Abstract
A bacterial primer set, known to produce a 542-bp amplicon specific for Bacteroides thetaiotaomicron, generated this product in PCR with 1 ng of extracted DNA from 92% of 25 human fecal samples, 100% of 20 sewage samples, and 16% of 31 dog fecal samples. The marker was not detected in 1 ng of fecal DNA from 61 cows, 35 horses, 44 pigs, 24 chickens, 29 turkeys, and 17 geese.
Fecal pollution of water is a problem of global concern, and procedures to determine host sources of pollution are critically needed (8, 11). Since the public health risk from contact with human feces may be greater than from contact with nonhuman feces (19, 22, 26), the presence/absence of human pollution is often a primary concern (10). This issue is especially important for urgent decisions such as those regarding beach closure and for instances of storm water overflow, with possible mixing of storm water and sewage. Many currently available methods for tracking pollution sources involve days to culture fecal indicator bacteria (such as Escherichia coli) and require reference libraries of known-host bacterial isolates (4, 13, 24). Intrinsic concerns about the representativeness of libraries, including questions of content (17, 27, 28) and geographic variation (12, 26) of enteric indicator bacteria, imply that a new database should be generated for each watershed studied. Libraries are intended to be collections of known-host enteric bacteria which are presumably host specific. We are aware of no means to distinguish enteric bacteria, which have a special niche in the intestinal tracts of particular hosts (21), from those which are transient or cosmopolitan (existing in multiple hosts) in nature (20, 23, 25); thus, many fecal isolates may be misclassified in host source libraries.
Culture- and library-independent source tracking methods are, therefore, a current focus of attention, since they have the potential to be rapid and less costly than numerous methods presently used to distinguish between human and nonhuman pollution sources (4, 13, 22). Bacteroides spp. are prospective alternative indicators of the host source of fecal pollution (7), since they are the most abundant bacteria in the human intestine (16) and have a much smaller presence in nonhuman hosts (9, 18). Total counts of fecal indicator bacteria have been reported as more useful when considered in tandem with human-specific Bacteroides-Prevotella markers or assay for enterovirus to predict the presence of human waste in water (3). Bacteroides markers, based on species composition differences in Bacteroides-Prevotella intestinal populations in humans, have reportedly also shown promise for application, since they were previously found in DNA of 11 of 13 (84.6%) human fecal samples and each of three sewage samples tested (2). A PCR assay for this human marker, using deliberately prepared test samples of feces-polluted water, also accurately detected the presence of human waste (6).
Bacteroides thetaiotaomicron is a particular candidate for association with humans, since it is a predominant species in human feces and is present in a much higher percentage of fecal samples from humans than from nonhumans (16, 18). A universal bacterial primer set (RW01 and DG74A) was recently reported to generate an amplicon (in addition to the expected 362-bp product) unique to B. thetaiotaomicron type strain VPI 5482 (27). To determine the relative specificity of this recently reported marker, we assayed for the 542-bp amplicon in human feces, sewage, and feces from nonhuman hosts. We also compared its specificity to a benchmark human Bacteroides primer set (2). Our intent was to gather information to predict the utility of the new marker, for a genomic site other than the 16S rRNA gene, as an indicator of human fecal pollution in water.
Preliminary validation of B. thetaiotaomicron marker.
The following isolates were obtained from the American Type Culture Collection: B. fragilis (ATCC 25285), B. vulgatus (ATCC 8482), B. ovatus (ATCC 8483), B. uniformis (ATCC 8492), and B. thetaiotaomicron (ATCC 29741). Whole-cell suspensions of Bacteroides cultures were lysed with Lyse-N-Go PCR reagent (Pierce Chemical Co., Rockford, IL). B. thetaiotaomicron primers (B.thetaF and B.thetaR) were constructed as previously described (27). PCR was performed with 1 ng of DNA from each strain under previously described conditions (27). The B. thetaiotaomicron marker was detected only in that particular species.
Collection of human and nonhuman fecal samples.
Numbers and sources of samples of human feces, sewage, and feces from dogs (house pets), beef cattle, dairy cattle, horses, pigs, chickens, turkeys, and geese are listed in Table 1. Human specimens were submitted directly by donors. Sewage was collected from inflow to waste treatment plants and sewage lines in six Missouri cities and towns. Dog samples were collected from private house pets and pets being boarded at a kennel. Farm animal, poultry, and goose samples were collected as certainly as possible from separate animals. Fresh fecal material was placed into plastic bags, kept cold during transport to the laboratory, and stored at −20°C until the time of DNA extraction.
TABLE 1.
Sources of fecal samples
| Host species or source | No. of specimens | No. of individuals | Sampling site(s) or method |
|---|---|---|---|
| Humans | 25 | 25 | Direct collection |
| Sewage | 20 | Unknown | 6 locations |
| Dogs | 31 | 31 | 4 locations |
| Beef cattle | 35 | 35a | 3 farms |
| Dairy cattle | 26 | 26 | 2 farms |
| Horses | 35 | 35 | 3 farms |
| Swine | 44 | 44 | 3 farms |
| Chickens | 24 | 24a | 1 farm |
| Turkeys | 29 | 29a | 2 farms |
| Geese | 17 | 17a | 3 locations |
Each fecal specimen could not be attributed to an individual animal with certainty.
Specificity of the B. thetaiotaomicron marker.
DNA was extracted from fecal samples with the BIO 101 FastDNA spin kit for soil (Q-Biogene, Inc., Carlsbad, CA) by using a modification of the manufacturer's instructions. Sterile swabs of fecal material were added directly to the lysing tube to begin processing. Forty-milliliter aliquots of sewage were first concentrated by centrifugation at 2,052 × g for 15 min at 4°C. The supernatant was discarded, except for 2 ml, which was used to resuspend the pellet. Four hundred microliters of resuspended sewage concentrate was then added to the lysing tube. Lysis was accomplished by placement of the tube into a Disruptor Genie (Scientific Industries, Inc., Bohemia, NY) for 1 min. Extracted DNA was quantitated in a BioSpec-mini DNA/RNA/protein analyzer (Shimadzu Scientific Instruments, Inc., Columbia, MD). B. thetaiotaomicron primers were constructed as previously described (27). Universal bacterial primers (5) and universal Bacteroides primers (1) were constructed according to published sequences and used in PCR to ensure the presence of adequate fecal DNA templates. All primer sequences used in this study are shown in Table 2. PCR mixtures, fecal DNA targets, and conditions for each primer set used separately were optimized to ensure target detection and determine the detection limits. Feces were assayed for markers associated with bacteria, Bacteroides spp., and B. thetaiotaomicron. The PCR to confirm the presence of Bacteroides was performed with incremental quantities of fecal DNA from 2 to 100 ng. Fifty nanograms of DNA proved to be the smallest quantity which would provide a sufficiently visible PCR product as proof of the presence of a universal Bacteroides target in all host species. Fecal DNA samples that were negative at 50 ng were subsequently examined by PCR with universal bacterial primers. Human waste is considered to contain higher levels of B. thetaiotaomicron per unit volume than nonhuman feces (9, 16). To reflect these different values, we used levels of fecal DNA ranging from 1 to 25 ng to determine the practical limits of B. thetaiotaomicron detection by PCR in feces from individuals representing various human and nonhuman hosts. Based on results of these experiments, we chose to use 1 ng of fecal DNA as the PCR template with the B. thetaiotaomicron (B.thetaF and B.thetaR) primers. The PCR for B. thetaiotaomicron was performed as described previously (27) with the following modifications. Eppendorf Master Mix 2.5× (Brinkman Instruments, Inc., Westbury, NY) was used for this PCR and all of the following PCR methods according to the manufacturer's instructions. The universal Bacteroides PCR was performed as described previously (1) with the following modifications. A single round of 25 cycles was performed at 94°C for 30 s, 53°C for 45 s, 72°C for 90 s, and 72°C for 6 min. The universal bacterial PCR was performed as described previously (5) except that the annealing temperature was 45°C. All PCR procedures were done with an Eppendorf Mastercycler or Mastercycler gradient PCR instrument (Brinkman Instruments, Inc., Westbury, NY). PCR products (10 μl) were visualized in a 1% agarose gel (Agarose Low EEO; Fisher Scientific, Pittsburgh, PA) stained with ethidium bromide. Conditions of electrophoresis were 100 V for 45 min, and examination was done using UV light.
TABLE 2.
Primers used in this studya
| Primer | Sequence (5′-3′) | Target | Reference |
|---|---|---|---|
| B.thetaF | AACAGGTGGAAGCTGCGGA | B. thetaiotaomicron | 25 |
| B.thetaR | AGCCTCCAACCGCATCAA | B. thetaiotaomicron | 25 |
| Bac32F | AACGCTAGCTACAGGCTT | Bacteroides-Prevotella | 2 |
| Bac708R | CAATCGGAGTTCTTCGTG | Bacteroides-Prevotella | 2 |
| HF183F | ATCATGAGTTCACATGTCCG | Human HF8 cluster, HF74 | 2 |
| 1070F | ATGGCTGTCGTCAGCT | Bacterial 16S RNA | 5 |
| Univ1392R | ACGGGCGGTGTGTAC | Universal | 5 |
HF183F and Bac32F were paired with Bac708R, 1070F was paired with Univ1392R, and B.thetaF was paired with B.thetaR.
Sensitivity study.
This experiment was intended to establish an estimate of the lowest quantity of actual target DNA sufficient for marker detection. PCR was performed with 0.5 to 0.0002 ng of B. thetaiotaomicron DNA. PCR was also performed using the same range of quantities of fecal DNA from two human samples and two sewage samples.
Comparison to a benchmark human Bacteroides marker.
The B. thetaiotaomicron marker was compared with the HF183F and HF654R benchmark human Bacteroides primer set (2) with respect to specificities for their respective targets. HF183 human marker PCR was performed using two rounds as previously reported (1), with 1 ng of fecal DNA as the template.
Application to environmental water samples.
Twenty-three water samples were collected at two sites downstream from the effluent discharge from a wastewater treatment plant in a major metropolitan area. Filtration and DNA extraction were performed by modification of previously reported methods (14, 15). Two hundred milliliters of water from each sample was filtered through a 0.40-μM polycarbonate filter (Millipore Corp., Billerica, MA), with a 0.45-μM mixed cellulose ester backing filter used to create a tight seal. The polycarbonate filter was placed into a 15-ml conical tube with beads from the previously described DNA extraction kit. All extraction reagents were described above under “Specificity of the B. thetaiotaomicron marker.” The tube was vortexed for 2 min, and the filter was removed. Further processing was as described in the kit manual.
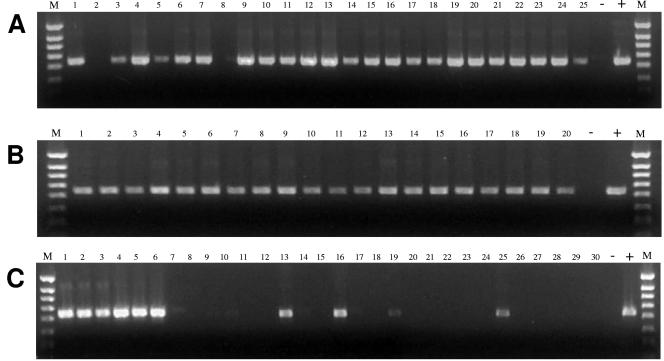
Results of the marker specificity study, with respect to the number of human fecal, sewage, and nonhuman fecal samples positive for each of the three markers used in this study, are shown in Table 3. The universal Bacteroides primer set generated PCR product visible on agar gels with DNA from all human fecal samples, all sewage samples, all dog, cattle, and swine samples, and many poultry, horse, and goose samples. One nanogram of fecal DNA was sufficient for the subject B. thetaiotaomicron primers to generate a strong PCR product for all 20 sewage samples and 23 of 25 human fecal samples (Table 3). The associated PCR products were represented by intense bands on the ethidium bromide-stained gels after electrophoresis (Fig. 1A and B). A distinct product was also produced with fecal DNA from 5 of 31 dogs (Table 3), possibly indicative of sharing of enteric bacteria between humans and dogs living in close association (16, 23). Five nanograms of fecal DNA produced no additional positive reactions for the human samples, but samples from 12 additional dogs, 2 of 24 chickens, 10 of 29 turkeys, and 2 of 44 pigs also became faintly positive. Figure 1C shows sample subsets (examples) of three isolates for each of the 10 host categories and includes the faint marker bands visible in lanes 13 (dog), 16 (chicken), 19 (turkey), and 25 (pig). Twenty nanograms of fecal DNA was required to generate even faintly visible PCR products from 12 of 61 cattle samples. At 25 ng of fecal DNA, there were still no positive PCRs for any of the samples from horses and geese. Note the relatively greater intensity of the marker bands in lanes 1 to 3 (sewer) and 4 to 6 (human). This template-dependent contrast between human and nonhuman samples reflects the relative numbers of B. thetaiotaomicron in these host classes.
TABLE 3.
Number of human fecal, sewage, and nonhuman fecal samples positive for the three markers used in this study
| Source | No. of positive samples/total no. of samples
|
||
|---|---|---|---|
| Bacteroidesa | B. thetaiotaomicronb | Human markerc | |
| Human | 25/25 | 23/25 | 12/25 |
| Sewage | 20/20 | 20/20 | 20/20 |
| Dog | 31/31 | 5/31 | 4/31 |
| Beef cattle | 35/35 | 0/35 | 0/6+ |
| Dairy cattle | 26/26 | 0/26 | 0/6+ |
| Chicken | 17/24 | 0/24 | 5/6+ |
| Turkey | 18/29 | 0/29 | 2/6+ |
| Horse | 29/35 | 0/35 | 0/6+ |
| Swine | 44/44 | 0/44 | 0/6+ |
| Goose | 9/17 | 0/17 | 0/6+ |
Primers Bac32F and Bac708R; 50 ng of fecal DNA.
Primers B.thetaF and B.thetaR; 1 ng of fecal DNA.
Primers HF183F and Bac708R; 1 ng of fecal DNA. +, random subset of six samples.
FIG. 1.
Gel electrophoresis of PCR products from reactions with B. thetaiotaomicron-specific primers B.thetaF and B.thetaR and DNA from human and nonhuman fecal sources. (A) PCR products generated with 1 ng of DNA from each of the human fecal samples, (B) PCR products generated with 1 ng of DNA from each of the sewage samples, and (C) PCR performed with 5 ng of fecal DNA from example subsets of three isolates from each of the 10 sources studied. Sources: sewer, lanes 1, 2, and 3; human, lanes 4, 5, and 6; dairy cattle, lanes 7, 8, and 9; beef cattle, lanes 10, 11, and 12; dog, lanes 13, 14, and 15; chicken, lanes 16, 17, and 18; turkey, lanes 19, 20, and 21; goose, lanes 22, 23, and 24; pig, lanes 25, 26, and 27; horse, lanes 28, 29, and 30. Lanes M have 1,000-bp ladders. Lanes +, positive control with B. thetaiotaomicron DNA. Lanes −, negative control without DNA.
Sensitivity assays reflected variation in B. thetaiotaomicron DNA content of the samples examined. The subject marker could be detected in as little as 0.0002 ng of “pure” B. thetaiotaomicron DNA. The detection limits for the two samples of human feces were 0.005 ng DNA for one and 0.0005 ng DNA for the other; both sewage samples had detection limits of 0.005 ng DNA.
Both the B. thetaiotaomicron primer set and the benchmark primer set amplified their markers in all 20 sewage samples at the level of 1 ng of DNA. At this same DNA level, the B. thetaiotaomicron marker was detected in 23 of 25 human fecal samples; the marker associated with the benchmark primers was detected in only 12 of 25 human samples. Both primer sets performed comparably by detecting their respective markers in 4 or 5 of the 31 dog fecal samples. The B. thetaiotaomicron marker, as described above, was not usually detected in samples of nonhuman fecal DNA at the 1-ng level. By contrast, the benchmark human Bacteroides primer set, used with random subsets of six fecal DNA samples from each category of production animals, horses, poultry, and geese (total of 42 samples), was strongly positive at 1 ng for five chicken and two turkey samples (Table 3). By this measure, the B. thetaiotaomicron primer set appeared to be a more effective indicator of human feces than the benchmark human primer set.
Ten of the 23 environmental water samples were positive for B. thetaiotaomicron. Three of these were positive when 1 ng of extracted DNA was used in the PCR, six were positive when 5 ng DNA was used, and one required 10 ng of DNA to generate a strong PCR product. This brief study was performed simply as an exercise in initial application of the subject method. No attempt was made to predict host association.
The disparity in relative presence of B. thetaiotaomicron in human and nonhuman enteric flora is key to the subject assay. The highest levels of B. thetaiotaomicron were reportedly found in human feces, with 10- to 100-fold-lower levels in dog and cat feces, and samples from farm animals had 103- to 105-fold-lower levels than those from house pets (9). In another study (18), nearly 80% of humans were reported to have high levels of B. thetaiotaomicron as opposed to about 15% of house pets. Nevertheless, interpretation of results of the B. thetaiotaomicron test as evidence of human fecal contamination should be done with the knowledge that animal sources can be responsible for false-positive identification.
Currently, we can only speculate on the successful application of the subject test, since our experience with application of the test is very limited. Initial work was done with fecal samples collected only in Missouri, and further evaluation will require expansion to a wider study area to determine whether the B. thetaiotaomicron marker is geographically stable. We furthermore expect that unknowns regarding source waters will include extent of fecal dilution, distance from pollution source to sampling site, and amount of B. thetaiotaomicron DNA degradation that may have occurred prior to testing. Nevertheless, we suggest that a strongly positive B. thetaiotaomicron test, done with a small quantity (e.g., 1 to 5 ng) of total DNA extracted from a water sample, would be primarily indicative of human-associated pollution. Strongly (or even faintly) positive tests, requiring larger DNA quantities, would justify additional consideration of domestic pets as the pollution source, followed by other possible animal hosts. It is expected that multiple and serial water samples, plus a knowledge of most-probable host sources, will be useful in interpretation of the B. thetaiotaomicron test results.
Since marker detection extended to the levels of 0.0002 ng of B. thetaiotaomicron DNA and 0.0005 ng of fecal DNA, the assay was considered to show promise of good sensitivity even after substantial target dilution. The B. thetaiotaomicron marker appeared to be a more accurate indicator for human feces than the benchmark at the 1-ng level. Furthermore, the B. thetaiotaomicron marker requires one PCR cycle, while the benchmark (2) requires two. The B. thetaiotaomicron assay can be performed in 5 h from the time of sample submission to the laboratory. This “same-day” analysis could help to support decisions on swimming-beach closure when used in combination with other source tracking assays. The procedure can be done inexpensively by laboratory personnel with basic skills and without highly specialized laboratory equipment. Such a library-independent approach also avoids the potentially confounding inclusion of cosmopolitan and transient bacteria that can diminish the validity of host-specific reference libraries.
Future studies will center on application of the subject method for detection of B. thetaiotaomicron DNA in environmental water samples. The method reported here is essentially a qualitative (presence/absence) analysis. Therefore, it is logical to consider its extension to a quantitative procedure using real-time PCR. We will examine freshwater and saltwater samples spiked with feces from human and nonhuman hosts and submitted to our laboratory “blindly.” We intend to include a study of environmental water samples from areas known to be impacted by waste from particular host species. Length of detection time for the bacterial DNA in the environment, under various water and climatic conditions, will also be of concern. Concurrent use of complementary microbial source tracking methods is always the suggested practice. Results of the present study indicate that the B. thetaiotaomicron test has the potential to become a valuable addition to the fecal source tracking “toolbox.” In this context, the utility and eventual role of this assay will be determined by further evaluation.
Acknowledgments
Funding was provided by the Food and Agriculture Policy Research Institute at the University of Missouri, the U.S. Geological Survey, and University of Missouri Outreach and Extension.
We are grateful for the assistance of Burton G. Schauf and Dwayne L. Miller in specimen procurement and logistical support. Thanks also to Ellen E. Swanson for technical assistance.
REFERENCES
- 1.Bernhard, A. E., and K. G. Field. 2000. Identification of nonpoint sources of fecal pollution in coastal waters by using host-specific 16S ribosomal DNA genetic markers from fecal anaerobes. Appl. Environ. Microbiol. 66:1587-1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bernhard, A. E., and K. G. Field. 2000. A PCR assay to discriminate human and ruminant feces on the basis of host differences in Bacteroides-Prevotella genes encoding 16S rRNA. Appl. Environ. Microbiol. 66:4571-4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boehm, A. B., J. A. Fuhrman, R. D. Mrse, and S. B. Grant. 2003. Tiered approach for identification of a human fecal pollution source at a recreational beach: case study at Avalon Bay, Catalina Island, California. Environ. Sci. Technol. 37:673-680. [DOI] [PubMed] [Google Scholar]
- 4.Carson, C. A., B. L. Shear, M. R. Ellersieck, and A. Asfaw. 2001. Identification of fecal Escherichia coli from humans and animals by ribotyping. Appl. Environ. Microbiol. 67:1503-1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ferris, M. J., G. Muyzer, and D. M. Ward. 1996. Denaturing gradient gel electrophoresis profiles of 16S rRNA-defined populations inhabiting a hot spring microbial mat community. Appl. Environ. Microbiol. 62:340-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Field, K. G., E. C. Chern, L. K. Dick, J. Fuhrman, J. Griffith, P. A. Holden, M. G. LaMontagne, J. Le, B. Olson, and M. T. Simonich. 2003. A comparative study of culture-independent genotypic methods of fecal source tracking. J. Water Health 1:181-194. [PubMed] [Google Scholar]
- 7.Fiksdal, L., J. S. Maki, S. J. LaCroix, and J. T. Staley. 1985. Survival and detection of Bacteroides spp., prospective indicator bacteria. Appl. Environ. Microbiol. 49:148-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fleisher, J. M., D. Kay, R. L. Salmon, F. Jones, M. D. Wyer, and A. F. Godfree. 1996. Marine waters contaminated with domestic sewage: nonenteric illnesses associated with bather exposure in the United Kingdom. Am. J. Public Health 86:1228-1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Geldreich, E. E. 1978. Bacterial populations and indicator concepts in feces, sewage, stormwater and solid wastes, p. 51-96. In G. Berg (ed.), Indicators of viruses in water and food. Ann Arbor Science Publishers, Inc., Ann Arbor, Mich.
- 10.Griffin, D. W., K. A. Donaldson, J. H. Paul, and J. B. Rose. 2003. Pathogenic human viruses in coastal waters. Clin. Microbiol. Rev. 16:129-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Griffith, J. F., S. B. Weisburg, and C. D. McGee. 2003. Evaluation of microbial source tracking methods using mixed fecal sources in aqueous test samples. J. Water Health 1:141-151. [PubMed] [Google Scholar]
- 12.Hartel, P. G. J. D. Summer, J. L. Hill, J. V. Collins, J. A. Entry, and W. I. Segars. 2002. Geographic variability of Escherichia coli ribotypes from animals in Idaho and Georgia. J. Environ. Qual. 31:1273-1278. [DOI] [PubMed] [Google Scholar]
- 13.Harwood, V. J., J. Whitlock, and V. Withington. 2000. Classification of antibiotic resistance patterns of indicator bacteria by discriminant analysis: use in predicting the source of fecal contamination in subtropical waters. Appl. Environ. Microbiol. 66:3698-3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haugland, R. A., J. L. Heckman, and L. J. Wymer. 1999. Evaluation of different methods for the extraction of DNA from fungal conidia by quantitative competitive PCR analysis. J. Microbiol. Methods 37:165-176. [DOI] [PubMed] [Google Scholar]
- 15.Haugland, R. A., N. Brinkman, and S. J. Vesper. 2002. Evaluation of rapid DNA extraction methods for the quantitative detection of fungi using real-time PCR analysis. J. Microbiol. Methods 50:319-323. [DOI] [PubMed] [Google Scholar]
- 16.Holdeman, L. V., I. J. Good, and W. E. C. Moore. 1976. Human fecal flora: variation in bacterial composition within individuals and a possible effect of emotional stress. Appl. Environ. Microbiol. 31:359-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson, L. K., M. B. Brown, E. A. Carruthers, J. A. Ferguson, P. E. Dombek, and M. J. Sadowsky. 2004. Sample size, library composition, and genotypic diversity among natural populations of Escherichia coli from different animals influence accuracy of determining sources of fecal pollution. Appl. Environ. Microbiol. 70:4478-4485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kreader, C. A. 1995. Design and evaluation of Bacteroides DNA probes for the specific detection of human fecal pollution. Appl. Environ. Microbiol. 61:1171-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krumperman, P. H. 1983. Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of fecal contamination of foods. Appl. Environ. Microbiol. 46:165-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McLellan, S. L., A. D. Daniels, and A. K. Salmore. 2003. Genetic characterization of Escherichia coli populations from host sources of fecal pollution by using DNA fingerprinting. Appl. Environ. Microbiol. 69:2587-2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McLellan, S. L. 2004. Genetic diversity of Escherichia coli isolated from urban rivers and beach water. Appl. Environ. Microbiol. 70:4658-4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moe, C. 2002. Waterborne transmission of infectious agents, pp. 184-204. In C. J. Hurst, R. L. Crawford, G. R. Knudsen, M. J. McInerney, and L. D. Stetzenbach (ed.), Manual of environmental microbiology, 2nd ed. ASM Press, Washington, D.C.
- 23.Myoda, S. P., C. A. Carson, J. J. Fuhrmann, B.-K. Hahm, P. G. Hartel, H. Yampara-Iquise, L. A. Johnson, R. L. Kuntz, C. H. Nakatsu, M. J. Sadowsky, and M. Samadpour. 2003. Comparison of genotypic-based microbial source tracking methods requiring a host origin database. J. Water Health 1:167-180. [PubMed] [Google Scholar]
- 24.Parveen, S., K. M. Portier, K. Robinson, L. Edmiston, and M. L. Tamplin. 1999. Discriminant analysis of ribotype profiles of Escherichia coli for differentiating human and nonhuman sources of fecal pollution. Appl. Environ. Microbiol. 65:3142-3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rodrigues, J., C. M. Thomazini, C. A. M. Lopes, and L. O. Dantas. 2004. Concurrent infection in a dog and colonization in a child with a human enteropathogenic Escherichia coli clone. J. Clin. Microbiol. 42:1388-1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scott, T. M., J. B. Rose, T. M. Jenkins, S. R. Farrah, and J. Lukasik. 2002. Microbial source tracking: current methodology and future directions. Appl. Environ. Microbiol. 68:5796-5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Teng, L.-J., P.-R. Hsueh, Y.-H. Huang, and J.-C. Tsai. 2004. Identification of Bacteroides thetaiotaomicron on the basis of an unexpected specific amplicon of universal 16S ribosomal DNA PCR. J. Clin. Microbiol. 42:1727-1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wiggins, B. A., P. W. Cash, W. S. Creamer, S. E. Dart, P. P. Garcia, T. M. Gerecke, J. Han, B. L. Henry, K. B. Hoover, E. L. Johnson, K. C. Jones, J. G. McCarthy, J. A. McDonough, S. A. Mercer, M. J. Noto, H. Park, M. S. Phillips, S. M. Purner, B. M. Smith, E. N. Stevens, and A. K. Varner. 2003. Use of antibiotic resistance analysis for representativeness testing of multiwatershed libraries. Appl. Environ. Microbiol. 69:3399-3405. [DOI] [PMC free article] [PubMed] [Google Scholar]