Abstract
A mosaic genomic island comprising Shigella resistance locus (SRL) sequences flanked by segments of Escherichia coli O157:H7 strain EDL933 O islands 43, 81, and 82 was identified in sorbitol-fermenting (SF) enterohemorrhagic Escherichia coli (EHEC) O157:H− strain 493/89. This mosaic island is absent from strain EDL933. PCR targeting the SRL-related sequence is a useful tool to distinguish SF EHEC O157:H− from EHEC O157:H7.
Sorbitol-fermenting (SF) enterohemorrhagic Escherichia coli (EHEC) O157:H− have been implicated in outbreaks, as well as sporadic cases of diarrhea and hemolytic-uremic syndrome (1, 9). Multilocus enzyme electrophoresis and sequence typing indicate that SF EHEC O157:H− are closely related to EHEC O157:H7 and to E. coli O55:H7 (16). SF EHEC O157:H− also possess a complete gene cluster encoding flagella, but loss of motility in these strains is caused by a 12-bp in-frame deletion in flhC that is required for transcriptional activation of genes involved in flagellin biosynthesis (11). The genome sequencing of EHEC O157:H7 strains EDL933 and Sakai (6, 13) demonstrated numerous islands of inserted DNA (O islands or SpLES) in both E. coli O157 pathogens that are absent from nonpathogenic E. coli K-12 strain MG1655 (3). To identify strain-specific genomic differences, suppression subtractive hybridization (SSH) has been used as a highly effective method (7, 15). This technique allows the identification of sequences that are present on one genome (“the tester”) but not the other genome (“the driver”) (7, 15). SSH between Shiga toxin-producing E. coli O91:H21 patient isolate and a nonpathogenic E. coli strain identified sequences from the E. coli O91:H21 virulence plasmids that were homologous to Shigella flexneri (15). The latter organism also contains a cluster of genes known as the Shigella resistance locus (SRL) that encodes resistance to streptomycin, ampicillin, chloramphenicol, and tetracycline (19). SRL is located within a 66-kb pathogenicity island (designated SRL PAI) in S. flexneri 2a strain YSH6000 (19); this PAI also carries a ferric-dicitrate uptake system (fec) (10).
SSH.
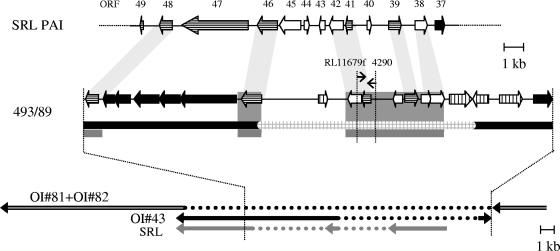
Using SSH between SF EHEC O157:H− strain 493/89 and EHEC O157:H7 strain EDL933 as described previously (7) and subsequent sequence analysis, several genes that are not present in E. coli O157:H7 strain EDL933 were identified. Some of them encoding potential virulence factors such as EHEC factor for adherence (efa1), cytolethal distending toxin (cdt-V), and Sfp fimbriae (sfp) have been characterized in our previous studies (4, 7, 8). The other genes detected by SSH and sequence analysis, which were present in strain 493/89 but absent from strain EDL933, demonstrated 96 to 98% homology to the respective genes of E. coli K-12. They included rspB (GenBank accession number AE000254) encoding starvation sensing protein, ycgZ (GenBank accession number AE000215) and ydfK (GenBank accession number AE000252), which encode hypothetical proteins. This strategy also provided evidence for the presence of a fragment with homology to the SRL PAI of S. flexneri, which is absent from E. coli K-12. Therefore, we sequenced (primers supercos1fwd and supercos1rwd) (7) one clone from a cosmid library of strain 493/89 (7), which contained this fragment. Sequences were analyzed (DNASIS; Hitachi Software) and compared to sequences from the NCBI database. The distribution of the SRL PAI-related sequences among 196 E. coli O157:H7 or H− and 12 E. coli O55:H7 strains isolated between 1987 and 2003 in five different countries (Table 1) was investigated by using PCR with primers RL11679f (5′-GTAGATATTCGGATGACACA-3′) and 4290 (5′-CAGACAACCTTATCCCATCG-3′) derived from the SRL sequence of strain 493/89 (Fig. 1). PCR was performed in 30 cycles of denaturing (94°C, 30 sec), annealing (55°C, 1 min), and extension (72°C, 1 min), followed by a final extension (72°C, 5 min). The absence of the SRL-related sequence spanned by the primers RL11679f and 4290 in PCR-negative strains was confirmed by Southern blot hybridization with an ECL Direct Nucleic Acid Labeling and Detection Systems kit (Amersham Pharmacia Biotech, Freiburg, Germany). This method was also used to test the presence of fecA, iha, ter, and ure genes. For this purpose, digoxigenin-labeled fecA probe derived from S. flexneri strain YSH6000 (GenBank accession no. AF326777) with primers Fec1 (5′-TGCCTTTGTTGTTGTCGTCA-3′) and Fec3 (5′-GAGACGCACAACCTGATGGT-3′), and iha, terC, and ureC probes derived from strain EDL933 by PCR with primers Iha-I (5′-CAG TTC AGT TTC GCA TTC ACC-3′) and Iha-II (5′-GTA TGG CTC TGA TGC GAT G-3′), TerC1 and TerC2 (2), and UreC-f and UreC-r (5), respectively, were used. Antibiotic susceptibilities were tested by using a standard disk diffusion method.
TABLE 1.
Distribution of SRL-related sequences among E. coli O157 and O55:H7 strains
| Serotypea | Country of origin | No. of strains | No. of strains from patients withb:
|
Sorbitol fermentation (no. of strains) | No. of strains with:
|
||||
|---|---|---|---|---|---|---|---|---|---|
| HUS | D | A | eae | stx | SRLc | ||||
| O157:H− | Austria, Czech Republic, Germany | 88 | 69 | 16 | 3 | 88 | 88 | 88d | 88 |
| O157:H− | Czech Republic, Germany | 7 | 1 | 6 | 0 | 7 | 7 | 0 | 7 |
| O157:H7 | Austria, Czech Republic, Denmark, Germany, United States | 101 | 79 | 20 | 2 | 0 | 101 | 101e | 0 |
| O55:H7 | Germany, United States | 12 | 0 | 12 | 0 | 12 | 12 | 0 | 0 |
| Total | 208 | 149 | 54 | 5 | 107 | 208 | 189 | 95 | |
H−, nonmotile.
HUS, hemolytic-uremic syndrome; D, diarrhea without HUS; A, asymptomatic.
The SRL-related fragment from strain 493/89 detected by PCR with primer pair RL11679f and 4290 (Fig. 1).
All strains had the stx2 genotype.
stx genotypes: stx1 (2 strains); stx2 (42 strains), stx1 stx2 (5 strains); stx2c (9 strains), stx1 stx2c (5 strains), and stx2 stx2c (38 strains).
FIG. 1.
Genetic organization of the mosaic genomic island in the SF EHEC O157:H− strain 493/89 in comparison to E. coli O157:H7 strain EDL933 and to the SRL PAI of Shigella flexneri 2a. At the top the SRL PAI sequence is shown, which contains the respective homologue ORFs. In the middle, the mosaic genomic island of strain 493/89 is demonstrated. Black ORFs demonstrate homologues to the EDL933 sequence, vertically striped ORFs are associated with transposase sequences, and horizontally striped ORFs are homologous to bacteriophage P4 sequences. White ORFs do not have any similarities in data banks. The black line represents homology to EDL933, whereas the square gray line represents the specific 493/89 sequence. Areas with similarities to the SRL PAI sequence are shown in dark gray. At the bottom the putative surrounding of the fragment as determined by cosmid shotgun sequencing is depicted. Both ends of the cosmid insert are homologous to three different O islands of the strain EDL933 (OI-41, OI-81, and OI-82). The sequence of contig 3 of 3, sections 44 to 46 of 290 of EDL933 (GenBank accession numbers AE005425 to AE005427, part of O islands 81 and 82) is inserted by contig 1 of 3, sections 100 and 101 of 155 of EDL933 (GenBank accession numbers AE005276 and AE005277, part of O island 43). Similarities to the SRL PAI sequence are indicated by gray arrows. The dotted lines mark the parts of the respective sequences of EDL933 and of SRL PAI, which are absent from strain 493/89. In addition, positions of PCR primers RL11679f and 4290 used to target the SRL-related sequence of SF EHEC O157:H− strains are shown.
Identification of an SRL-related sequence-containing mosaic genomic island in SF EHEC O157:H− strain 493/89.
A 19.9-kb fragment from a cosmid clone of SF E. coli O157:H− strain 493/89 was completely sequenced. Both ends of the cosmid insert are homologous to the genome of strain EDL933 and flank an 8.8-kb fragment (GenBank accession no. AJ534392) that is absent from EDL933 (Fig. 1). This DNA sequence is highly homologous to SRL of S. flexneri strain YSH6000 (GenBank accession no. AF326777) (19). Figure 1 demonstrates the organization of the 19.9-kb fragment and of its putative flanking regions, which was deduced by “shotgun” sequencing. The 8.8-kb region, which exhibits no homology to EDL933, is flanked by stretches of the O island (OI)-43 of EDL933 chromosome (GenBank accession numbers AE005276 and AE005277), which are ordered in opposite direction to each other. These stretches encode a transposase-associated protein and proteins with unknown function. They are furthermore flanked by fragments of the EDL933 genome (GenBank accession numbers AE005425 to AE005427), which encode a putative outer membrane receptor for iron or colicin (OI-82), a penicillin-binding protein, an exonuclease, and metabolic and hypothetical proteins. Seven open reading frames (ORFs) of the E. coli 493/89 cosmid sequence are 88 to 97% homologous to ORFs 38, 39, 41, 42, 46, and 48 of the SRL PAI. The order and distance of these ORFs in strain 493/89 are comparable with those in the SRL PAI (Fig. 1). Five of these ORFs are located within the 8.8-kb 493/89 fragment which is absent from EDL933 (Fig. 1). For only two of these SRL PAI ORFs (ORF 41 and ORF 46) a putative function has been published (3, 14). ORFs 39 and 48 are bacteriophage P4 related; ORFs 46 and 48 exhibit, in addition, similarity to prophage 933L (19).
The iha, ure, and ter genes of E. coli O157:H7 strain EDL933 are absent from SF EHEC O157:H− strain 493/89.
While nonvirulence loci (GenBank accession numbers AE005276 and AE005277) are present on the OI-43 stretches found within the mosaic genomic island of SF EHEC O157:H− strain 493/89, the iha, ure, and ter genes present on OI-43 of strain EDL933 (13, 18), which encode a putative adhesin Iha (IrgA homologue adhesin) (18), urease (5), and tellurite resistance (2), have not been identified on this mosaic island. Moreover, as demonstrated by Southern blot hybridization with the respective probes, they are absent from the genome of SF EHEC O157:H− strain 493/89 (data not shown).
Absence of multiple antibiotic resistance and fecA from SF EHEC O157:H− strain 493/89.
Strain 493/89 is susceptible to ampicillin, streptomycin, and tetracycline and resistant to chloramphenicol. Thus, the multiple antibiotic resistance encoded by the SRL PAI in S. flexneri (19) is absent from strain 493/89, a finding consistent with the absence of the antibiotic resistance-loci in this strain. fecA probe hybridized to a 10-kb HindIII DNA fragment from S. flexneri 2a strain MT90 but not to DNA from strain 493/89 (data not shown).
Distribution of SRL-related sequences among E. coli O157.
PCR targeting the SRL-related sequence of strain 493/89 (Fig. 1) demonstrated that this region was present in each of 88 SF EHEC O157:H− but in none of 101 EHEC O157:H7 of different stx genotypes (Table 1). It was also absent from each of 12 E. coli O55:H7 strains that are proposed to be ancestral to E. coli O157:H7 and SF E. coli O157:H− (16). The presence of the SRL-related sequences in seven stx-negative SF E. coli O157:H− (Table 1) confirms that these strains are closely related to stx-positive SF E. coli O157:H−, as previously demonstrated by their spectrum of putative virulence genes (4, 9). The absence of the SRL-related 493/89 sequence from E. coli O157:H7 strains was confirmed by Southern blot hybridization with digoxigenin-labeled probe derived from strain 493/89 with primers RL11679f and 4290. Taken together, the experiments on the distribution of the SRL-related sequences demonstrate that within members of EHEC 1 group consisting of E. coli O55:H7, E. coli O157:H7, and SF E. coli O157:H−, SRL-related sequences were only present in SF EHEC O157:H− and their stx-negative derivatives.
The identification of the genomic mosaic island in SF EHEC O157:H− strain 493/89 extends recent reports on the presence of EDL933 OI mosaics in non-O157:H7 EHEC strains (12, 17). However, in comparison to the mosaic islands reported in EHEC O26 (12) and EHEC O113 (17), which contain putative virulence genes (12, 17), none of the ORFs of the 493/89 mosaic genomic island encodes a product currently known to be associated with virulence. Specifically, the OI-43 iha gene encoding the Iha putative adhesin (18) is absent from the OI-43 segment present in strain 493/89. Moreover, this OI-43 segment lacks genes of ter and ure operons located on OI-43 of EDL933 (13), which encode tellurite resistance and urease, respectively, in E. coli O157:H7 (2, 5). This is consistent with a general absence of ter genes (and, consequently, tellurite resistance), and ure genes from SF EHEC O57:H− clinical isolates as demonstrated in our previous studies (2, 5).
In a study on the distribution of SRL among members of the family Enterobacteriaceae (20), SRL PAI has been widespread among Shigella isolates resistant to the four respective antibiotics (20). Among 24 pathogenic E. coli strains investigated, including enteropathogenic E. coli, enteroinvasive E. coli, enteroaggregative E. coli, enterotoxigenic E. coli, and EHEC, some contained single markers of the SRL PAI (int, orf58, and fecA) but showed no SRL-encoded antibiotic resistance, suggesting that the SRL PAI is not present in these strains (20).
All EHEC O157:H7 and SF EHEC O157:H− possess rfbO157, fliC encoding the H7 antigen, eae-γ, and largely (O157:H7) or obligatorily (SF O157) contain stx2 genes (9; the present study). Phenotypic markers may not reliably distinguish EHEC O157:H7 from SF EHEC O157:H− strains because some O157:H7 strains ferment sorbitol rapidly (within 24 to 48 h) and SF O157:H− strains can display delayed sorbitol fermentation (after 24 h) (H. Karch, unpublished data). Also, the intensity of the color reaction indicating sorbitol fermentation on sorbitol MacConkey agar substantially varies among SF EHEC O157:H− strains and, moreover, it rapidly decreases during prolonged incubation or storage at 4°C (H. Karch, unpublished data). Therefore, among EHEC O157 which harbor stx2 but not stx1, SRL-related sequences detected by PCR developed in the present study represent a valuable marker for distinguishing O157:H7 from SF O157:H− strains. Moreover, the conservation of the SRL sequences in all 82 SF EHEC O157:H− strains investigated indicates a clonal origin of such strains and suggests a considerable stability of this element.
In the SRL PAI sequence, a similar organization and orientation exists as in the prophage P4 genome. This infers a common origin of sequences of parts of phage P4 and the SRL PAI. P4 could be a prophage of E. coli O157:H−, as well as of Shigella, and hence be responsible for the partial similar structures of both pathovars. The presence of several markers of the SRL PAI in SF E. coli O157:H− might indicate that these strains diverged from the evolutionary pathway of EHEC O157:H7 at an early stage and acquired SRL. It was reported that SRL can undergo both integrase-mediated and non-integrase-mediated excision in the same strain (19). Functional analysis of the SRL-related ORFs is under way in an attempt to explain a mode of acquisition and/or deletion of parts of this DNA segment by SF E. coli O157:H−.
The sequence of the 8,846-bp SRL-related fragment from strain 493/89 has been deposited in GenBank (accession no. AJ534392).
Acknowledgments
This study was supported by grants from the Bundesministerium für Bildung und Forschung Project Network of Competence Pathogenomics Alliance “Functional Genomic Research on Enterohemorrhagic Escherichia coli” (BD 119523) and from the First European Graduate College “Gene Regulation in and by Microbial Pathogens.”
We thank Phillip Tarr (Washington University School of Medicine, St. Louis, Mo.) for critical reading of the manuscript and stimulating discussions.
REFERENCES
- 1.Ammon, A., L. R. Peterson, and H. Karch. 1999. A large outbreak of hemolytic-uremic syndrome caused by an unusual sorbitol-fermenting strain Escherichia coli O157:H−. J. Infect. Dis. 179:1274-1277. [DOI] [PubMed] [Google Scholar]
- 2.Bielaszewska, M., P. I. Tarr, H. Karch, W. Zhang, and W. Mathys. 2005. Phenotypic and molecular analysis of tellurite resistance among enterohemorrhagic Escherichia coli O157:H7 and sorbitol-fermenting O157:NM clinical isolates. J. Clin. Microbiol. 43:452-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 4.Friedrich, A. W., K. V. Nierhoff, M. Bielaszewska, A. Mellmann, and H. Karch. 2004. Phylogeny, clinical associations, and diagnostic utility of the pilin subunit gene (sfpA) of sorbitol-fermenting, enterohemorrhagic Escherichia coli O157:H−. J. Clin. Microbiol. 42:4697-4701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Friedrich, A. W., R. Köck, M. Bielaszewska, W. Zhang, H. Karch, and W. Mathys. 2005. Distribution of the urease gene cluster among and urease activities of enterohemorrhagic Escherichia coli O157 isolates from humans. J. Clin. Microbiol. 43:546-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hayashi, T., K. Makino, M. Ohnishi, K. Kurokawa, K. Ishii, K. Yokoyama, C. G. Han, E. Ohtsubo, K. Nakayama, T. Murata, M. Tanaka, T. Tobe, T. Iida, H. Takami, T. Honda, C. Sasakawa, N. Ogasawara, T. Yasunaga, S. Kuhara, T. Shiba, M. Hattori, and H. Shinagawa. 2001. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 8:11-22. [DOI] [PubMed] [Google Scholar]
- 7.Janka, A., M. Bielaszewska, U. Dobrindt, and H. Karch. 2002. Identification and distribution of the enterohemorrhagic Escherichia coli factor for adherence (efa1) gene in sorbitol-fermenting Escherichia coli O157:H−. Int. J. Med. Microbiol. 292:207-214. [DOI] [PubMed] [Google Scholar]
- 8.Janka, A., M. Bielaszewska, U. Dobrindt, L. Greune, M. A. Schmidt, and H. Karch. 2003. The cytolethal distending toxin (cdt) gene cluster in enterohemorrhagic Escherichia coli O157:H− and O157:H7: characterization and evolutionary considerations. Infect. Immun. 71:3634-3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karch, H., and M. Bielaszewska. 2001. Sorbitol-fermenting Shiga toxin-producing Escherichia coli O157:H− strains: epidemiology, phenotypic and molecular characteristics, and microbiological diagnosis. J. Clin. Microbiol. 39:2043-2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luck, S. N., S. A. Turner, K. Rajakumar, H. Sakellaris, and B. Adler. 2001. Ferric dicitrate transport system (Fec) of Shigella flexneri 2a YSH6000 is encoded on a novel pathogenicity island carrying multiple antibiotic resistance genes. Infect. Immun. 69:6012-6021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Monday, S. R., S. A. Minnich, and P. C. Feng. 2004. A 12-base-pair deletion in the flagellar master control gene flhC causes nonmotility of the pathogenic German sorbitol-fermenting Escherichia coli O157:H− strains. J. Bacteriol. 186:2319-2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morabito, S., R. Tozzoli, E. Oswald, and A. Caprioli. 2003. A mosaic pathogenicity island made up of the locus of enterocyte effacement and a pathogenicity island of Escherichia coli O157:H7 is frequently present in attaching and effacing E. coli. Infect. Immun. 71:3343-3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perna, N. T., G. Plunket III, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 14.Polo, S., T. Sturniolo, G. Deho, and D. Ghiotti. 1996. Identification of a phage-coded DNA-binding protein that regulates transcription from late promoters in bacteriophage P4. J. Mol. Biol. 257:745-755. [DOI] [PubMed] [Google Scholar]
- 15.Pradel, N., S. Leroy-Setrin, B. Joly, and V. Livrelli. 2002. Genomic subtraction to identify and characterize sequences of Shiga toxin-producing Escherichia coli O91:H21. Appl. Environ. Microbiol. 68:2316-2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reid, S. D., C. J. Herbelin, A. C. Bumbaugh, R. K. Selander, and T. S. Whittam. 2000. Parallel evolution of virulence in pathogenic Escherichia coli. Nature 406:64-67. [DOI] [PubMed] [Google Scholar]
- 17.Shen, S., M. Mascarenhas, K. Rahn, J. B. Kaper, and M. A. Karmali. 2004. Evidence for a hybrid genomic island in verocytotoxin-producing Escherichia coli CL3 (serotype O113:H21) containing segments of EDL933 (serotype O157:H7) O islands 122 and 48. Infect. Immun. 72:1496-1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tarr, P. I., S. S. Bilge, J. C. Vary, S. Jelacic, R. L. Habeeb, T. R. Ward, M. R. Baylor, and T. E. Besser. 2000. Iha: a novel Escherichia coli O157:H7 adherence-conferring molecule encoded on a recently acquired chromosomal island of conserved structure. Infect. Immun. 68:1400-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Turner, S. A., S. N. Luck, H. Sakellaris, K. Rajakumar, and B. Adler. 2001. Nested deletions of the SRL pathogenicity island of Shigella flexneri 2a. J. Bacteriol. 183:5535-5543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Turner, S. A., S. N. Luck, H. Sakellaris, K. Rajakumar, and B. Adler. 2003. Molecular epidemiology of the SRL pathogenicity island. Antimicrob. Agents Chemother. 47:727-734. [DOI] [PMC free article] [PubMed] [Google Scholar]