and sterile 0.85% saline solution and then isolated on EMB agar. E. coli isolates were randomly analyzed.
FIG. 1.
Map showing location of the study site in the El Yunque tropical rain forest in Luquillo, Puerto Rico.
PFGE.
The conditions used for typing E. coli by PFGE were obtained from a standard methodology for tracking E. coli O157:H7 outbreaks (5). Briefly, E. coli isolates were subcultured on EMB at 37°C for 16 to 18 h. From the overnight E. coli culture, a single colony was obtained and incubated on tryptic soy agar overnight at 37°C. Then, single colonies were suspended in 3 ml of TE buffer (100 mM Tris, 100 mM EDTA, pH 8.0) to a transmittance between 13 and 15%. Plugs were formed by mixing 0.2 ml of cell suspension of proteinase K solution (20 μg enzyme/ml H2O; Sigma Chemicals Company, MO) and 0.2 ml of agarose solution (1.6% pulsed-field certified agarose [Bio-Rad Laboratories, CA]-1.0% sodium dodecyl sulfate in 10 mM Tris-1 mM EDTA, pH 8.0). The mixture of cell-agarose was pipetted into plug molds designed for PFGE (Bio-Rad Laboratories, CA). Solidified plugs were transferred to 1.5 ml of lysis buffer (50 mM Tris, 50 mM EDTA [pH 8.0], 1% Sarcosine, 0.5 mg/ml proteinase K) and incubated for 16 to 20 h in a shaker water bath at 50°C. The lysis buffer was removed and the plugs washed six times with 15 ml of sterile TE buffer (10 mM Tris, 1 mM EDTA, pH 8.0) for 20 min in a 50°C shaker water bath. Two 1-mm-thick slices were cut from the plugs with a sterile razor blade. Then the sliced plugs were incubated in 100 μl of restriction enzyme solution for 3 h at 37°C. The enzyme solution consists of 30 U XbaI in 100 μl of 1× enzyme buffer (Promega Corporation, WI). The PFGE was done in 1.2% of pulsed-field certified agarose (Bio-Rad Laboratories, CA) in a 0.5× Tris-borate-EDTA running buffer. Electrophoresis was performed using a CHEF Mapper (Bio-Rad Laboratories, CA). The running time was 20 h, with a linear ramping from 2.16 s to a 54.17-s switch time at an angle of 120° (60°/−60°) at 6.0 V/cm and 14°C. The gels were stained for 30 min with 600 μg of ethidium bromide in 500 ml of sterile water and washed with distilled water for 30 min. Gel analysis was performed using the Diversity Database software (Bio-Rad Laboratories, CA). E. coli K12 (ATCC 29425) was used as the control.
Statistical analysis.
For the cluster analysis and discriminant analysis, the bands were identified as binary variables such as presence or absence of the band in the fingerprint (1 and 0, respectively). For the cluster analysis of binary data, Systat recommends using normalized percent disagreement metric distance, that is, the percentage of comparisons resulting in disagreement in two profiles (20). The cluster method used was the complete linkage which computed the between-cluster distances using the most distant pair of objects in two clusters (20). The discriminant analysis was performed stepwise, using a tolerance of 0.001 which measured the correlation of a candidate variable with the variables included in the model. The probability to remove or enter a variable in the model was set at 0.15. The scores of the first two canonical variables (CVs), which are the linear combinations of variables that discriminate among groups, were obtained and used to show the relations of the isolates in a plot. For cross validation of the classification results, separate analyses were run, the isolates were grouped randomly, and the random groups were identified using letters (a, b, c, d) in repeating sequences from a to d. The results of the classification of isolates with their respective groups are presented in a classification matrix (each case is classified into a group, though when computing the classification functions using all cases, sometimes results are more optimistic) and jackknife classification matrix (which compute the classification, leaving out one case at a time). The assigned group similarities were calculated with the between-groups F matrix using Mahalanobis D2 statistics that calculate the distance between the centroids of the groups.
The effect of the number of isolates versus the number of variables in the fidelity of discriminant analysis was analyzed by the creation of different artificial data sets with variation in the proportion in the number of samples and number of variables. The use of an artificial data set is important because it ensures that no relationships develop within any particular group. The artificial data sets were created using Bernoulli distribution with a success probability set at 0.25 in Minitab 12 software (Minitab Inc.). The proportion of the number of samples versus the number of variables is presented in Table 3.
TABLE 3.
Testing the effect of the n × p range in the classification results of data generated randomlya
| n × p | % Overall classification
|
|
|---|---|---|
| Correct | Jackknife | |
| 86 × 65 | 55 | 39 |
| 86 × 130 | 100 | 21 |
| 86 × 254 | 100 | 12 |
| 300 × 254 | 73 | 51 |
| 300 × 65 | 29 | 23 |
The artificial data set was created using Bernoulli distribution with a success probability set at 0.25 in Minitab 12 software (Minitab Inc.). The artificial data sets have variation in sample numbers and variable numbers.
RESULTS
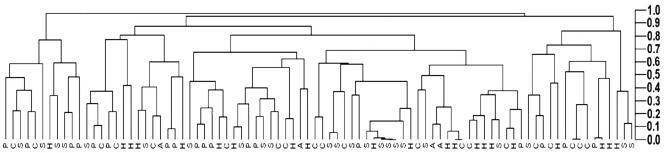
Eighty-six E. coli isolates were analyzed with XbaI digestion followed by PFGE. The number of isolates analyzed, although it may seem small, may be typical of a study involving routine source tracking in the environment by using PFGE. Fragment sizes were from 30 kbp to 600 kbp. The cluster analysis of the different band patterns placed almost every E. coli isolate in a different cluster (Fig. 2). In the dendrogram, the isolates were not clearly clustered with the source of origin, although some isolates from the same sources were clustered together. This could be attributed to a possible clonal replication of the isolates. The overall cluster formation does not indicate a clear relationship between sources and isolate.
FIG. 2.
Cluster analysis of the PFGE results from 86 E. coli isolates. The letters represent the source of origin, as follows: A, animal; H, human; S, soil; P, pristine waters; R, recreational waters.
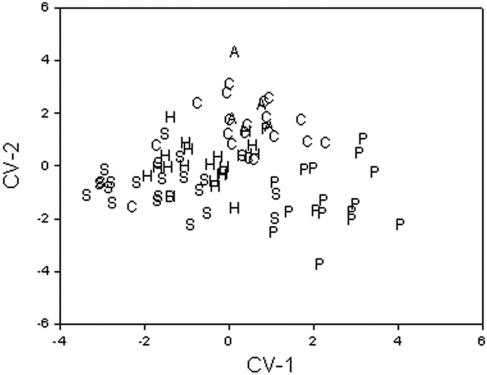
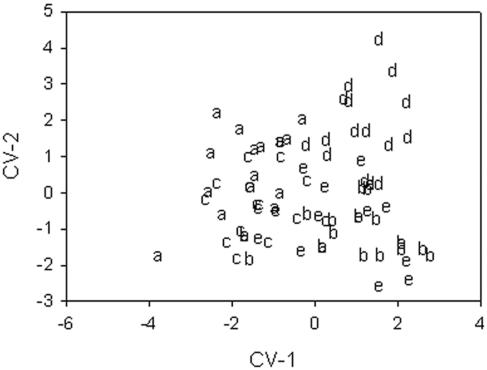
The results of the classification matrix are presented in Table 1. The cases are classified into columns. The overall correct classification average was 84% with the classification matrix and 57% with the jackknife classification matrix. The sources with the highest correct classification were animal samples with 100% and pristine samples with 94%. The results obtained with the jackknife approach show the pristine isolates with 71% and soil isolates with 70% of the isolates. Figure 3 shows two canonical function variables (CV-1, CV-2). CV-1 explains 42.3% of the dispersion, and CV-2 explains 25.6%. The pristine samples were localized in the positive values of CV-1, and soil samples were localized in the negative values of CV-1. The samples of the contaminated and recreational water are localized in the positive values of CV-2. The human isolates were dispersed between the soil and pristine- and contaminated-water isolates. The between-groups F-matrix results show that the human source isolates and the animal source isolates are the closest to each other (f = 1.53) and that the soil source isolates and pristine-water-source isolates are the groups farthest from each other. The results of the random grouping of the samples were 63% for the classification matrix and 43% for the jackknife classification matrix (Table 2). Figure 4 shows two canonical function variables for the validation. CV-1 explains 49.7% of the dispersion, and CV-2 explain, 40.5%. Random group d was localized in the positive values of CV-1, and random group a was localized in the negative values of CV-1. Figure 4 also shows that ordinations of the isolates are similar to ordinations of the isolates observed in Fig. 3.
TABLE 1.
Classification table for the 86 E. coli isolates with the source of origina
| Source | Animal | Human | Pristine | Soil | Contaminatedb | % Correct |
|---|---|---|---|---|---|---|
| Animal | 4 (1) | 0 (1) | 0 (0) | 0 (1) | 0 (1) | 100 (25) |
| Human | 0 (2) | 16 (9) | 0 (1) | 3 (4) | 2 (4) | 76 (43) |
| Pristine | 0 (2) | 0 (1) | 16 (12) | 0 (0) | 1 (2) | 94 (71) |
| Soil | 0 (0) | 3 (4) | 2 (2) | 18 (16) | 0 (1) | 78 (70) |
| Contaminated | 0 (2) | 2 (5) | 0 (1) | 1 (2) | 18 (11) | 86 (52) |
| Total | 4 (7) | 21 (20) | 18 (16) | 22 (24) | 21 (19) | 84 (57) |
Cases are classified into columns. Numbers in parentheses are the results obtained in the jackknife classification matrix.
These are for isolates from contaminated and recreational waters.
FIG. 3.
Dot plot of the two first canonical variables obtained for 86 E. coli isolates analyzed by PFGE. The letters represent the source of origin, as follows: A, animal; H, human; S, soil; P, pristine waters; R, recreational waters.
TABLE 2.
Classification table of the 86 E. coli isolates grouped randomlya
| Random group | a | b | c | d | e | % Correct |
|---|---|---|---|---|---|---|
| a | 12 (9) | 1 (1) | 4 (7) | 1 (1) | 0 (0) | 67 (50) |
| b | 0 (1) | 10 (6) | 2 (2) | 0 (2) | 5 (6) | 59 (35) |
| c | 6 (7) | 1 (2) | 9 (7) | 0 (0) | 1 (1) | 53 (41) |
| d | 0 (4) | 0 (2) | 0 (0) | 14 (9) | 3 (2) | 82 (53) |
| e | 0 (0) | 3 (6) | 4 (4) | 0 (1) | 10 (6) | 59 (35) |
| Total | 18 (21) | 15 (17) | 19 (20) | 15 (13) | 19 (15) | 64 (43) |
Cases are classified into columns. Numbers in parentheses are the results obtained in the jackknife classification matrix.
FIG. 4.
Dot plot of the first two canonical variables of E. coli PFGE data grouped randomly. The symbols (a, b, c, d, e) represent the random groups.
The results observed by analyzing artificial data 1 sets with discriminant analysis are shown in Table 3. The data sets with the same proportion of the numbers of samples versus the numbers of variables (n × p) as the data obtained in our experiment show 55% correct classification with the classification matrix and 39% with the jackknife classification matrix. Overall, the classification rates increase when the numbers of variables are higher than the number of samples. In the data set with 86 samples and 254 variables, 100% correct classification was obtained. In this case, this proportion of n × p does not influence the jackknife classification matrix. However, when the sample and variable numbers are high, the jackknife classification analysis could give false-positive classification results. In the data set with 300 samples and 254 variables, the classification with the jackknife matrix was 51% correct.
DISCUSSION
The advantage of using a standardized protocol for PFGE is the robustness, as evidenced by the highly reproducible gels. Duplicate experiments using the same isolates displayed identical XbaI-PFGE profiles, demonstrating the robustness of the method, as well as genetic heterogeneity in E. coli populations (data not shown). Various studies have successfully typed E. coli isolates by pulsed-field gel electrophoresis using the XbaI restriction enzyme exclusively (26, 27, 29). Two isolates are considered to be closely or possibly related if their PFGE patterns have almost the same number of bands (25). In this study, PFGE failed to reveal differences among the sources of E. coli isolates. Neither the environmental nor the fecal strains shared band patterns among themselves, which would have allowed us to separate them into discrete groups.
Cluster analysis did not show any relations between samples and their environment. Only the discriminant function analysis grouped the samples with the source of origin. The discrepancies observed between the cluster analysis and discriminant function analysis rely on the mathematical bases of the two analyses. The mathematical calculation of the discriminant analysis maximizes the variability between groups, removing all variables that do not increase or account for that variability (18). To validate the results, it is recommended that the analysis be run several times, assigning the groups randomly. Our validation analyses indicate artifact possibility in the classification results. We observed differences in the classification results obtained with the jackknife classification matrix. The classification matrix and the jackknife classification matrix have differences in the calculation of the classification function. The classification matrix results are more optimistic than those obtained with the jackknife matrix. If the results obtained with the jackknife classification matrix are lower than those obtained with the classification matrix, it may indicate that there are too many variables in the model (24). Therefore, the high degree of genetic heterogeneity observed in these E. coli populations results in too many different RFLP band patterns that increase the number of variables and make source tracking analysis more difficult.
The results obtained with the randomly generated RFLP data show that when the number of bands analyzed is greater than the number of isolates, the probability of a false-positive classification increases. Discriminant analysis is a useful method to select the variables that increase or account for any difference between groups. When many variables are used in the analysis, it is more likely to group the observations with a particular model, making the final significant model of the groups not valid (13). However, it has been observed that an increase in the number of variables may lead to better results when trying to classify isolates according to different sources. For example, Leung et al. (16) compared the use of AFLP and ERIC-PCR for the discrimination of E. coli from different animal sources. In their comparison, they had 63 isolates and a total of 390 bands produced by AFLP and they reported 100% correct grouping when using discriminant analysis. Using the ERIC-PCR with the same isolates but considerably lower total numbers of bands (18 bands), they reported a 33% average correct classification result. Seurinck et al. (21) reported that when two fingerprint analyses (BOX-PCR and 16-23S rRNA intergenic spacer region PCR) were used together, they correctly grouped the isolates as to the sources. However, the same grouping was lower when only one fingerprint analysis was used. These reported results describe the same phenomenon that we obtained using our artificial data set. Using a large number of variables allowed us to obtain greater correct classification rates in the discriminant analysis; however, these statistically significant classification rates may be not biologically valid and thus the fidelity of the analysis decreases. Therefore, the results obtained with artificial data sets, as we carried out, demonstrated the importance of reducing the number of variables in order to increase the fidelity of the results when stepwise discrimination methods are used to classify the isolates in source tracking.
Our results demonstrated the genetic heterogeneity in environmental and human gut populations of E. coli. This heterogeneity is clearly observed in the dendrograms obtained for the PFGE of these isolates (Fig. 2). The classification and the canonical variance analysis resolve the pristine- and contaminated-recreational-water and soil isolates. However, the relationship between the human isolates is not clearly observed. The heterogeneity observed in the human isolates could be the result of a large within-host variability.
The heterogeneity of the E. coli populations is also observed in other recently published articles using different techniques for ribotyping and fingerprinting (4, 9, 21). Although the objective of those articles was to demonstrate the application of molecular techniques to identify the source of fecal contamination, they also demonstrated the diversity of E. coli populations in different hosts and environments. The diversity of E. coli populations was also shown to be high in closed environments such as bovine feed lots (28). Due to the E. coli genetic diversity, there is a need for intensive sampling and for an enormously large number of isolates for source tracking to be successful.
The success of source tracking methods depends on the geographical structure, host specificity, and stability through time of the species being monitored. Gordon (10) described how the characteristics of the E. coli populations invalidate the use of source tracking methods to identify the source. First, the clonal composition shows little temporal stability in the E. coli populations obtained from wild animal hosts (14). In gulls and cows, a clonal dominance was described in one individual and a high variability in E. coli populations between members of the same host species (19). Second, the geographical structure or host specificity accounts for little of the observed genetic diversity in E. coli populations (23).
We have shown that background E. coli populations in the environment are genetically heterogeneous. In our laboratory, we observed constant changes in the pristine-environment E. coli population genetic patterns (15). Therefore, the genetic heterogeneity as well as the possible temporal diversity of E. coli environmental populations should be considered in cases when this genus and species is used to track fecal contamination. Finally, this study also cautions the user of different statistical analyses for source tracking purposes. Our results demonstrate that different results can be obtained when different statistical analyses are used. For example, to differentiate between environmental and clinical E. coli isolates, the cluster analysis does not indicate a clear relation between sources and isolates. However, with the discriminant analysis, the correct classification average was 84%, and with a jackknife classification matrix, it was 57%. The fidelity of discriminant analysis for source tracking may be improved by removing the clonal isolates and by the addition of similarity value thresholds and quality factor thresholds, such as the ones described by Hassan et al. (12). These similarity value thresholds and quality factor thresholds, when they are used as published previously (12) in discriminant analysis, increase the correct assignment of isolates to a source, but the percentage of isolates classified onto a source drastically decreases and thus the possibility of incorrectly classifying an isolate to any given source also decreases. Previous studies have grouped E. coli isolates into possible source types; however, since grouping and reliability depend on the statistical analysis used, the analyst should be aware of possible unreliable groupings.
Acknowledgments
We thank Luis A. Perichi from the Department of Mathematics, University of Puerto Rico, for assisting us with the statistical analysis programs.
This study was funded by RCMI, RISE, and by a grant from the WRRI at the University of the Virgin Islands.
REFERENCES
- 1.American Public Health Association. 1988. Standard methods for the examination of water and wastewater, 17th ed. American Public Health Association/American Water Works Association/Water Environment Federation, Washington, D.C.
- 2.Bermúdez, M., and T. C. Hazen. 1988. Phenotypic and genotypic comparison of Escherichia coli from pristine tropical waters. Appl. Environ. Microbiol. 54:979-983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carrillo, M., E. Estrada, and T. C. Hazen. 1985. Survival and enumeration of the fecal indicators Bifidobacterium adolescentis and Escherichia coli in a tropical rain forest watershed. Appl. Environ. Microbiol. 50:468-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carson, C. A., B. L. Shear, M. R. Ellersieck, and A. Asfaw. 2001. Identification of fecal Escherichia coli from humans and animals by ribotyping. Appl. Environ. Microbiol. 67:1503-1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. 1997. Standardized molecular subtyping of Escherichia coli O157:H7 by pulsed-field gel electrophoresis: a training manual. Foodborne and Diarrheal Diseases Branch, Division of Bacterial and Mycotic Diseases, National Center for Infectious Diseases, Centers for Disease Control and Prevention, Atlanta, Ga.
- 6.Dombek, P. E., L. K. Johnson, S. T. Zimmerley, and M. J. Sadowsky. 2000. Use of repetitive DNA sequences and the PCR to differentiate Escherichia coli isolates from human and animal sources. Appl. Environ. Microbiol. 66:2572-2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Field, K. G. 2002. Fecal source tracking with Bacteroides. U.S. EPA workshop on microbial source tracking, Irvine, Calif. [Online.] ftp://ftp.sccwrp.org/pub/download/PDFs/Micro_source_tracking_wkshop/04_Field.pdf.
- 8.Finney, M. 1993. Pulsed-field gel electrophoresis, p. 2.5.9-2.5. 17. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology, vol. 1. Greene-Wiley, New York, N.Y. [Google Scholar]
- 9.Geornaras, I., J. W. Hastings, and A. von Holy. 2001. Genotypic analysis of Escherichia coli strains from poultry carcasses and their susceptibilities to antimicrobial agents. Appl. Environ. Microbiol. 67:1940-1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gordon, D. M. 2001. Geographical structure and host specificity in bacteria and the implications for tracing the source of coliform contamination. Microbiology 147:1079-1085. [DOI] [PubMed] [Google Scholar]
- 11.Hardina, C. M., and R. S. Fujioka. 1991. Soil: the environmental source of Escherichia coli and enterococci in Hawaii's streams. Environ. Toxicol. Water Qual. Int. J. 6:185-195. [Google Scholar]
- 12.Hassan, W. N., S. Y. Wang, and R. D. Ellender. 2005. Methods to increase fidelity of repetitive extragenic palindromic PCR fingerprint-based bacterial source tracking efforts. Appl. Environ. Microbiol. 71:512-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.James, M. 1985. Feature selection-variable selection, p. 127-149. In Classification algorithms. Collins Professional and Technical Books, William Collins Sons & Co. Ltd., London, United Kingdom.
- 14.Jenkins, M. B., P. G. Hartel, T. J. Olexa, and J. A. Stuedemann. 2003. Putative temporal variability of Escherichia coli ribotypes from yearling steers. J. Environ. Qual. 32:305-309. [DOI] [PubMed] [Google Scholar]
- 15.Lasalde, C., R. Rodriguez, H. Smith, and G. A. Toranzos. Heterogeneity of uidA gene in environmental Escherichia coli populations. J. Water Health, in press. [DOI] [PubMed]
- 16.Leung, K. L., R Mackereth, Y Tien, E Topp. 2004. A comparison of AFLP and ERIC-PCR analyses for discriminating Escherichia coli from cattle, pig and human sources. FEMS Microbiol. Ecol. 47:111-119. [DOI] [PubMed] [Google Scholar]
- 17.López-Torres, A., T. Hazen, and G. A. Toranzos. 1987. Distribution and In situ survival and activity of Klebsiella pneumoniae and Escherichia coli in tropical rain forest watershed. Curr. Microbiol. 15:213-218. [Google Scholar]
- 18.Manly, B. 1986. Discriminant function analysis, p. 86-99. In Multivariate statistical methods: a primer. Chapman and Hall, New York, N.Y.
- 19.McLellan, S. L., A. D. Daniels, and A. K. Salmore. 2003. Genetic characterization of Escherichia coli populations from host sources of fecal pollution by using DNA fingerprinting. Appl. Environ. Microbiol. 69:2587-2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rivera, S. C., T. C. Hazen, and G. A. Toranzos. 1988. Isolation of fecal coliforms from pristine sites in a tropical rain forest. Appl. Environ. Microbiol. 54:513-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seurinck, S., W. Verstraete, and S. D. Siciliano. 2003. Use of 16S-23S rRNA intergenic spacer region PCR and repetitive extragenic palindromic PCR analyses of Escherichia coli isolates to identify nonpoint fecal sources. Appl. Environ. Microbiol. 69:4942-4950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Solo-Gabriele, H. M., M. A. Wolfert, T. R. Desmarais, and C. J. Palmer. 2000. Sources of Escherichia coli in a coastal subtropical environment. Appl. Environ. Microbiol. 66:230-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Souza, V., M. Rocha, A. Valera, and L. E. Eguiarte. 1999. Genetic structure of natural populations of Escherichia coli in wild hosts on different continents. Appl. Environ. Microbiol. 65:3373-3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Systat 9. 1999. Statistics I. SPSS Inc., Chicago, Ill.
- 25.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vali, L., K. A. Wisely, M. C. Pearce, E. J. Turner, H. I. Knight, A. W. Smith, S. G. Amyes. 2004. High-level genotypic variation and antibiotic sensitivity among Escherichia coli O157 strains isolated from two Scottish beef cattle farms. Appl. Environ. Microbiol. 70:5947-5954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Valverde, A., T. M. Coque, M. P. Sanchez- Moreno, A. Rollan, F. Baquero, and R. Canton. 2004. Dramatic increase in prevalence of fecal carriage of extended-spectrum beta-lactamase-producing Enterobacteriaceae during nonoutbreak situations in Spain. J. Clin. Microbiol. 42:4769-4775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang, H. H., R. T. Vinopal, D. Grasso, and B. F. Smets. 2004. High diversity among environmental Escherichia coli isolates from a bovine feedlot. Appl. Environ. Microbiol. 70:1528-1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao, S., S. Qaiyumi, S. Friedman, R. Singh, S. L. Foley, D. G. White, P. F. McDermott, T. Donkar, C. Bolin, S. Munro, E. J. Baron, R. D. Walker. 2003. Characterization of Salmonella enterica serotype Newport isolated from humans and food animals. J. Clin. Microbiol. 41:5366-5371. [DOI] [PMC free article] [PubMed] [Google Scholar]