Abstract
The pathways for degradation of aromatic hydrocarbons are constantly modified by a variety of genetic mechanisms. Genetic studies carried out with Pseudomonas stutzeri OX1 suggested that the tou operon coding for toluene o-xylene monooxygenase (ToMO) was recently recruited into a preexisting pathway that already possessed the ph operon coding for phenol hydroxylase (PH). This apparently resulted in a redundancy of enzymatic activities, because both enzymes are able to hydroxylate (methyl)benzenes to (methyl)catechols via the intermediate production of (methyl)phenols. We investigated the kinetics and regioselectivity of toluene and o-xylene oxidation using Escherichia coli cells expressing ToMO and PH complexes. Our data indicate that in the recombinant system the enzymes act sequentially and that their catalytic efficiency and regioselectivity optimize the degradation of toluene and o-xylene, both of which are growth substrates. The main product of toluene oxidation by ToMO is p-cresol, the best substrate for PH, which catalyzes its transformation to 4-methylcatechol. The sequential action of the two enzymes on o-xylene leads, via the intermediate 3,4-dimethylphenol, to the exclusive production of 3,4-dimethylcatechol, the only dimethylcatechol isomer that can serve as a carbon and energy source after further metabolic processing. Moreover, our data strongly support a metabolic explanation for the acquisition of the ToMO operon by P. stutzeri OX1. It is possible that using the two enzymes in a concerted fashion confers on the strain a selective advantage based on the ability of the microorganism to optimize the efficiency of the use of nonhydroxylated aromatic hydrocarbons, such as benzene, toluene, and o-xylene.
Several bacterial strains use specific hydrocarbon substrates as their primary sources of carbon and energy, and the number of such strains is continuously increasing (13, 28, 30, 34). The wide range of substrates that can be transformed by these microorganisms make them a powerful tool for the bioremediation of environmentally harmful substances. Degradation of aromatic hydrocarbons by aerobic bacteria is generally carried out through catabolic routes usually divided into an upper pathway, which produces dihydroxylated aromatic intermediates by the action of monooxygenases, and a lower pathway, which transforms these intermediates into molecules that enter the citric acid cycle (26). Molecular elucidation of these catabolic pathways has indicated that new pathways are continually generated by a variety of genetic mechanisms, such as gene transfer, mutational drift, genetic recombination, and transposition (30, 31, 35, 37). Pseudomonas stutzeri OX1 is one microorganism that is capable of growth on highly toxic aromatic compounds, such as benzene, toluene, o-xylene, and dimethylphenols, each of which can serve as a sole source of carbon and energy (3). This strain is an interesting model system for studying hydrocarbon catabolism because of its peculiar ability to transform a wide range of these molecules compared with other similar microorganisms.
In P. stutzeri, (methyl)benzenes are initially activated by sequential introduction of two adjacent hydroxyl groups to form (methyl)phenols and, eventually, (methyl)catechols (Fig. 1) (5). These metabolites are subsequently cleaved into 2-hydroxymuconic semialdehyde derivatives, which, upon further processing, are transformed into citric acid cycle intermediates (1). We have recently demonstrated (8, 9) that the initial hydroxylation steps are carried out by two evolutionarily distinct bacterial multicomponent monooxygenases (BMMs), toluene o-xylene monooxygenase (ToMO), belonging to the family consisting of four-component aromatic/alkene monooxygenases (group 2 BMMs) (6, 9), and phenol hydroxylase (PH), belonging to the group consisting of toluene 2-monooxygenases (T2MO)/phenol hydroxylases (group 1 BMMs) (8). The ring cleavage step that allows aromatic hydrocarbons access to the lower metabolic pathway is catalyzed by a catechol 2,3-dioxygenase (C2,3O) (1).
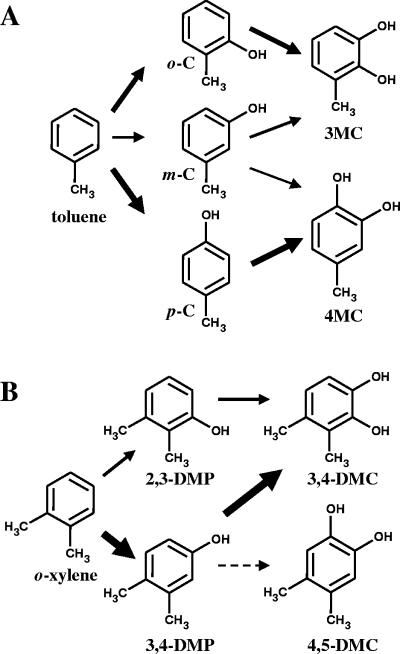
FIG. 1.
Proposed pathways for the conversion of toluene to methylcatechols (A) and for the conversion of o-xylene to dimethylphenols (B) catalyzed by ToMO and PH mixtures. The thickness of the arrows is roughly proportional to the relative abundance of each species. o-C, o-cresol; m-C, m-cresol; p-C, p-cresol.
Genetic studies carried out with P. stutzeri OX1 have suggested that the genes coding for ToMO were recently acquired by horizontal gene transfer and incorporated into a preexisting catabolic route (4). Thus, the acquisition of ToMO by P. stutzeri OX1 is an example of expansion of a catabolic pathway. The result of this expansion is an apparent redundancy in enzymatic activity, since ToMO and PH hydroxylate both benzene and phenol (1, 5, 8). To elucidate the metabolic significance of the coexpression of two multicomponent enzymes with very similar catalytic activities, we recently investigated the kinetics of benzene oxidation catalyzed by ToMO and PH (8). The data obtained strongly supported the hypothesis that ToMO and PH form a metabolic chain in which the depletion of phenol by PH enhances the use of benzene in P. stutzeri. However, this hypothesis needs to be extended to other substrates in order to establish, in general, whether the concerted use of the two different monooxygenases controls the metabolic flow of aromatic molecules that can be used by the microorganism as growth substrates which enter the lower pathway.
In the present work we investigated toluene and o-xylene oxidation using recombinant Escherichia coli cells expressing ToMO and PH complexes to collect data on how the biochemical properties of the two complexes could influence the efficiency of methylated aromatic utilization. Our results confirmed the general hypothesis that the two monooxygenases of P. stutzeri OX1 form a metabolic chain, thus expanding the catabolic potential of the microorganism by optimizing the degradation of benzene, toluene, and o-xylene.
MATERIALS AND METHODS
Materials.
Bacterial culturing, plasmid purification, and transformation were performed as described by Sambrook et al. (27). E. coli strain JM109 was obtained from Novagen. E. coli strain JM101 was purchased from Boehringer. The pGEM-3Z expression vector and Wizard SV gel and PCR clean-up system for elution of DNA fragments from the agarose gel were obtained from Promega. Enzymes and other reagents used for DNA manipulation were obtained from New England Biolabs. All other chemicals were obtained from Sigma. The methods used for expression and purification of recombinant C2,3O from P. stutzeri OX1 are described elsewhere (33).
Expression vectors.
Plasmids pBZ1260 (5) and pJSX148 (1) containing the ToMO and PH gene clusters, respectively, were kindly supplied by P. Barbieri (Dipartimento di Biologia Strutturale e Funzionale, Università dell'Insubria, Varese, Italy). Plasmid pBZ1260 containing the tou gene cluster cloned in the pGEM-3Z vector was used to express ToMO. For expression of PH, the DNA sequence coding for the PH complex from plasmid pJSX148 was subcloned into the vectors pGEM-3Z and pBZ1260. To do this, the DNA fragment coding for the PH cluster was excised from the pJSX148 vector using XbaI restriction endonuclease, purified by agarose gel electrophoresis, ligated with the pGEM-3Z and pBZ1260 vectors previously cut with the same enzyme, and used to transform JM101 competent cells. The resulting recombinant plasmids were designated pGEM-3Z/PH and pGEM-3Z/ToMO-PH, respectively. It should be noted that vector pGEM-3Z/ToMO-PH allows transcription of ToMO and PH open reading frames into a single mRNA under control of the lac promoter of the pGEM-3Z vector. Plasmids pGEM-3Z/PH and pGEM-3Z/ToMO-PH were used to express the PH complex alone and to coexpress the ToMO and PH complexes, respectively.
Whole-cell assays.
The assays were performed as described previously (8) using E. coli JM109 cells transformed with plasmids pGEM-3Z/PH, pBZ1260, and pGEM-3Z/ToMO-PH, which expressed PH, ToMO, and PH/ToMO complexes, respectively. The recombinant strains were routinely grown in Luria-Bertani medium (27) supplemented with 50 μg/ml ampicillin at 37°C to an optical density at 600 nm (OD600) of ∼0.5. Expression of the recombinant protein was induced with 0.4 mM isopropyl-β-d-thiogalactopyranoside at 37°C in the presence of 100 μM Fe(NH4)2(SO4)2. One hour after induction, cells were collected by centrifugation and suspended in M9 minimal medium containing 0.4% glucose. The enzymatic activity of cells was measured with phenol as the substrate by monitoring the production of catechol in continuous coupled assays with recombinant C2,3O from P. stutzeri OX1, as described previously (8). The specific activities of the cells ranged from 10 to 14 mU/OD600 for cells expressing PH and from 14 to 16 mU/OD600 for cells expressing ToMO. One milliunit was defined as the amount of catalyst that oxidized 1 nmol of phenol per min at 25°C.
Determination of apparent kinetic parameters.
All the kinetic parameters were determined using whole cells. The apparent kinetic parameters for phenol, o-, m-, and p-cresol, and 2,3-dimethylphenol (2,3-DMP) hydroxylation were determined by a continuous colorimetric assay coupled with C2,3O as described elsewhere (8). Induced cells were used at a concentration of 0.25 to 0.5 mU/ml. Substrates were added from stock solutions in water at a maximum concentration of 1 mM. The amounts of semialdehyde produced from catechol, 3-methylcatechol (3-MC), 4-MC, and 3,4-dimethylcatechol (3,4-DMC) were determined by measuring the increase in absorbance at 375 nm (ɛ375 = 29,100 M−1 cm−1), at 388 nm (ɛ388 = 6,100 M−1 cm−1), at 382 nm (ɛ382 = 21,550 M−1 cm−1), and at 324 nm (ɛ324 = 15,310 M−1 cm−1), respectively, for the different semialdehyde products.
The apparent kinetic parameters for benzene, toluene, o-xylene, and 3,4-DMP were determined by a discontinuous assay with cells suspended at a concentration of 0.5 to 1 mU/ml in 500 μl (final volume) of M9 minimal medium containing 0.4% glucose at 25°C. Reactions were started by addition to cell suspensions of different amounts of substrates (final concentration, up to 1 mM) in N,N-dimethylformamide. The reactions were stopped at different times by addition of 50 μl of 1 M HCl. Samples were centrifuged at 12,000 rpm for 20 min at 4°C. The soluble fractions were stored at −20°C until further analysis by high-pressure liquid chromatography (HPLC) as described below.
Maximum rates (Vmax) for all substrates were always normalized to the maximum rate measured for phenol in a parallel assay. This was done for each sample of induced cells, and the experiment was repeated for each substrate with at least three independent samples of cells. For each sample of induced cells, the levels of expression of ToMO or PH were measured as described below and used to calculate the apparent kcat for phenol. The amounts of ToMO and PH complexes were determined by densitometric scanning of Coomassie blue-stained sodium dodecyl sulfate-polyacrylamide gels (22) containing cell extracts. Five different amounts of samples of lysed cells were run on a single gel together with five different amounts of purified monooxygenases used as standards. ToMO and PH were expressed at similar levels.
Apparent kcat values for each substrate were calculated by determining the products of the apparent kcat for phenol and the individual substrate Vmax/phenol Vmax ratios determined as described above. Relative errors for the apparent kcat values were calculated by adding the relative error for the apparent kcat for phenol and the relative errors for the substrate Vmax/phenol Vmax ratios.
Apparent kinetic parameters were calculated with the program GraphPad Prism (GraphPad Software).
Identification of products.
Reaction products were identified with a HPLC system equipped with a Waters 1525 binary pump coupled to a Waters 2996 photodiode array detector. Mono- and dihydroxylated products were separated using an Ultrasphere C18 reverse-phase column (4.6 by 250 mm; pore size, 80 Å), and the absorbance of the eluate at 274 nm was monitored. Separation was carried out at a flow rate of 1 ml/min by using a two-solvent system comprising a 0.1% formic acid solution in water (solvent A) and a 0.1% formic acid solution in methanol (solvent B). Phenol and catechol were separated using 12 min of isocratic elution with 10% of solvent B, followed elution with a linear 10 to 30% solvent B gradient in 10 min and an isocratic 30% solvent B step. The retention times of phenol and catechol were 25.2 and 14 min, respectively. 2,3-DMP, 3,4-DMP, 3,4-DMC, and 4,5-DMC were separated by isocratic elution with 40% solvent B; the retention times for these compounds were 26.7, 24.4, 14, and 11.4 min, respectively.
Oxidation products obtained from toluene were separated using a two-solvent system comprising a 0.1% formic acid solution in water (solvent A) and a 0.1% formic acid solution in acetonitrile (solvent B). 4-Methylcatechol, 3-methylcatechol, and m-, p-, and o-cresols were separated using 5 min of isocratic elution with 15% solvent B, followed by elution with a linear 15 to 25% solvent B gradient in 5 min and then an isocratic 25% solvent B step. Under these conditions m- and p-cresols were not separated, and they eluted in a single peak at 22 min. The retention times of 4-methylcatechol, 3-methylcatechol, and o-cresol were 13.1, 14.1, and 23.3 min, respectively. The products were identified by comparing their HPLC retention times and UV-visible spectra with those of standard solutions. The amount of each product was determined by comparing the area of the peak with the areas obtained using known concentrations of standards.
The regiospecificity of toluene hydroxylation was determined by HPLC using an acetylated Cyclobond I 2000 column (Advanced Separation Technologies Inc.) and monitoring the absorbance of the eluate at 274 nm. The acetylated β-cyclodextrin-silica column allowed separation of the three cresol isomers without extraction, which is required by the usual gas chromatography analysis (23, 32). Separation was carried out at a flow rate of 1 ml/min using a two-solvent system comprising 20 mM ammonium acetate in water (pH 5.0) (solvent A) and methanol (solvent B). o- m- and p-cresols were separated using 5 min of isocratic elution with 20% solvent B, followed by elution with a linear gradient from 20 to 60% solvent B in 20 min. The retention times of o-, m-, and p-cresols were 12.8, 14.5, and 15.2 min, respectively.
Time course of toluene and o-xylene oxidation.
The rate of product formation by E. coli cells expressing PH, ToMO, or a mixture of the two proteins was measured by the HPLC discontinuous assay. Each assay mixture contained either 30 μM toluene or 20 μM o-xylene. All cells were used at a concentration of 1 mU/ml when toluene was used as substrate and at a concentration of 1.5 mU/ml when o-xylene was used.
Rate of (di)methylcatechol production as a function of ToMO concentration.
The rate of 3- and 4-MC production from toluene was measured as a function of the ToMO concentration at ToMO concentrations ranging from 0.29 to 2.32 mU/ml with a constant concentration of cells expressing PH (0.29 mU/ml) in the presence of 30 μM toluene. The reactions in aliquots (500 μl) were stopped at 5, 10, and 15 min by adding 50 μl of 1 M HCl, and the products were analyzed by the HPLC system as described above.
The rate of 3,4-DMC production from 40 μM o-xylene was measured as a function of the ToMO concentration at ToMO concentrations ranging from 0.3 to 6 mU/ml with a constant concentration of cells expressing PH (range, 0.36 to 1 mU/ml). 3,4-DMC was measured by the continuous coupled assay with C2,3O (3 U/ml). Experimental data were fitted to the following equation: rate = (Vmax × [ToMO])/(K + [ToMO]) where Vmax is the maximum rate and K is a constant corresponding to the ToMO concentration which gives a rate of Vmax/2.
The rate of 3,4-DMC production from 40 μM o-xylene was also measured by using E. coli cells harboring vector pGEM-3Z/ToMO-PH. The cell concentration corresponded to 1.17 mU/ml of ToMO and 0.5 mU/ml of PH. An assay with phenol was used to determine the total monooxygenase activity of cells. The enzymatic activity of ToMO was measured by assaying the conversion of 3,4-DMP to 4,5-DMC because this product is exclusively formed by ToMO (see below and Fig. 1). The activity of PH was calculated by determining the difference between the total activity and the ToMO activity.
RESULTS
Regioselectivity of ToMO and PH with toluene.
It has been demonstrated previously that ToMO is able to hydroxylate toluene, forming a mixture of o-, m-, and p-cresols (4-6). In addition, both ToMO and PH are able to further hydroxylate the three cresol isomers to catechols (Fig. 1A) (1). Data for the relative amounts of products are available only for the hydroxylation of toluene by ToMO (32).
The relative abundance of each isomer was determined by incubating E. coli cells expressing ToMO or PH with toluene or the three cresol isomers at concentrations ranging from 0.1 to 1 mM as described in Materials and Methods. The data in Table 1 indicate that p-cresol and o-cresol are the main products of toluene oxidation by ToMO and that there is slightly more of the para isomer. The hydroxylation of toluene by PH-expressing cells also resulted in a mixture of products, but in this case the ortho isomer accounted for about two-thirds of the cresol produced. Both enzymes were found to add the second hydroxyl group ortho to the hydroxyl group already present on the aromatic ring, thus producing only 3-MC from o-cresol and only 4-MC from p-cresol (data not shown). The oxidation of m-cresol catalyzed by ToMO produced almost exclusively 4-MC, whereas PH produced predominantly 3-MC (Table 1).
TABLE 1.
Regioselectivity of PH and ToMO with toluene, m-cresol, o-xylene, and 3,4-DMP
| Substrate | Product | % witha:
|
|
|---|---|---|---|
| PH | ToMO | ||
| Toluene | o-Cresol | 70 | 36 |
| m-Cresol | 7 | 19 | |
| p-Cresol | 23 | 45 | |
| o-Xylene | 2,3-DMP | 80 | 20 |
| 3,4-DMP | 20 | 80 | |
| m-Cresol | 3-MC | 96 | 5 |
| 4-MC | 4 | 95 | |
| 3,4-DMC | 3,4-DMC | 99 | 10 |
| 4,5-DMC | 1 | 90 | |
The error was ∼1%.
Kinetic parameters of ToMO and PH with toluene.
The kinetic parameters of ToMO and PH activities with toluene and cresols, as determined by whole-cell assays, are shown in Table 2. The specificity constants of ToMO for benzene and toluene are at least 1 order of magnitude higher than those measured for the same substrates when PH was used. In contrast, when hydroxylated substrates were used, the specificity constants for ToMO with phenols were found to be 2 to 30 times lower than those measured for PH.
TABLE 2.
Apparent kinetic constants of PH and ToMO
| Substrate | PH
|
ToMO
|
||||
|---|---|---|---|---|---|---|
| kcat (s−1)a | Km (μM)b | kcat/Km (s−1 μM−1) | kcat (s−1)a | Km (μM)b | kcat/Km (s−1 μM−1) | |
| Phenol | 1.02 | 0.6 | 1.69 | 1 | 2.18 | 0.46 |
| p-Cresol | 0.77 | 0.6 | 1.29 | 0.63 | 13.26 | 0.047 |
| o-Cresol | 0.946 | 3.13 | 0.3 | 0.9 | 6.0 | 0.15 |
| m-Cresol | 0.609 | 1.8 | 0.34 | 0.44 | 9.38 | 0.047 |
| 2,3-DMP | 0.9425 | 33.87 | 0.028 | 0.2 | 25.7 | 0.008 |
| 3,4-DMP | 0.771 | 3.37 | 0.228 | 0.38 | 7.7 | 0.049 |
| Benzene | 0.092 | 25 | 0.004 | 0.36 | 0.2 | 1.81 |
| Toluene | 0.148 | 22 | 0.007 | 0.42 | 3.93 | 0.11 |
| o-Xylene | 0.125 | 32.5 | 0.004 | 0.25 | 3.73 | 0.067 |
The errors for kcat values were about 26%. The errors for the kcat values for phenol were about 20%.
The errors for Km values were less than 10%. The error for the Km of ToMO for benzene was about 25%.
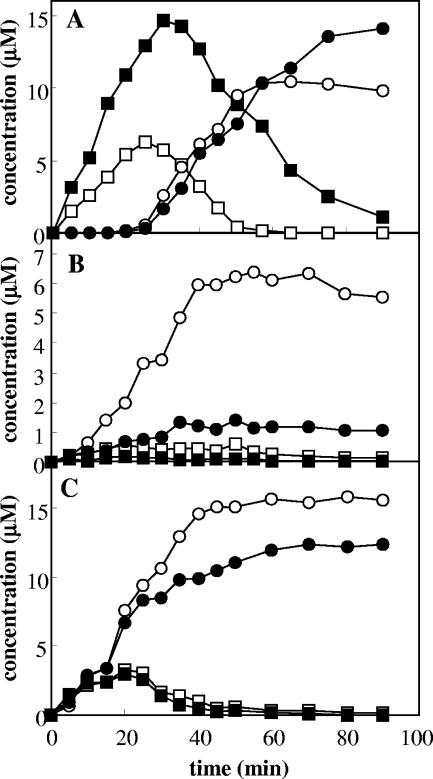
We also determined by using HPLC the production of cresols and methylcatechols from toluene as a function of time (Fig. 2). When E. coli cells expressing ToMO were incubated with 30 μM toluene, a concentration 7.5-fold higher than the ToMO Km value, cresols were produced at a linear rate, whereas methylcatechols were not detected for 30 min. Production of methylcatechols started only after about 60 to 70% of the toluene had been converted to cresols (Fig. 2A). When E. coli cells expressing PH were incubated with 30 μM toluene (Fig. 2B), a short lag period for production of catechols was observed. Moreover, toluene transformation by PH was not very efficient since only 25% of the toluene was converted into methylcatechols after 90 min. Most likely, this depended on the limited amount of cresols produced, which reached a constant, low value after 20 min of incubation. When a mixture of E. coli cells expressing PH and ToMO was incubated with 30 μM toluene, no lag phase for production of methylcatechols was observed, and the formation rates, as determined by the slopes of the plots shown in Fig. 2C, were fivefold higher than those recorded with cells expressing PH alone (Fig. 2B). The concentrations of cresols quickly reached constant, low values, and toluene was completely converted into 55% 3-MC and 45% 4-MC after 50 min. These percentages are very different from those obtained with cells expressing ToMO alone (62% 4-MC and 38% 3-MC) (Fig. 2A) or cells expressing PH alone (17% 4-MC and 83% 3-MC) (Fig. 2B). Interestingly, the values measured under these conditions are the values expected based on the hypothesis that ToMO catalyzes the first hydroxylation reaction and PH catalyzes the second hydroxylation reaction. In fact, according to the reaction scheme in Fig. 1A, after complete conversion of cresols to methylcatechols, the expected percentage of 3-MC based on the total amount of methylcatecols (3-MC%) is given by the following equation: 3-MC% = o-C% + f × m-C%, where o-C% and m-C% are the percentages of o- and m-cresols, respectively, produced by the enzyme that catalyzes the first hydroxylation step and f is the fraction of m-cresol converted to 3-MC by the enzyme that catalyzes the second hydroxylation reaction. If ToMO is the catalyst of the first reaction, then the o-C% and m-C% values are 36 and 19%, respectively (Table 1). Assuming that PH catalyzes the second reaction, the f value is 0.96 (Table 1). Substituting these values in the equation above, 3-MC% is 54.24%, a value which is identical, within the experimental error, to the experimentally measured value.
FIG. 2.
Kinetics of production of o-cresol (□), m- and p-cresols (▪), 3-methylcatechol (○), and 4-methylcatechol (•) by cells expressing ToMO at a concentration of 1 mU/ml (A), cells expressing PH at a concentration of 1 mU/ml (B), and a mixture of cells expressing ToMO (1 mU/ml) and cells expressing PH (1 mU/ml) (C) incubated with 30 μM toluene.
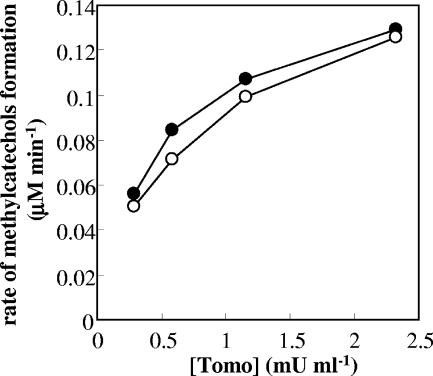
In order to investigate the effects of the relative amounts of ToMO- and PH-expressing cells, the initial rates of methylcatechol production were monitored by incubating toluene with different amounts of cells expressing ToMO (0.29 to 2.3 mU/ml) and/or a constant amount of cells expressing PH (0.29 mU/ml). Under these conditions, methylcatechols were produced at a rate that increased with a hyperbolic dependence on the concentration of ToMO-expressing cells (Fig. 3). Under the same experimental conditions, ToMO-expressing cells alone did not form a significant amount of methylcatechol from toluene at the concentrations tested, whereas only small amounts of methylcatechol were produced by cells expressing PH alone (data not shown).
FIG. 3.
Rates of formation of 3-MC (•) and 4-MC (○) by mixtures of cells expressing PH (0.29 mU/ml) and cells expressing ToMO (0.29 to 2.3 mU/ml) incubated with 30 μM toluene. The concentrations of 3-MC and 4-MC were measured by HPLC as described in Materials and Methods.
Regioselectivity of ToMO and PH with o-xylene.
The relative abundance of dimethylphenol and dimethylcatechol isomers was determined by incubating E. coli cells expressing ToMO (1 mU/ml) or PH (1 mU/ml) with o-xylene, 2,3-DMP, or 3,4-DMP at concentrations ranging from 0.25 to 1 mM. The results (Table 1) indicate that the two monooxygenases are able to catalyze both hydroxylation steps, but with very different regioselectivities. ToMO produces 20% 2,3-DMP and 80% 3,4-DMP from o-xylene, whereas PH produces 80% 2,3-DMP and 20% 3,4-DMP (Fig. 1B). The different regioselectivities of the two enzymes are even more evident in the second hydroxylation step. The hydroxylation of 3,4-DMP by ToMO yields a mixture of 3,4-DMC (10%) and 4,5-DMC (90%) (Table 1 and Fig. 1B), whereas the same substrate is transformed exclusively into 3,4-DMC by PH. 2,3-DMP is converted solely to 3,4-DMC by both enzymes.
Kinetic parameters of ToMO and PH with o-xylene.
The kinetic parameters of ToMO and PH with o-xylene for the first and second reaction steps were determined by whole-cell assays and are shown in Table 2. ToMO had a specificity constant for o-xylene that was 17 times higher than that of PH, whereas PH was about four times more active than ToMO with both 2,3-DMP and 3,4-DMP. In the second hydroxylation step both monooxygenases exhibited kcat/Km values for 3,4-DMP that were about 10 times higher than those measured for 2,3-DMP.
We also determined by HPLC the production of dimethylphenols and dimethylcatechols as a function of time (Fig. 4). When E. coli cells expressing ToMO (1.5 mU/ml) were incubated with 20 μM o-xylene, dimethylphenols were produced at a linear rate, whereas dimethylcatechols were detected only after about 15 min (Fig. 4A). In contrast, when E. coli cells expressing PH (1.5 mU/ml) were incubated with 20 μM o-xylene, only 2,3-DMP and 3,4-DMC were detected (Fig. 4B). The concentration of 2,3-DMP reached a low but constant value after about 30 min, whereas no 3,4-DMP was detected. 3,4-DMC accumulated at a low but constant rate after a short lag period (about 5 min). No 4,5-DMC was detected. When a mixture of E. coli cells expressing PH and ToMO (both at a concentration of 1.5 mU/ml) was incubated with 20 μM o-xylene, only 2,3-DMP and 3,4-DMC were detected (Fig. 4C), as was the case for cells expressing only PH, but 3,4-DMC was produced at a rate that was about 13 times higher.
FIG. 4.
Kinetics of production of 2,3-DMP (□), 3,4-DMP (▪), 3,4-DMC (○), and 4,5-DMC (•) by cells expressing ToMO at a concentration of 1.5 mU/ml (A), cells expressing PH at a concentration of 1.5 mU/ml (B), and a mixture of cells expressing ToMO (1.5 mU/ml) and cells expressing PH (1.5 mU/ml) (C).
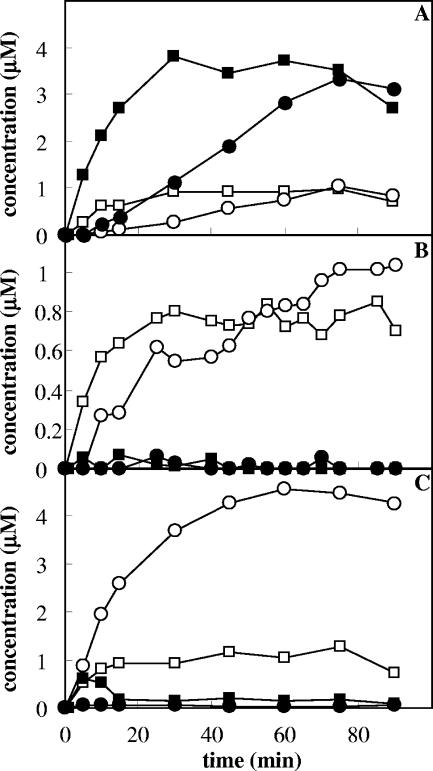
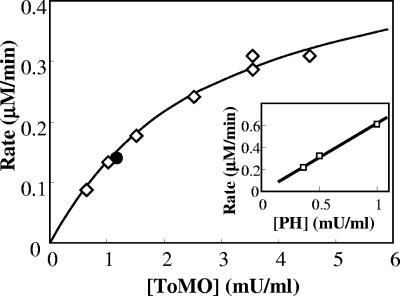
The interplay between the ToMO and PH complexes in o-xylene oxidation was investigated further by incubating different amounts of cells expressing ToMO (0.3 to 6 mU/ml) and a constant amount of cells expressing PH (0.36 mU/ml) with 40 μM o-xylene and measuring the rate of 3,4-DMC formation by a coupled assay with C2,3O. The assay was repeated at PH concentrations up to 1 mU/ml. Cells expressing ToMO or PH alone at the concentrations indicated above did not form significant quantities of 3,4-DMC (data not shown). When a mixture of the two types of recombinant cells was used, however, 3,4-DMC was produced at a rate that was proportional to the concentration of ToMO (Fig. 5). When the rate of 3,4-DMC formation at a constant PH concentration was plotted as a function of the ToMO concentration, a hyperbolic function was obtained (Fig. 5). Moreover, when constant amounts of ToMO were used, the rate of 3,4-DMC formation was found to be directly proportional to the PH concentration (Fig. 5, inset).
FIG. 5.
Kinetics of 3,4-DMC formation. Cells expressing PH were used at constant concentration (0.5 mU/ml), and the rate of 3,4-DMC formation was measured as a function of the concentration of cells expressing ToMO (⋄) in the presence of 40 μM o-xylene. The solid circle indicates the rate of 3,4-DMC production by cells coexpressing ToMO (1.17 mU/ml) and PH (0.5 mU/ml). The rate of 3,4-DMC production was measured by the continuous coupled assay with C2,3O as described in Materials and Methods. The inset shows the rate of 3,4-DMC formation as a function of the PH concentration at a constant concentration of ToMO (4.5 mU/ml).
In order to verify that the expression of ToMO and PH in different cells could influence the degradation of o-xylene carried out under the experimental conditions described above, o-xylene oxidation was also studied using E. coli cells coexpressing the two complexes. The clone E. coli JM109/pGEM-3Z/ToMO-PH was found to express ToMO and PH with a ratio of specific activities of 2.3:1. When 40 μM o-xylene was incubated with an amount of cells corresponding to 1.17 mU/ml of ToMO and 0.5 mU/ml of PH, the rate of 3,4-DMC formation was found to be identical, within the experimental error, to the rate measured using cells expressing the two complexes separately but corresponding to the same ToMO and PH activity (Fig. 5).
DISCUSSION
The throughput of a metabolic route depends upon several factors. Some of these factors are the specificity and regioselectivity of the enzymes in the pathway and their catalytic efficiencies. For degradation of aromatic molecules by P. stutzeri OX1, two different enzymes, ToMO and PH, catalyze the obligatory hydroxylation of the substrate (1, 5, 8). The existence of these two seemingly similar enzymes in P. stuzteri suggests either that they act independently with different substrates or that they comprise a metabolic chain. We have suggested previously (8) that the latter possibility is supported by recent experimental data for benzene hydroxylation by these enzymes which suggest that the different catalytic efficiencies of ToMO and PH appear to be finely tuned to optimize the use of benzene as a growth substrate. In order to establish whether the concerted use of ToMO and PH is a general control system in P. stuzteri for optimal use of aromatic molecules as growth substrates, the kinetic parameters and the regioselectivity of the two enzymes for the transformation of toluene and o-xylene were determined.
Toluene metabolism.
ToMO and PH are able to hydroxylate both toluene and cresols, confirming that the conversion of toluene into methylcatechol is a two-step process that requires cresol intermediates (Fig. 1A) (1). This is also true for the conversion of benzene to catechol by these enzymes (8, 9). The regioselectivities of ToMO and PH, however, are different. Cresols are produced by ToMO with a relaxed regioselectivity, preferentially yielding the ortho and para isomers. Such an unusual relaxed regioselectivity has been reported previously only for benzene monooxygenase from Pseudomonas aeruginosa JI104 (19), whereas most of the other characterized monooxygenases belonging to the family of four-component aromatic/alkene monooxygenases exhibit more restricted regioselectivity (15-17, 24, 34).
PH, which belongs to the large family of three-component T2MO/PHs (8), yields the three cresol isomers, but there is a preference for the ortho isomer, which accounts for 70% of the product. Thus, PH is more restricted in its regioselectivity than ToMO, but it is more relaxed than other group 1 BMMs, like T2MO, which produce only o-cresol (16, 17, 24).
It should also be noted that PH and ToMO still have opposite regioselectivities when they act on a hydroxylated substrate. For example, 4-MC is the major product of ToMO reactivity with m-cresol, whereas 3-MC is the predominant product of PH catalysis with the same substrate.
The catalytic constants of the two monooxygenases provide insight into their substrate specificities (Table 2). The specificity constant of ToMO for toluene is about 16-fold lower than that for benzene. This considerable difference is essentially due to the Km with toluene, which is about 20-fold higher than that with benzene, whereas the kcat values are very similar for these two substrates. Thus, ToMO is a more efficient catalyst for benzene oxidation. The Km of ToMO with toluene is very similar to that of toluene 4-monooxygenase (T4MO) (23); however, its kcat and kcat/Km values are sevenfold lower. These data imply that T4MO is a better catalyst for toluene transformation than ToMO. On the other hand, a different picture emerges when the data for benzene transformation are compared. T4MO has a specificity constant for benzene (25) that is lower than that previously reported for ToMO for the same substrate (8). Thus, benzene is an optimal substrate for ToMO, whereas toluene is transformed better by T4MO. This conclusion agrees with the metabolic features of Pseudomonas mendocina KR1 and P. stutzeri OX1. P. mendocina KR1 is highly specialized for growth on toluene (34), while P. stutzeri OX1, which is more versatile, can grow efficiently on both benzene and toluene (3, 5; P. Barbieri, personal communication).
When the values of the kinetic constants of PH with toluene are compared to those measured with benzene, a different picture emerges than the picture obtained for ToMO. All the kinetic constants for PH are very similar for both substrates (Table 2), suggesting that PH does not discriminate between benzene and toluene. Moreover, the specificity constant for toluene is 16-fold lower than that of ToMO, and this indicates that PH is a less efficient catalyst than ToMO with both of these hydrophobic substrates.
The specificity constants of ToMO for p- and m-cresols are identical to each other and are threefold lower than that for toluene, whereas the specificity constant for o-cresol is very similar to that for toluene. Since o-cresol accounts for only 36% of the products of the first hydroxylation reaction, we concluded that ToMO has a higher catalytic efficiency in the first step of toluene hydroxylation than in the second step, even if the difference is less pronounced than that observed in the case of benzene hydroxylation (Table 2). On the other hand, the specificity constants of PH for o- and m-cresols are similar and are about 4-5 fold higher than those for toluene. The kcat/Km value with p-cresol is about 190 times higher than that with toluene and is very similar to that with phenol. Therefore, PH shows greater catalytic efficiency in the second step of hydroxylation, and p-cresol and phenol are the preferred substrates. Taken together, these results suggest that in P. stutzeri ToMO could act as a toluene-oxidizing enzyme and PH could act as a cresol-oxidizing enzyme. The time-dependent conversion of toluene to methylcatechol reinforces this hypothesis (Fig. 2) because in the recombinant system that we used the combination of the two monooxygenases converted toluene to methylcatechol much more efficiently than ToMO or PH alone. Moreover, the percentages of 3- and 4-MC that we measured are almost identical to the values that can be calculated by assuming that ToMO catalyzes the first hydroxylation step and PH catalyzes the second hydroxylation step (see above).
o-Xylene metabolism.
A large number of bacteria are able to use m- and p-xylenes as growth substrates (2, 10, 20, 21, 36), but few of these bacteria are known to grow on o-xylene; the bacteria that can grow on o-xylene include P. stutzeri OX1 (1, 3, 5), Corynebacterium sp. strain C125 (29), and some Rhodococcus strains (7, 11, 18). Catabolism of m- and p-xylenes usually occurs through the progressive oxidation of a methyl group (2, 10), whereas o-xylene catabolism occurs by direct hydroxylation of the aromatic ring (12, 14), as is the case in P. stutzeri (1, 5).
o-Xylene hydroxylation by ToMO and PH produces 3,4- and 4,5-DMC via the intermediate formation of 2,3- and 3,4-DMP (Fig. 1B). Only 3,4-DMC, however, can be oxidized by C2,3O, the enzyme that opens the gate to the lower pathway. 4,5-DMC is not a substrate for this enzyme (1) and is therefore a dead-end product of o-xylene metabolism that results in the loss of carbon atoms from the growth substrate and in the useless depletion of NADH used by ToMO and PH in the hydroxylation reactions.
Several studies have shown that only 2,3-DMP and 3,4-DMC can be detected in the culture medium of o-xylene-oxidizing bacteria (7, 11, 18, 29), including P. stutzeri (3), for which monooxygenase-mediated o-xylene catabolism has been reported. Thus, the absence of other dimethylphenol or dimethylcatechol isomers suggests that a restricted regiospecificity of the monooxygenases acting on o-xylene would channel this substrate to the exclusive formation of 2,3-DMP in the first hydroxylation step and to 3,4-DMC in the second hydroxylation step (7, 11, 18, 29).
The data that we have collected indicate that both ToMO and PH alone are able to transform o-xylene into dimethylcatechols through a two-step process (Table 2), in which dimethylphenol intermediates are further hydroxylated to dimethylcatechols (Fig. 1A) (4). However the regioselectivities of the two enzymes are different in both reaction steps. ToMO converts o-xylene predominantly to 3,4-DMP, whereas PH produces predominantly 2,3-DMP (Table 1). Both monooxygenases convert 2,3-DMP to 3,4-DMC, but they exhibit different regioselectivities with 3,4-DMP. ToMO oxidizes 3,4-DMP mainly to 4,5-DMC, which cannot be further metabolized (5). PH converts 3,4-DMP exclusively to 3,4-DMC. Thus, our data indicate that only PH can produce a dimethylcatechol which can be further metabolized through the lower pathway.
The catalytic constants of the two enzymes reported in Table 2 clearly indicate that ToMO is a more efficient catalyst than PH in the first hydroxylation step and has a 17.7-fold-higher specificity constant. On the other hand, PH is more efficient than ToMO in the second hydroxylation step and has specificity constants for 2,3- and 3,4-DMP that are about three- and fourfold higher, respectively, than those determined for ToMO. Our kinetic data indicate that 3,4-DMP is transformed into products more efficiently than 2,3-DMP, especially by PH, which has a kcat/Km that is fivefold higher than that of ToMO. The regioselectivity and the kinetic constants of ToMO are in agreement with the finding that when E. coli cells expressing ToMO are incubated with o-xylene, only 4,5-DMC is produced after a long lag phase (Fig. 4A). On the other hand, E. coli cells expressing PH produce 3,4-DMC, but at a very low rate, likely because of the poor conversion of o-xylene to dimethylphenol in the first step (Fig. 4B).
A mixture of E. coli cells expressing PH and ToMO produced only 2,3-DMP and 3,4-DMC (Fig. 4C). However, in this case, no lag phase was observed in the production of 3,4-DMC, and the rate of 3,4-DMC formation was 13-fold greater than the rate recorded with cells expressing PH alone. 2,3-DMP, on the other hand, was produced at a low and constant level (Fig. 4C). The fact that 4,5-DMC was not detected in this mixture is consistent with the conclusion that the second hydroxylation step is performed by PH. 2,3-DMP accumulated in these reactions because it is not a good substrate either for PH or for ToMO, whereas 3,4-DMP was not detected likely because it is the preferred substrate for PH.
The rate of 3,4-DMC formation catalyzed by mixtures of different amounts of cells expressing ToMO and a constant amount of cells expressing PH was found to be proportional to the concentration of ToMO (Fig. 5), and there was a hyperbolic dependence on the ToMO concentration. According to the conclusion described above that ToMO catalyzes the first step of the reaction by producing dimethyphenols, which in turn are hydroxylated to 3,4-DMC by PH, low concentrations of ToMO would be limiting for the system; hence, the rate of 3,4-DMC formation would increase when the ToMO concentration increased. On the other hand, higher ToMO concentrations would provide PH with a saturating amount of DMP isomers. Under these conditions, the PH concentration would be the rate-limiting factor for production of 3,4-DMC, whose rate of formation should be independent of the ToMO concentration. This behavior is exactly what was experimentally observed (Fig. 5). Moreover, when ToMO was used at a constant concentration, the maximum rate of 3,4-dimethylcatechol formation increased linearly with increasing PH concentrations (Fig. 5, inset).
The rate of 3,4-DMC formation was also measured using E. coli cells coexpressing ToMO and PH and was found to be almost identical to the rate measured using the same amounts of ToMO and PH expressed in different cells (Fig. 5). Therefore, the presence of ToMO and PH in different cellular compartments does not influence either the percentages of the products of the conversion of o-xylene to dimethylcatechols or their rate of formation.
In conclusion, the data for the oxidation of toluene and o-xylene, together with the data reported previously for the transformation of benzene (8), strongly support a general hypothesis concerning the metabolism of aromatic compounds in P. stutzeri OX1. ToMO and PH act sequentially and allow efficient conversion of nonhydroxylated aromatic hydrocarbons to substituted catechols. Moreover, this coupling is particularly important in the case of o-xylene degradation because the sequential action leads to production of 3,4-DMC, the sole dimethylcatechol that can enter the lower pathway and lead to the production of precursors for biosynthetic reactions. This hypothesis also provides a metabolic explanation for the acquisition of the ToMO operon by P. stutzeri OX1 (4). The use of the two enzymes in a concerted fashion confers on the strain a selective advantage based on the ability to maximize the efficiency of exploitation of nonhydroxylated aromatic hydrocarbons, such as benzene, toluene, and o-xylene.
Acknowledgments
We are indebted to Giuseppe D'Alessio, Department of Structural and Functional Biology, University of Naples Federico II, and to Gennaro Marino, Department of Organic Chemistry and Biochemistry, University of Naples Federico II, for critically reading the manuscript.
This work was supported by grants PRIN/2000 and PRIN/2002 from the Ministry of University and Research.
REFERENCES
- 1.Arenghi, F. L., D. Berlanda, E. Galli, G. Sello, and P. Barbieri. 2001. Organization and regulation of meta cleavage pathway gene for toluene and o-xylene derivative degradation in Pseudomonas stutzeri OX1. Appl. Environ. Microbiol. 67:3304-3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Assinder, S. J., and P. A. Williams. 1990. The TOL plasmids: determinants of the catabolism of toluene and xylenes. Adv. Microb. Physiol. 31:1-69. [DOI] [PubMed] [Google Scholar]
- 3.Baggi, G., P. Barbieri, E. Galli, and S. Tollari. 1987. Isolation of a Pseudomonas stutzeri strain that degrades o-xylene. Appl. Environ. Microbiol. 53:2129-2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barbieri, P., F. L. Arenghi, G. Bertoni, F. Bolognese, and E. Galli. 2001. Evolution of catabolic pathways and metabolic versatility in Pseudomonas stutzeri OX1. Antonie Leeuwenhoek 79:135-140. [DOI] [PubMed] [Google Scholar]
- 5.Bertoni, G., F. Bolognesi, E. Galli, and P. Barbieri. 1996. Cloning of the genes for and characterization of the early stages of toluene catabolism in Pseudomonas stutzeri OX1. Appl. Environ. Microbiol. 62:3704-3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bertoni, G., M. Martino, E. Galli, and P. Barbieri. 1998. Analysis of the gene cluster encoding toluene/o-xylene monooxygenase from Pseudomonas stutzeri OX1. Appl. Environ. Microbiol. 64:3626-3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bickerdike, S. R., R. A. Holt, and G. M. Stephens. 1997. Evidence for metabolism of o-xylene by simultaneous ring and methyl group oxidation in a new soil isolate. Microbiology 143:2321-2329. [DOI] [PubMed] [Google Scholar]
- 8.Cafaro, V., V. Izzo, R. Scognamiglio, E. Notomista, P. Capasso, A. Casbarra, P. Pucci, and A. Di Donato. 2004. Phenol hydroxylase and toluene/o-xylene monooxygenase from Pseudomonas stutzeri OX1: interplay between two enzymes. Appl. Environ. Microbiol. 70:2211-2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cafaro, V., R. Scognamiglio, A. Viggiani, V. Izzo, I. Passaro, E. Notomista, F. Dal Piaz, A. Amoresano, A. Casbarra, P. Pucci, and A. Di Donato. 2002. Expression and purification of the recombinant subunits of toluene/o-xylene monooxygenase and reconstitution of the active complex. Eur. J. Biochem. 269:5689-5699. [DOI] [PubMed] [Google Scholar]
- 10.Davey, J. F., and D. T. Gibson. 1974. Bacterial metabolism of para- and meta-xylene: oxidation of a methyl substituent. J. Bacteriol. 119:923-929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Di Gennaro, P., E. Rescalli, E. Galli, G. Sello, and G. Bestetti. 2001. Characterization of Rhodococcus opacus R7, a strain able to degrade naphthalene and o-xylene isolated from a polycyclic aromatic hydrocarbon-contaminated soil. Res. Microbiol. 152:641-651. [DOI] [PubMed] [Google Scholar]
- 12.Favaro, R., C. Bernasconi, N. Passini, G. Bertoni, G. Bestetti, E. Galli, and G. Deho. 1996. Organisation of the tmb catabolic operons of Pseudomonas putida TMB and evolutionary relationship with the xyl operons of the TOL plasmid pWW0. Gene 182:189-193. [DOI] [PubMed] [Google Scholar]
- 13.Fetzner, S., and J. R. van der Meer. 2000. Enzymes involved in the aerobic bacterial degradation of N-heteroaromatic compounds: molybdenum hydroxylases and ring-opening 2,4-dioxygenases. Naturwissenschaften 87:59-69. [DOI] [PubMed] [Google Scholar]
- 14.Franklin, F. C., M. Bagdasarian, M. M. Bagdasarian, and K. N. Timmis. 1981. Molecular and functional analysis of the TOL plasmid pWWO from Pseudomonas putida and cloning of genes for the entire regulated aromatic ring meta cleavage pathway. Proc. Natl. Acad. Sci. USA 78:7458-7462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson, G. R., and R. H. Olsen. 1997. Multiple pathways for toluene degradation in Burkholderia sp. strain JS150. Appl. Environ. Microbiol. 63:4047-4052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson, G. R., and R. H. Olsen. 1995. Nucleotide sequence analysis of genes encoding a toluene/benzene-2-monooxugenase from Pseudomonas sp. strain JS150. Appl. Environ. Microbiol. 61:3336-3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kahng, H. Y., J. C. Malinverni, M. M. Majko, and J. J. Kukor. 2001. Genetic and functional analysis of the tbc operons for catabolism of alkyl- and chloroaromatic compounds in Burkholderia sp. strain JS150. Appl. Environ. Microbiol. 67:4805-4816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim, D., Y. S. Kim, S. K. Kim, S. W. Kim, G. J. Zylstra, Y. M. Kim, and E. Kim. 2002. Monocyclic aromatic hydrocarbon degradation by Rhodococcus sp. strain DK17. Appl. Environ. Microbiol. 68:3270-3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kitayama, A., E. Suzuki, Y. Kawakami, and T. Nagamune. 1996. Gene organization and low regioselectivity in aromatic-ring hydroxylation of a benzene monooxygenase of Pseudomonas aeruginosa JI104. Ferment. Bioeng. 82:421-425. [Google Scholar]
- 20.Kunz, D. A., and P. Chapman. 1981. Catabolism of pseudocumene and 3-ethyltoluene by Pseudomonas putida (arvilla) mt-2: evidence for new functions of the TOL (pWWO) plasmid. J. Bacteriol. 146:179-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kunz, D. A., and P. J. Chapman. 1981. Isolation and characterization of spontaneously occurring TOL plasmid mutants of Pseudomonas putida HS1. J. Bacteriol. 146:952-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 23.Mitchell, K. H., J. M. Studts, and B. G. Fox. 2002. Combined participation of hydroxylase active site residues and effector protein binding in a para to ortho modulation of toluene 4-monooxygenase regiospecificity. Biochemistry 41:3176-3188. [DOI] [PubMed] [Google Scholar]
- 24.Newman, L. M., and L. P. Wackett. 1995. Purification and characterization of toluene 2-monooxygenase from Burkholderia cepacia G4. Biochemistry 34:14066-14076. [DOI] [PubMed] [Google Scholar]
- 25.Pikus, J. D., J. M. Studts, K. McClay, R. J. Steffan, and B. G. Fox. 1997. Changes in the regiospecificity of aromatic hydroxylation produced by active site engineering in the diiron toluene 4-monooxygenase. Biochemistry 36:9283-9289. [DOI] [PubMed] [Google Scholar]
- 26.Powlowski, J., and V. Shingler. 1994. Genetics and biochemistry of phenol degradation by Pseudomonas sp. CF600. Biodegradation 5:219-236. [DOI] [PubMed] [Google Scholar]
- 27.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 28.Sanseverino, J., B. M. Applegate, J. M. King, and G. S. Sayler. 1993. Plasmid-mediated mineralization of naphthalene, phenanthrene, and anthracene. Appl. Environ. Microbiol. 59:1931-1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schraa, G., B. M. Bethe, A. R. van Neerven, W. J. Van den Tweel, E. Van der Wende, and A. J. Zehnder. 1987. Degradation 1,2-dimethylbenzene by Corynebacterium strain C125. Antonie Leeuwenhoek 53:159-170. [DOI] [PubMed] [Google Scholar]
- 30.van der Meer, J. R. 1997. Evolution of novel metabolic pathways for the degradation of chloroaromatic compounds. Antonie Leeuwenhoek 71:159-178. [DOI] [PubMed] [Google Scholar]
- 31.van der Meer, J. R., W. M. de Vos, S. Harayama, and A. J. B. Zehnder. 1992. Molecular mechanisms of genetic adaptation to xenobiotic compounds. Microbiol. Rev. 56:677-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vardar, G., and T. K. Wood. 2004. Protein engineering of toluene-o-xylene monooxygenase from Pseudomonas stutzeri OX1 for synthesizing 4-methylresorcinol, methylhydroquinone, and pyrogallol. Appl. Environ. Microbiol. 70:3253-3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Viggiani, A., L. Siani, E. Notomista, L. Birolo, P. Pucci, and A. Di Donato. 2004. The role of conserved residues H246, H199 and Y255 in the catalysis of catechol 2,3-dioxygenase from Pseudomonas stutzeri OX1. J. Biol. Chem. 279:48630-48639. [DOI] [PubMed] [Google Scholar]
- 34.Whited, G. M., and D. T. Gibson. 1991. Toluene-4-monooxygenase, a three component enzyme system that catalyzes the oxidation of toluene to p-cresol in Pseudomonas mendocina KR1. J. Bacteriol. 173:3010-3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Williams, P. A., and J. R. Sayers. 1994. The evolution of pathways for aromatic hydrocarbon oxidation in Pseudomonas. Biodegradation 5:195-217. [DOI] [PubMed] [Google Scholar]
- 36.Williams, P. A., and M. J. Worsey. 1976. Ubiquity of plasmids in coding for toluene and xylene metabolism in soil bacteria: evidence for the existence of new TOL plasmids. J. Bacteriol. 125:818-828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wyndham, R. C., A. E. Cashore, C. H. Nakatsu, and M. C. Peel. 1994. Catabolic transposons. Biodegradation 5:323-342. [DOI] [PubMed] [Google Scholar]