Abstract
To investigate the responses of Baltic Sea wintertime bacterial communities to changing salinity (5 to 26 practical salinity units), an experimental study was conducted. Bacterial communities of Baltic seawater and sea ice from a coastal site in southwest Finland were used in two batch culture experiments run for 17 or 18 days at 0°C. Bacterial abundance, cell volume, and leucine and thymidine incorporation were measured during the experiments. The bacterial community structure was assessed using denaturing gradient gel electrophoresis (DGGE) of PCR-amplified partial 16S rRNA genes with sequencing of DGGE bands from initial communities and communities of day 10 or 13 of the experiment. The sea ice-derived bacterial community was metabolically more active than the open-water community at the start of the experiment. Ice-derived bacterial communities were able to adapt to salinity change with smaller effects on physiology and community structure, whereas in the open-water bacterial communities, the bacterial cell volume evolution, bacterial abundance, and community structure responses indicated the presence of salinity stress. The closest relatives for all eight partial 16S rRNA gene sequences obtained were either organisms found in polar sea ice and other cold habitats or those found in summertime Baltic seawater. All sequences except one were associated with the α- and γ-proteobacteria or the Cytophaga-Flavobacterium-Bacteroides group. The overall physiological and community structure responses were parallel in ice-derived and open-water bacterial assemblages, which points to a linkage between community structure and physiology. These results support previous assumptions of the role of salinity fluctuation as a major selective factor shaping the sea ice bacterial community structure.
Sea ice covers vast areas of earth's surface annually and serves as a habitat for diverse microbial communities, which makes it one of the largest biomes known (12). The sea ice internal environment, the brine channel system, is characterized by low temperature, relatively high salinity, and elevated concentrations of dissolved nutrients and dissolved organic carbon. Natural variations in sea ice salinity are extreme; under non-ice conditions, only estuarine organisms experience salinity changes of this magnitude (34). However, unlike in estuaries, salinity variation inside sea ice also extends above the ambient seawater salinity.
Upon ice formation, bacteria incorporated into the newly forming ice must cope with severe physicochemical stress caused by salinity extremes and ice nucleation (41). Immediately after ice formation, a strong reduction in bacterial metabolic activity occurs as the water column bacteria decline. Subsequently, metabolic activity is restored as the psychrophilic population becomes established (5, 20, 21, 29). A gradual transition occurs in bacterial community composition from dominant psychrotolerant to obligate psychrophilic bacteria as sea ice formation progresses from the initial stages to a consolidated ice sheet (11, 23, 41). Low temperature alone or the ability to utilize available substrates at low temperatures does not account for the selective enrichment of psychrophiles (38, 43, 44). Sea ice bacterial assemblages have to be able to cope with constant and irregular salinity fluctuations, as brine salinity and chemical composition change over short time scales along with ice temperature changes (14, 19). Therefore, frequency, magnitude, and rate of salinity variation are believed to be major selective factors shaping the ice bacterial community (27, 41). Also, in open-water ecosystems, salinity fluctuations have been shown to significantly alter bacterioplankton community composition (1, 41).
In addition to the polar oceans, annual sea ice formation occurs in adjacent temperate sea areas as in the Baltic Sea. The northern Baltic Sea is annually ice covered for up to 6 months (45). Despite the brackish parent water, the sea ice contains a fully developed brine channel system and hosts diverse and active organism assemblages comparable to those of polar sea ice (29). Measured brine salinity in the Baltic Sea ice varies between 6 and 30 practical salinity units (psu) (26, 28, 32, 37). Even though brine salinity in polar sea ice may be substantially higher during winter, it is likely to be lower in spring and summer because of the flushing effect of ice melt and saline depletion resulting from the period of high-salinity brine drainage over winter (14, 23, 36). The spring-summer period is accompanied by maximal heterotrophic activity in sea ice (8, 22, 31), and polar sea ice bacteria may have preferential adaptation to the lower end of the salinity range, i.e., below the salinity of oceanic water (40). Nichols et al. (40) suggested that information on the physiological response of psychrophilic bacteria to combined temperature-salinity stress is very crucial to an understanding of the entire bacterial sea ice community. However, studies on the effects of salinity variations on sea ice natural bacterial assemblages have been lacking up to now. The aim of this study was to describe the effects of increasing salinity on the physiology and structure of the Baltic Sea ice and open-water natural bacterial assemblages. An experimental approach, designed to cover the salinity range from the ambient Baltic seawater in the study area to the Baltic Sea ice brine environment, was applied.
MATERIALS AND METHODS
Sampling and experimental setup.
In this study, two successive experiments with identical experimental setups were conducted. Water and ice samples for the experiments were collected at a coastal location in southwest Finland (59°51′N, 23°16′E) in the northern Baltic Sea. The water samples were taken at 10-m depths from the ice edge of an open-water area on 15 March (experiment 1) and under the seasonal ice cover on 23 March (experiment 2) in 2001. The ice samples for both experiments had been obtained 2 or 3 days earlier from 40-cm-thick level ice of a semienclosed bay, approximately 20 km from the water-sampling site. The ice samples were taken with a Mark II-type power auger (Kovacs Enterprises, Lebanon, NH) and cut into 10-cm sections, and the lowermost two sections were placed in clean plastic containers. The ice samples were then allowed to melt in the dark at 5°C.
For each of the two successive experiments, two separate eight-unit series were prepared, one containing only the open-water bacterial community (hereafter referred to as the “W” [water] series) and the other containing the open-water community and an addition of ice bacteria (hereafter referred to as the “I” [ice] series). Polycarbonate bottles of 1,200 ml (NalgeNunc International, Rochester, NY) were filled with 660 ml of seawater sample that was filtered through a 20-μm net to remove larger predators of bacteria and 540 ml of the same sample water filtered through a 0.45- plus 0.2-μm Sartobran 300-capsule filter (Sartorius, Göttingen, Germany) to remove bacteria. The W series contained only the open-water bacterial community already present in 20-μm-filtered inoculum. The I series was amended with sea ice bacteria that had been concentrated by centrifugation from the melted ice sample prefiltered through a 1-μm polycarbonate membrane filter (Millipore, Billerica, MA). That procedure resulted in the abundance of the ice bacteria which was 10% of that of the open-water bacteria already present in the experimental units (bacterial abundance was determined as described below). Salinity of the two experimental series was adjusted with Tropic Marin synthetic sea salt (Dr. Biener GMHB, Wartenberg, Germany), resulting in four salinity levels (5, 12, 19, and 26 psu), each in duplicate units in both series. To ensure sufficient bacterial growth, all units were amended with 1 mg liter−1 sucrose C and 40 μg liter−1 PO4−3 P. All units were incubated at 0°C in the dark for 17 or 18 days, and sampling was performed at 1- to 4-day intervals. Evolution of the total bacterial number, bacterial average cell volume, and total incorporation of thymidine and leucine were followed over the course of the experiment. The leucine and thymidine incorporation were measured from both duplicate units. Since the standard deviations between duplicate units of the leucine and thymidine incorporation were generally very small (see Fig. 2), the bacterial abundance and average cell volume measurements as well as community DNA collection were done from only one of the duplicate units.
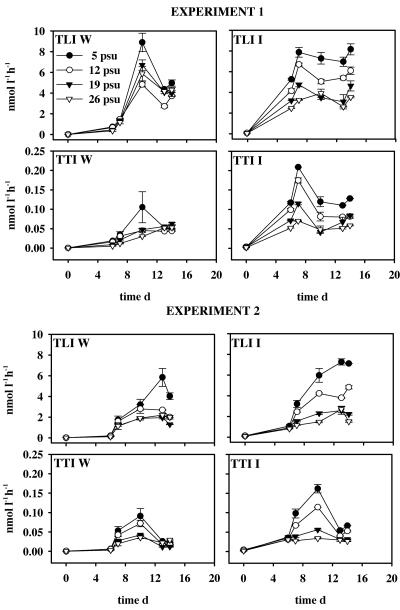
FIG. 2.
Total leucine incorporation (TLI) and total thymidine incorporation (TTI) in the open-water (W) and ice (I) series of experiments 1 and 2; error bars denote standard deviations between the duplicate units. d, day.
Physiological responses.
For determination of bacterial abundance and average cell volume, 20-ml samples were fixed with 25% electron microscopy grade glutaraldehyde (1% final concentration; Sigma-Aldrich Company, St. Louis, MO) and stored in a cool, dark place prior to analysis. A 5-ml subsample was then filtered onto 0.2-μm-pore-size black polycarbonate filters (Poretics, Livermore, CA) and stained with 0.02% acridine orange (25). The same filter was used for both bacterial abundance and average bacterial cell volume measurements, with variation coefficients between repetitive measurements being 6.4 and 3.3%, respectively. The total bacterial numbers were counted using a Leitz Aristoplan (Leica Microsystems, Bensheim, Germany) epifluorescence microscope equipped with Leitz a I3 filter and PL Fluotar 100 × 12.5/20× oil immersion objective giving a total magnification of ×1,000. The total bacterial number was calculated by counting at least 200 cells recorded in a minimum of 20 random fields of a New Porton E11 counting grid (Graticules Ltd., Ebenbridge, Kent, United Kingdom). The average bacterial cell volume was determined by image analysis (35). At least 200 bacteria were recorded from each filter using a Photometrics CH250/A charge-coupled-device camera (Princeton Instruments, Monmouth Junction, NJ) connected to a Leitz Aristoplan epifluorescence microscope and PMIS image acquisition software (GKR Computer Consulting, Unterföhring, Germany). The digital images obtained were analyzed with LabView (National Instruments, Austin, TX)-based LabMicrobe software (DiMedia, Kvistgård, Denmark).
Specific growth rates (μ) of bacteria were calculated according to the following formula (equation 1):
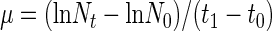 |
(1) |
where Nt and N0 are the cell numbers at the end and beginning of a sampling interval (i.e., between two successive samplings), respectively; t1 − t0 is the time (days) between these samplings.
The bacterial relative DNA and protein synthesis activities were measured using [3H]thymidine to determine the total thymidine incorporation (15, 16) and [14C]leucine for the total leucine incorporation (30) (dual labeling) as a proxy. Three 10-ml aliquots and a formaldehyde-killed (0.5% final concentration) absorption blank were amended with l-[U-14C]leucine (specific activity, 315 mCi mmol−1; Amersham Biosciences, Buckinghamshire, United Kingdom) and [methyl-3H]thymidine (specific activity, 20 Ci mmol−1; NEN, Boston, MA). The concentrations used, 14 nM for thymidine and 166 nM for leucine, were tested to be above the saturating concentration. The samples were incubated at 0°C for 2 h, and incorporation was terminated with formaldehyde. The macromolecules were collected using the standard cold-trichloroacetic acid extraction technique, and samples were counted with a Wallac WinSpectral 1414 liquid scintillation counter (Perkin-Elmer, Wellesley, MA [formerly Wallac, Turku, Finland]).
Community structure responses.
For bacterial community structure analysis, community DNA was isolated at the start of the experiment from the original melted ice samples, and the sample water was used for the W and I units. The bacterial cells were collected from all 16 units on day 10 for experiment 1 and on day 13 for experiment 2 by filtering the maximum possible amount of sample water (80 to 500 ml) onto a 47-mm-diameter Supor 200 polyethersulfone filter (Pall Corp., East Hills, NY). The filters were stored in a lysis buffer consisting of 40 mM EDTA, 400 mM NaCl, 750 mM sucrose, and 50 mM Tris HCl (pH 8.3) (18) at −80°C. In the laboratory, DNA was extracted following the hot-phenol method as described previously (18), except for precipitation of DNA, which was performed by adding a 1/10 volume of 3 M sodium acetate and 2 volumes of 94.6% ethyl alcohol and letting the sample precipitate at −20°C for 2 to 24 h. After centrifugation (16,000 × g, 20 min) the pellet was rinsed with 70% ethyl alcohol and centrifuged (16,000 × g, 10 min), resuspended in 60 μl of Tris-EDTA buffer, and stored at −20°C. The DNA extracts obtained were subsequently purified using a Prep-A-Gene DNA purification kit according to the manufacturer's instructions (Bio-Rad Laboratories, Hercules, CA).
Partial 16S rRNA genes were amplified from the community DNA with the general eubacterial primers F984GC and R1378 (24). The 25-μl PCRs contained 3 μl of the DNA extract as template, 0.3 μM of each primer, 200 μM of each deoxynucleoside triphosphate (Finnzymes, Espoo, Finland), 0.3 U DyNAzyme II polymerase (Finnzymes), 1× DyNAzyme II polymerase buffer, 700 μM MgCl2, and 1.0 M betaine. The PCR cycling consisted of a 3-min denaturation step at 94°C, followed by 35 cycles at 94°C for 1 min, 53°C for 1 min, and 72°C for 1 min, with a final extension step of 10 min at 72°C. The PCR products were checked in 1.5% (wt/vol) agarose gels in 0.5× TAE buffer (20 mM Tris-acetate, 0.5 mM EDTA [pH 8.9]).
Denaturing gradient gel electrophoresis (DGGE) was used to study the composition of and changes in the bacterial communities between the start and end (day 10 or 13) of the experiments. The PCR products were loaded onto 6% acrylamide-bis-acrylamide (37.5:1) DGGE gels with a vertical denaturing gradient from 38% to 45% (100% denaturant consisting of 7 M urea and 40% formamide). The running conditions were 150 V at 60°C in 1× TAE buffer for 4.5 h. The gels were stained with GelStar nucleic acid stain (Rockland, ME) for 40 min and photographed under UV light. Some of the most prominent bands with interesting positions in the gels were then excised for sequencing. The excised bands were stored in 50 μl of Tris-EDTA buffer at −20°C.
For sequencing, the partial 16S rRNA genes from the DGGE bands were amplified as described above, using 5 μl of the DGGE band sample as a template. To enhance liberation of the template from the gel, the melted sample was kept in a lab bench shaker for 30 min at room temperature. The PCR products were purified for sequencing with Ultrafree-DA columns (Millipore, Billerica, MA). The sequencing was carried out with an ABI Prism 310 Genetic Analyzer (PE Applied Biosystems, Foster City, CA) with the ABI Prism BigDye Terminator Cycle Sequencing Ready Reaction kit according to the manufacturer's recommendations (PE Applied Biosystems). The above-mentioned primers were used for sequencing reactions, and the amplicons were resolved in both directions. The sequences from each band were checked by aligning them with the PILEUP program of GCG package version 10.1 (Accelcrys, San Diego, CA [formerly Genetics Computer Group, WI]) and manually edited using the GeneDoc multiple sequence alignment editor.
In order to enhance detection of bands in the DGGE gels, all lanes of the gel images were scanned with public domain imageJ software (available at http://rsb.info.nih.gov/ij/), and the bands were detected from intensity histograms. To reveal similarities between different communities, band presence/absence data were subsequently used for cluster analysis (hierachical clustering, Ward linkage, and percent distance) using Systat 7.0 software (SPSS; Munich, Germany).
Nucleotide sequence accession numbers.
The sequences described above were deposited in GenBank under accession numbers AY271857 to AY271864.
RESULTS
Total bacterial number and bacterial cell volumes.
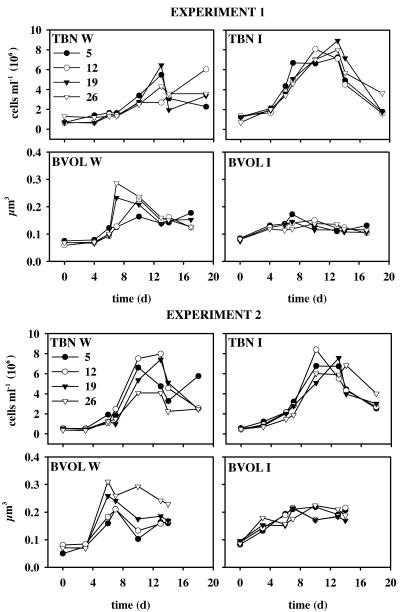
Increase in cell numbers began in the W units mainly after day 3 or 4, and cell density maximum was attained around days 10 or 13 in all units with the exception of the W 12-psu unit in experiment 1 (Fig. 1). In the I units, bacterial number increased immediately after the start of the experiment, with bacterial abundance being 1.9 to 4.9 times the initial values at day 6 (Fig. 1). Bacterial maximum abundances were of the same magnitude in I and W series in both experiments, and only the W 26-psu unit showed a lower maximum abundance than the I 26-psu unit. No clear differences in the temporal evolution of bacterial abundance or bacterial maximum abundance could be seen between the different salinity treatments in either I or W series (Fig. 1). In both experiments, development of the average bacterial cell volume differed clearly in the W and I series (Fig. 1). In the W series, the increase in average bacterial cell volume was pronounced after day 4, and a 3.4- to 4.4-fold increase in comparison to the initial value was seen in the two units with the highest salinity. In the 5- and 12-psu units, cell size maximum was attained 1 to 2 days later, and the increase was 2.1- to 3.8-fold. In the I series, the average cell volume increased 1.4- to 2.7-fold in all units 4 to 7 days after the start of the experiments and then remained rather steady throughout the rest of the course of the experiments. Maximal growth rates averaged for all salinities were (mean ± standard deviation) 0.30 ± 0.08 and 0.37 ± 0.06 in experiment 1 and 0.42 ± 0.10 and 0.38 ± 0.05 in experiment 2 W and I series, respectively.
FIG. 1.
Total bacterial number (TBN) and average bacterial cell volume (BVOL) in the open-water (W) and ice (I) series of experiments 1 and 2. d, day.
Leucine and thymidine incorporation.
Temporal development of the leucine and thymidine incorporation differed in the I and W series (Fig. 2). The initial leucine incorporation values on day 0 were low (0.002 to 0.13 nmol liter−1 h−1) although 8 and 15 times higher on average in the I series than in the W series in experiments 1 and 2, respectively. Temporal development of leucine incorporation in I series showed a similar pattern in both experiments, with incorporation values being significantly higher in the 12- and 5-psu units than in the 16- and 29-psu units (P < 0.016 [separate Mann-Whitney U tests for days 6 to 13]) and with maximum values decreasing along with the salinity increase. In W series, incorporation maximum was attained on days 10 to 13, with the unit with ambient salinity of 5 psu showing higher values than units with elevated salinity. The initial thymidine incorporation values were on average five and six times higher in the I unit than in the W unit in experiments 1 and 2, respectively. In the W series, the thymidine incorporation maximum was seen only in 5-psu or 5- and 12-psu units on day 10 in experiments 1 and 2, respectively. In the I series, temporal development of thymidine incorporation differed from that of leucine incorporation, having a clear peak on days 7 and 10 in experiments 1 and 2, respectively (Fig. 2).
Similar to leucine incorporation, thymidine incorporation was significantly higher in the two lower-salinity units than in the two higher-salinity units (P < 0.016 [separate Mann-Whitney U tests for days 7 to 13]).
Community structure.
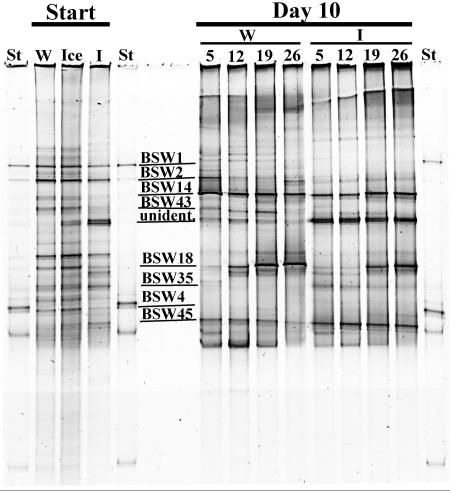
In the DGGE band pattern intensity histograms of experiments 1 and 2, 23 and 32 bands were detected in the initial water samples and 30 and 24 bands were detected in the ice samples, respectively (Fig. 3 [experiment 1 DGGE band pattern is shown]). The water and ice communities were initially rather similar; however, certain bands, including a multiple band marked “unidentified” (Fig. 3), were present in the ice community only. During the course of the experiment, the community structures changed (Fig. 3). Bands present in the starting communities, such as BSW1 and BSW2, either were not found in the communities on day 10 or were less intense. Alternatively, bands such as BSW14 were more intense on day 10 than at the start. Differences in the band patterns between the W and I communities on day 10 were also detected. The band BSW43 showed higher intensity in the W community, whereas the bands BSW35 and BSW45 and the unidentified band showed higher intensities in the I series (Fig. 3). The most explicit differences in band intensities in both experiments along the salinity gradient were observed in band BSW18, which showed the highest intensities in the highest salinities (19 and 26 psu).
FIG. 3.
DGGE band pattern of PCR-amplified partial 16S rRNA genes of eubacteria from the initial (Start), open-water (W), and ice (I) series communities of experiment 1 with the melted ice sample (Ice) and those from the W and I communities on day 10 of the experiment 1 (Day 10) grown with salinities of 5, 12, 19, and 26 psu. The lanes with standards are marked with St and the sequenced bands are indicated with lines and codes on the gel below, with the exception of the not-sequenced multiple band marked as “unident.”
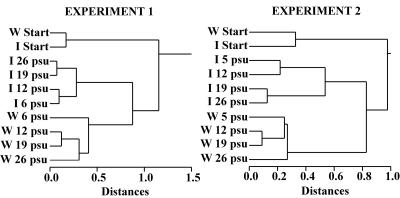
The main results of the DGGE band pattern cluster analysis were alike in both experiments (Fig. 4). Initial communities differed more from the end communities than from each other. The W series end communities clustered apart from the I end communities. There was more variability along the salinity gradient in the W than I end communities, as I series end communities formed two separate clusters, whereas three clusters or branches were formed in the W series.
FIG. 4.
Cluster analysis dendrograms of the open-water (W) and ice (I) series initial (Start) and end communities based on band presence/absence data in DGGE gel images of experiments 1 and 2.
Eight individual bands were amplified and sequenced from the DGGE gels (Fig. 3 and Table 1). The lengths of the resolved partial 16S rRNA sequences varied from 311 to 422 nucleotides. A BLAST search (September 2004) against our sequences resulted in sequences showing maximum similarities (98 to 100%) with our sequences (Table 1).
TABLE 1.
Partial 16S rRNA gene sequences derived from Baltic Sea water and ice communities and their closest matches in GenBank
| Division | Partial 16S rRNA gene sequence and GenBank accession no. (length [bp]) | Identical nucleotides/total (% similarity) | Closest matches in GenBank (accession no.) | Geographical origin | Reference or source |
|---|---|---|---|---|---|
| α-Proteobacteria | BSW1, AY271857 (422) | 375/375 (100) | Uncultured bacterium, clone AS 76 (AJ550432) | Archipelago Sea, the Baltic Sea | J. Sipura, unpublished |
| α-Proteobacteria | BSW2, AY271858 (418) | 371/371 (100) | Uncultured bacterium, clone AS 32 (AJ550431) | Archipelago Sea, the Baltic Sea | Sipura, unpublished |
| β-Proteobacteria | BSW35, AY271862 (418) | 401/404 (99) | Beta proteobacterium Wuba72 (AF336361) | Karstic aquifer, Wulfbach River resurgence, Germany | J. Bohnert et al., unpublished |
| 400/404 (99) | Glacier bacterium FJS31 (AY315178) | Sediments and ice from two Southern Hemisphere glaciers | J. Foght et al., unpublished | ||
| 400/404 (99) | Uncultured beta proteobacterium, clone NE83 (AJ575698) | Surface waters of humic Lake Grosse, Fuchskuhle, Germany | 9 | ||
| γ-Proteobacteria | BSW14, AY271860 (407) | 403/407 (99) | Psychromonas sp. strain IC004 (U85849) | Antarctic sea ice | 3 |
| γ-Proteobacteria | BSW18, AY271861 (407) | 401/401 (100) | Uncultured bacterium clone, ARKICE-28 (AF468310) | Arctic sea ice | 6 |
| 401/401 (100) | Colwellia sp. strain ANT9207 (AY167299) | Antarctic sea ice | 6 | ||
| γ-Proteobacteria | BSW45, AY271864 (419) | 408/408 (100) | Uncultured bacterium clone, ARKIA-102 (AF468282) | Arctic sea ice | 6 |
| 408/408 (100) | Shewanella sp. strain GA-22 (AJ563805) | Antarctic | 17 | ||
| 408/408 (100) | Shewanella livingstonis LMG19866T (AJ300834) | Antarctic | 4 | ||
| 408/408 (100) | Shewanella frigidimarina LMG19867 (AJ300833) | Antarctic | 4 | ||
| Flavobacteria | BSW43, AY271863 (311) | 306/311 (98) | Antarctic bacterium R-9003 (AJ441000) | Microbial mats from Antarctic lakes | 46 |
| 305/311 (98) | Uncultured flavobacterium SIC.ARCTIC.156 (AF277542) | Arctic sea ice | 7 | ||
| Actinobacteria | BSW4, AY271859 (361) | 341/341 (100) | Uncultured bacterium, clone AS 35 (AJ550436) | Archipelago Sea, the Baltic Sea | Sipura, unpublished |
Each of the sequenced bands showed the highest similarities with GenBank sequences of bacteria originating from either the Baltic Sea or the polar regions (Table 1). The bands/sequences BSW1, BSW2, and BSW4, which were present mainly in the initial communities (Fig. 3), had 100% sequence similarities to uncultured bacterial clones from the Baltic Sea. BSW14, which was mainly present in the day 10 communities (Fig. 3), showed 99% partial 16S rRNA gene sequence similarity to Psychromonas sp. sequences from the Antarctic sea ice (Table 1). Band BSW18 was most pronounced in the higher salinities of the day 10 communities (Fig. 3). It showed 100% sequence similarity to an uncultured bacterial clone, ARKICE-28, from the Arctic sea ice and Colwellia sp. ANT9207 from the Antarctic sea ice (Table 1). Band/sequence BSW35 was found mainly in the ice communities, showing the highest similarities (99%) to β-proteobacteria from a river resurgence and a humic lake in Germany and to a bacterium from glaciers in the Southern Hemisphere (Table 1). Band/sequence BSW43 was most pronounced in the initial water and ice communities and in the water communities on day 10, except in those with the highest salinity. It showed the highest similarity (98%) with sequences from the Antarctic bacterium R-9003 from microbial mats and with an uncultured Flavobacterium sp. from the Arctic sea ice. BSW45, which was present in all lanes and most pronounced in the ice communities on day 10, had 100% similarities with several bacteria from both the Arctic and Antarctic, which were mainly from the genus Shewanella. In addition to the bands that were sequenced, a multiple band that could not be sequenced was seen in the DGGE gel. This band (marked as “unidentified”) was absent in the initial water communities and most pronounced in the day 10 ice communities.
DISCUSSION
Initial communities.
We examined the responses of wintertime Baltic Sea bacterial communities to salinity change over a lower salinity range in comparison to salinity of oceanic water. This 5- to 26-psu range corresponds not only to the Baltic Sea ice brine salinity range but also to conditions that polar sea ice bacterial assemblages encounter during the spring-summer period. The community structure of the initial water and ice bacterial assemblages were rather similar, and the initial communities in W and I series clustered together in both experiments (Fig. 4). All eight bands sequenced were closely related to sequences from the Baltic Sea or various polar habitats (Table 1). No obvious resemblance to the only existing published report on Baltic Sea ice bacterial community structure from the Kiel Bight (42) was found.
The close association between our phylotypes from the Baltic Sea and from both polar areas corresponds with the recent findings of Brinkmeyer et al. (6) on phylogenetic convergence between Arctic and Antarctic sea ice bacterial communities. As found in previous studies of polar sea ice (3, 6, 7, 27), the majority of sequenced clones belonged to the γ- and α-proteobacteria and the Cytophaga-Flavobacterium-Bacteroides group. BSW35 was the only exception, showing 99% sequence similarity with environmental clones of β-proteobacteria from glaciers from the Southern Hemisphere and 98% sequence similarity with an environmental clone from Arctic sea ice (6).
Ice-derived bacterial communities.
In contrast to the water column bacterial communities, the major part of the sea ice bacterial community is believed to be metabolically active and cultivable (6). Sea ice bacteria added to the I units were metabolically active, which was shown by the higher initial leucine and thymidine incorporation rates observed in the I series than those observed in the W series. It was further assumed that this active 10% subpopulation of sea ice bacteria was able to overgrow the more slowly growing open-water-derived bacterial assemblage in the I units as described previously (13), leading to formation of an ice-derived bacterial assemblage in the I series. This assumption is supported by the earlier onset of increase of bacterial cell abundance and leucine and thymidine incorporation observed in the I series compared with that of the W series (Fig. 1 and 2).
Ice-derived bacterial communities were able to adapt to salinity change without major changes in growth rates or maximal bacterial cell numbers, which is consistent with the results from experimental work with a single sea ice bacterial species (38, 40). The significantly lower leucine and thymidine incorporation rates in the two units with higher salinity, however, suggest that the sea ice-derived assemblage could grow more efficiently in the upper end of the salinity range used. The physiological differences between low- and high-salinity units in the I series may have resulted from changes in community structure (Fig. 3). In I series, two separate groups corresponding to the significant differences observed in the leucine and thymidine incorporation were found in the cluster analysis. The most notable response of an organism to the salinity change was observed in band BSW18 associated with Colwellia spp. (Table 1). The intensity of the band increased along with increasing salinity in both W and I series (Fig. 3). The low intensity of BSW18 at low salinity may be due to the sea salt requirement of Colwellia spp., with a reported lower salinity limit of 25% of oceanic water (9 psu) for growth (2). The occurrence of band BSW18 in the initial open-water community at the depth of 10 m and ambient salinity of 5.3 psu remains unexplained.
Open-water bacterial communities.
In all the W series units, average bacterial cell volume increased sharply prior to bacterial abundance, with the increase being pronounced in the two units with the highest salinity (Fig. 1). This bacterial cell volume increase and lower maximal cell numbers in the unit with the highest salinity may be related to salinity stress. The overall physiological responses of the open-water assemblages (W series) were markedly different from those of the I series. This may be a reflection of different salinity adaptation strategies in these two adjacent wintertime bacterial communities. Current knowledge on the phylogenetic succession of bacterial assemblages in aquatic ecosystems and the transition mechanisms between phylogenetically distinct assemblages is limited. However, severe physiological stress to the bacteria is assumed to be accompanied by phylogenetic succession (10, 39). In comparison to the I series, the W series end communities showed a more variable clustering pattern along with the salinity increase (Fig. 4), which is parallel to the physiological responses observed and may have resulted from salinity stress encountered by the open-water bacterial assemblage. All end communities clustered apart from the initial communities, which probably reflected the effects of the experimental conditions. Clearly, conditions in batch cultures differ from those in the natural environment and are thus to some degree selective; however, all end communities were subsets of the initial communities and thus represent major components of the initial communities, as only the prominent species present in a bacterial community are assumed to be visible in PCR-DGGE. The advantage of this type of batch experiment is that it allows testing of specific factors, in this case, salinity, while other conditions are kept constant (33).
Conclusions.
In this study, we present the first results on physiological and community structure responses of sea ice bacterial communities to salinity change. The results show that under experimental conditions, the sea ice-derived natural bacterial communities were able to maintain balanced growth over the salinity range used with small changes in community structure and physiology especially within the two groups observed, which points to effective adaptation to salinity fluctuations. The open-water bacterial assemblages seemed to suffer from osmotic stress along with the salinity rise and responded to increased salinity with greater changes in community structure and physiology. The parallel physiological and community structure responses in both the ice and open-water bacterial assemblages point to a clear linkage between the phylogenetic structure and physiological responses of these bacterial communities, as suggested previously by Del Giorgio and Bouvier (10) for estuarine bacterioplankton. Adaptation to salinity fluctuations is crucial for sea ice organisms living in a constantly changing salinity environment, while open-water assemblages tend to respond with changes in community structure. Results of this study well support the earlier assumptions on the role of salinity fluctuation as a major selective factor shaping the sea ice bacterial community structure.
Acknowledgments
This work was financially supported by the Walter and Andrée de Nottbeck Foundation (H.K.) and grants (48004-FIBRE program, 201576) from the Academy of Finland to K.S. and the Finnish Institute of Marine Research (H.K. and M.L.).
The assistance of Tvärminne Zoological Station staff in logistics and sampling is gratefully acknowledged.
REFERENCES
- 1.Bouvier, T. C., and P. A. del Giorgio. 2002. Compositional changes in free-living bacterial communities along a salinity gradient in two temperate estuaries. Limnol. Oceanogr. 47:453-470. [Google Scholar]
- 2.Bowman, J. P., J. J. Gosink, S. A. McCammon, T. E. Lewis, D. S. Nichols, J. H. Skerratt, J. T. Staley, and T. A. McMeekin. 1998. Colwellia demingiae sp. nov., Colwellia hornerae sp. nov., Colwellia psychrotropica sp. nov.: psychrophilic Antarctic species with the ability to synthetize docosahexaenoic acid (22:6ω3). Int. J. Syst. Bacteriol. 48:1171-1180. [Google Scholar]
- 3.Bowman, J. P., S. A. McCammon, M. V. Brown, D. S. Nichols, and T. A. McMeekin. 1997. Diversity and association of psychrophilic bacteria in Antarctic sea ice. Appl. Environ. Microbiol. 63:3068-3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bozal, N., M. J. Montes, E. Tudela, F. Jimenez, and J. Guinea. 2002. Shewanella frigidimarina and Shewanella livingstonensis sp. nov. isolated from Antarctic coastal areas. Int. J. Syst. Evol. Microbiol. 52:195-205. [DOI] [PubMed] [Google Scholar]
- 5.Brierley, A. S., and D. N. Thomas. 2002. Ecology of southern ocean pack ice. Adv. Mar. Biol. 43:171-276. [DOI] [PubMed] [Google Scholar]
- 6.Brinkmeyer, R., K. Knittel, J. Juergens, H. Weyland, R. Amann, and E. Helmke. 2003. Diversity and structure of bacterial communities in Arctic versus Antarctic sea ice: a comparison. Appl. Environ. Microbiol. 69:6610-6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown, M. V., and J. P. Bowman. 2001. A molecular phylogenetic survey of sea-ice microbial communities (SIMCO). FEMS Microbiol. Ecol. 35:267-275. [DOI] [PubMed] [Google Scholar]
- 8.Bunch, J. N., and R. C. Harland. 1990. Bacterial production in the bottom surface of sea ice in the Canadian Subarctic. Can. J. Fish Aquat. Sci. 47:1986-1995. [Google Scholar]
- 9.Burkert, U., F. Warnecke, D. Babenzien, E. Zwirnmann, and J. Pernthaler. 2003. Members of a readily enriched β-proteobacterial clade are common in the surface waters of a humic lake. Appl. Environ. Microbiol. 69:6550-6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Del Giorgio, P. A., and T. C. Bouvier. 2002. Linking the physiology and phylogenetic successions in free-living bacterial communities along an estuarine salinity gradient. Limnol. Oceanogr. 47:471-486. [Google Scholar]
- 11.Delille, D. 1992. Marine bacterioplankton at the Weddell Sea ice edge, distribution of psychrophilic and psychrotrophic populations. Polar Biol. 12:205-210. [Google Scholar]
- 12.Dieckmann, G. S., and H. H. Hellmer. 2003. The importance of sea ice: an overview, p. 1-21. In D. N. Thomas and G. S. Dieckmann (ed.), Sea ice: an introduction to its physics, chemistry, biology and geology. Blackwell Science, Oxford, United Kingdom.
- 13.Ducklow, H. 2000. Bacterial production and biomass in the oceans, p. 85-120. In D. Kirchman (ed.), Microbial ecology of the oceans. Wiley-Liss, New York, N.Y.
- 14.Eicken, H. 2003. From the microscopic to the macroscopic, to the regional scale: growth, microstructure and properties of sea ice, p. 22-81. In D. N. Thomas and G. S. Dieckmann (ed.), Sea ice: an introduction to its physics, chemistry, biology and geology. Blackwell Science, Oxford, United Kingdom.
- 15.Fuhrman, J. A., and F. Azam. 1980. Bacterioplankton secondary production estimates for coastal waters of British Columbia, Antarctica, and California. Appl. Environ. Microbiol. 39:1085-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fuhrman, J. A., and F. Azam. 1982. Thymidine incorporation as a measure of heterotrophic bacterioplankton production in marine surface waters: evaluation and field results. Mar. Biol. 66:109-120. [Google Scholar]
- 17.Gentile, G., V. Bonasera, C. Amico, L. Giuliano, and M. M. Yakimov. 2003. Shewanella sp. GA-22, a psychrophilic hydrocarbonoclastic Antarctic bacterium producing polyunsaturated fatty acids. J. Appl. Microbiol. 95:1124-1133. [DOI] [PubMed] [Google Scholar]
- 18.Giovannoni, S. J., E. F. DeLong, T. M. Schmidt, and N. R. Pace. 1990. Tangential flow filtration and preliminary phylogenetic analysis of marine picoplankton. Appl. Environ. Microbiol. 56:2572-2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Granskog, M. A., H. Kaartokallio, and K. Shirasawa. 2003. Nutrient status of Baltic Sea ice: evidence for control by snow-ice formation, ice permeability, and ice algae. J. Geophys. Res. 108:3253. [Online.] doi: 10.1029/2002JC001386. [DOI] [Google Scholar]
- 20.Grossmann, S. 1994. Bacterial activity in sea ice and open waters of the Weddell Sea, Antarctica: a microautoradiographic study. Microb. Ecol. 28:1-18. [DOI] [PubMed] [Google Scholar]
- 21.Grossmann, S., and M. Gleitz. 1993. Microbial responses to experimental sea-ice formation: implications for the establishment of Antarctic sea-ice communities. J. Exp. Mar. Biol. Ecol. 173:273-289. [Google Scholar]
- 22.Haecky, P., and A. Andersson. 1999. Primary and bacterial production in sea ice in the northern Baltic sea. Aquat. Microb. Ecol. 20:107-118. [Google Scholar]
- 23.Helmke, E., and H. Weyland. 1995. Bacteria in sea ice and underlying seawater of the eastern Weddell Sea in midwinter. Mar. Ecol. Prog. Ser. 117:269-287. [Google Scholar]
- 24.Heuer, H., M. Krsek, P. Baker, K. Smalla, and E. M. H. Wellington. 1997. Analysis of actinomycete communities by specific amplification of genes encoding 16S rRNA and gel-electrophoretic separation in denaturing gradients. Appl. Environ. Microbiol. 63:3233-3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hobbie, J. E., R. J. Daley, and S. Jasper. 1977. Use of Nuclepore filters for counting bacteria by fluorescence microscopy. Appl. Environ. Microbiol. 33:1225-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ikävalko, J., and H. A. Thomsen. 1997. The Baltic Sea ice biota (March 1994): a study of the protistan community. Eur. J. Protistol. 33:229-243. [Google Scholar]
- 27.Junge, K., F. Imhoff, J. Staley, and J. W. Deming. 2002. Phylogenetic diversity of numerically important Arctic sea-ice bacteria cultured at subzero temperature. Microb. Ecol. 43:315-328. [DOI] [PubMed] [Google Scholar]
- 28.Kaartokallio, H. 2001. Evidence for active microbial nitrogen transformations in sea ice (Gulf of Bothnia, Baltic Sea) in midwinter. Polar Biol. 24:21-28. [Google Scholar]
- 29.Kaartokallio, H. 2004. Food web components and physical and chemical properties of Baltic Sea ice. Mar. Ecol. Prog. Ser. 273:49-63. [Google Scholar]
- 30.Kirchman, D., E. K'Nees, and R. Hodson. 1985. Leucine incorporation and its potential as a measure of protein synthesis by bacteria in natural aquatic systems. Appl. Environ. Microbiol. 49:599-607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kottmeier, S. T., S. M. Grossi, and C. W. Sullivan. 1987. Sea ice microbial communities. VIII. Bacterial production in annual sea ice of McMurdo sound, Antarctica. Mar. Ecol. Prog. Ser. 35:175-186. [Google Scholar]
- 32.Laamanen, M. 1996. Cyanoprokaryotes in the Baltic Sea ice and winter plankton. Algol. Stud. 83:423-433. [Google Scholar]
- 33.Langenheder, S., V. Kisand, J. Wikner, and L. J. Tranvik. 2003. Salinity as a structuring factor for the composition and performance of bacterioplankton degrading riverine DOC. FEMS Microbiol. Ecol. 45:189-202. [DOI] [PubMed] [Google Scholar]
- 34.Lizotte, M. P. 2003. The microbiology of sea ice, p. 184-210. In D. N. Thomas and G. S. Dieckmann (ed.), Sea ice: an introduction to its physics, chemistry, biology and geology. Blackwell Science, Oxford, United Kingdom.
- 35.Massana, R., J. M. Gasol, P. K. Bjørnsen, N. T. Blackburn, Å. Hagström, S. Hietanen, B. H. Hygum, J. Kuparinen, and C. Pedrós-Alió. 1997. Measurement of bacterial size via image analysis of epifluorescence preparations: description of an inexpensive system and solutions to some of the most common problems. Sci. Mar. 61:397-407. [Google Scholar]
- 36.Melnikov, I. A. 1997. The Arctic sea ice ecosystem. Gordon and Breach Scientific Publishers, Amsterdam, The Netherlands.
- 37.Mock, T., K. M. Meiners, and H. C. Giesenhagen. 1997. Bacteria in sea ice and underlying brackish water at 54° 26′ 5" N (Baltic Sea). Mar. Ecol. Prog. Ser. 158:23-40. [Google Scholar]
- 38.Nedwell, D. B. 1999. Effect of low temperature on microbial growth: lowered affinity for substrates limits growth at low temperature. FEMS Microbiol. Ecol. 30:101-111. [DOI] [PubMed] [Google Scholar]
- 39.Nichols, D. S., A. R. Greenhill, C. T. Shadbolt, T. Ross, and T. A. McMeekin. 1999. Physicochemical parameters for growth of the sea ice bacteria Glaciecola punicea ACAM 611T and Gelidibacter sp. strain IC158. Appl. Environ. Microbiol. 65:3757-3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nichols, D. S., J. Olley, H. Garda, R. R. Brenner, and T. A. McMeekin. 2000. Effect of temperature and salinity stress on growth and lipid composition of Shewanella gelidimarina. Appl. Environ. Microbiol. 66:2422-2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nichols, D. S., P. D. Nichols, and T. A. McMeekin. 1995. Ecology and physiology of psychrophilic bacteria from Antarctic saline lakes and sea-ice. Sci. Prog. 78:311-347. [Google Scholar]
- 42.Petri, R., and J. F. Imhoff. 2001. Genetic analysis of sea-ice bacterial communities of the western Baltic Sea using an improved double gradient method. Polar Biol. 24:252-257. [Google Scholar]
- 43.Pomeroy, L. R., and W. J. Wiebe. 2001. Temperature and substrates as interactive limiting factors for marine heterotrophic bacteria. Aquat. Microb. Ecol. 23:187-204. [Google Scholar]
- 44.Reay, D. S., D. B. Nedwell, J. Priddle, and J. C. Ellis-Evans. 1999. Temperature dependence of inorganic nitrogen uptake: reduced affinity for nitrate at suboptimal temperatures in both algae and bacteria. Appl. Environ. Microbiol. 65:2577-2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seinä, A., and J. Peltonen. 1991. Duration of the ice season and statistics of fast ice thickness along the Finnish coast 1961-1990. Finnish Marine Research 258:1-46.
- 46.Van Trappen, S., J. Mergaert, S. Van Eygen, P. Dawyndt, M. C. Cnockaert, and J. Swings. 2002. Diversity of 746 heterotrophic bacteria isolated from microbial mats from ten Antarctic lakes. Syst. Appl. Microbiol. 25:603-610. [DOI] [PubMed] [Google Scholar]