Abstract
Glyphosate has been used globally as a safe herbicide for weed control. It inhibits 5-enolpyruvylshikimate-3-phosphate (EPSP) synthase (AroA), which is a key enzyme in the aromatic amino acid biosynthetic pathway in microorganisms and plants. A Pseudomonas putida strain, 4G-1, was isolated from a soil heavily contaminated by glyphosate in China. Its AroA-encoding gene (aroA) has been cloned, sequenced, and expressed in Escherichia coli. Phylogenetic analysis revealed that this AroA belongs neither to class I nor to class II AroA enzymes. When compared with E. coli AroA, 4G-1 AroA shows similar values for Km[PEP], Km[S3P], and specific enzyme activity. Moreover, 4G-1 AroA exhibits high tolerance to glyphosate, which indicates a protein with a high potential for structural and functional studies of AroA in general and its potential usage for the generation of transgenic crops resistant to the herbicide.
Biotechnology offers a sustainable approach to many industrial and environmental applications of economic benefit. Microbial biodiversity, particularly in extreme or polluted ecosystems, offers an opportunity to extract novel genes and proteins for industrial and environmental applications. China offers an extensive source of such biodiversity. Glyphosate is a broad-spectrum herbicide that blocks plant growth by inhibiting 5-enolpyruvylshikimate-3-phosphate (EPSP) synthase (AroA) (8, 29). AroA is a key enzyme involved in aromatic amino acid biosynthesis. It catalyzes an unusual reaction between shikimate-3-phosphate (S3P) and phosphoenolpyruvate (PEP), with transfer of the carboxyvinyl moiety from PEP to S3P to form EPSP and inorganic phosphate (11). Glyphosate inhibits AroA in a slowly reversible reaction, which is competitive versus PEP and noncompetitive versus S3P (2, 30). Glyphosate binds to the binding site for PEP and forms a stable ternary complex with the enzyme and S3P (AroA-S3P-glyphosate) (23). This ternary complex represents the actual enzyme-bound form of glyphosate, which is responsible for its herbicide activity (26).
Two classes of AroA have been reported (1). Class I includes AroA enzymes from Escherichia coli, Aeromonas salmonicida, Petunia petunia, and Arabidopsis thaliana. Class I enzymes are naturally sensitive to glyphosate. Their mutants with tolerance to glyphosate can be generated by different methods (4, 10, 20). For example, highly glyphosate tolerant AroA has been produced by a G96A substitution in the aroA gene of Escherichia coli (20). However, glyphosate tolerance in class I enzymes is often paralleled by a decrease in the affinity of AroA synthase for PEP (1, 26). Class II AroA enzymes, which share less than 30% amino acid identity with class I AroA enzymes, have been identified from Agrobacterium tumefaciens CP4, Bacillus subtilis, and Pseudomonas sp. strain PG2982. In contrast to class I enzymes, class II enzymes usually have not only high affinity for PEP but also natural tolerance to glyphosate (1).
In this study, we have retrieved a novel AroA-encoding gene (aroA) from a Pseudomonas putida strain isolated from a soil heavily contaminated by glyphosate in China. Sequence analysis shows that this novel enzyme has less than 30% amino acid identity to both class I and class II AroA enzymes. This enzyme has similar Km[S3P] and Km[PEP] values, but a much higher Ki[glyphosate] value, compared with Escherichia coli AroA.
MATERIALS AND METHODS
Medium and chemicals.
S3P (barium) was a gift from Nikolaus Amrhein (ETH Zürich, Zürich, Switzerland). Glyphosate (free acid form) and PEP were purchased from Sigma. All chemicals were of analytical grade. The minimal medium used for the glyphosate tolerance test was M63 (16) supplemented with 0.4% glucose as a carbon source plus 100 μg/ml vitamin B1 and an appropriate antibiotic.
Isolation of microorganisms.
Isolation of the glyphosate-tolerant strains was undertaken using an inoculum of heavily contaminated soil from a glyphosate-producing factory in Hebei province, China. Soil samples were suspended in 0.9% (wt/vol) NaCl solution, diluted, and spread onto M63 plates containing 60 mM glyphosate. Dozens of colonies were isolated after 2 days of incubation at 28°C and were further screened on M63 plates in the presence of higher concentrations (120 mM and 200 mM) of glyphosate. One strain, 4G-1, grew well in the presence of 200 mM glyphosate and was chosen for further studies.
Isolation of the gene involved in glyphosate tolerance.
Chromosomal DNA was isolated from strain 4G-1 and was subsequently partially digested with Sau3AI to produce 30- to 40-kb fragments. These DNA fragments were dephosphorylated and ligated with the SuperCos 1 cosmid vector (Stratagene). The resulting ligation mixture was packaged with GIGAPACK III packaging extract (Stratagene), and thus a genomic library was constructed as described by the manufacturer. Escherichia coli XL1-Blue MR colonies carrying the cosmid library were spread onto M63 agar plates containing 120 mM glyphosate and were incubated at 37°C overnight. One glyphosate-resistant colony was obtained, which contained a cosmid with an insert of approximately 35 kb and was designated pBD1. The pBD1 cosmid DNA was prepared, partially digested with Sau3AI, and ligated into the pUC18 vector (18). The ligation mixtures were transformed to the Escherichia coli aroA mutant strain AB2829 (22) and screened on M63 agar plates supplemented with 120 mM glyphosate. One colony harboring a plasmid (designed pBD2) with a ∼2.0-kb insert fragment was selected. The insertion fragment contained in pBD2 was subsequently sequenced.
Expression of His-tagged AroA from Pseudomonas putida 4G-1 and N-terminal amino acid sequencing.
A DNA fragment containing the entire coding region of AroA was obtained by PCR using pBD2 as a template with the following two primers based on pBD2: primer 1 (5′-GCTCTAGAAGTGTTGGAACAATATG-3′) and primer 2 (5′-GTTACTCGAGTGAGAAATTAAATTGATGGTTT-3′) (the introduced restriction enzyme sites are underlined). Primer 2 contains a mutation where the stop codon TGA was replaced with CTC. The fragment was digested with XbaI and XhoI, cloned into the corresponding restriction sites of pET-28a (Novagen, Inc.), and confirmed by DNA sequencing. The plasmid was transformed to Escherichia coli BL21(DE3) (Novagen, Inc.) to express the His-tagged AroA. The protein was purified using a HisTrap HP kit (Amersham Biosciences) according to the manufacturer's instructions. The protein was further purified by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and blotted onto a polyvinylidene difluoride membrane for N-terminal protein sequencing using an Applied Biosystems Procise sequencer.
Construction of recombinant plasmids.
Primers 3 (5′-CGGGATCCTAAGTAAGTGAAAGTAACAATACAGC-3′) and 4 (5′-CGGGATCCCTTCTTCGGACAATGACAGAC-3′), specific for aroA, were designed based on the sequence of pBD2; primers 5 (5′-CGGGATCCGTTAATGCCGAAATTTTGCTTAATC-3′) and 6 (5′-CGGGATCCAGGTCCGAAAAAAAACGCCGAC-3′), specific for the aroA gene of Escherichia coli, were designed based on the sequence available in GenBank. BamHI sites were introduced into the forward and reverse primers (one into each primer) in order to ligate fragments with the pACYC184 vector (3). When pBD2 and Escherichia coli ET8000 genomic DNA (15) were used as templates, the aroA gene fragments from Pseudomonas putida 4G-1 and Escherichia coli were PCR amplified, BamHI digested. and ligated into the pACYC184 vector. Plasmids with the aroA gene and Tetr promoter in the same direction were chosen in order to ensure that the aroA genes would be driven by the Tetr promoter of pACYC184 (designated pACYC-aroAP. putida and pACYC-aroAE. coli, respectively). In addition, a glyphosate-tolerant mutant (G96A) (20) of aroAE. coli (designated pACYC-aroAE. coli-G96A) was constructed using the method of PCR overlap extension (12).
Cell growth in the presence of glyphosate.
The Escherichia coli aroA mutant strain AB2829 harboring either pACYC-aroAP. putida, pACYC-aroAE. coli, or pACYC-aroAE. coli-G96A was grown with shaking at 37°C in liquid M63 minimal medium supplemented with glyphosate at various concentrations ranging from 0 to 100 mM. Cell densities were measured by attenuance at 600 nm (D600).
Preparation of crude AroA.
The Escherichia coli aroA mutant strain AB2829 containing either pACYC-aroAP. putida, pACYC-aroAE. coli, or pACYC-aroAE. coli-G96A was grown in 300 ml LB broth. When the D600 reached 2.5, the cells were collected by centrifugation and resuspended in 30 ml 50 mM Tris-HCl buffer (pH 7.0)-0.2 mM dithiothreitol. The cell suspension was lysed using a French press. The crude homogenate was clarified by centrifugation at 8,000 rpm and 4°C for 10 min. Ammonium sulfate was added to the supernatants to 35% saturation, and the resulting precipitate was removed by centrifugation. The supernatant was brought to 70% saturation with ammonium sulfate and the precipitate collected by centrifugation. The precipitate was dissolved in 50 mM Tris-HCl (pH 7.0)-0.1 mM dithiothreitol. The samples were dialyzed overnight in 50 mM Tris-HCl (pH 7.0)-0.1 mM dithiothreitol.
Enzyme assay.
AroA activity was measured by the production of inorganic phosphate using the malachite green dye assay method (14). The standard reaction was carried out at 28°C in a final volume of 50 μl containing 50 mM HEPES (pH 7.0), 1 mM S3P, 1 mM PEP, and the crude enzyme. After incubation for 1 to 5 min, 800 μl of malachite green-ammonium molybdate colorimetric solution was added, and 1 min later, 0.1 ml of a 34% sodium citrate solution was added to stop the reaction. After a 30-min incubation at room temperature, absorbances of samples were measured at 660 nm. In this case, the same reaction solution without S3P was used as the zero control.
Nucleotide sequence accession number.
The sequence of the ∼2.0-kb insert of pBD2 is available from EMBL under accession number AJ812018.
RESULTS
Isolation and identification of the glyphosate-tolerant strain 4G-1.
A total of 48 strains that can grow on M63 plates containing 60 mM glyphosate have been isolated from heavily contaminated soil (see Materials and Methods); 13 of these can grow in the presence of 120 mM glyphosate. One, strain 4G-1, grows well on M63 plates in the presence of 200 mM glyphosate. 4G-1 is a gram-negative bacterium, and Biolog profiling showed that it was most similar to Pseudomonas putida. Its 16S rRNA gene sequence (1,501 bp) confirmed that isolate 4G-1 was strongly related to Pseudomonas putida (99.86%). Therefore, strain 4G-1 was assigned to Pseudomonas putida.
Cloning of the gene involved in glyphosate tolerance.
To isolate the gene involved in glyphosate tolerance from Pseudomonas putida 4G-1, a genomic library was constructed from this strain and further screened (see Materials and Methods). Of approximately 3,000 ampicillin-resistant Escherichia coli XL1-Blue MR colonies, 1 colony was observed to have the ability to grow on an M63 agar plate supplemented with 120 mM glyphosate. The cosmid DNA isolated from this colony was subcloned into the pUC18 vector, and one of the resulting plasmid subclones (pBD2) could support the growth of the Escherichia coli aroA mutant strain AB2829 on M63 agar plates supplemented with 120 mM glyphosate. This result suggests that pBD2 may encode an aroA gene that may also be involved in glyphosate tolerance.
Sequence and functional analysis of the aroA gene of Pseudomonas putida 4G-1.
The ∼2.0-kb insert of pBD2 was sequenced and analyzed. One open reading frame (ORF), designated ORF431, was deduced; it contains a single peptide of 431 amino acids that starts with a possible GUG start codon. It shares 28% identity with AroA of Escherichia coli. Therefore, ORF431 encodes an AroA protein, and it was designated aroA.
In order to eliminate the possibility that the flanking regions of ORF431 in pBD2 may also be required for glyphosate tolerance, a PCR experiment was carried out to amplify only the sequence encoding ORF431 from pBD2. The resulting DNA fragment was subsequently cloned into pACYC184, resulting in plasmid pACYC184-aroAP. putida. When this plasmid was transformed into the Escherichia coli aroA mutant strain AB2829, it was able to complement its aroA mutation for growth on M63 minimal medium supplemented with 120 mM glyphosate.
To determine whether ORF431 was translated, AroAH6 P. putida, containing an 8-amino-acid C-terminal extension terminating with His6, was expressed and purified. The molecular size of the predicted product of ORF431 (47 kDa) agreed well with that of the purified AroAH6 P. putida (47 kDa by sodium dodecyl sulfate-polyacrylamide gel electrophoresis) (data not shown). The N-terminal amino acid sequence of AroAH6 P. putida was further determined as M-K-V-T-I-Q-P-G-D-L by automated Edman degradation. The result confirmed that the initiator codon of AroAP. putida is GUG, which is used as a start codon in less than 10% of bacterial genes (13).
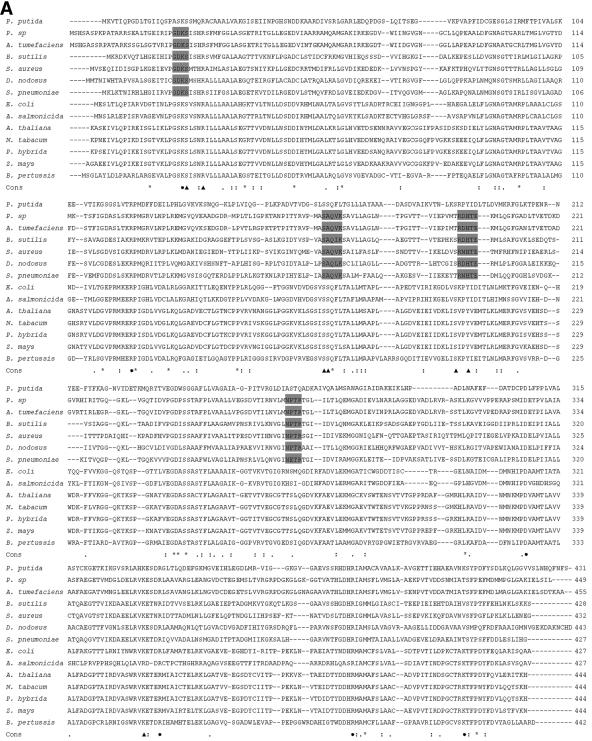
Multiple sequence alignment showed that AroAP. putida has only <30% identity to both class I and class II AroA enzymes (Fig. 1). However, the residues involved in PEP and S3P binding are all highly conserved in AroAP. putida and class I AroA enzymes, whereas these residues are not so conserved in class II AroA enzymes. The domains that are important for glyphosate tolerance and maintenance of productive PEP binding in class II AroA enzymes were not found in AroAP. putida (1) (Fig. 1A). This indicates that AroAP. putida is more closely related to class I AroA enzymes, though phylogenetic analysis revealed that AroAP. putida belongs neither to class I nor to class II (Fig. 1B).
FIG. 1.
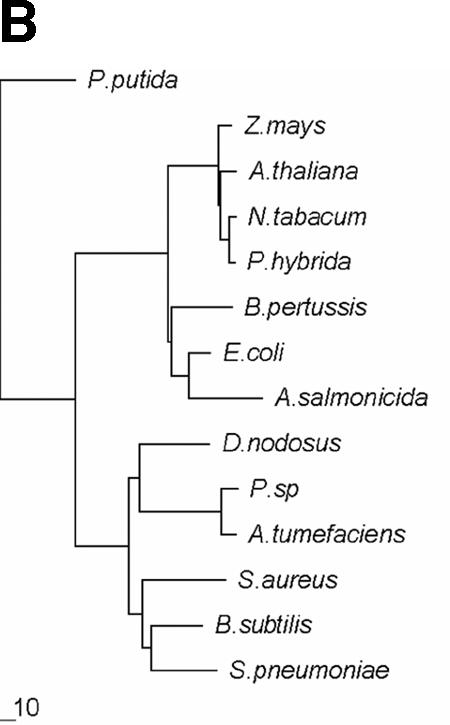
Sequence alignment and phylogenetic analysis of AroAP. putida with class I and class II AroA proteins. (A) Multiple sequence alignment performed using T-Coffee (19) with minor manual adjustments. Asterisks, identical residues; periods, conserved residues; colons, strongly conserved residues. Triangles and circles, residues important for S3P binding and PEP binding, respectively, in AroAE. coli (23, 25). Domains important for glyphosate tolerance and maintenance of productive PEP binding in class II AroA are shaded (1). (B) Phylogenetic tree of AroA constructed using DNASTAR based on the results of alignment with T-Coffee. The tree suggests that AroAP. putida (P. putida; AJ812018) has less than 30% identity to class I and class II AroA proteins. Class I AroA proteins shown are from E. coli (P07638), Aeromonas salmonicida (Q03321), Arabidopsis thaliana (P05466), Nicotiana tabacum (P23981), Petunia hybrida (P11043), Zea mays (CAA44974), and Bordetella pertussis (P12421). Class II AroA proteins shown are from Pseudomonas sp. strain PG2982 (P56952), Agrobacterium tumefaciens CP4 (Q9R4E4), Bacillus subtilis (P20691), Staphylococcus aureus (Q05615), Dichelobacter nodosus (Q46550), and Streptococcus pneumoniae (Q9S400).
Structure of Pseudomonas putida AroA.
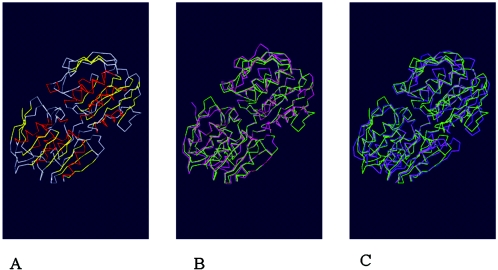
The structure of AroAP. putida was modeled by SWISS-MODEL and drawn with Swiss-PdbViewer (9, 24). It is very similar to the known structure of AroA (21, 23, 28). The protein folds to two similar globular inside-out α-β-barrel domains, which are connected by a two-stranded hinge (Fig. 2). AroAP. putida has 28% amino identity to Escherichia coli AroA and 27% amino identity to Streptococcus pneumoniae AroA (Fig. 1); however, the structure of AroAP. putida is much more similar to that of Escherichia coli AroA than to that of Streptococcus pneumoniae AroA (Fig. 2). The results also suggested that AroAP. putida is more closely related to class I AroA enzymes.
FIG. 2.
Backbone traces of AroA proteins from P. putida 4G-1, E. coli, and S. pneumoniae. (A) Backbone Cα atom trace of AroAP. putida. The structure was modeled by SWISS-MODEL based on the structures of AroAE. coli and AroAS. pneumoniae (9, 21, 23) and was drawn with Swiss-PdbViewer (24, 28). (B) Backbone Cα atom traces of AroAP. putida (green) and AroAE. coli (pink) (9, 23). (C) Backbone Cα atom traces of AroAP. putida (green) and AroAS. pneumoniae (purple) (21).
Growth of cells in the presence of glyphosate.
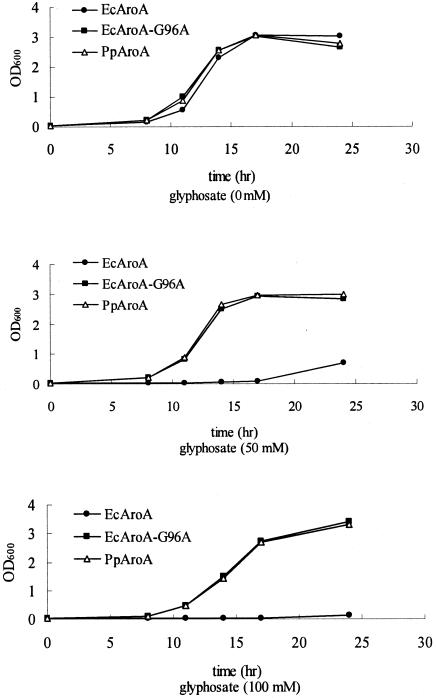
The growth curves of the Escherichia coli aroA mutant AB2829 harboring plasmid pACYC-aroAE. coli, pACYC-aroAE. coli-G96A, or pACYC-aroAP. putida are shown in Fig. 3. The cells were grown in liquid M63 minimal medium containing various concentrations of glyphosate. The results show that all cells grew well in M63 medium without glyphosate. The growth of cells harboring pACYC-aroAE. coli was strongly inhibited at 50 or 100 mM glyphosate. In contrast, cells harboring pACYC-aroAE. coli-G96A or pACYC-aroAP. putida grew well at 50 or 100 mM glyphosate. The result suggested that aroAP. putida encodes a highly glyphosate tolerant AroA.
FIG. 3.
Growth of the E. coli aroA mutant strain AB2829 harboring either pACYC-aroAE. coli (EcAroA), pACYC-aroAE. coli-G96A (EcAroA-G96A), or pACYC-aroAP. putida (PpAroaA) in liquid M63 minimal medium supplemented with glyphosate at the concentrations indicated.
Kinetic properties of AroA variants.
Protein extracts were prepared from Escherichia coli aroA mutant AB2829 cells containing different plasmids harboring different aroA genes and were assayed for AroA activity. As a control, only traces of EPSP synthase activity were detected in crude extracts of Escherichia coli aroA mutant AB2829 host cells with the pACYC184 vector. Compared with AroAE. coli, AroAP. putida showed a 300-fold-higher Ki[glyphosate] value and a similar Km[S3P] value (Table 1). Its Km[PEP] was similar to that of AroAE. coli and 23-fold less than that of AroAE. coli-G96A (Table 1). The purified AroAE. coli and AroAP. putida with the His tag showed similar kinetic parameters as crude enzymes (data not shown). These results further exemplify the natural tolerance of AroAP. putida to glyphosate and its high affinity for PEP.
TABLE 1.
Kinetic properties of AroAE. coli, AroAE. coli-G96A, and AroAP. putidaa
| Enzyme | Sp actb (nkat/mg protein) | Km[PEP]c (μM) | Km[S3P]d (μM) | Ki[glyphosate]f (mM) |
|---|---|---|---|---|
| AroAE. coli | 7.43 ± 0.12 | 9.43 ± 1.98 | 27.45 ± 5.73 | (0.608 ± 0.151) × 10−3 |
| AroAE. coli-G96A | 1.25 ± 0.04 | 234.86 ± 7.79 | NDe | 2.525 ± 1.062 |
| AroAP. putida | 9.36 ± 0.27 | 10.36 ± 4.12 | 19.50 ± 1.18 | 0.183 ± 0.084 |
The results presented are averages of two sets of experiments done in triplicate.
Determined at 1.0 mM PEP and 1.0 mM S3P.
Determined by the Lineweaver-Burk method. The PEP concentration was varied from 20 to 400 μM, while the S3P concentration was fixed at 1 mM.
Determined by the Lineweaver-Burk method. The S3P concentration was varied from 20 to 250 μM, while the PEP concentration was fixed at 1 mM.
ND, not determined.
Competitive inhibition by glyphosate with respect to PEP was demonstrated by lines converging on the x axes of Lineweaver-Burk plots.
DISCUSSION
In this study, a novel aroA gene was retrieved from a Pseudomonas putida strain, 4G-1, isolated from an extremely polluted environment in China; its product (AroAP. putida) has natural glyphosate tolerance. AroAP. putida shows only 28% identity to Pseudomonas putida KT2440 AroA (17) (data not shown). The 1,914-bp fragment contained in pBD2 has a 44.5% G+C content, in contrast to the 61.6% G+C content of the Pseudomonas putida KT2440 genome (17). Differences in G+C content have been suggested to imply lateral gene transfers (7). Pseudomonas putida has been reported to have acquired genes via lateral gene transfer among soil microbial communities (27, 31). It may be the case that Pseudomonas putida 4G-1 acquired this novel aroA gene from other organisms recently via horizontal gene transfer.
Sequence analysis and structure modeling had shown that AroAP. putida does not belong to class II AroA enzymes, which have natural glyphosate tolerance. Though AroAP. putida is related to class I AroA proteins, it did not have the motifs that confer glyphosate tolerance in class I AroA (Fig. 1A) (4, 20). Inhibition of AroA by glyphosate has been shown to proceed through the formation of a stable but noncovalent AroA-S3P-glyphosate ternary complex (23). It has been reported that the glyphosate binding site is identical to the binding site for PEP (6, 23). However, the Gly96-to-Ala mutation of Escherichia coli AroA confers glyphosate insensitivity without changing the PEP binding site (20). This is due to an additional methyl group in position 96 of AroAE. coli that has an indirect effect on glyphosate binding (5). Compared with AroAE. coli, AroAP. putida has an identical PEP binding site and even an identical S3P binding site (Fig. 1). AroAP. putida probably has some residues that exert an indirect effect on glyphosate binding, leading to glyphosate tolerance. Since AroAP. putida is more closely related to class I AroA enzymes, studies on the mechanism of glyphosate tolerance of AroAP. putida may help us to obtain a new type of class I AroA proteins that not only show high tolerance to glyphosate but also have high affinity for PEP.
The special structure of AroAP. putida is expected to facilitate the study of the mechanism and function of AroA. Mutagenesis of AroAP. putida to study the different characteristics of this novel AroA is under way. The high glyphosate tolerance of AroAP. putida may also be invaluable for the generation of transgenic crops resistant to glyphosate.
Acknowledgments
We thank Jin-Dong Zhao and Wei-Qun Shen (Peking University) for protein N-terminal sequencing, Zhi-Ting Li for advice and critical reading of the manuscript, Nikolaus Amrhein (ETH Zürich, Zürich, Switzerland) for sending us the S3P substrate, and the Escherichia coli Genetic Stock Center, Yale University, for providing the Escherichia coli aroA mutant strain AB2829.
This research was supported by grants from the Ministry of Science and Technology of China (The National Transgenic Research and Industrialization Special Foundation of China, grants J00-A-010 and JY03-A-05), the National Natural Science Foundation of China (grant 39925017), the Higher Education Authority of Ireland PRTLI programme, and EU contracts QLK3-CT2000-00164 and QLK3-CT-2001-00101.
REFERENCES
- 1.Barry, G. F., G. M. Kishore, and S. R. Padgette. March 1992. DNA encoding class II 5′-enolpyruvyl shikimate-3-phosphate synthase—for producing plants and bacteria tolerant to glyphosate herbicides. World Patent WO 9,204,449.
- 2.Boocock, M. R., and J. R. Coggins. 1983. Kinetics of 5-enolpyruvylshikimate-3-phosphate synthase inhibition by glyphosate. FEBS Lett. 154:127-133. [DOI] [PubMed] [Google Scholar]
- 3.Chang, A. C., and S. N. Cohen. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 134:1141-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Comai, L., L. C. Sen, and D. M. Stalker. 1983. An altered aroA gene product confers resistance to the herbicide glyphosate. Science 221:370-371. [DOI] [PubMed] [Google Scholar]
- 5.Eschenburg, S., M. L. Healy, M. A. Priestman, G. H. Lushington, and E. Schonbrunn. 2002. How the mutation glycine 96 to alanine confers glyphosate insensitivity to 5-enolpyruvyl shikimate-3-phosphate synthase from Escherichia coli. Planta 216:129-135. [DOI] [PubMed] [Google Scholar]
- 6.Eschenburg, S., W. Kabsch, M. L. Healy, and E. Schonbrunn. 2003. A new view of the mechanisms of UDP-N-acetylglucosamine enolpyruvyl transferase (MurA) and 5-enolpyruvylshikimate-3-phosphate synthase (AroA) derived from X-ray structures of their tetrahedral reaction intermediate states. J. Biol. Chem. 278:49215-49222. [DOI] [PubMed] [Google Scholar]
- 7.Folkesson, A., A. Advani, S. Sukupolvi, J. D. Pfeifer, S. Normark, and S. Lofdahl. 1999. Multiple insertions of fimbrial operons correlate with the evolution of Salmonella serovars responsible for human disease. Mol. Microbiol. 33:612-622. [DOI] [PubMed] [Google Scholar]
- 8.Franz, J. E., M. K. Mao, and J. A. Sikorski. 1997. Glyphosate: a unique global herbicide. American Chemical Society monograph 189. Oxford University Press, New York, N.Y.
- 9.Guex, N., and M. C. Peitsch. 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714-2723. [DOI] [PubMed] [Google Scholar]
- 10.He, M., Z. Y. Yang, Y. F. Nie, J. Wang, and P. Xu. 2001. A new type of class I bacterial 5-enopyruvylshikimate-3-phosphate synthase mutants with enhanced tolerance to glyphosate. Biochim. Biophys. Acta 1568:1-6. [DOI] [PubMed] [Google Scholar]
- 11.Herrmann, K. M., and L. M. Weaver. 1999. The shikimate pathway. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50:473-503. [DOI] [PubMed] [Google Scholar]
- 12.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 13.Kozak, M. 1999. Initiation of translation in prokarytotes and eukaryotes. Gene 234:187-208. [DOI] [PubMed] [Google Scholar]
- 14.Lanzetta, P. A., L. J. Alvarez, P. S. Reinach, and O. A. Candia. 1979. An improved assay for nanomole amounts of inorganic phosphate. Anal. Biochem. 100:95-97. [DOI] [PubMed] [Google Scholar]
- 15.MacNeil, T., D. MacNeil, and B. Tyler. 1982. Fine-structure deletion map and complementation analysis of the glnA-glnL-glnG region in Escherichia coli. J. Bacteriol. 150:1302-1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 17.Nelson, K. E., C. Weinel, I. T. Paulsen, R. J. Dodson, H. Hilbert, V. A. Martins dos Santos, D. E. Fouts, S. R. Gill, M. Pop, M. Holmes, L. Brinkac, M. Beanan, R. T. DeBoy, S. Daugherty, J. Kolonay, R. Madupu, W. Nelson, O. White, J. Peterson, H. Khouri, I. Hance, P. C. Lee, E. Holtzapple, D. Scanlan, K. Tran, A. Moazzez, T. Utterback, M. Rizzo, K. Lee, D. Kosack, D. Moestl, H. Wedler, J. Lauber, D. Stjepandic, J. Hoheisel, M. Straetz, S. Heim, C. Kiewitz, J. A. Eisen, K. N. Timmis, A. Dusterhoft, B. Tummler, and C. M. Fraser. 2002. Complete genome sequence and comparative analysis of the metabolically versatile Pseudomonas putida KT2440. Environ. Microbiol. 4:799-808. [DOI] [PubMed] [Google Scholar]
- 18.Norrander, J., T. Kempe, and J. Messing. 1983. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene 26:101-106. [DOI] [PubMed] [Google Scholar]
- 19.Notredame, C., D. G. Higgins, and J. Heringa. 2000. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302:205-217. [DOI] [PubMed] [Google Scholar]
- 20.Padgette, S. R., D. B. Re, C. S. Gasser, D. A. Eichholtz, R. B. Frazier, C. M. Hironaka, E. B. Levine, D. M. Shah, R. T. Fraley, and G. M. Kishore. 1991. Site-directed mutagenesis of a conserved region of the 5-enolpyruvylshikimate-3-phosphate synthase active site. J. Biol. Chem. 266:22364-22369. [PubMed] [Google Scholar]
- 21.Park, H., J. L. Hilsenbeck, H. J. Kim, W. A. Shuttleworth, Y. H. Park, J. N. Evans, and C. Kang. 2004. Structural studies of Streptococcus pneumoniae EPSP synthase in unliganded state, tetrahedral intermediate-bound state and S3P-GLP-bound state. Mol. Microbiol. 51:963-971. [DOI] [PubMed] [Google Scholar]
- 22.Pittard, J., and B. J. Wallace. 1966. Distribution and function of genes concerned with aromatic biosynthesis in Escherichia coli. J. Bacteriol. 91:1494-1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schonbrunn, E., S. Eschenburg, W. A. Shuttleworth, J. V. Schloss, N. Amrhein, J. N. Evans, and W. Kabsch. 2001. Interaction of the herbicide glyphosate with its target enzyme 5-enolpyruvylshikimate 3-phosphate synthase in atomic detail. Proc. Natl. Acad. Sci. USA 98:1376-1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schwede, T., J. Kopp, N. Guex, and M. C. Peitsch. 2003. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 31:3381-3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shuttleworth, W. A., M. E. Pohl, G. L. Helms, D. L. Jakeman, and J. N. Evans. 1999. Site-directed mutagenesis of putative active site residues of 5-enolpyruvylshikimate-3-phosphate synthase. Biochemistry 38:296-302. [DOI] [PubMed] [Google Scholar]
- 26.Sikorski, J. A., and K. J. Gruys. 1997. Understanding glyphosate's molecular mode of action with EPSP synthase: evidence favoring an allosteric inhibitor model. Acc. Chem. Res. 30:2-8. [Google Scholar]
- 27.Sinclair, M. I., P. C. Maxwell, B. R. Lyon, and B. W. Holloway. 1986. Chromosomal location of TOL plasmid DNA in Pseudomonas putida. J. Bacteriol. 168:1302-1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stallings, W. C., S. S. Abdel-Meguid, L. W. Lim, H. S. Shieh, H. E. Dayringer, N. K. Leimgruber, R. A. Stegeman, K. S. Anderson, J. A. Sikorski, S. R. Padgette, and G. M. Kishore. 1991. Structure and topological symmetry of the glyphosate target 5-enolpyruvylshikimate-3-phosphate synthase: a distinctive protein fold. Proc. Natl. Acad. Sci. USA 88:5046-5050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Steinrucken, H. C., and N. Amrhein. 1980. The herbicide glyphosate is a potent inhibitor of 5-enolpyruvyl-shikimic acid-3-phosphate synthase. Biochem. Biophys. Res. Commun. 94:1207-1212. [DOI] [PubMed] [Google Scholar]
- 30.Steinrucken, H. C., and N. Amrhein. 1984. 5-Enolpyruvylshikimate-3-phosphate synthase of Klebsiella pneumoniae. 1. Purification and properties. Eur. J. Biochem. 143:341-349. [DOI] [PubMed] [Google Scholar]
- 31.Williams, P. A., and K. Murray. 1974. Metabolism of benzoate and the methylbenzoates by Pseudomonas putida (arvilla) mt-2: evidence for the existence of a TOL plasmid. J. Bacteriol. 120:416-423. [DOI] [PMC free article] [PubMed] [Google Scholar]